Abstract
The pseudoresponse regulators (PRRs) participate in the progression of the circadian clock in Arabidopsis thaliana. The founding member of the family, TIMING OF CAB EXPRESSION1 (TOC1), is an essential component of the transcriptional network that constitutes the core mechanism of the circadian oscillator. Recent data suggest a role in circadian regulation for all five members of the PRR family; however, the molecular function of TOC1 or any other PRRs remains unknown. In this work, we present evidence for the involvement of PRR3 in the regulation of TOC1 protein stability. PRR3 was temporally coexpressed with TOC1 under different photoperiods, yet its tissue expression was only partially overlapping with that of TOC1, as PRR3 appeared restricted to the vasculature. Decreased expression of PRR3 resulted in reduced levels of TOC1 protein, while overexpression of PRR3 caused an increase in the levels of TOC1, all without affecting the amount of TOC1 transcript. PRR3 was able to bind to TOC1 in yeast and in plants and to perturb TOC1 interaction with ZEITLUPE (ZTL), which targets TOC1 for proteasome-dependent degradation. Together, our results indicate that PRR3 might function to modulate TOC1 stability by hindering ZTL-dependent TOC1 degradation, suggesting the existence of local regulators of clock activity and adding to the growing importance of posttranslational regulation in the design of circadian timing mechanisms in plants.
INTRODUCTION
The Arabidopsis thaliana circadian oscillator is constructed from a network of multiple interlocking transcriptional feedback loops (Salome and McClung, 2004; Mas, 2005; Gardner et al., 2006; Ueda, 2006). At the core of the oscillator, the key components identified to date are the MYB transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LONG HYPOCOTYL (LHY), which are positively regulated by the pseudoresponse regulator (PRR) TIMING OF CAB EXPRESSION1 (TOC1) and the MYB transcription factor LUX ARRYTHMO (LUX) during the night. In turn, CCA1 and LHY act as repressors of TOC1 and LUX expression during the day (Wang et al., 1997; Wang and Tobin, 1998; Strayer, 2000; Alabadi et al., 2002; Mizoguchi et al., 2002; Hazen et al., 2005b). Therefore, the reciprocal regulation of TOC1, LUX, CCA1, and LHY forms a negative feedback loop that is crucial for maintaining circadian rhythmicity.
The regulation of transcription is not the only mechanism that controls clock progression, as modulation of protein stability has emerged as an important aspect of the circadian clockwork (Millar, 2000; Harms et al., 2004; Shu and Hong-Hui, 2004; Liu, 2005; Ivleva et al., 2006; Dardente and Cermakian, 2007; Farre and Kay, 2007; Gallego and Virshup, 2007). ZEITLUPE (ZTL), the founding member of the protein family characterized by a LIGHT, OXYGEN, or VOLTAGE (LOV) domain, an F-box motif, and kelch repeats, mediates the degradation of TOC1 via the formation of a multiprotein Skp/Cullin1/F-box complex that catalyzes the ubiquitylation of proteins destined for proteasomal degradation (Kiyosue and Wada, 2000; Somers et al., 2000, 2004; Schultz et al., 2001; Han et al., 2004; Fukamatsu et al., 2005; Kevei et al., 2006). In the absence of ZTL, the pace of the clock is delayed as a result of increased TOC1 stability (Mas et al., 2003a). ZTL turnover is also proteasome-dependent and is regulated by the clock in a phase-specific manner, the latter being a critical issue in shaping ZTL expression profile, as ZTL mRNA does not oscillate robustly (Somers et al., 2000; Kim et al., 2003a). Similarly, oscillation of LHY protein in light/dark conditions was observed even upon constitutive expression of LHY, since the protein is degraded by the proteasome during the night (Kim et al., 2003a; Song and Carre, 2005).
TOC1 belongs to a family of PRRs along with four other members: PRR9, PRR7, PRR5, and PRR3 (Matsushika et al., 2000). Expression of the PRRs is regulated by the circadian clock, and their transcripts are partially overlapping but peak at different times of the day: PRR9 mRNA in the morning, PRR7 and PRR5 mRNA at ∼8 h after dawn, and PRR3 and TOC1 mRNA in the evening (Matsushika et al., 2000). These proteins feature a pseudo receiver domain at the N terminus and a CCT (for CONSTANS, CONSTANS-LIKE, and TOC1) domain at the C terminus (Strayer, 2000). The pseudo receiver domain shows high similarity to receiver domains of two-component response regulators but lacks the key Asp residue that accepts a phosphoryl group to modulate the activity of the protein. CCT domain–containing proteins have been implicated in many aspects of plant physiology and are thought to be involved in protein–protein interaction (Kurup et al., 2000; Wenkel et al., 2006).
Mutations in TOC1 are known to shorten the period of many clock-controlled processes by severely affecting the expression of core clock components to the point of causing arrhythmia under certain conditions (Somers et al., 1998; Strayer, 2000; Alabadi, 2001; Mas et al., 2003a). This last effect was also observed as a result of increased levels of TOC1 under all conditions tested, evidence that clock progression is sensitive not only to the level but also to the cyclic profile of TOC1 expression (Mas et al., 2003a).
In addition to TOC1, loss- and gain-of-function studies have suggested a role in the circadian clockwork for most of the PRRs. PRR single mutants were found to cause a mild effect on period length, whereas some double and triple mutant combinations result in a severe clock phenotype: the double mutant prr9 prr7 displayed a longer period than either of the single mutants, and the prr5 prr7 double mutant phenotype was reported to exhibit very short to arrhythmic oscillations that became further compromised in the prr9 prr7 prr5 triple mutant (Farre et al., 2005; Fujimori et al., 2005; Nakamichi et al., 2005a, 2005b; Salome and McClung, 2005). Interestingly, combining the clock phenotype of the short-period prr5 mutation with the long-period prr9 mutation resulted in a wild-type period (Eriksson et al., 2003). Hence, despite their molecular similarity, not all PRRs are functionally equivalent, although some may have overlapping functions.
