Abstract
The eukaryotic defense response posttranscriptional gene silencing (PTGS) is directed by short-interfering RNAs and thwarts invading nucleic acids via the RNA slicing activity of conserved ARGONAUTE (AGO) proteins. PTGS can be counteracted by exogenous or endogenous suppressors, including the cytoplasmic exoribonuclease XRN4, which also degrades microRNA (miRNA)-guided mRNA cleavage products but does not play an obvious role in development. Here, we show that the nuclear exoribonucleases XRN2 and XRN3 are endogenous PTGS suppressors. We also identify excised MIRNA loops as templates for XRN2 and XRN3 and show that XRN3 is critical for proper development. Independently, we identified the nucleotidase/phosphatase FIERY1 (FRY1) as an endogenous PTGS suppressor through a suppressor screen in a hypomorphic ago1 genetic background. FRY1 is one of six Arabidopsis thaliana orthologs of yeast Hal2. Yeast hal2 mutants overaccumulate 3′-phosphoadenosine 5′-phosphate, which suppresses the 5′→3′ exoribonucleases Xrn1 and Rat1. fry1 mutant plants recapitulate developmental and molecular characteristics of xrn mutants and likely restore PTGS in ago1 hypomorphic mutants by corepressing XRN2, XRN3, and XRN4, thus increasing RNA silencing triggers. We anticipate that screens incorporating partially compromised silencing components will uncover additional PTGS suppressors that may not be revealed using robust silencing systems.
INTRODUCTION
RNA silencing, at both the transcriptional and posttranscriptional levels, regulates endogenous and foreign gene expression in eukaryotes (Baulcombe, 2004; Kawasaki and Taira, 2005; Wassenegger, 2005; Zaratiegui et al., 2007). Posttranscriptional gene silencing (PTGS) is directed in a sequence-specific manner by short-interfering RNAs (siRNAs) to repress gene expression via RNA slicing activity, which is performed by conserved ARGONAUTE (AGO) proteins (Zamore and Haley, 2005; Ambros and Chen, 2007; Peters and Meister, 2007). We now know that thousands of unique siRNA sequences exist in plants, highlighting their potential to influence the activity of many endogenous gene networks (Lu et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007). Another class of small RNAs, microRNAs (miRNAs), is distinguished from siRNAs based on their biogenesis, but, like siRNAs, directs gene repression via AGO proteins (Bartel, 2004). miRNAs are essential for normal animal and plant development (Carrington and Ambros, 2003; Mallory and Vaucheret, 2006) and appear to be the most abundant small RNAs that exist in plants.
PTGS was first recognized as a plant immune response to virus infection, where viral RNAs trigger PTGS and then are targeted for elimination (Voinnet, 2001; Waterhouse et al., 2001; Ding and Voinnet, 2007). Many viruses have evolved to escape this immune response by producing proteins that interfere with key components of the silencing machinery (Roth et al., 2004). For example, the tombusvirus protein p19, the closterovirus protein p21, and the potyvirus protein HC-Pro prevent productive silencing by binding to siRNAs (Lakatos et al., 2006), the cucumovirus protein 2b interacts directly with AGO1 and inhibits its slicing activity (Zhang et al., 2006), and the polerovirus protein P0 suppresses silencing by interacting with and destabilizing AGO1 (Pazhouhandeh et al., 2006; Baumberger et al., 2007; Bortolamiol et al., 2007).
In addition to the numerous viral suppressors of gene silencing, endogenous silencing suppressors have been reported in plants. The first of these suppressors, the Nicotiana calmodulin-related silencing suppressor rgs-CAM, was found in a screen for proteins interacting with the viral suppressor HC-Pro (Anandalakshmi et al., 2000). rgs-CAM suppresses both potato virus X–induced gene silencing and sense transgene–mediated PTGS (S-PTGS) when expressed at high levels in Nicotiana (Anandalakshmi et al., 2000). The Arabidopsis thaliana exoribonuclease XRN4 suppresses PTGS, likely by degrading decapped messages, which are plausible templates for RNA-dependent RNA polymerases (RdRPs) (Gazzani et al., 2004). Indeed, xrn4 mutations promote RdRp-dependent silencing (Gazzani et al., 2004) and also lead to the overaccumulation of miRNA-generated cleavage products (Souret et al., 2004). Furthermore, mutations in ENHANCED SILENCING PHENOTYPE (ESP) genes involved in RNA processing and 3′ end formation enhance the RNA silencing of a potato virus X phytoene desaturase amplicon, presumably by allowing RNAs with aberrant 3′ termini to enter the PTGS pathway (Herr et al., 2006). In addition, the Arabidopsis putative RNase L inhibitor RLI2 was shown to suppress transgene-mediated silencing when expressed at high levels transiently in Nicotiana benthamiana (Sarmiento et al., 2006).
