Abstract
Positive signaling by nitrate in its assimilation pathway has been studied in Chlamydomonas reinhardtii. Among >34,000 lines generated by plasmid insertion, 10 mutants were unable to activate nitrate reductase (NIA1) gene expression and had a Nit− (no growth in nitrate) phenotype. Each of these 10 lines was mutated in the nitrate assimilation–specific regulatory gene NIT2. The complete NIT2 cDNA sequence was obtained, and its deduced amino acid sequence revealed GAF, Gln-rich, Leu zipper, and RWP-RK domains typical of transcription factors and transcriptional coactivators associated with signaling pathways. The predicted Nit2 protein sequence is structurally related to the Nin (for nodule inception) proteins from plants but not to NirA/Nit4/Yna proteins from fungi and yeast. NIT2 expression is negatively regulated by ammonium and is optimal in N-free medium with no need for the presence of nitrate. However, intracellular nitrate is required to allow Nit2 to activate the NIA1 promoter activity. Nit2 protein was expressed in Escherichia coli and shown to bind to specific sequences at the NIA1 gene promoter. Our data indicate that NIT2 is a central regulatory gene required for nitrate signaling on the Chlamydomonas NIA1 gene promoter and that intracellular nitrate is needed for NIT2 function and to modulate NIA1 transcript levels.
INTRODUCTION
Nitrate is not only a substrate for the nitrate assimilation pathway but is also a key signal regulating several metabolic, developmental, and cellular differentiation processes. Plants have developed mechanisms to sense nitrate and to regulate expression of the enzymes required for nitrate assimilation in response to cellular levels of reduced nitrogenous compounds (Crawford, 1995; Crawford and Glass, 1998; Daniel-Vedele et al., 1998; Forde, 2000). Plants also respond to nitrate levels by regulating their metabolic carbon allocation (Scheible et al., 1997a), their roots–shoots balance (Scheible et al., 1997b), and their state of root development (Zhang and Forde, 1998) and stomatal opening (Guo et al., 2003). In Chlamydomonas reinhardtii, gametogenesis is also modulated by the presence or absence of nitrate (Pozuelo et al., 2000).
Microarray analysis of nitrate-responsive genes in Arabidopsis thaliana has shown the importance of nitrate as a molecular signal (Wang et al., 2003). Over 1000 genes were found to respond to low levels of nitrate after only 20 min, and new connections between nitrate levels and glycolysis, trehalose-6-P synthesis, iron metabolism, and sulfate uptake and reduction have been uncovered. Wide transcriptome analysis for >1400 putative transcription factors has been performed to identify processes affected by long-term nitrogen deprivation or short-term nitrate nutrition in Arabidopsis (Scheible et al., 2004). Multiple genes for nitrate assimilation were induced within 30 min, and there was a coordinate induction of genes assigned to RNA synthesis and processing. Many genes from different families responded to changes in N nutrition (Scheible et al., 2004). However, specific roles for these genes are still to be defined in most cases.
Nitrate, as a positive signal, plays an essential role in activating expression of the genes involved in its assimilation, initially at the level of transcription (Crawford, 1995; Crawford and Glass, 1998; Galván and Fernández, 2001). Ammonium, or compounds derived from ammonium, such as Gln, provides a negative signal, leading to the repression of genes encoding nitrate transporters and enzymes of the pathway (Cove, 1979; Crawford, 1995; Quesada et al., 1997; Vidmar et al., 2000; Galván and Fernández, 2001). Nevertheless, how nitrate is sensed to regulate the expression of nitrate-regulated genes is unknown in photosynthetic eukaryotes.
On the other hand, the fungal regulatory genes for nitrate assimilation are well characterized. The expression of nitrate assimilation genes is regulated by two main transcription factors, AREA and NIRA in Aspergillus and the homologous genes NIT2 and NIT4 in Neurospora (Crawford and Arst, 1993; Marzluf, 1997). In the yeast Hansenula polymorpha, two NIRA/NIT4 homologous genes have been identified (YNA1 and YNA2) (Siverio, 2002). NIRA/NIT4/YNA genes from Aspergillus, Neurospora, and Hansenula, respectively, are pathway-specific genes involved in nitrate induction, corresponding to GAL4-like Cys6/Zn2 binuclear zinc cluster proteins (Marzluf, 1997; Siverio, 2002). AREA, a GATA family transcription factor involved in the utilization of alternative nitrogen sources, and NIRA act synergistically. The DNA binding of NIRA depends on nitrate concentration and on the presence of a functional AREA gene (Narendja et al., 2002).
Among photosynthetic eukaryotes, NIT2 of Chlamydomonas is the only positively acting regulatory gene for the nitrate assimilation pathway identified to date. The ANR1 MADS box factor that was linked to nitrate regulation of root differentiation, but not to the nitrate assimilation pathway, was originally proposed to respond to nitrate in Arabidopsis roots (Zhang and Forde, 1998). It has subsequently been shown to respond instead to N deprivation (Gan et al., 2005). Chlamydomonas nit2 mutants are unable to grow on nitrate as the sole nitrogen source, and they do not express many of the genes required for nitrate assimilation (NIA1, NRT2.1, NRT2.2, NII1, NAR2, and NAR1.1) (Galván and Fernández, 2001). NIT2 was cloned by transposon tagging, and its expression was shown to be repressed in ammonium-grown cells (Schnell and Lefebvre, 1993). However, the protein encoded by NIT2 has not been characterized, nor is it known whether Chlamydomonas uses additional regulatory genes for nitrate assimilation.
Previous studies, using the arylsulfatase reporter gene under the control of the nitrate reductase (NIA1) promoter region as a nitrate sensor (Ohresser et al., 1997; Loppes et al., 1999; Llamas et al., 2002), have been used to understand nitrate signaling in Chlamydomonas. Nitrate sensing to control NIA1 gene promoter activity occurs intracellularly and is directly dependent on the activity of the nitrate transporters. The high-affinity nitrate transporter (HANT) system I (Nrt2.1/ Nar2) is responsible for the entry into the cell of very small amounts of nitrate still present in theoretically nitrogen-free medium (Llamas et al., 2002). In addition, two short regions in the NIA1 promoter were found to be required for the activation and repression of the NIA1 gene (Loppes and Radoux, 2002).
In this work, we examine the regulatory genes essential for nitrate assimilation in Chlamydomonas, the structural characteristics of Nit2p, and the mechanism of nitrate sensing. We find that NIT2 is an essential regulatory gene for nitrate assimilation, and only mutants affected at this gene, whose deficiency led to the lack of nitrate induction of NIA1, could be identified among >34,000 insertional mutants. The deduced NIT2 amino acid sequence reveals motifs characteristic of signaling transcription factors and transcription coactivators in plants. NIT2 expression is regulated posttranscriptionally and requires intracellular nitrate to activate NIA1 gene expression. Nit2p binds specifically to the short regions previously identified in the NIA1 promoter, explaining their role in the activation and repression of this gene.
RESULTS
Search for New Genes Involved in the Regulation of Nitrate-Responsive Genes
Insertional mutagenesis was used to identify new genes mediating the transcriptional regulation of NIA1 and other nitrate assimilation genes. First, the 4.2-kb plasmid pSP124S conferring bleomycin resistance (Lumbreras et al., 1998) was used to transform Chlamydomonas strain 704. This strain is wild type for nitrate assimilation and contains in its genome the reporter gene ARS encoding Arylsulfatase (Ars) under the control of the NIA1 gene promoter (Ohresser et al., 1997). Using expression of this reporter gene as a genetic screen, mutants lacking the function of a gene needed for positive regulation by nitrate were identified. These mutants would also be expected to grow poorly on nitrate or nitrite medium and be unable to express the pNIA1:ARS reporter gene in nitrate-containing medium.