In this work, we focus on the function of PRR3, whose temporal expression largely overlaps with that of TOC1, and present evidence for the involvement of PRR3 in the modulation of TOC1 stability through interference with the binding of ZTL to TOC1. This provides the first insight into a molecular function of one of the PRRs and indicates additional fine-tuning of the level of regulation, acting on the stability of the core clock component TOC1. In addition, we show that PRR3 expression is detected mainly in the vasculature of the leaves, where altered expression of this gene appears to have a strong effect on clock-regulated genes that are specifically expressed in the vascular tissues. This observation suggests the existence of mechanisms that fine-tune the plant clock in different tissues in plants, similar to what is well known in animal systems.
RESULTS
PRR3 Is Coregulated with TOC1 under Different Photoperiods, but Their Expression Pattern Is Only Partially Overlapping
In order to gain insight into the time of day when PRR3 functions in the clock, we analyzed PRR3 temporal expression by diurnal time course array data that were obtained from Diurnal (http://diurnal.cgrb.oregonstate.edu). PRR3 transcription exhibits a circadian profile in constant white light (Matsushika et al., 2000), and we observed that PRR3 transcript also peaked at dusk under long-day and short-day conditions. Interestingly, the PRR3 circadian profile was remarkably similar to that of TOC1 even under different photoperiods, indicating that the temporal expression of these genes was highly correlated (Figures 1A and 1B).
Figure 1.
The Temporal Expression of the Core Clock Components TOC1 and PRR3 Is Coregulated, and the PRR3 Vasculature Expression Pattern Partially Overlaps with That of TOC1.
(A) and (B) Diurnal time course array data from Diurnal (http://diurnal.cgrb.oregonstate.edu). Relative expression of PRR3 (open circles) and TOC1 (open squares) under long days (LD; 16-h light/8-h dark cycles) (A) and short days (SD; 8 h-light/16-h dark cycles) (B).
(C) and (D) Expression pattern of PRR3 and TOC1. Two-week-old plants were entrained in photocycles (12 h of light/12 h of dark), and GUS activity was detected for PRR3:GUS (C) and TOC1:GUS (D). Bars = 1 mm.
To further explore the relationship between TOC1 and PRR3, we also analyzed the spatial expression patterns of PRR3 and TOC1 using promoter-driven β-glucuronidase (GUS) gene constructs (PPRR3:GUS and PTOC1:GUS). Interestingly, PRR3:GUS activity was observed mainly in the vascular tissues of cotyledons and leaves (Figure 1C). No GUS activity was detected in the roots. PTOC1:GUS was broadly expressed, and TOC1 promoter activity was detected in cotyledons, whole leaves, and roots (Figure 1D).
Together, these data indicate that PRR3 and TOC1 expression are temporally correlated but spatially overlapping only in the vasculature.
Alteration of PRR3 Gene Expression Affects Clock Progression in the Vasculature
Next, we investigated whether a clock phenotype would result from a decrease in PRR3 levels by testing a prr3 mutant and PRR3 RNA interference (RNAi) plants using the well-characterized bioluminescent circadian reporters CCR2:LUC and CAB2:LUC (Millar et al.,1995; Strayer, 2000). PRR3RNAi plants were constructed by cloning a fragment of 480 bp that is unique to the PRR3 cDNA in the sense and antisense orientations into an RNAi vector and transforming the construct into the CCR2:LUC background. Real-time quantitative RT-PCR (RT-qPCR) analyses confirmed a strong reduction in PRR3 mRNA in several lines (down to 12% of wild-type levels; see Supplemental Figure 1 online). Bioluminescent analyses of CCR2 rhythms revealed that both PRR3RNAi lines had a shorter period (PRR3RNAi plant 25, 24.3 ± 0.1 h; PRR3RNAi plant 26, 23.8 ± 0.1 h; PRR3RNAi plant 58, 24.4 ± 0.1 h) than the wild type (24.9 ± 0.1 h) (P < 0.01) in free-running conditions (Figure 2A). In addition, prr3-1, a T-DNA insertion line from the Salk collection (see Methods), was transformed with the same reporter that was observed to cause a similar effect (prr3-1, 23.8 ± 0.0 h; wild type, 24.6 ± 0.1 h) (P < 0.01) (see Supplemental Figure 2 online). Similar results were obtained when transforming the PRR3RNAi construct into the CAB2:LUC background (PRR3RNAi plant 14, 23.0 ± 0.2 h; PRR3RNAi plant 19, 23.3 ± 0.6 h; wild type, 23.5 ± 0.2 h) (P < 0.05) (Figure 2B).
Figure 2.
Effects of Decreased PRR3 Levels on Different Circadian Reporters.
(A) CCR2:LUC bioluminescence rhythms from the wild type and PRR3RNAi in constant light (left), relative amplitude error (Rel Amp) graph (center), and bar graph of the period length (right).
(B) CAB2:LUC bioluminescence rhythms from the wild type and PRR3RNAi in constant light (left), relative amplitude error graph (center), and bar graph of the period length (right).
(C) PRR9:LUC bioluminescence rhythms from the wild type and PRR3RNAi in constant light (left), relative amplitude error graph (center), and bar graph of the period length (right).
Traces represent an average of 20 to 40 seedlings for CCR2:LUC and 6 to 12 seedlings for CAB2:LUC and PRR9:LUC. The experiments were repeated three times with similar results. Values significantly different from the wild type are indicated (* P < 0.01, ** P < 0.05).
(D) to (F) Expression profiles of CCA1 (D), LHY (E), and CDF1 (F) analyzed by RT-qPCR. Seedlings were entrained in 12 h of light and 12 h of dark for 7 d and then transferred in constant light. After 2 d, samples were harvested every 4 h at the indicated Zeitgeber times (ZT). Open symbols, wild type; closed symbols, PRR3RNAi lines 25 and 58.
This modest effect on period length suggests a role for PRR3 in the plant clock; however, the confined spatial expression pattern of PRR3 led us to consider whether the ubiquitous expression of CCR2 and the broad expression of CAB2 might not accurately portray the circadian function of PRR3 (Carpenter et al., 1994; Thain et al., 2002). To test this possibility, we constructed a PRR9:LUC promoter fusion as a circadian reporter, as we could observe an intense activity of the PRR9 promoter in the vasculature of PRR9:GUS plants (see Supplemental Figure 3 online).