Genetic and biochemical studies have identified several effectors of plant RNA silencing pathways, including Dicer-like RNaseIII proteins (DCL), RNA polymerases (NRPD and RDR), double-stranded RNA binding proteins (DRB), an RNA methylase (HEN1), an RNA-protecting protein (SGS3), and small RNA-guided RNA slicer proteins (AGO) (Matzke and Birchler, 2005; Brodersen and Voinnet, 2006; Vaucheret, 2006; Henderson and Jacobsen, 2007). AGO proteins are conserved in all systems capable of RNA silencing and have been described as essential RNA silencing components (Carmell et al., 2002). The Arabidopsis AGO1 protein binds both siRNAs and miRNAs and, through the direction of these small RNAs, slices homologous transcripts during posttranscriptional silencing (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005).
Although several suppressors of the PTGS pathway have been identified, we reasoned that the contribution of some silencing suppressors could be masked in the robust transgenic silencing systems that are used generally and that a global view of endogenous suppressor proteins could be revealed in a genetic background in which an essential player in silencing, such as AGO1, is suppressed partially. To uncover novel PTGS suppressors, we performed a genetic screen in which two hypomorphic ago1 mutants, ago1-27 and ago1-33, which are 99 and 97% defective for RNA silencing at the L1 transgene locus, respectively (Morel et al., 2002; Sorin et al., 2005), were mutagenized and the M2 progeny were screened for second site mutations that compensate AGO1 deficiency. The hypomorphic ago1-27 and ago1-33 mutants used in this screen were identified previously in a separate screen for mutants defective for PTGS of the L1 35S:GUS (for β-glucuronidase) locus (Morel et al., 2002; Sorin et al., 2005). Line L1 accumulates high levels of GUS siRNAs and GUS expression is silenced during development (Elmayan et al., 1998; Boutet et al., 2003). Our previous forward genetic screen for mutants impaired in L1 silencing identified some of the first endogenous plant components of RNA silencing (RDR6, SGS3, AGO1, and HEN1) (Elmayan et al., 1998; Mourrain et al., 2000; Morel et al., 2002; Boutet et al., 2003), demonstrating the utility of the L1 locus as a model for endogenous silenced loci. Both L1/ago1-27 and L1/ago1-33 mutants accumulate high GUS mRNA and protein levels and show a strong reduction in GUS siRNA accumulation. In addition to releasing siRNA-mediated silencing (Boutet et al., 2003), these mutants exhibit mild increases in miRNA target mRNA levels (Vaucheret et al., 2004), owing to mutations within the RNaseH-like PIWI domain that likely reduce slicer activity, as reported for ago1-25 (Baumberger and Baulcombe, 2005), but they accumulate miRNAs at wild-type levels. The mild defects in the miRNA pathway lead to moderate developmental abnormalities in homozygous ago1-27 and ago1-33 mutants, but importantly, unlike null ago1 mutants, these hypomorphic mutants are fertile (Morel et al., 2002; Sorin et al., 2005), allowing a screen for the restoration of silencing in the progeny of mutagenized seeds.