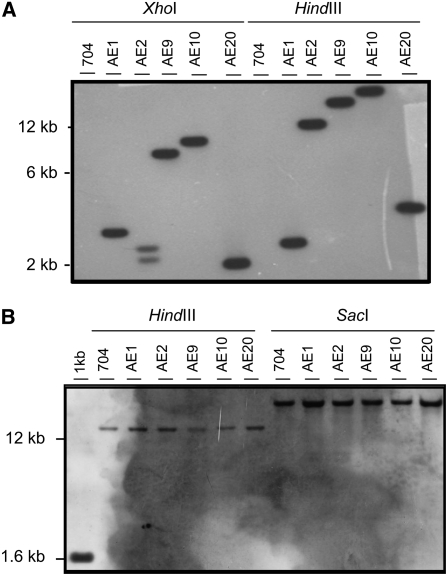
A total of 12,090 bleomycin-resistant (BleR) transformants were isolated, 20 of which were unable to grow in nitrate medium. Five strains—AE1, AE2, AE9, AE10, and AE20—lacked Ars activity in nitrate medium and thus were candidates to be defective in a nitrate signaling gene. A DNA gel blot of DNA digests from these strains using the bleomycin resistance gene as a probe showed that each strain had undergone a unique single integration of the selectable marker DNA (Figure 1A). However, genetic crosses with the wild-type strain 6145c gave Nit+ BleR and Nit− BleS (for bleomycin-susceptible) segregants in about equal numbers as Nit− BleR and Nit+ BleS segregants, indicating that the mutation responsible for the Nit phenotype is independent of the BleR gene insertion. In addition, genetic crosses of these five strains with the nit2 mutant strain resulted in 100% Nit− segregants. DNA gel blot analysis of the NIT2 gene region showed no change in the expected size of the digestion bands in the AE mutants compared with the parental strain (Figure 1B). Moreover, in vivo genetic complementation of the isolated AE Nit− strains was tested using mutant strains 305 (nia1), S16 (nrt2.1, nrt2.2, and nar2), D2 (nia1, nrt2.1, nrt2.2, and nar2), and nit2. These complementation results indicated that all five of the AE mutant strains were allelic with the nit2 mutation. Transformation of these strains with a plasmid carrying the complete NIT2 gene (Schnell and Lefebvre, 1993) resulted in the recovery of wild type (Nit+) colonies (Table 1). These data indicate that these five BleR mutants contain mutations in the NIT2 gene unrelated to the bleomycin marker insertion, probably due to the mutagenic effects of bleomycin as discussed below.
Figure 1.
Detection by DNA Gel Blot of the Bleomycin Resistance Marker and the NIT2 Gene in the Parental Strain 704 and AE Mutants.
Genomic DNA (5 μg) of the indicated strains was digested with the indicated restriction enzymes and hybridized with a 1.2-kb XhoI fragment from plasmid pSP124S (BLE) (A) or with a 1.5-kb HindIII-SalI fragment of plasmid pMN68 of the NIT2 gene (B).
Table 1.
Features of Insertional Mutants
| No. of Transformants
|
Complementation with Strains
|
||||||
|---|---|---|---|---|---|---|---|
| Mutants | Plasmid | + | − | 305(nia1) | nit2 | Insertion Localization | Distance to NIT2 |
| AE1 | pMN68 | 84 | 0 | + | − | n.d. | n.d. |
| AE2 | pMN68 | 69 | 0 | + | − | n.d. | n.d. |
| AE9 | pMN68 | 36 | 0 | + | − | n.d. | n.d. |
| AE10 | pMN68 | 104 | 0 | + | − | n.d. | n.d. |
| AE20 | pMN68 | 31 | 0 | + | − | n.d. | n.d. |
| 89.87 | pNit2 | 42 | 0 | + | − | 9(324200↓) | 0 |
| 177.1 | pNit2 | 79 | 0 | + | − | 9(329046⊣) | 2.5 |
| pMN68 | 78 | 0 | |||||
| 229.13 | pNit2 | 5 | 0 | + | − | 9(323980↓) | 0 |
| 257.67 | pNit2 | 6 | 0 | + | − | 9(304248⊢) | 15 |
| 257.92 | |||||||
The five AE mutants were isolated after insertional mutagenesis using the pBle marker DNA (Lumbreras et al., 1998), and the other five mutants were isolated using the pAphVIII marker (Sizova et al., 2001). Transformation of mutants with the indicated plasmid DNA bearing the complete NIT2 gene was performed with 1 μg of DNA (+) or with no DNA (−) as a control (Kindle, 1990). Genetic complementation with the nit2 mutant was performed as detailed in Methods; −, no complementation. The insertion of the bleomycin resistance gene is unlinked to the Nit− phenotype and was not determined (n.d.). Localizations of the paromomycin resistance gene insertions for the indicated mutants are indicated according to version 3 of the JGI at the Chlamydomonas genome server as follows: first number, number of scaffold; numbers in parentheses, position in the scaffold of the insertion; symbols ↓,⊢, and ⊣, insertion positions of the inserted plasmid DNA within the NIT2 gene, toward the right, or toward the left, respectively. Mutants 257.67 and 257.92 correspond to an identical insertion event. GenBank National Center for Biotechnology Information accession numbers for genomic sequences adjacent to the marker insertion are as follows: DQ385508, mutant 89.87; AY704202, mutant 177.1; DQ385509, mutant 229.13; and AY704210, mutant 257.67.
New mutants insensitive to nitrate were further isolated from an ordered mutant library of Chlamydomonas (González-Ballester et al., 2005b). This collection of 22,000 mutant strains was produced by transformation of the 704 strain with the paromomycin resistance gene APHVIII (Sizova et al., 2001). Five Nit− mutants identified from the mutant library also displayed a nitrate-insensitive phenotype (i.e., no expression of the pNIA1:ARS reporter gene). In each of these mutants, which carried a single insertion of the marker gene, the region adjacent to the insertion was identified by restriction enzyme site-directed amplification PCR (González-Ballester et al., 2005a) (Table 1). Two of these mutants (257.67 and 257.92) had insertions at the same nucleotide. Since these two mutants were isolated from the same selection plate, they were probably sibling strains produced by a mitotic event that occurred after transformation but before selection. These mutants contained insertions either within the NIT2 gene (mutants 89.87 and 229.13) or in the adjacent regions, accompanied by deletions of all or a portion of the NIT2 gene (mutants 177.1 and 257.67) (Table 1). Each of these mutants was shown to be allelic with nit2 in complementation tests using stable diploids. Growth on nitrate medium and expression of the pNIA1:ARS reporter upon nitrate induction could be rescued by transforming the insertional mutants with plasmids containing the NIT2 gene (Table 1). We conclude that these five mutants are nit2 alleles.
The chlorate-resistant mutants RP3 and DC2III were previously shown to have a Nit− phenotype similar to nit2. These two strains were found to be affected within the promoter and the transcript NIT2 regions, respectively (see Supplemental Figure 1 online).
Characteristics of the NIT2 Sequence
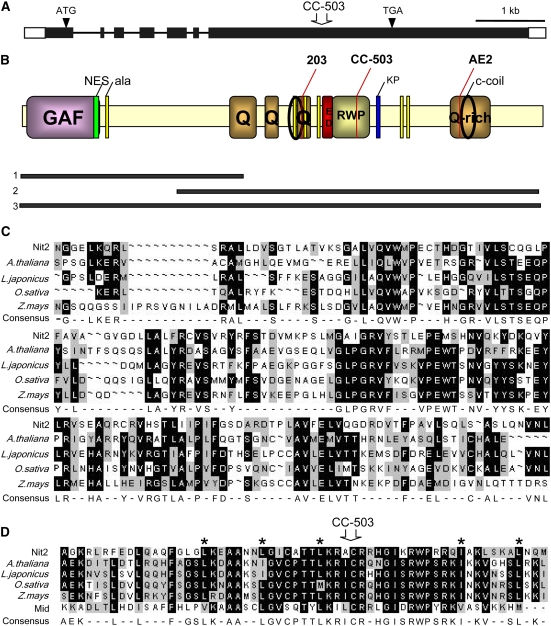
A full-length NIT2 cDNA was isolated and sequenced. The open reading frame of 3591 bp has a 5′ leader sequence containing stop codons in all three reading frames and an unusually long 3′ untranslated region of 1952 bp. From comparison of this cDNA and genomic sequences from Chlamydomonas available from the Joint Genome Institute (JGI; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), we conclude that NIT2 has six introns, all located in the 5′ half of the gene (Figure 2A). Further confirmation that this sequence is from NIT2 comes from the fact that the Chlamydomonas strain used by the JGI to obtain the sequence of the nuclear genome (JGI strain CC-503) carries spontaneous mutations in both NIT1 and NIT2 (Harris, 1989). In comparing our sequence with the sequence of the entire genome, the nit2 mutation in strain CC-503 is caused by a C-to-A transversion at position 2555 of the cDNA within the last exon of the gene, resulting in a stop codon (open arrow in Figure 2A).