PRR3RNAi was transformed into the PRR9:LUC background, and bioluminescence analyses exhibited a shortening in period length significantly greater than that observed on CCR2 rhythms (PRR3RNAi plant 39, 22.9 ± 0.1 h; PRR3RNAi plant 44, 23.1 ± 0.1 h; PRR3RNAi plant 56, 23.2 ± 0.2 h; wild type, 24.6 ± 0.1 h) (P < 0.01) (Figure 2C).
In addition, we examined the expression profile of CYCLING DOF FACTOR1 (CDF1) in wild-type and PRR3RNAi plants. CDF1 expression was previously shown to be controlled by the circadian clock and restricted to the vasculature (Imaizumi et al., 2005). After 2 d in constant conditions, CDF1 mRNA peaked considerably earlier (6 to 8 h) in PRR3RNAi lines than in the wild-type plants, while the peak of expression of CCA1 and LHY, two ubiquitously expressed morning genes (J. Pruneda-Paz and S.A. Kay, unpublished results), showed only a modest advance (Figures 2D to 2F). These results indicate that decreased levels of PRR3 had a stronger effect on the expression of clock-regulated genes that are specifically expressed in the vascular tissues.
These phenotypes support the notion that PRR3 participates in the regulation of the Arabidopsis circadian clock and might play an important role in clock progression in the vascular tissue.
TOC1 Protein Accumulation Is Reduced in the prr3-1 Mutant and PRR3RNAi Lines and Increased in P35S:PRR3 Lines
A decrease in PRR3 expression affects the pace of the clock in a similar manner as a decrease in TOC1 expression. These observations led us to the following hypotheses: (1) PRR3 and TOC1 have overlapping functions; or (2) PRR3 might influence the stability or function of TOC1.
To test the first hypothesis, we constructed a toc1 prr3 double mutant and also transformed the PRR3RNAi construct into the toc1 mutant background. We reasoned that if TOC1 and PRR3 functions are partially redundant, the double mutant should have a stronger phenotype than either single mutant. The clock phenotypes of the toc1 prr3 and toc1 PRR3RNAi double mutants were indistinguishable from that of the toc1 mutant alone, indicating that TOC1 was epistatic to PRR3 (see Supplemental Figures 4A and 4B online). In order to test the possibility that PRR3 might regulate TOC1 stability, we set out to monitor TOC1 protein and mRNA levels in prr3-1 mutant and PRR3RNAi lines. Transgenic plants expressing TOC1 fused to yellow fluorescent protein (YFP) under the control of theTOC1 native promoter (designated TOC1 Mini Gene [TMG]) were crossed to the prr3-1 mutant or transformed with the PRR3RNAi construct, and the amount of TOC protein was detected on protein gel blots.
In addition, transgenic plants expressing a transcriptional fusion of the PRR3 coding region with the strong, constitutive cauliflower mosaic virus 35S promoter were generated to assess the effect of increased levels of PRR3 on TOC1. The P35S:PRR3 construct was also transformed into the CCR2:LUC reporter background, in which higher PRR3 expression caused a 1- to 1.5-h increase in the free-running period of CCR2:LUC (see Supplemental Figure 2B online).
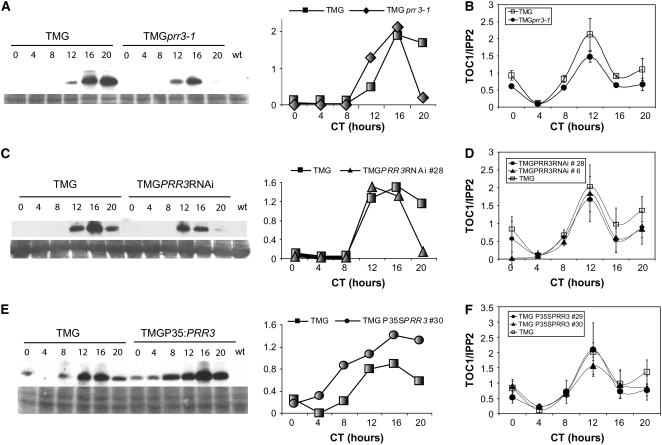
Time-course analyses revealed that the pattern of TOC1 protein expression was altered in all backgrounds. In prr3-1, TOC1 levels were reduced significantly at the end of the dark period (at circadian time 20), and a similar effect was observed in the PRR3RNAi background (Figures 3A and 3C). Increased levels of PRR3 caused a dramatic change in the TOC1 expression profile, as TOC1 protein was detected at all time points in the PRR3-overexpressing background (Figure 3E). Interestingly, rhythmic oscillation of TOC1 protein was still observed in the P35S:PRR3 plants.
Figure 3.
TOC1 Protein Levels Are Affected by Increased or Decreased Levels of PRR3 Independently of Transcription.
Immunodetection and quantification of TOC1-YFP protein using anti-GFP antibody in TMG and in prr3-1 TMG (A), PRR3RNAi TMG (line 6) (C), and P35S:PRR3 TMG (line 30) (E). The experiments were repeated three times. TOC1 mRNA levels analyzed with RT-qPCR in TMGprr3-1 (B), PRR3RNAi TMG (lines 6 and 28) (D), and P35S:PRR3 TMG (lines 29 and 30) (F). Seedlings of each genotype were entrained in 12-h light/12-h dark cycles and harvested every 4 h after dawn for 24 h before undergoing protein extraction at the indicated circadian times (CT). Immunodetection of TOC1-YFP protein was performed using anti-GFP antibody. The experiments were repeated three times.
In order to establish whether the changes in TOC1 protein profiles in the prr3 mutant, PRR3RNAi, and the PRR3-overexpressor might be due to changes in TOC1 mRNA levels, RT-qPCR analyses were performed to monitor TOC1 transcript in all of the genotypes. In photocycles (12 h of light/12 h of dark), TOC1 mRNA expression maintained a rhythmic pattern with similar levels in all lines tested, indicating that PRR3 does not affect the TOC1 transcription profile (Figures 3B, 3D, and 3F). Together, these data indicate that disruption or decreased levels of PRR3 resulted in lower TOC1 protein, while increased levels of PRR3 had the opposite effect of making TOC1 protein more abundant, all without affecting TOC1 transcription.
As we have shown above, PRR3 is involved in a molecular mechanism that affects the levels of TOC1 protein. As PRR3 expression is restricted to the vasculature, we set out to examine whether the abundance of TOC1 protein would be different in the tissues in which TOC1 and PRR3 are coexpressed.