Our search for endogenous silencing suppressors revealed that the 5′→3′ exoribonucleases XRN2 and XRN3 and the 3′(2′),5′-bisphosphate nucleotidase/inositol polyphosphate 1-phosphatase FIERY1 (FRY1) individually act as PTGS suppressors. FRY1 (also known as SAL1 and HOS2) was reported previously to act during both sulfur assimilation and phosphoinositide signaling (Quintero et al., 1996; Xiong et al., 2001, 2004) and to negatively regulate abscisic acid and stress signaling, including responses to cold and osmotic stress (Xiong et al., 2001). In L1/ago1/fry1 plants, GUS mRNA accumulation was reduced and GUS siRNA accumulation was elevated compared with L1/ago1 plants, and the accumulation of these PTGS hallmarks in L1/ago1/fry1 plants resembled their accumulation in the original L1-silenced line, demonstrating that L1-PTGS was reestablished in a classical fashion. Consistent with the role of FRY1 during S-PTGS, fry1 mutants are hyperresistant to Cucumber mosaic virus (CMV), which is targeted by PTGS. It is likely that fry1 mutants restore PTGS in ago1 hypomorphic mutants by corepressing the exoribonucleases XRN2, XRN3, and XRN4, which increases RNA silencing triggers. Indeed, fry1 mutants overaccumulate miRNA target cleavage products, which are templates of XRN4 (Souret et al., 2004), and MIRNA loops, which are templates of both XRN2 and XRN3 (this work). We anticipate that screens incorporating weakly silenced systems or partially compromised silencing components will uncover additional PTGS suppressors that may not be revealed using robust silencing systems.
RESULTS
Identification of Second Site Mutations That Restore PTGS in ago1 Hypomorphs
To identify second site mutations that compensate AGO1 deficiency, M2 populations derived from mutagenized ago1-27 and ago1-33 seeds were screened first for the restoration of wild-type development. We were unable to identify plants exhibiting a wild-type phenotype. Therefore, M2 populations were screened for the restoration of RNA silencing at the L1 locus. Two and five mutants with restored RNA silencing at the L1 locus were identified in the progeny of mutagenized ago1-27 and ago1-33 seeds, respectively. The presence of second site suppressor mutations that do not affect the integrity of either the 35S-GUS transgene at the L1 locus or the mutant ago1-27 or ago1-33 endogenous locus was established based on the following two results: (1) after self-fertilization, 100% of the progeny of the seven restored mutants expressed GUS at high levels 1 week after germination and silenced GUS later during development, similar to L1, indicating that the 35S-GUS transgene was functional; and (2) after outcrossing to L1 plants followed by self-fertilization, 3 of 16 of the F2 plants exhibited the ago1-27 or ago1-33 phenotype and expressed GUS at high levels, similar to the original ago1-27 and ago1-33 mutants, indicating that the second site mutations were not linked to the mutant ago1-27 or ago1-33 locus.
Map-Based Cloning Identifies fry1 as a Suppressor of ago1 Hypomorphs
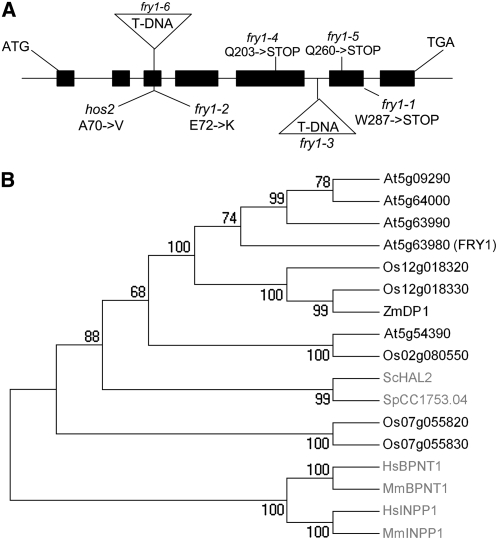
Genetic complementation tests and map-based cloning revealed that two mutants recovered in the ago1-27 and ago1-33 mutagenesis both had mutations in the conserved FRY1/HOS2/SAL1 gene (Figure 1) (Quintero et al., 1996; Xiong et al., 2001, 2004), indicating the capacity of this gene to regulate RNA silencing in two different hypomorphic ago1 backgrounds. The two mutants, ago1-27 fry1-4 and ago1-33 fry1-5, each had single point mutations in FRY1 that changed different Gln codons to stop codons, truncating the protein at positions 203 and 260, respectively (Figure 1A). The two mutants exhibited similar developmental defects, which included crinkly leaves, rounded leaf margins, and delayed flowering, compared with the original ago1-27 and ago1-33 mutants (Figure 2; data not shown). fry1-4 and fry1-5 plants, in which ago1-27 and ago1-33 mutations had been segregated away, exhibited developmental defects (Figure 2) similar to that of a complete loss-of-function fry1-6 mutant, in which a T-DNA insertion truncates the protein at position 71 (Figure 1A), indicating that the developmental defects and RNA silencing restoration resulted from the impairment of a single gene.