Figure 2.
NIT2 Gene Structure and Nit2 Protein Domains.
(A) NIT2 gene structure (accession number DQ311647). Arrowheads show initiation (ATG) and termination (TGA) codons. Black boxes show the exon regions. The open arrow indicates the C-to-A transversion at position 2555 of the cDNA in strain CC-503, used for the genome sequence initiative (JGI).
(B) Nit2 protein structure. GAF domain, residues 9 to 175. Green bar, Leu-rich NES (residues 166 to 175) in an RPT_1 domain. Q, Gln-rich regions (residues 475 to 523, 539 to 558, 623 to 646, and 985 to 1068). Yellow bars (ala), groupings of approximately six to eight Ala residues. Circles, coiled-coil regions (residues 614 to 638 and 1016 to 1050). Red bar, acidic domain (residues 681 to 703). RWP, RWP-RK domain (residues 720 to 778). Blue bar, four KP grouped repetitions. The three black bars below the scheme represent the lengths of the Nit2 protein residues expressed in E. coli: bar 1, Nt Nit2p fragment of 503 amino acids; bar 2, *Nit2p of 831 residues; bar 3, complete Nit2. Mutations in strains 203, Nit2 (CC-503), and AE2 are indicated with long red lines.
(C) Alignment of GAF domains from Nit2 and several Nin plant proteins: Arabidopsis thaliana (NP_195547), Lotus japonicus (CAE30325), Oryza sativa (BAD81274), and Zea mays (AAV64196).
(D) Alignment of the RWP-RK domain from Nit2 and several Nin plant proteins (the same as in [C]) and the Cr Mid protein (AAC49753), with stars showing Leu heptal repetitions. The position from which the deletion in strain Nit2 (CC-503) occurs in also indicated with an open arrow.
The deduced Nit2 protein of 1196 amino acids (Figure 2B) contains several domains characteristic of transcription factors and transcription coactivators in other systems. No similarity is seen in comparing the Chlamydomonas Nit2p sequence with the nitrogen regulatory proteins from Neurospora crassa (Nit2 and Nit4) and Aspergillus nidulans (AreA and NirA). At the N terminus, Nit2p contains a GAF domain similar to those found in phytochromes (Aravind and Ponting, 1997) and cGMP-specific phosphodiesterases (Rybalkin et al., 2003), and in NifA (Little and Dixon, 2003), a transcriptional factor required for the activation of molybdenum nitrogenase for nitrogen fixation in Azotobacter vinelandii. Nit2p also contains a RWP-RK motif within a putative Leu zipper DNA binding domain. This RWP-RK arrangement is found in a Chlamydomonas protein involved in the dominance of the minus mating-type locus (minus dominance or mid) (Ferris et al., 2002). This motif is also found in plant proteins involved in nitrogen-controlled development, such as nodule inception proteins or Nin and Nin-like gene products (Schauser et al., 1999, 2005) (Figure 2D). Nevertheless, although several Nin-like proteins carry both GAF and RWP-RK domains (Figures 2C and 2D), the first Nin protein described (Schauser et al., 1999) lacks the first conserved region present in Nit2p and the other proteins. Database searching showed that plant proteins in different organisms contained one or both domains, with intermediate degrees of conservation with Nit2p. Additional motifs in the protein were four Gln-rich domains, Ala groupings of approximately six to eight residues, two coiled-coil regions, an acidic domain close to the RWP-RK domain, and a nuclear export sequence (NES; la Cour et al., 2003) at the end of the GAF domain (Figure 2B).
Some mutant alleles have been sequenced in an attempt to identify point mutations important for Nit2p function (Figure 2B). Mutant 203 has a very low reversion rate and has been widely used in genetic, biochemical, and molecular studies of nitrate assimilation. This mutant is impaired in the expression of genes for nitrate and nitrite assimilation (Fernández and Matagne, 1986; Prieto and Fernández, 1993; Quesada et al., 1993). The NIT2 gene in this strain was found to have two mutations: at positions 2192 and 2195, both being G-to-T transversions resulting in the changes Q634H and M635I, respectively, in Nit2p. These point mutations fall within the third Gln-rich region containing Ala repeats, highlighting the importance of this domain for Nit2p function. In mutant AE2, deletion of an A nucleotide at position 3275 of the cDNA was found. This deletion occurs within the fourth Gln-rich domain of the protein, as indicated in Figure 2, causing a frameshift of the protein from position 995. This frameshift elongates the protein by 172 residues unrelated to the Nit2p sequence. Even though Nit2p in mutant AE2 still contains most domains (GAF, Leu zipper, and RWP-RK), it is not functional, indicating that most of the protein is not sufficient to drive expression of the endogenous NIA1 gene and that this C-terminal domain is important for Nit2p structure or function.
NIT2 and Nitrate Signaling
The reporter gene pNIA1:ARS was transferred into a nit2 mutant background (203 mt–derived) by a genetic cross between the strains nit2− and 704 (pNIA1:ARS). Fifty percent of the segregants from this cross were incapable of growing in nitrate medium and scored as nit2. None of these Nit− strains had detectable Ars activity when assayed on agar plates. Thus, the presence of a functional NIT2 gene product is essential for the activity of the NIA1 gene promoter. Segregants containing the pNIA1:ARS reporter gene were identified by complementation with strains containing the wild-type NIT2 allele. Four strains that carried both the reporter gene and the nit2 mutation were randomly chosen. When these strains were grown in ammonium and transferred to liquid medium containing nitrate to induce pNIA1:ARS expression, Nia1:Ars activity was undetectable.
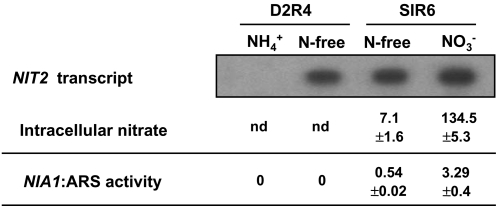
NIT2 gene expression is subject to repression by ammonium and induction in nitrogen-free medium (Schnell and Lefebvre, 1993). However, N-free medium can contain very low concentrations of nitrate sufficient to accumulate in the cell using high-affinity nitrate transport systems and thus to signal positive NIA1 gene expression (Llamas et al., 2002). To determine whether NIT2 is subject to positive control by nitrate or only to repression by ammonium, two isogenic strains, D2R4 and SIR6, were used (Figure 3). Strain D2R4 has a deletion of the NRT2.2, NRT2.1, NAR2, NIA1, and NAR1 genes; thus, it lacks HANT systems I and II and is unable to accumulate nitrate intracellularly from the minor concentrations present in nitrogen-free medium (Llamas et al., 2002). In addition, strain D2R4 contains the low-affinity system III, capable of transporting nitrate at millimolar concentrations (Rexach et al., 1999). Strain SIR6 contains HANT system I in the genetic background of strain D2R4. These two strains were cultivated in ammonium and high CO2 and transferred to ammonium, nitrate, or nitrogen-free medium, and transcript accumulation of NIT2 was analyzed. Strain D2R4 accumulated NIT2 transcripts in nitrogen-free medium even though nitrate did not accumulate intracellularly, indicating that nitrate is not essential for NIT2 gene expression (Figure 3). In addition, even though NIT2 transcripts were present, intracellular nitrate is also needed to stimulate NIA1 expression. As shown in Figure 3, in N-free medium, pNia1:Ars activity was detected only in strain SIR6 (which can accumulate nitrate) but not in strain D2R4 (which cannot).
Figure 3.
RNA Gel Blot Analysis of NIT2 Expression, Intracellular Nitrate, and Ars Activity in D2R4 and SIR6 Strains.
Ammonium-grown cells were transferred to medium containing 8 mM NH4Cl or 4 mM KNO3 or to N-free medium for 1.5 h of incubation. Isolated poly(A) mRNA (10 μg) of the indicated strains and conditions was loaded onto the gel, and a 1-kb PstI-KpnI fragment from the pMN68 plasmid was used as a probe for NIT2 detection. Intracellular nitrate concentrations (expressed as nanomoles of nitrate per milligram of protein) and Ars activity (milliunits per milligram of protein) in these strains were determined. nd, not detectable.