In order to visualize TOC1 protein, we fused TOC1 to the GUS gene under the control of the TOC1 native promoter (TMG:GUS). TOC1 protein clearly localized to the nucleus, and it was detected in all of the tissues in which the TOC1 gene was found to be expressed (Figures 1 and 4). Yet, the GUS activity appeared to be more intense in the veins of the leaves compared with the epidermis (Figure 4). These data indicate that the levels of TOC1 protein might be higher in the vasculature and strengthen the hypothesis of a local increase of TOC1 stability, possibly mediated by PRR3.
Figure 4.
TOC1 Protein Is More Abundant in the Vasculature of the Leaves.
Expression patterns of TOC1 protein. Two-week-old plants were entrained in photocycles (12 h of light/12 h of dark), and GUS activity was detected for TMG:GUS. The localization of TOC1 to the nuclei is indicated by arrowheads.
(A) True leaf (left), and enlarged detail of the area of vasculature (right) enclosed in the dotted rectangle. Bar = 1 mm.
(B) Close-up of the epidermis of a true leaf (left), and enlarged detail (right) of the area enclosed in the dotted rectangle. Bar = 0.1 mm.
PRR3 Binds TOC1 but Not ZTL and Interferes with the Binding of TOC1 to ZTL in Yeast
To investigate the molecular mechanism underlying the effect of decreased and increased levels of PRR3 on TOC1, we asked whether PRR3 was able to interact physically with TOC1. PRR3 fused to the DNA binding domain of GAL4 (bait) was coexpressed with TOC1 fused to the activation domain of GAL4 (prey) in yeast, and the interaction between these proteins was assayed by monitoring β-galactosidase (β-gal) activity. Strong β-gal activity in yeast colonies that expressed both TOC1 and PRR3 indicated a physical interaction between these proteins (no β-gal activity was detected in the control) (Figure 5A). The interaction between TOC1 and PRR3 was also verified in vivo by transient expression of PRR3 and TOC1 in tobacco (Nicotiana bethamiana) leaves. Tagged versions of both PRR3 and TOC1 were coexpressed in tobacco, and TOC1-GFP (for green fluorescent protein) was immunoprecipitated with an anti-GFP antibody. Subsequent detection of hemagglutinin (HA)-PRR3 with an anti-HA antibody revealed a protein of ∼70 kD (consistent with the predicted size of the PRR3 protein) only in the sample in which both PRR3 and TOC1 were present, indicating that PRR3 is able to bind TOC1 in planta (Figure 5B).
Figure 5.
PRR3 Interacts with TOC1 in Yeast and in Plants and Is Able to Disrupt TOC1 Binding to ZTL.
(A) Yeast two-hybrid assay. β-Gal activity reveals the interaction of full-length PRR3-BD and TOC1-AD. The combination of PRR3 and the empty vector expressing the activation domain of GAL4 was used as a control.
(B) Protein gel blot analysis of TOC1 immunoprecipitation with anti-GFP antibody and detection of PRR3 with anti-HA antibody. Agrobacterium tumefaciens strains containing P35S:HAPRR3, P35S:TOC1-GFP, and P35S:HA19 were infiltrated into tobacco leaves, and TOC1 was immunoprecipitated using anti-GFP antibody. The asterisk indicates an unspecific band.
(C) Yeast three-hybrid assay. Yeast liquid cultures coexpressing TOC1-BD, ZTL-AD, and free PRR3 were grown overnight in the presence (black bars) or absence (gray bars) of Met. The O-nitrophenyl-β-d-galactopyraniside assay was performed on colonies from three distinct transformation events, and the results are expressed in β-gal units. The error bars represent sd.
As we observed that PRR3 binds TOC1 in vitro and in vivo and that increased levels of PRR3 resulted in higher levels of TOC1 protein, we speculated that the binding of PRR3 to TOC1 might prevent the formation of a TOC1-ZTL complex that leads to TOC1 degradation (Mas et al., 2003b).
In order to test this hypothesis, we performed a yeast three-hybrid assay. TOC1 was fused to the DNA binding domain of GAL4 (bait) and coexpressed with ZTL fused to the activation domain of GAL4 (prey) in yeast. The bridge protein PRR3 was then conditionally expressed under the control of the MET25 promoter, whose activity is repressed in the presence of Met, and β-gal activity was monitored by a liquid culture assay (Tirode et al., 1997). In the absence of PRR3, TOC1 and ZTL were able to interact, while in the presence of PRR3, the formation of the TOC1-ZTL complex was disturbed, as indicated by a 50% decrease in β-gal activity (Figure 5C).
In addition, we coexpressed PRR3 fused to the DNA binding domain of GAL4 with the full-length ZTL and the LOV domain fused to the activation domain of GAL4 in yeast. No β-gal activity was detected in yeast colonies that expressed either ZTL or the LOV domain and PRR3, indicating that PRR3 and ZTL are not likely to interact physically in yeast (Figure 5A).
In summary, these data indicate that PRR3 interacts physically with TOC1 and acts as a competitor to modulate the extent of the interaction between TOC1 and ZTL.
Increased Levels of PRR3 Partially Rescue the TOC1RNAi Clock Phenotype
The data presented thus far suggest that PRR3 regulates TOC1 stability by physically binding to TOC1 and preventing ZTL-dependent degradation. This model presents a number of testable predictions. First, PRR3 overexpression should ameliorate the short-period phenotype observed upon the reduction of TOC1 protein levels but, critically, should have no effect on the period length in a toc1 null allele. Second, increased levels of PRR3 should weaken the strong clock phenotype of a ZTL-overexpressor line.