Figure 1.
Structure and Conservation of FRY1.
(A) FRY1 (At5g63980) gene structure. Exons are represented by black boxes. The positions of the ATG start and TGA stop codons are indicated. The location of fry1 T-DNA insertions (triangles), nonsense mutations, and missense mutations with the corresponding amino acid changes are indicated above or below the gene structure.
(B) Phylogeny of the FRY1 protein. In black are plants (At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays), and in gray are fungi (yeast; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe) and mammals (Mm, Mus musculus; Hs, Homo sapiens).
Figure 2.
Developmental Abnormalities of ago1, fry1, and xrn Mutants.
(A) and (B) ago1-27 and ago1-33 mutant lines used for mutagenesis. Plants are dwarf and have serrated leaves.
(C) to (F) fry1-4 and fry1-5 mutants recovered from the mutagenesis screen are shown in the ago1 mutant backgrounds ([C] and [D]) or alone ([E] and [F]). Plants are dwarf and have crinkly leaves, rounded leaf margins, and delayed flowering compared with wild-type plants.
(G) A Columbia (Col) accession wild-type plant.
(H) fry1-6 T-DNA insertion mutants display developmental anomalies identical to fry1-4 and fry1-5 mutants.
(I) xrn2-1 mutants do not display obvious developmental defects.
(K) Hypomorphic xrn3-3 mutants display crinkly leaves and delayed flowering.
(M) xrn4-6 mutants display slightly serrated leaves.
(J), (L), and (N) xrn2-1 xrn3-3 (J), xrn2-1 xrn4-6 (L), and xrn3-3 xrn4-6 (N) double mutants show additive developmental defects, and xrn2-1 xrn3-3 mutants (J) resemble fry1-4, fry1-5, and fry1-6 single mutants ([E], [F], and [H], respectively).
Transgene mRNA and siRNA Accumulation Is Restored in ago1 fry1 Mutants
Several features of RNA silencing were analyzed to characterize the effects of the fry1 mutations. GUS mRNAs accumulated at high levels in ago1-27 and ago1-33 mutants, whereas they accumulated at low levels in L1 controls, fry1-4 and fry1-5 plants, and ago1-27 fry1-4 and ago1-33 fry1-5 mutant plants, owing to the restoration of silencing (Figure 3A). Reciprocally, GUS siRNAs accumulated at high levels in L1, fry1-4, and fry1-5 mutant plants, whereas they were below detectable levels in ago1-27 and ago1-33 mutants (Figure 3B). GUS siRNA accumulation was restored in both ago1-27 fry1-4 and ago1-33 fry1-5 mutants (Figure 3A), and maximal GUS silencing was achieved by 18 d in ago1-33 fry1-4 mutants, similar to L1 control plants, but occurred later in development in ago1-27 fry1-5 mutants (Figure 3; data not shown). This difference suggested that RNA silencing was more easily restored in ago1-33 than in ago1-27 and provided a plausible explanation for the higher number of suppressor loci recovered from the ago1-33 mutagenesis compared with the ago1-27 mutagenesis.
Figure 3.
ago1 fry1 Double Mutants Posttranscriptionally Silence L1.
RNA gel blot analysis of GUS mRNA (A) and GUS siRNA (B) accumulation in L1 control and ago1, fry1, and ago1 fry1 mutant 18-d-old seedlings.