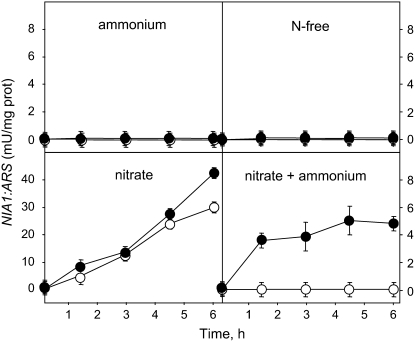
These data suggest that NIT2 may require some sort of activation by intracellular nitrate to mediate its positive signal on the NIA1 gene promoter. To test this hypothesis, ARS activity expression from the NIA1 promoter was analyzed in strain D2R4 in different nitrogen sources (Figure 4). Nia1:Ars activity was determined at different times and compared with that observed in cells previously preincubated during 3 h in nitrogen-free medium to allow NIT2 transcript accumulation. This preincubation of strain D2R4 did not allow the expression of NIA1:ARS in ammonium medium or in nitrogen-free medium, as shown above. However, in nitrate or nitrate plus ammonium medium, the preincubation allowed NIA1:ARS expression, resulting in a greater induction in nitrate medium and in an appreciable Nia1:Ars activity in nitrate plus ammonium medium. During the first 30 min, this activity reached 40% of the levels observed using medium containing only nitrate, with little subsequent increase.
Figure 4.
Nia1:Ars Activity Expression under Different Nitrogen Sources in Strain D2R4.
Ammonium-grown cells were transferred, either directly (open symbols) or after a 3-h incubation in N-free medium (closed symbols), to medium containing 5 mM (NH4)2SO4, 4 mM KNO3, or 5 mM (NH4)2SO4 plus 4 mM KNO3 or N-free medium and bubbled with 5% CO2. At the indicated times, Ars activity was determined.
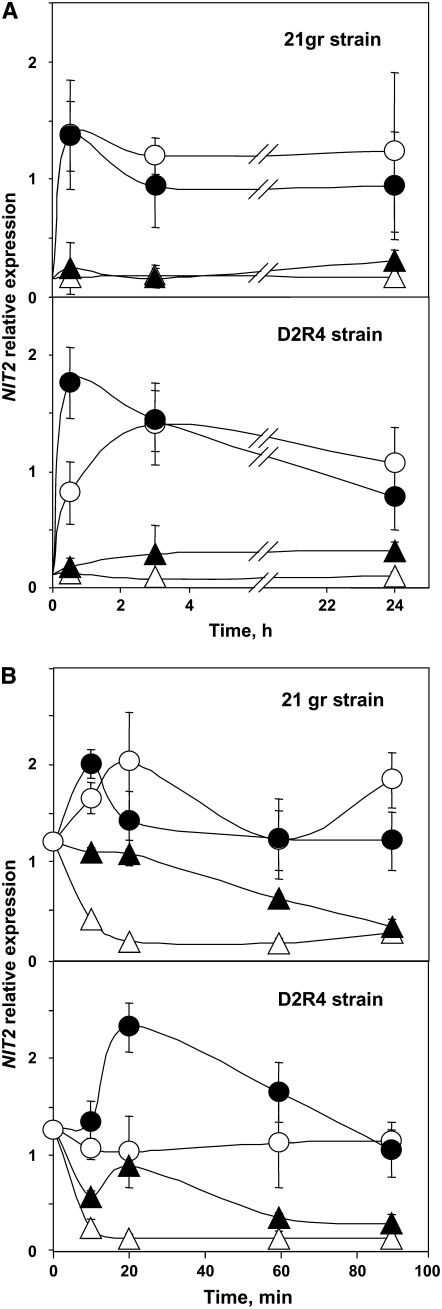
Nit2 expression was quantified by real-time PCR in the wild-type strain 21gr and in strain D2R4. As shown in Figure 5A, NIT2 transcripts were scarce but detectable in ammonium medium and in similar amounts in both strains. Transfer to nitrate or N-free medium resulted in a rapid increase (10 to 20 times) of the transcript levels. Although nitrate is not required for optimal expression of NIT2 transcript, it showed a slight positive effect. This effect of nitrate was more evident in a strain (D2R4) unable to assimilate nitrate. Different levels of NIT2 transcript accumulation in these experiments may be explained by potential differences in transcript stability under the conditions studied. These effects on NIT2 transcript abundance were studied in cells from the wild type and the mutant D2R4 incubated for 3 h in N-free medium (Figure 5B). After transfer to ammonium medium, NIT2 transcripts turned over rapidly in both strains. In 15 to 20 min, transcript levels decreased to the low levels present in this medium. The presence of nitrate (in nitrate plus ammonium–containing medium) appeared to stabilize the transcripts, so that elevated transcript levels were maintained for 90 min. In a true N-free medium condition (strain D2R4), NIT2 transcripts were maintained in constant amounts and nitrate appeared to stabilize transcripts relative to N-free medium. However, in the wild-type strain, NIT2 transcripts were maintained at high levels in both N-free and nitrate-containing media (Figure 5B). When these experiments were repeated with amounts of actinomycin D sufficient to block transcription (20 μg/mL) (Harris, 1989), the high and stable levels of NIT2 transcripts in N-free medium decreased to the basal amounts in ammonium in ∼90 min. However, the presence of actinomycin D did not affect the decrease in NIT2 transcript levels in ammonium or ammonium plus nitrate–containing medium. These results suggest that ammonium inhibits NIT2 transcription and nitrate stabilizes NIT2 transcripts.
Figure 5.
Effect of the Nitrogen Source on NIT2 Transcript Amounts in the Wild-Type 21gr and Mutant D2R4.
Ammonium-grown cells from the indicated strains were either transferred directly (A) or incubated for 3 h in N-free medium and then transferred (B) to medium containing 5 mM (NH4)2SO4 (open triangles), 4 mM KNO3 (closed circles), or 5 mM (NH4)2SO4 plus 4 mM KNO3 (closed triangles) or N-free medium (open circles) and bubbled with 5% CO2. At the indicated times, the cells were processed as detailed in Methods, and the amount of NIT2 transcripts was determined by real-time PCR.
Binding of Nit2p to the NIA1 Promoter
To study Nit2p binding to the NIA1 promoter, the recombinant full-length Nit2 protein was expressed in Escherichia coli. Similarly, the *Nit2p fragment of 831 amino acids at the Nit2 C terminus lacking the GAF domain fragment (Figure 2B, bar 2) and an N-terminal fragment of 503 amino acids (Figure 2B, bar 1) were expressed. It is noteworthy that this fragment contains in its first 175 residues the GAF domain bound to 328 other residues of the protein containing a Gly-rich region and almost the first Gln-rich region. Whether the GAF domain in this N-terminal fragment of Nit2p could correspond to a nitrate binding site was also determined. Using procedures described by Nagore et al. (2006), we were unable to detect binding to nitrate, suggesting that the GAF domain in the N terminus of Nit2p might bind another kind of small molecule.
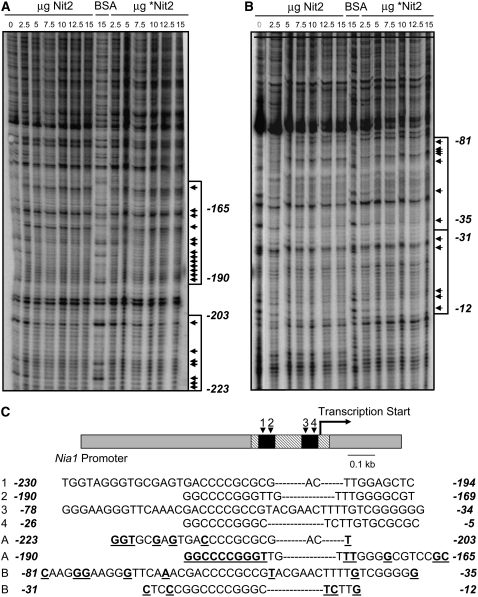
Footprinting experiments were performed with each of the preparations of Nit2p indicated above (Figure 2B). For complete Nit2p and *Nit2p without the GAF domain, proteins were purified from the soluble fraction. In footprinting experiments, Nit2p and *Nit2p-GAF modified the pattern of bands at the same positions in the NIA1 promoter using primers from both orientations (Figures 6A and 6B). These modified bands, highlighted in Figure 6C, were located within the region previously identified by Loppes and Radoux (2002) as essential for activation and repression of the NIA1 gene in Chlamydomonas. However, the Nt Nit2p fragment, which was able to bind to different regions of the 1059-bp NIA1 promoter (data not shown), did not produce any consistent or reproducible pattern of binding in footprinting experiments. Thus, Nit2p appears to bind to specific regions of the NIA1 promoter by means of sequences involving regions of the protein outside of the GAF domain.