In order to test the first prediction (i.e., that elevated levels of PRR3 could partially rescue the clock phenotype due to decreased levels of TOC1 by stabilizing TOC1 protein), a TOC1RNAi line (previously designated TOC1RNAi-24 in Mas et al., 2003a) was transformed with the P35S:PRR3 construct. The TOC1RNAi P35S:PRR3 lines displayed an increase in the period length of CAB2:LUC (TOC1RNAiP35S:PRR3 line 52, 21.1 ± 0.2 h; line 77, 21.0 ± 0.2 h) compared with the parental TOC1RNAi line (19.5 ± 0.1 h) (P < 0.01) (Figure 6A). Hence, increased levels of PRR3 caused a slight but reproducible increase of the TOC1RNAi period toward that of the wild type. Importantly, no change in period was seen after introducing P35S:PRR3 in the toc1-4 mutant background (toc1-4P35S:PRR3 line 68, 19.3 ± 0.4 h; line 69, 19.6 ± 0.2 h; line 71, 19.7 ± 0.3 h; line 77, 19.2 ± 0.3 h; toc1-4, 19.5 ± 0.1 h) (P ≥ 0.5). The toc1-4 allele is a point mutation that creates a premature stop close to the N terminus of the protein, resulting in a null mutation (Hazen et al., 2005a) (Figure 6B). Together, these results support the notion that PRR3 stabilizes TOC1 protein in vivo.
Figure 6.
Increased Levels of PRR3 Partially Rescue the TOC1RNAi Clock Phenotype but Not a TOC1 Null (toc1-4) Phenotype.
(A) CAB2:LUC bioluminescence rhythms for the wild type, TOC1RNAi line 24, and P35S:PRR3 TOC1RNAi T2 lines (left) and relative amplitude (Rel Amp) graph (right).
(B) CAB2:LUC bioluminescence rhythms for the wild type, toc1-4, and toc1-4 P35S:PRR3 (the trace for toc1-4 P35S:PRR3 represents the average of four independently transformed lines [lines 68, 69, 71, and 77]) (left) and relative amplitude graph (right).
The traces represent an average of 12 seedlings. The experiments were repeated three times with similar results. The error bars were omitted for clarity.
Increased Levels of PRR3 Rescue the Severe Clock Phenotype Due to Constitutive Expression of ZTL
We demonstrated that PRR3 is able to compete with ZTL for TOC1 binding in yeast. Next, we set out to investigate whether PRR3 was able to interfere with ZTL action on TOC1 stability in vivo. It was reported previously that increased levels of ZTL shortened the period length in a dosage-dependent manner and caused arrhythmicity at the highest doses (Somers et al., 2004). As this phenotype is likely due to the extreme instability of the TOC1 protein in such a background, increased levels of PRR3 might be able to partially ameliorate the ZTL mutant phenotype by binding to TOC1 and preventing ZTL from interacting with TOC1.
To generate a ZTL-overexpressor (P35S:ZTL), ZTL coding sequence was expressed under the control of the 35S promoter in a CAB2:LUC background and fast Fourier transform nonlinear least square (FFT-NLLS) analyses confirmed a severe loss of circadian rhythmicity, since no rhythm could be fitted for CAB2:LUC traces from most P35S:ZTL seedlings and the few fitted rhythms exhibited a relative amplitude error > 0.6 (see Methods) (Figure 7). These plants were transformed with the P35S:PRR3 construct, and several independent transgenic lines recovered rhythmic oscillation of the CAB2:LUC reporter, as exemplified by P35S:ZTLP35S:PRR3 lines 89 and 99: 96.7% of P35S:ZTLP35S:PRR3 line 89 seedlings (n = 30) and 90% of P35S:ZTLP35S:PRR3 line 99 seedlings (n = 30) exhibited relative amplitude errors < 0.6 when analyzed using FFT-NLLS (Figure 7). In addition, these P35S:ZTLP35S:PRR3 lines displayed a period of CAB2 that was close to that of the wild type (wild type, 23.06 ± 0.09 h; P35S:ZTLP35S:PRR3 line 89, 22.70 ± 0.1 h; P35S:ZTLP35S:PRR3 line 99, 22.36 ± 0.12 h). This indicates that PRR3 protein counteracts ZTL action in vivo. Together, our results suggest that PRR3 might interfere with the formation of the ZTL-TOC1 complex through binding to TOC1, resulting in the stabilization of TOC1.
Figure 7.
Increased Levels of PRR3 Rescue the P35S:ZTL Clock Phenotype.
CAB2:LUC bioluminescence rhythms for the wild type, P35S:ZTL, and two lines of P35S:PRR3 P35S:ZTL (lines 89 and 99) and relative amplitude (Rel Amp) graph (bottom). Traces for the wild type and P35S:ZTL represent an average of 12 seedlings, and traces for P35S:ZTL P35S:PRR3 represent an average of 29 seedlings. The experiments were repeated three times with similar results. The error bars represent se.
DISCUSSION
PRR3 Plays a Role in Clock Progression in the Vasculature
We started our analysis by examining the temporal and spatial patterns of PRR3 expression. Surprisingly, the PRR3 promoter is active mainly in the vascular tissue of cotyledons and leaves, although we cannot exclude the possibility that PRR3 might be expressed below detection levels in other tissues. Such a restricted expression pattern might explain the apparently modest clock phenotype observed in mutants with decreased or disrupted expression of PRR3, as most of the commonly used clock assays can only reflect global alterations of the circadian system. By combining tissue-specific bioluminescence and transcriptional assays, we were able to show that PRR3 plays an important role in the progression of the circadian clock in the vasculature, as decreased levels of PRR3 caused an evident shortening of the period of the clock-controlled genes that are expressed mainly in the same tissue. Together with previous observations (Thain et al., 2000, 2002), our findings suggest the existence of a tissue-specific circadian clock mechanism. Moreover, one of the most important clock output pathways for time measurement in photoperiodic responses has been shown to act through tissue-specific factors (An et al., 2004; Imaizumi and Kay, 2006). To date, no other factors have been implicated in a local clock machinery, and analyzing PRR3 function might represent the entry point for exploring the properties of the circadian clockwork that differ among plant tissues.
Tissue-Specific Regulation of TOC1: An Additional Level of Complexity
Under different photoperiods, PRR3 mRNA peaks at dusk. A similar temporal expression profile was observed for TOC1, raising the question of whether PRR3 might have overlapping functions with TOC1. Instead, we found that PRR3 regulates the levels of TOC1 protein and that TOC1 abundance appears to be increased in the vasculature, where PRR3 and TOC1 are coexpressed.
TOC1 abundance was reported previously to be under the control of a diurnal mechanism, as the protein was found to be unstable in the dark and stable in the light (Mas et al., 2003b). In addition, robust cycling of TOC1 protein in light/dark conditions was observed even in the transgenic plants constitutively and ubiquitously expressing TOC1 mRNA (A. Para and S.A. Kay, unpublished results). Our data suggest that the regulation of TOC1 protein levels might have not only a temporal component but also a spatial component. This finding expands and adds complexity to the molecular mechanisms that underlie the regulation of a critical core clock element.