(A) L1 control and L1/fry1-4 and L1/fry1-5 mutant plants are silenced and accumulate low GUS mRNA levels. ago1-27 and ago1-33 mutants release L1 silencing and accumulate high levels of GUS mRNA. Both fry1-4 and fry1-5 restore L1 silencing in ago1 mutant backgrounds and accumulate low levels of GUS mRNA, although L1 silencing is restored to a lesser extent in L1/ago1-27/fry1-4 mutants (4.2 versus 1.3). Normalized values of GUS mRNA to ethidium bromide–stained 25S RNA (bottom panel), with L1 levels set at 1.0, are indicated.
(B) RNA gel blot analysis of GUS siRNA accumulation. L1 control, L1/fry1-4, L1/fry1-5, L1/ago1-27/fry1-4, and L1/ago1-33/fry1-5 mutant plants are silenced and accumulate GUS siRNAs. ago1-27 and ago1-33 mutants release L1 silencing and do not accumulate detectable GUS siRNAs. Blots were stripped and rehybridized with a probe complementary to U6 snRNA as a loading control. Normalized values of GUS siRNAs to U6 small nuclear RNA (snRNA), with L1 levels set at 1.0, are indicated. ND, not detected.
fry1 Mutants Overaccumulate miRNA Target Cleavage Products
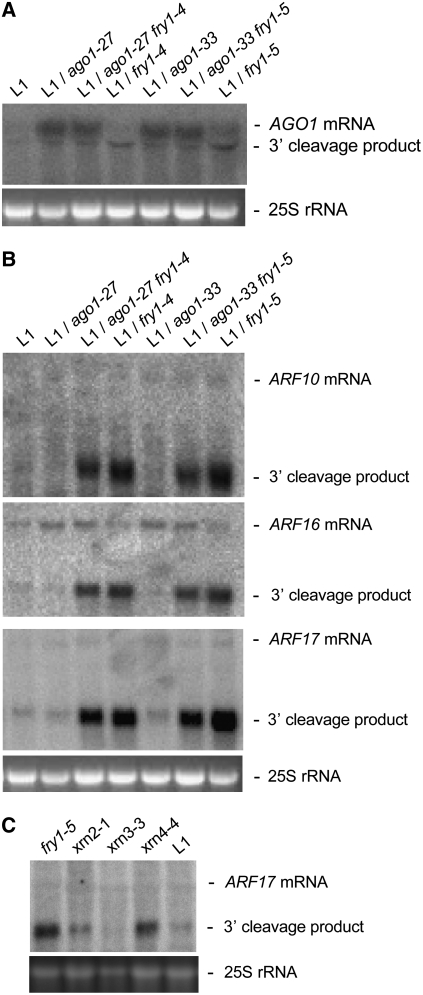
AGO1 mRNA overaccumulated in ago1-27 fry1-4, ago1-33 fry1-5, ago1-27, and ago1-33 mutants, which have reduced slicer activity. Conversely, AGO1 mRNA levels were similar in fry1-4, fry1-5, and control L1 plants, whereas miR168-guided AGO1 3′ cleavage products overaccumulated in fry1-4 and fry1-5 compared with control L1 plants (Figure 4). A dramatic increase in the accumulation of miR160-guided ARF10, ARF16, and ARF17 3′ cleavage products was observed in fry1-4 and fry1-5 mutants compared with control L1 plants as well as in ago1-27 fry1-4 and ago1-33 fry1-5 compared with ago1-27 and ago1-33 controls (Figures 4B and 4C). This overaccumulation was similar to that previously reported for xrn4 mutants (Figure 4C) (Souret et al., 2004). By contrast, the accumulation of full-length ARF10, ARF16, and ARF17 mRNAs was similar among ago1-27, ago1-33, ago1-27 fry1-4, and ago1-33 fry1-5 and among fry1-4, fry1-5, and control L1 plants (Figure 4B). Together, these results indicate that fry1 mutations affect the accumulation of miRNA target cleavage products but not that of miRNA targeted full-length mRNAs.
Figure 4.
fry1 Mutants Overaccumulate miRNA Target 3′ Cleavage Products.
RNA gel blot analysis of AGO1 full-length mRNA and miR168 generated 3′ AGO1 cleavage products, and ARF10, ARF16, and ARF17 full-length RNA and miR160 generated 3′ ARF cleavage products in L1 control and ago1, fry1, ago1 fry1, xrn2, xrn3, and xrn4 mutant 18-d-old seedlings.