Figure 6.
DNase I Footprinting of Nit2 Protein on the NIA1 Promoter Region.
(A) Patterns of fragments resulting from digestion with DNase I of the 5′ end 32P-labeled DNA fragment upon binding with no protein, with different amounts of full-length Nit2p or Nit2 protein without GAF domain (*Nit2), or with BSA. Sequences protected from DNase I are indicated by arrows (band identification was performed by sequencing; data not shown).
(B) Patterns of fragments resulting from digestion with DNase I of the 3′ end 32P-labeled DNA fragment. The regions of DNA protected from DNase I digestion by Nit2 or truncated *Nit2 proteins are indicated by arrowheads.
(C) NIA1 gene promoter scheme. Oblique hatching shows the DNA fragment used in footprinting assays. Black boxes indicate two regions of the promoter that are essential to full expression of the gene (Loppes and Radoux, 2002). The palindromic sequences proposed as sequence signatures for the DNA binding sites in experiments A and B are compared with those present in the NIA1 gene promoter. Bases showing a clear modified pattern in DNase I digestions are shown in boldface and underlined.
DISCUSSION
The regulatory genes and mechanisms controlling nitrate assimilation are mostly unknown in algae and plants, in contrast with the well-characterized regulatory pathways in fungi (Crawford and Arst, 1993; Marzluf, 1997; Siverio, 2002). The only regulatory gene for the nitrate assimilation pathway identified to date in photosynthetic eukaryotes is NIT2 from Chlamydomonas (Schnell and Lefebvre, 1993; Galván and Fernández, 2001). However, the protein encoded by NIT2 and the mechanism by which Nit2p regulates nitrate assimilation have not been described previously.
In this report, >34,000 potential insertional mutants were examined for defects in the regulation of nitrate assimilation, and only 10 new mutants were found, all of which were nit2 alleles. Interestingly, the five bleomycin-resistant/nitrate-sensing mutants were unrelated to the marker insertion and might have been caused by mutagenesis derived from the action of bleomycin as a mutagen (Hecht, 2000). By contrast, all five paromomycin-resistant/nitrate-sensing mutants resulted from insertion of the marker DNA into the NIT2 gene, demonstrating the usefulness of this library for insertional mutagenesis (González-Ballester et al., 2005b). Since mutants were selected in ammonium medium for antibiotic resistance and screened afterward for nitrate-sensing defects, it is unlikely that our selection protocol prevented the isolation of any particular class of mutants, such as mutants with a lethal phenotype on nitrate medium. In addition, two previously isolated mutant strains, RP3 and DC2III, with a Nit− phenotype related to NIT2 were proposed to be affected either at NIT2 or at the closely linked gene NIT9 (Prieto and Fernández, 1993; Rexach et al., 1999; Navarro et al., 2005). Now, these mutants are shown to be affected within the NIT2 locus, promoter, and transcribed regions, so they are allelic to the other nit2 mutants isolated (see Supplemental Figure 1 online). Thus, NIT2 is the only essential regulatory gene identified and characterized so far in Chlamydomonas.
The predicted Nit2 protein sequence shows typical elements of a transcription factor, and its sequence is unrelated to the known fungal regulatory proteins for nitrate assimilation (Crawford and Arst, 1993; Marzluf, 1997; Siverio, 2002). Nit2p appears to be a Leu zipper protein, probably acting as a dimer and containing a putative RWP-RK DNA binding domain. This domain is found in several Arabidopsis proteins, such as NLP, in the Nin protein of Lotus japonicus (Schauser et al., 1999) and other plants (Schauser et al., 2005) (Figure 2), and in the minus dominance (Mid) protein of Chlamydomonas (Ferris et al., 2002; Lin and Goodenough, 2007). Both Nin and Mid proteins have been proposed to act as transcription factors, and both are involved in nitrogen-controlled developmental processes. The structural similarities between Nit2p, Nin, and Mid proteins may have functional significance and reflect a selection during evolution for a family of transcriptional factors mediating nitrogen signals. Deletion of the C-terminal moiety of the protein or point mutations at the third Gln-rich domain led to an inability of Nit2p to promote NIA1 transcription, as did a frameshift mutation at the fourth Gln-rich domain. These Gln-rich domains are thought to provide appropriate protein–protein interactions for the buildup of the transcriptional machinery (Owens et al. 2003). In addition, motifs named GAF domains, for their presence in cGMP-regulated cyclic nucleotide phosphodiesterases, certain adenylyl cyclases, and the bacterial transcription factor FhlA, regulate protein function by binding ligands such as nucleotides and small molecules. Examples of small ligands binding to proteins with GAF domains include 2-oxoglutarate binding to the NifA protein (Little and Dixon, 2003) and nitric oxide binding to the NorR protein (Busch et al., 2005). An interesting possibility is that Nit2p or an associated protein functions as a sensing protein similarly to NorR (Busch et al., 2005; D'Autreaux et al., 2005). It seems that the Nit2p fragment containing the GAF domain does not bind nitrate, so the effect of this anion on Nit2 activity might occur at another level. Alternatively, this GAF domain could bind cGMP, as described for other proteins with GAF domains from different organisms (Ho et al., 2000; Bridges et al., 2005).
Previous studies showed that nitrate regulation of the NIA1 gene promoter in Chlamydomonas occurs through intracellular sensing of nitrate levels (Llamas et al., 2002). Conclusions derived from data on the expression of nitrate assimilation genes in fungal NIA1 mutants were puzzling, since these genes were highly expressed in medium with no added nitrate, suggesting an autoregulation mechanism mediated by NIA1 itself (Cove and Pateman, 1969; Crawford and Arst, 1993; Marzluf, 1997). Similar results were found in Chlamydomonas and the yeast Hansenula, in which it was subsequently shown that this overexpression is the result of the accumulation of intracellular nitrate from minor concentrations present in N-free medium and is dependent on HANT activity (Llamas et al., 2002; Navarro et al., 2003). Thus, Chlamydomonas strains lacking HANTs have been critically important in demonstrating the role of intracellular nitrate in gene regulation.
Recently, it was shown in A. nidulans that AreA regulates NirA through the nitrate transporters. Nuclear import of NirA triggered by nitrate is an essential step in NirA activation as a transcription factor (Berger et al., 2006), with an additional regulatory checkpoint of nitrate effects mediated by the nuclear export machinery of NirAp (Bernreiter et al., 2007). Our work (Figure 2) shows that NIT2 does not require nitrate for its expression, because its transcripts accumulated at maximum levels in N-free medium in a mutant (D2R4) devoid of HANT system I (Rexach et al., 1999). Intracellular nitrate, however, is required for NIT2 to promote transcription of the NIA1 gene. Only when both intracellular nitrate and NIT2 transcripts were present (i.e., in the SIR6 strain) did we observe expression from the Ars reporter construct. The precise mechanism of Nit2 activation by nitrate remains to be determined in Chlamydomonas. Nuclear import and additional export of Nit2p might play an important regulatory role, similar to the mechanism described for the fungal nitrate transcription factors (Berger et al., 2006). In fact, Nit2p has a typical NES (Figure 2B). Although activation mechanisms might be shared between the fungal/yeast and algal/plant systems, the nature of the regulatory transcriptional factors is very different. In Chlamydomonas, Nit2p is a RWP-RK domain–containing protein within a Leu zipper motif. Thirteen additional RWP proteins are present in the Chlamydomonas genome, although only Nit2p shows phylogenetic proximity to Mid from Chlamydomonas or NLP and Nin from plants (Lin and Goodenough, 2007).
Our data support the conclusion that Nit2 is a DNA binding protein. Nit2p and *Nit2 without the GAF domain bind to specific sequences in the NIA1 promoter DNA, indicating that the GAF domain does not participate in DNA binding. Because the NES is so close to the GAF domain, a possibility is that the NES activity is modulated by the GAF through the binding of small molecules. The sequences that bind to Nit2p reside within a NIA1 promoter region identified previously by Loppes and Radoux (2002) as essential for positive and negative control of NIA1 expression.