PRR3 Competes with ZTL for Binding to TOC1
A crucial role in the regulation of TOC1 stability is played by ZTL, which is able to mediate TOC1 degradation through the proteasome pathway (Mas et al., 2003b). TOC1 and ZTL proteins show similar expression profiles, but TOC1 levels decrease only at the end of the dark period, suggesting that additional factors and/or posttranslational modifications might be required for targeting TOC1 to degradation (Kim et al., 2003b). PRR3 might bind TOC1 at the transition from light to dark and into the night, coincident with the rising ZTL levels, therefore modulating the interaction between TOC1 and ZTL. However, PRR3 does not bind to ZTL in yeast, indicating that the binding of PRR3 to TOC1 could mask the protein–protein interaction domains or the docking sites for ZTL, resulting in an increase of TOC1 stability. Thus, both the change in TOC1 protein stability in relation to the levels of PRR3 and the significant weakening of the ZTL-overexpressor phenotype due to the constitutive expression of PRR3 hint at an additional molecular mechanism that participates in the fine-tuning of the Arabidopsis circadian clock.
Functional Implications for the PRR3-TOC1 Complex
In the presence of low levels of PRR3, the TOC1 protein profile was affected mainly in the night, while elevated levels of PRR3 resulted in an increase in the accumulation of TOC1 protein.
As shown previously, TOC1 is likely to be an indirect regulator of CCA1 and LHY expression (Alabadi, 2001). No change in the expression level/phase of these two genes was observed when the levels of PRR3 were altered (data not shown). However, we have shown that PRR3 is expressed mainly in the vasculature; therefore, the effect on the TOC1 protein might be restricted to this tissue. Thus, it is conceivable that, if a reduction in TOC1 protein levels would result in a change in CCA1 and LHY levels and/or expression profiles, such an alteration is likely to occur only in the tissues in which TOC1 and PRR3 are coexpressed. As the commonly used transcriptional profiling methods can only detect global alterations of gene expression, a test of this idea will have to await the development of tools that will allow us to study the levels of specific factors in the vascular tissue.
In the case of increased levels of PRR3, either the extent of the increase in TOC1 protein stability was not enough to detect an effect or TOC1 might need other factors whose levels or activity are not affected by PRR3. Similarly, no increase in CCA1 levels was observed in the ztl mutant background, in which TOC1 is more stable (Mas et al., 2003a; Somers et al., 2004). Then again, some TOC1 targets might be more sensitive to the levels of the proteins than others, or the contribution of TOC1 to CCA1 mRNA levels might be buffered by other components of the network of interlocking loops (Ueda, 2006).
It is also possible that the PRR3-TOC1 complex has other functional implications. It has been reported that ZTL can bind CRY1 and phyB in vitro (Jarillo, 2001). This observation led to the hypothesis that ZTL might participate in a gating mechanism, since it could convey light signals to the central oscillator through the interaction with these photoreceptors, and the cycling changes in ZTL protein abundance would trigger the degradation of ZTL targets in a circadian manner. Overexpression of ZTL and PRR3 caused hyposensitivity to red light during deetiolation (Murakami et al., 2004; Somers et al., 2004). Furthermore, mutations in ZTL and overexpression of PRR3 delayed flowering under long days but not short days, indicating that ZTL and PRR3 might be involved in daylength responses (Somers et al., 2000; Murakami et al., 2004). The circadian clock plays a major role in the measurement of daylength, a key factor in the determination of flowering time (Imaizumi and Kay, 2006; Hotta et al., 2007), and the vasculature of the leaves has been shown to be where photoperiodic cues are perceived and integrated to initiate flowering (Takada and Goto, 2003; Endo et al., 2005, 2007). Therefore, the modulation of TOC1 binding to ZTL through the PRR3-TOC1 complex might have an important role in specific clock output, such as flowering and/or the modulation of light perception through TOC1 stability.
Together, the data presented here contribute to our current understanding of the circadian clockwork by proposing an unexpected molecular function for a member of a known protein family involved in the progression of the Arabidopsis circadian clock.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown as reported previously (Mas et al., 2003b). Ecotype Col-0 harboring the CCR2:LUC reporter was transformed with either the PRR3RNAi or P35S:PRR3 construct. The prr3-1 (SALK_090261) mutant was obtained from the Salk collection through the ABRC. TOC1RNAi-24 was described previously, as were the toc1-2 and toc1-4 alleles (Strayer, 2000; Mas et al., 2003a; Hazen et al., 2005a). TMG construction was reported elsewhere (Mas et al., 2003b).
Plasmid Construction
PPRR3:GUS and PPRR9:GUS constructs were created by cloning the genomic sequences −1.8kb/+123bp and −1.3kb/+81bp, respectively, into pMDC63 (Curtis and Grossniklaus, 2003). For TOC1:GUS, the TOC1 genomic sequence −1.7kb/+27bp was cloned into pBI101 (Clontech). The PRR9:LUC construct was obtained by cloning the genomic sequence −1.3kb/−1bp into pZPXomegaLuc (Michael and McClung, 2003).
To make the PRR3RNAi construct, 480 bp was amplified from a cDNA pool using primers PRR3RNAi F (5′-AACGGGAGTGGAACTCAGAGT-3′) and PRR3RNAi R (5′-caccAGCCTTCTTAGCAGAAGTAGC-3′) and cloned into the pH7WG2 binary vector (Karimi et al., 2002). The HA-PRR3 coding sequence was amplified with the forward primer 5′-caccATGTACCCATACGATGTTCCAGATTACGCTATGTGTTTTAATAACATTGA-3′ and the reverse primer 5′-CAATTGTCTTCACTTCCTG-3′ (uppercase letters indicate genomic sequence, lowercase letters indicate the sequence required for Gateway cloning, and the underlined sequence indicates the HA sequence) and cloned into the pH7WG2 binary vector to make the P35S:PRR3 construct (Karimi et al., 2002). The forward primer contains the sequence for the HA tag.
To overexpress ZTL, the ZTL coding sequence was cloned into pRTL2 vector, and the expression cassette was then cloned into pBI221 binary vector (Hajdukiewicz et al., 1994).