Interestingly, NIT2 is expressed constitutively in low amounts, and when ammonium is removed from the medium, NIT2 transcript levels increase by ∼15 fold. In A. nidulans, AreA acts to facilitate the expression of genes involved in metabolizing alternative nitrogen sources. A signaling mechanism involving the regulated degradation of AreA transcripts in response to ammonium and Gln provides the first direct means of monitoring nitrogen signaling in this fungus (Morozov et al., 2001). Our observations indicate that, as with AreA, regulation of NIT2 transcript levels would ensure that cells preferentially use ammonium rather than nitrate. While in the fungal or yeast systems two regulatory genes are required for nitrate induction of the nitrate pathway (Crawford and Arst, 1993; Marzluf, 1997; Siverio, 2002), based on the data presented here it is likely that NIT2 is the only gene in Chlamydomonas required for NIA1 induction in response to changes in nitrate concentrations. Now that the complete Chlamydomonas genomic DNA sequence is available (Merchant et al., 2007), it is clear that NIT2 is a single-copy gene.
NIT2 also provides a negative signal in the presence of nitrate for the expression of Cr AMT1.1, a putative ammonium transporter, thus connecting the pathway for ammonium uptake with that of nitrate assimilation (González-Ballester et al., 2004). This dual role of Nit2p in gene expression, dependent on intracellular nitrate, positive for nitrate assimilation genes and negative for AMT1.1 expression, reinforces the central role of NIT2 in the nitrogen metabolism in the alga. Thus, we propose that Chlamydomonas Nit2p is a transcriptional factor mediating nitrogen status of the cell.
METHODS
Strains and Growth Conditions
Chlamydomonas reinhardtii strain 704 (mt+ cw15 arg7+ pnia1:ars+) was kindly provided by Roland Loppes (University of Liege, Belgium) and characterized as described elsewhere (Ohresser et al., 1997; Loppes et al., 1999). Strains 6145c mt− and 21gr mt+ (wild type), 305 (mt−nia1), nit2 (mt−nit2) D2R4 (mt−Δ[nia1, nar1, nar2, nrt2.1, nrt2.2]∷pnia1:ars+), S16 (mt−Δ[nrt2.1, nrt2.2, nar2]), D2 (mt−Δ [nia1, nrt2.1, nrt2.2, nar2]), and SIR6 (mt−D2R4∷pT2CO-5 [NRT2.1, NAR2]) have been described elsewhere (Harris, 1989; Llamas et al., 2002). Mutants 203 (Fernández and Matagne, 1986), RP3 mt− (nit2*) (Prieto and Fernández, 1993), and DC2III (mt−Δ [nia1, nrt2.1, nrt2.2, nar2] nit9) (Rexach et al., 1999) were characterized previously. Cells were grown at 25°C under continuous light with 5% (v/v) CO2-enriched air in minimal medium containing 8 mM ammonium chloride (Harris, 1989). Cells were collected at the midexponential phase of growth by centrifugation (4000g, 5 min), washed twice with 10 mM potassium phosphate, pH 7, and incubated in the indicated medium.
Insertional Mutagenesis of Chlamydomonas Cells
Cells were washed in N-free medium and concentrated to ∼108 cells/mL. Transformation using the glass bead method (Kindle, 1990) was performed using 100 ng of pSP124S, conferring bleomycin resistance (Lumbreras et al., 1998), or 100 ng of pSI104, conferring paromomycin resistance (Sizova et al., 2001), as detailed elsewhere (González-Ballester et al., 2005b).
Genetic Analyses and Complementation Tests
Genetic crosses were performed using the random spore-plating method (Levine and Ebersold, 1960; Harris, 1989). Segregants were analyzed for their ability to grow on medium containing nitrate by replica plating. In vivo complementation was performed by isolating stable vegetative diploids according to a previously described method (Ebersold, 1967; Fernández and Matagne, 1986).
Ars Assays
Cells were collected by centrifugation at 3000g and disrupted by freezing/thawing in 250 μL of 0.4 M Gly-NaOH buffer, pH 9. After centrifugation, 2.5 μL of 1 M imidazole and 50 μL of the enzyme substrate α-naphthylsulfate were added to 100 μL of the extract. The samples were incubated for 30 min at 37°C. The reaction was stopped with 50 μL of 20% SDS, 200 μL of sodium acetate buffer, pH 4.8, and 50 μL of 10 mg/mL tetrazotized σ-dianisidine. The absorbance was determined at 540 nm (Ohresser et al., 1997). Ars activity is expressed as milliunits, corresponding to nanomoles of α-naphthol produced per minute of assay. The data presented in this work correspond to mean values and sd from at least three independent experiments. Ars activity was also determined in situ by assaying the activity on the agar plates after removing cells from the agar surface with a razor blade and following the procedure reported previously (Ohresser et al., 1997).
Nitrate Determination
To determine intracellular nitrate accumulation, cells were collected by centrifugation at 3000g, washed with ice-cold 75 mM potassium phosphate buffer, pH 7.0, and disrupted by freezing/thawing in 20 mM potassium phosphate buffer, pH 4.0, at a dilution of 1:10 (w/v). After eliminating the cell debris by centrifugation at 10,000g, nitrate concentrations in the supernatant were determined by HPLC with a Partisil SA ion-exchange column (10 μm) using the same extraction buffer, pH 4.0, at a flow rate of 0.4 mL/min (Llamas et al., 2002).
Isolation of DNA and RNA from Chlamydomonas and Hybridization Analysis
Isolation of genomic DNA and DNA gel blot analysis were performed according to previously described methods (Sambrook et al., 1989). Conditions for hybridization were as described by Schloss et al. (1984), and washes were performed at 65°C with 0.2× SSC solution (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) containing 0.2% SDS. Total RNA isolation was performed as reported previously (Schloss et al., 1984). The PolyTtract mRNA isolation systems kit from Promega was used following the instructions of the manufacturer. Poly(A) RNA was fractionated on 1.6% agarose gels containing 17.5% formaldehyde and transferred onto nylon membranes (Nytran-N2; Schleicher and Schuell) in 10× SSC. Conditions for hybridization were as reported (Schloss et al., 1984) at 42°C and 50% formamide. Washes were performed at 65°C with 0.2× SSC solution containing 0.2% SDS. Radioactive probes were labeled by the random primer method (Feinberg and Vogelstein, 1984) using [α-32P]dCTP for both RNA and DNA gel blot analyses. The hybridization probes used were as follows: the 1-kb PstI-KpnI fragment of the NIT2 gene from plasmid pMN68 (Schnell and Lefebvre, 1993), the 2.2-kb fragment nit2-52 obtained by PCR amplification at the NIT2 5′ end, and the 2.4-kb fragment nit2-32 fragment obtained by PCR amplification from the NIT2 3′ end (see Supplemental Figure 1 online). Digoxigenin labeling was performed according to the instructions of the manufacturer (Boehringer Mannheim).
PCR Procedures
PCR was performed in a final volume of 25 μL with the following components: 5 pmol of each primer, 0.2 mM of each nucleotide triphosphate, 0.5 units of Taq DNA polymerase from Biotools (B&M Labs), 2.5 μL of specific buffer (containing 2.5 mM MgCl2), 50 to 100 ng of DNA, and 1% DMSO. Primers for restriction enzyme site-directed amplification PCR were as follows: T1RP (5′-CGCATTGCTGCTCGTTCA-3′) and T2RP (5′-TCGGGCAACATTCTCCGCTTT-3′) in strain RP3 and T1Bas (5′-GCAGCTTCCATGAACAACAA-3′) and T2Bas (5′-GGCGACGCGGGGGATGAC-3′) in strain DC2III; for specific amplification of the inserted DNA in strain DC2III, BasInsUpper (5′-GGGATGCACATGGGTGTCA-3′) and BasInsLower (5′-GGCGACTGGAGGGGATGCT-3′) were used.
Synthesis of cDNA
cDNA synthesis was performed with 3 μg of total RNA by reverse transcription amplification using the SuperScript II kit (Invitrogen), following the instructions of the manufacturer, in the presence of an RNase inhibitor, RNAguard (Amersham Pharmacia Biotech), and QT primer (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTT-3′) as well as specific primers 5RTUpperNit2 (5′-GCGCCATGCGAGGGGTTCACA-3′), ATGNit2Upper (5′-CTTCGAATGAATACAGGGATGGCTTAC-3′), BasInsUpper (5′-GGGATGCACATGGGTGTCA-3′), LowerHindIII (5′-CAAGCTTATCGCGCGCTGATGCCGGCCA-3′), 4RTNit2Lower (5′-GTGCAGGTTGCCGGTGGACA-3′), and 23T1Bas (5′-GCAGCTTCCATGAACAACAA-3′).