All Arabidopsis transformations were achieved by Agrobacterium tumefaciens infiltration using the floral dip method. For tobacco (Nicotiana bethamiana) infiltration, TOC1 genomic sequence (4500 bp downstream of the ATG) was cloned into pMDC83 and transformed into Agrobacterium.
GUS Staining
Seedlings were grown for 2 weeks in photocycles and then stained as described by Imaizumi et al. (2005). Photographs were taken with a Diagnostic Instruments SPOT-RT Slider camera mounted on a Nikon SMZ800 stereoscope equipped with Dyna Light 150 fiber optics.
Bioluminescence Analyses
Seeds were germinated on selective medium and entrained in 12-h light/12-h dark photocycles. One-week-old seedlings were transferred to plates without selection and placed in continuous light (60 μmol·m−2·s−1). Bioluminescence was detected and the data were analyzed as described (Millar et al., 1995; Somers et al., 1998). FFT-NLLS was used to estimate period length and relative amplitude error to assess rhythm robustness. The relative amplitude error value of 0.6 was used as a cutoff to identify rhythmic traces within the circadian range
Yeast Two- and Three-Hybrid Assays
For yeast two-hybrid assay, BP reactions via the Gateway LR system (Invitrogen) were performed to clone PRR3 coding sequence into the pASGW vector to fuse it with the GAL4 DNA binding domain and to clone full-length TOC1, full-length ZTL, and the ZTL LOV domain into pACTGW vector to fuse them with GAL4 activation domain (Nakayama et al., 2002). For yeast three-hybrid assay, the PCR product NotI-PRR3cDNA-BamHII was cloned into MCSII in pBridge (Clontech) downstream of the MET25 promoter and XmaI-TOC1cDNA-SalI was cloned into MCSI in the same vector. As prey, ZTL coding sequence was cloned into pACTGW vector. These construct were used to transform the suitable yeast strains, and the selection and β-gal assays were conducted following the manufacturer's recommendations (Clontech Yeast Protocols Handbook).
Tobacco Infiltration
Agrobacterium containing P35S:TOC1, P35S:HAPRR3, and P35S:HA19 was grown overnight and resuspended in infiltration buffer (10 mM MgCl2 and 0.15 μM acetosyringone) to reach a concentration of 0.8 OD600. This suspension was pressure-infiltrated into tobacco leaves using a syringe. After 2 d, the tissue was infiltrated with 0.50 μM MG132 and harvested after 4 h.
Protein Gel Blots and Coimmunoprecipitation
Proteins were extracted from 0.2 g of plant tissue and ground in 0.5 mL of grinding buffer (50 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, 2 mM DTT, 5 mM EDTA, 0.2% polyvinylpolypyrrolidone, 50 μM MG132, and protease inhibitor cocktail [Roche]). Protein concentration was determined using the Bradford method (Bio-Rad), and ∼60 to 80 μg of total protein was loaded per lane to separate them on a 10% acrylamide/bisacrylamide gel. Homogeneous protein transference to nitrocellulose membranes was confirmed by Ponceau red staining. Horseradish peroxidase–conjugated anti-GFP antibody (Molecular Probes) was used to detect TOC1-YFP.
For coimmunoprecipitation, proteins were extracted in IP buffer (50 mM Na-P, pH 7.4, 100 mM NaCl, 10% glycerol, 0.1% Triton X-100, 5 mM DTT, 5 mM EDTA, 0.5% polyvinylpolypyrrolidone, 50 μM MG132, phosphatase inhibitor cocktail [Sigma-Aldrich], and protease inhibitor cocktail [Roche]), and the immunoprecipitation was performed at 4°C using Dynabeads Protein G (Invitrogen) coated with anti-GFP antibody (Invitrogen). PRR3 detection was performed using anti-HA antibody 3F10 (Roche), and TOC1 was detected using horseradish peroxidase–conjugated anti-GFP antibody (Molecular Probes).
RNA Extraction and RT-qPCR
Total RNA from 10-d-old seedlings was isolated with the RNeasy plant mini kit (Qiagen). First-strand cDNA was produced from 1.5 μg of total RNA using the iScript Select cDNA synthesis kit (Bio-Rad). The product was diluted 1:5 with water, and 2 μL was used as template for RT-qPCR amplification on a MyiQ iCylcer (Bio-Rad) with the following primers and probes: for TOC1, forward, 5′-TCTTCGCAGAATCCCTGTGAT-3′; reverse, 5′-GCTGCACCTAGCTTCAAGCA-3′; probe, 5′-ATGATGTCGAGGCAAGACGAAGTCCC-3′; for PRR3, forward, 5′-TCCCGTTATCACTCCCACCATCTTGG-3′; reverse, 5′-GTGGGAGTAGTGGTGGTTTGAGTA-3′; probe, 5′-TTTGTCCAAGAACTCTGAGTTCCA-3′. Primers and probes for CCA1 and CDF1 were described previously (Farre et al., 2005; Imaizumi et al., 2005). Each RT-qPCR experiment was repeated three times.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At5g60100 (PRR3), At5g61380 (TOC1), At5g57360 (ZTL), At5g62430 (CDF1), At2g46830 (CCA1), and At1g01060 (LHY).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PRR3 mRNA Levels in PRR3RNAi and P35S:PRR3 Lines.
Supplemental Figure 2. Effects of the Disruption of the PRR3 Gene in prr3-1 and of the Increased Expression of PRR3 on CCR2:LUC.
Supplemental Figure 3. Expression Pattern of PRR9:GUS.
Supplemental Figure 4. TOC1 Is Epistatic to PRR3.