Quantification of Gene Expression by Real-Time PCR
PCR was performed on the LightCycler instrument (Bio-Rad iCycler iQ real-time PCR detection system) using Taq DNA polymerase with its corresponding buffer (Biotools Biotechnological and Medical Laboratories) in a 25-μL final volume with 0.2 mM of each nucleotide triphosphate and 5 pmol of each primer. The specific primer set (forward, 5′-GCGCCATGCGAGGGGTTCACA-3′; reverse, 5′-CGAGCTCAAGCTTTTTTTTTTTTTTTGTA-3′) amplified a 132-bp Nit2 cDNA fragment. Each reaction was performed on 3 μL of a 1:5 (v/v) dilution of the first cDNA strand, synthesized as described above. As fluorescent dye, SYBR Green I diluted 1:10,000 (v/v) in water (Molecular Probes) was used. The reaction was incubated at 96°C for 3 min, followed by 40 cycles of 30 s at 96°C, 30 s at 60°C, 20 s at 72°C, and 10 s at 87°C, where fluorescence was measured to avoid primer-dimer and background signals. The specificity of PCR amplifications was checked by a melting curve program (60 to 95°C with a heating rate of 0.5°C/s and a continuous fluorescence measurement) and analyzed by electrophoresis on a 1.6% agarose gel. The data are expressed as relative values with respect to the constitutive expression of the ubiquitin ligase gene used as a control (González-Ballester et al., 2004).
Expression and Purification of Nit2
The full-length Nit2 protein and the N-terminal Nit2 fragment of 503 amino acids (Figure 2B) were expressed from pQ80L in Escherichia coli strain TP1000 for 3 h at 37°C in Luria-Bertani medium containing 1 mM isopropyl β-thiogalactoside at A600 = 0.5. Constructions corresponding to 6×His-tagged full-length Nit2p resulted in the expression of minor amounts of soluble protein, since most localized in the inclusion bodies at the different temperatures (28 to 45°C) and isopropyl β-d-thiogalactoside concentrations (1 to 500 μM) used. This fact is not surprising considering that the protein has an isoelectric point of 6.85 and a high proportion of hydrophobic residues with long stretches of Ala and Gln residues. The truncated Nit2 protein, *Nit2 (Figure 2B, bar 2), lacking the first 365 amino acids, was expressed from pET30b(+) in E. coli strain BL21 using the same conditions as full-length Nit2. Harvested cells were extracted and purified using 1 mL of nickel-nitrilotriacetic acid superflow matrix per 2-liter culture (Qiagen). His-tagged Nit2 protein was purified according to previously reported methods (Schwarz et al., 1997).
DNase I Footprinting
The DNase I protection assay was performed as described previously (Frías et al., 2000). DNA fragment used in DNase I footprinting assays was obtained by PCR amplification using NIA1 promoter as template (primers Upper, 5′-CCTTGGGAAGCGTTGGACTATAT-3′; and Lower, 5′-ATTGGTCGGTTCCTGTATCGTCTT-3′) and 5′ end-labeled with polynucleotide kinase (Promega).
DNA Sequencing and Sequence Analysis
DNA sequencing was performed at the Servicio Central de Apoyo a la Investigación (University of Córdoba) with fluorescence-labeled terminators for an automatic sequencer (model ABI 377; Applied Biosystems Perkin-Elmer), according to the manufacturer's instructions, or at the Advanced Genome Analysis Center of the University of Minnesota. Sequences were analyzed using DNAstar software version 4.05 (Lasergene Navigator), the Bioedit Sequence Alignment Editor version 5.0.9 (Department of Microbiology North Carolina State University), the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/), and the Chlamydomonas JGI server (http://www.chlamy.org). Domain predictions were analyzed with the Smart server (http://smart.embl-heidelberg.de/).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NIT2 cDNA sequence, DQ311647; genomic sequences adjacent to the marker insertion for mutant 89.87, DQ385508; mutant 177.1, AY704202; mutant 229.13, DQ385509; and mutant 257.67, AY704210; DNA inserted (419 bp) at the NIT2 gene in mutant DC2III, DQ835537.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Analysis of the NIT2 Genomic Region in Mutants RP3 and DC2III.
Supplementary Material
Acknowledgments
This work was funded by the Ministerio de Educación y Ciencia, Spain (Grants BMC2002-03325 and BFU2005-07521), the European Commission (Grant HPRN-CT-2002-00247), the Junta de Andalucía, Spain (Grant CVI-0128), and by National Institute of General Medical Sciences Grant GM-34437 (to P.A.L.). A.C. and J.J.H. thank the Ministerio de Educación y Ciencia, Spain, for PhD fellowships. We thank M. Isabel Macías and Raquel González for laboratory support and A. Valladares, A. Herrero, and E. Flores for help in footprinting experiments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Emilio Fernández (bb1feree@uco.es).
Online version contains Web-only data.
References
- Aravind, L., and Ponting, C.P. (1997). The GAF domain: An evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22 458–459. [DOI] [PubMed] [Google Scholar]
- Berger, H., Pachlinger, R., Morozov, I., Goller, S., Narendja, F., Caddick, M., and Strauss, J. (2006). The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol. Microbiol. 59 433–446. [DOI] [PubMed] [Google Scholar]
- Bernreiter, A., Ramon, A., Fernandez-Martinez, J., Berger, H., Araujo-Bazan, L., Espeso, E.A., Pachlinger, R., Gallmetzer, A., Anderl, I., Scazzocchio, C., and Strauss, J. (2007). Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol. Cell. Biol. 27 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, D., Fraser, M.E., and Moorhead, G.B. (2005). Cyclic nucleotide binding proteins in the Arabidopsis thaliana and Oryza sativa genomes. BMC Bioinformatics 6 6 10.1186/1471-2105-6-6. [DOI] [PMC free article] [PubMed]
- Busch, A., Strube, K., Friedrich, B., and Cramm, R. (2005). Transcriptional regulation of nitric oxide reduction in Ralstonia eutropha H16. Biochem. Soc. Trans. 33 193–194. [DOI] [PubMed] [Google Scholar]
- Cove, D.J. (1979). Genetic studies of nitrate assimilation in Aspergillus nidulans. Biol. Rev. Camb. Philos. Soc. 54 291–327. [DOI] [PubMed] [Google Scholar]
- Cove, D.J., and Pateman, J.A. (1969). Autoregulation of the synthesis of nitrate reductase in Aspergillus nidulans. J. Bacteriol. 97 1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M. (1995). Nitrate: Nutrient and signal for plant growth. Plant Cell 7 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M., and Arst, N.H.J. (1993). The molecular genetics of nitrate assimilation in fungi and plants. Annu. Rev. Genet. 27 115–146. [DOI] [PubMed] [Google Scholar]
- Crawford, N.M., and Glass, A.D.M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3 389–395. [Google Scholar]
- Daniel-Vedele, F., Filleur, S., and Caboche, M. (1998). Nitrate transport: A key step in nitrate assimilation. Curr. Opin. Plant Biol. 1 235–239. [DOI] [PubMed] [Google Scholar]
- D'Autreaux, B., Tucker, N.P., Dixon, R., and Spiro, S. (2005). A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437 769–772. [DOI] [PubMed] [Google Scholar]
- Ebersold, W.T. (1967). Chlamydomonas reinhardtii: Heterozygous diploid strains. Science 157 447–449. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1984). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137 266–267. [DOI] [PubMed] [Google Scholar]
- Fernández, E., and Matagne, R.F. (1986). In vivo complementation analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr. Genet. 10 397–403. [DOI] [PubMed] [Google Scholar]
- Ferris, P.J., Armbrust, E.V., and Goodenough, U.W. (2002). Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde, B.G. (2000). Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 1465 219–235. [DOI] [PubMed] [Google Scholar]
- Frías, J.E., Flores, E., and Herrero, A. (2000). Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol. Microbiol. 38 613–625. [DOI] [PubMed] [Google Scholar]
- Galván, A., and Fernández, E. (2001). Eukaryotic nitrate and nitrite transporters. Cell. Mol. Life Sci. 58 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Y., Filleur, S., Rahman, A., Gotensparre, S., and Forde, B.G. (2005). Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222 730–742. [DOI] [PubMed] [Google Scholar]