Supplementary Material
Acknowledgments
We thank Samuel Hazen, Ghislain Breton, Dimitri Nusinow, and Marco Gallio for critical reading of the manuscript and all members of the Kay laboratory for useful discussions. We are also grateful to Aura de Schopke and Kathryn Spencer for technical assistance. This work was supported by National Institutes of Health Grants GM-56006 and GM-67837 (to S.A.K.). T.I. was supported by National Institutes of Health Grant GM-079712. A.P. was supported by a postdoctoral fellowship from Wenner-Gren Stiftelserna. This is manuscript No. 19089 of the Scripps Research Institute.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Steve A. Kay (skay@ucsd.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alabadi, D. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Alabadi, D., Yanovsky, M.J., Mas, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12 757–761. [DOI] [PubMed] [Google Scholar]
- An, H., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Carpenter, C.D., Kreps, J.A., and Simon, A.E. (1994). Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 104 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardente, H., and Cermakian, N. (2007). Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol. Int. 24 195–213. [DOI] [PubMed] [Google Scholar]
- Endo, M., Mochizuki, N., Suzuki, T., and Nagatani, A. (2007). CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M., Nakamura, S., Araki, T., Mochizuki, N., and Nagatani, A. (2005). Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M.E., Hanano, S., Southern, M.M., Hall, A., and Millar, A.J. (2003). Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218 159–162. [DOI] [PubMed] [Google Scholar]
- Farre, E.M., Harmer, S.L., Harmon, F.G., Yanovsky, M.J., and Kay, S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15 47–54. [DOI] [PubMed] [Google Scholar]
- Farre, E.M., and Kay, S.A. (2007). PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J. 52 548–560. [DOI] [PubMed] [Google Scholar]
- Fujimori, T., Sato, E., Yamashino, T., and Mizuno, T. (2005). PRR5 (PSEUDO-RESPONSE REGULATOR 5) plays antagonistic roles to CCA1 (CIRCADIAN CLOCK-ASSOCIATED 1) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 69 426–430. [DOI] [PubMed] [Google Scholar]
- Fukamatsu, Y., Mitsui, S., Yasuhara, M., Tokioka, Y., Ihara, N., Fujita, S., and Kiyosue, T. (2005). Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol. 46 1340–1349. [DOI] [PubMed] [Google Scholar]
- Gallego, M., and Virshup, D.M. (2007). Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8 139–148. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Hubbard, K.E., Hotta, C.T., Dodd, A.N., and Webb, A.A. (2006). How plants tell the time. Biochem. J. 397 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Han, L., Mason, M., Risseeuw, E.P., Crosby, W.L., and Somers, D.E. (2004). Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J. 40 291–301. [DOI] [PubMed] [Google Scholar]
- Harms, E., Kivimae, S., Young, M.W., and Saez, L. (2004). Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 19 361–373. [DOI] [PubMed] [Google Scholar]
- Hazen, S.P., Borevitz, J.O., Harmon, F.G., Pruneda-Paz, J.L., Schultz, T.F., Yanovsky, M.J., Liljegren, S.J., Ecker, J.R., and Kay, S.A. (2005. a). Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 138 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005. b). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta, C.T., Gardner, M.J., Hubbard, K.E., Baek, S.J., Dalchau, N., Suhita, D., Dodd, A.N., and Webb, A.A. (2007). Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 30 333–349. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., and Kay, S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11 550–558. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Ivleva, N.B., Gao, T., LiWang, A.C., and Golden, S.S. (2006). Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. USA 103 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo, J.A. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410 487–490. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kevei, E., et al. (2006). Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 140 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y., Song, H.R., Taylor, B.L., and Carre, I.A. (2003. a). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W.Y., Geng, R., and Somers, D.E. (2003. b). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. USA 100 4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., and Wada, M. (2000). LKP1 (LOV kelch protein 1): A factor involved in the regulation of flowering time in Arabidopsis. Plant J. 23 807–815. [DOI] [PubMed] [Google Scholar]
- Kurup, S., Jones, H.D., and Holdsworth, M.J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21 143–155. [DOI] [PubMed] [Google Scholar]
- Liu, Y. (2005). Analysis of posttranslational regulations in the Neurospora circadian clock. Methods Enzymol. 393 379–393. [DOI] [PubMed] [Google Scholar]
- Mas, P. (2005). Circadian clock signaling in Arabidopsis thaliana: From gene expression to physiology and development. Int. J. Dev. Biol. 49 491–500. [DOI] [PubMed] [Google Scholar]
- Mas, P., Alabadi, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003. a). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, P., Kim, W.Y., Somers, D.E., and Kay, S.A. (2003. b). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41 1002–1012. [DOI] [PubMed] [Google Scholar]
- Michael, T.P., and McClung, C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J. (2000). Clock proteins: Turned over after hours? Curr. Biol. 10 R529–R531. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2 629–641. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Yamashino, T., and Mizuno, T. (2004). Characterization of circadian-associated APRR3 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 45 645–650. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T., and Mizuno, T. (2005. a). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46 609–619. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Yamashino, T., and Mizuno, T. (2005. b). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46 686–698. [DOI] [PubMed] [Google Scholar]
- Nakayama, M., Kikuno, R., and Ohara, O. (2002). Protein-protein interactions between large proteins: Two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res. 12 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome, P.A., and McClung, C.R. (2004). The Arabidopsis thaliana clock. J. Biol. Rhythms 19 425–435. [DOI] [PubMed] [Google Scholar]
- Salome, P.A., and McClung, C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, T.F., Kiyosue, T., Yanovsky, M., Wada, M., and Kay, S.A. (2001). A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y., and Hong-Hui, L. (2004). Transcription, translation, degradation, and circadian clock. Biochem. Biophys. Res. Commun. 321 1–6. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Kim, W.Y., and Geng, R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A.R., Pearson, M., and Kay, S.A. (1998). The short-period mutant toc1-1 alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125 485–494. [DOI] [PubMed] [Google Scholar]
- Song, H.R., and Carre, I.A. (2005). DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol. Biol. 57 761–771. [DOI] [PubMed] [Google Scholar]
- Strayer, C.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain, S.C., Hall, A., and Millar, A.J. (2000). Functional independence of circadian clocks that regulate plant gene expression. Curr. Biol. 10 951–956. [DOI] [PubMed] [Google Scholar]
- Thain, S.C., Murtas, G., Lynn, J.R., McGrath, R.B., and Millar, A.J. (2002). The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 130 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode, F., Malaguti, C., Romero, F., Attar, R., Camonis, J., and Egly, J.M. (1997). A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J. Biol. Chem. 272 22995–22999. [DOI] [PubMed] [Google Scholar]
- Ueda, H.R. (2006). Systems biology flowering in the plant clock field. Mol. Syst. Biol. 2 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wenkel, S., Turck, F., Singer, K., Gissot, L., Le Gourrierec, J., Samach, A., and Coupland, G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.