- González-Ballester, D., Camargo, A., and Fernández, E. (2004). Ammonium transporter genes in Chlamydomonas: The nitrate-specific regulatory gene Nit2 is involved in Amt1;1 expression. Plant Mol. Biol. 56 863–878. [DOI] [PubMed] [Google Scholar]
- González-Ballester, D., de Montaigu, A., Galván, A., and Fernández, E. (2005. a). Restriction enzyme site-directed amplification PCR: A tool to identify regions flanking a marker DNA. Anal. Biochem. 340 330–335. [DOI] [PubMed] [Google Scholar]
- González-Ballester, D., de Montaigu, A., Higuera, J.J., Galván, A., and Fernández, E. (2005. b). Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 137 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F.Q., Young, J., and Crawford, N.M. (2003). The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. (1989). The Chlamydomonas Sourcebook. (New York: Academic Press). [DOI] [PubMed]
- Hecht, S.M. (2000). Bleomycin: New perspectives on the mechanism of action. J. Nat. Prod. 63 158–168. [DOI] [PubMed] [Google Scholar]
- Ho, Y.S., Burden, L.M., and Hurley, J.H. (2000). Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19 5288–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle, K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour, T., Gupta, R., Rapacki, K., Skriver, K., Poulsen, F.M., and Brunak, S. (2003). NESbase version 1.0: A database of nuclear export signals. Nucleic Acids Res. 31 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, R.P., and Ebersold, W.T. (1960). The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 14 197–216. [DOI] [PubMed] [Google Scholar]
- Lin, H., and Goodenough, U.W. (2007). Gametogenesis in the Chlamydomonas reinhardtii mating type is controlled by two genes, NID and MTD1. Genetics 176 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, R., and Dixon, R. (2003). The amino-terminal GAF domain of Azotobacter vinelandii NifA binds 2-oxoglutarate to resist inhibition by NifL under nitrogen-limiting conditions. J. Biol. Chem. 278 28711–28718. [DOI] [PubMed] [Google Scholar]
- Llamas, A., Igeño, M.I., Galván, A., and Fernández, E. (2002). Nitrate signaling on the nitrate reductase gene promoter depends directly on the activity of the nitrate transport systems in Chlamydomonas. Plant J. 30 261–271. [DOI] [PubMed] [Google Scholar]
- Loppes, R., Ohresser, M., Radoux, M., and Matagne, R. (1999). Transcriptional regulation of the Nit1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: Effect of various environmental factors on the expression of a reporter gene under the control of the Nit1 promoter. Plant Mol. Biol. 41 701–711. [DOI] [PubMed] [Google Scholar]
- Loppes, R., and Radoux, M. (2002). Two short regions of the promoter are essential for activation and repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Mol. Genet. Genomics 268 42–48. [DOI] [PubMed] [Google Scholar]
- Lumbreras, V., Stevens, D.R., and Purton, S. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii by an endogenous intron. Plant J. 14 441–447. [Google Scholar]
- Marzluf, G.A. (1997). Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov, I.Y., Galbis-Martinez, M., Jones, M.G., and Caddick, M.X. (2001). Characterization of nitrogen metabolite signaling in Aspergillus via the regulated degradation of AreA mRNA. Mol. Microbiol. 42 269–277. [DOI] [PubMed] [Google Scholar]
- Nagore, D., Sanz, B., Soria, J., Llarena, M., Llama, M.J., Calvete, J.J., and Serra, J.L. (2006). The nitrate/nitrite ABC transporter of Phormidium laminosum: phosphorylation state of NrtA is not involved in its substrate binding activity. Biochim. Biophys. Acta 1760 172–181. [DOI] [PubMed] [Google Scholar]
- Narendja, F., Goller, S.P., Wolschek, M., and Strauss, J. (2002). Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol. Microbiol. 44 573–583. [DOI] [PubMed] [Google Scholar]
- Navarro, F.J., Perdomo, G., Tejera, P., Medina, B., Machin, F., Guillen, R.M., Lancha, A., and Siverio, J.M. (2003). The role of nitrate reductase in the regulation of the nitrate assimilation pathway in the yeast Hansenula polymorpha. FEMS Yeast Res. 4 149–155. [DOI] [PubMed] [Google Scholar]
- Navarro, M.T., Mariscal, V., Macías, M.I., Fernández, E., and Galván, A. (2005). Chlamydomonas reinhardtii strains expressing nitrate reductase under control of the cabII-1 promoter: Isolation of chlorate resistant mutants and identification of new loci for nitrate assimilation. Photosynth. Res. 83 151–161. [DOI] [PubMed] [Google Scholar]
- Ohresser, M., Matagne, R., and Loppes, R. (1997). Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr. Genet. 31 264–271. [DOI] [PubMed] [Google Scholar]
- Owens, B.M., Zhu, Y.-X., Suen, T.-C., Wang, P.-X., Greenblatt, J.F., Goss, P.E., and Hawley, R.G. (2003). Specific homeodomain-DNA interactions are required for HOX11-mediated transformation. Blood 101 4966–4974. [DOI] [PubMed] [Google Scholar]
- Pozuelo, M., Merchán, F., Macías, M.I., Beck, C.F., Galván, A., and Fernández, E. (2000). The negative effect of nitrate on gametogenesis is independent of nitrate assimilation in Chlamydomonas reinhardtii. Planta 211 287–292. [DOI] [PubMed] [Google Scholar]
- Prieto, R., and Fernández, E. (1993). Toxicity and mutagenesis by chlorate are independent of nitrate reductase activity in Chlamydomonas reinhardtii. Mol. Gen. Genet. 237 429–438. [DOI] [PubMed] [Google Scholar]
- Quesada, A., Galván, A., Schnell, R.A., Lefebvre, P.A., and Fernández, E. (1993). Five nitrate assimilation-related loci are clustered in Chlamydomonas reinhardtii. Mol. Gen. Genet. 240 387–394. [DOI] [PubMed] [Google Scholar]
- Quesada, A., Krapp, A., Trueman, L.J., Daniel-Vedele, F., Fernández, E., Forde, B.G., and Caboche, M. (1997). PCR-identification of a Nicotiana plumbaginifolia cDNA homologous to the high-affinity nitrate transporters of the crnA family. Plant Mol. Biol. 34 265–274. [DOI] [PubMed] [Google Scholar]
- Rexach, J., Montero, B., Fernández, E., and Galván, A. (1999). Differential regulation of the high affinity nitrite transport systems III and IV in Chlamydomonas reinhardtii. J. Biol. Chem. 274 27801–27806. [DOI] [PubMed] [Google Scholar]
- Rybalkin, S.D., Rybalkina, I.G., Shimizu-Albergine, M., Tang, X.B., and Beavo, J.A. (2003). PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. EMBO J. 22 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Wieloch, W., and Stougaard, J. (2005). Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60 229–237. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R., González-Fontes, A., Lauerer, M., Muller-Reber, B., Caboche, M., and Stitt, M. (1997. a). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., Lauerer, M., Schulze, E.D., Caboche, M., and Stitt, M. (1997. b). Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J. 11 671–691. [Google Scholar]
- Scheible, W.R., Morcuende, R., Czechowski, T., Fritz, C., Osuna, D., Palacios-Rojas, N., Schindelasch, D., Thimm, O., Udvardi, M.K., and Stitt, M. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, J., Silflow, C.D., and Rosenbaum, J.L. (1984). mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol. Cell. Biol. 4 424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell, R.A., and Lefebvre, P.A. (1993). Isolation of the Chlamydomonas regulatory gene nit2 by transposon tagging. Genetics 194 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, G., Boxer, D.H., and Mendel, R.R. (1997). Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 272 26811–26814. [DOI] [PubMed] [Google Scholar]
- Siverio, J.M. (2002). Assimilation of nitrate by yeasts. FEMS Microbiol. Rev. 26 277–284. [DOI] [PubMed] [Google Scholar]
- Sizova, I., Fuhrmann, M., and Hegemann, P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277 221–229. [DOI] [PubMed] [Google Scholar]
- Vidmar, J.J., Zhuo, D., Siddiqi, M.Y., and Glass, A.D. (2000). Isolation and characterization of HvNRT2.3 and HvNRT2.4, cDNAs encoding high-affinity nitrate transporters from roots of barley. Plant Physiol. 122 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Okamoto, M., Xing, X., and Crawford, N.M. (2003). Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and Forde, B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.