(A) AGO1 full-length mRNA levels are increased in ago1 and ago1/fry1 mutants, whereas AGO1 3′ cleavage products, but not AGO1 full-length mRNA, overaccumulate in fry1 single mutants.
(B) and (C) fry1 mutants and ago1 fry1 double mutants overaccumulate ARF10, ARF16, and ARF17 3′ cleavage products (B), similar to xrn4 mutants (C). Ethidium bromide–stained 25S RNA is shown as a loading control.
fry1 Mutants Overaccumulate Excised MIRNA Loops
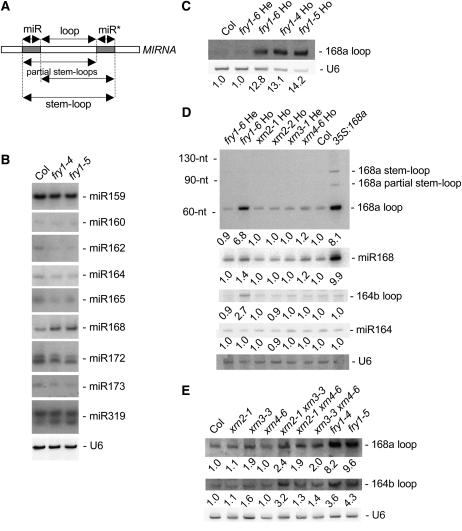
miRNA accumulation either was unchanged or was decreased slightly in fry1 mutants, except for miR168 accumulation, which was increased (Figures 5B and 5D). Thus, the dramatic increase in miRNA target cleavage products cannot be explained by changes at the miRNA level. Because fry1 mutations affect the accumulation of miRNA target cleavage products but not that of miRNA target full-length mRNAs, we hypothesized that FRY1 could function during the degradation of intermediates or nonfunctional end products of small RNA pathways. Therefore, we analyzed the accumulation of stem-loop and partially processed stem-loop intermediates and excised loop end products derived from miRNA precursors during DCL1-mediated maturation (Figure 5A) in fry1 mutants. Among several miRNAs tested, only MIR164b- and MIR168a-derived products were detected (Vaucheret et al., 2006). Excised MIR164b- and MIR168a-derived loops, but not stem-loops or partially processed stem-loops, overaccumulated in fry1 compared with control L1 plants (Figures 5C to 5E).
Figure 5.
MIRNA Loops Overaccumulate in fry1 and xrn2 xrn3 Double Mutants.
(A) Diagram of the various intermediates and end products generated during MIRNA maturation. The stem-loop is the RNA sequence flanked at each end by the miRNA and miRNA* sequences; partial stem-loops are generated after either the 3′ end of the miRNA or the 5′ end of miRNA* is defined. The positions of the 21-nucleotide miRNA (miR) and miRNA* (miR*) species are indicated by gray boxes (the miR* is the 21-nucleotide RNA species that is opposite of and paired to the miRNA in the MIRNA hairpin). The loop sequence is between the miRNA and the miRNA* and is liberated during miRNA maturation.
(B) RNA gel blot analysis of miRNA accumulation in Col control and fry1-4 and fry1-5 mutant inflorescences. miRNA accumulation is either unchanged or slightly reduced in fry1 mutants except for miR168, which slightly overaccumulates. Blots were stripped and rehybridized with a probe complementary to U6 RNA as a loading control.
(C) RNA gel blot analysis showing that a homozygous fry1-6 T-DNA insertion mutant and fry1-4 and fry1-5 ethyl methanesulfonate mutants overaccumulate MIR168a loops in inflorescence tissues. U6 hybridization is shown as a loading control. Normalized values of the MIR168a loop RNA to U6 snRNA, with Col levels set at 1.0, are indicated.
(D) RNA gel blot analyses of MIR168a and MIR164b maturation intermediates and end products in inflorescence tissues of controls (Col and MIR168a-overexpressing plants; 35S:168a), hemizygous and homozygous fry1 mutants, two homozygous xrn2 mutants, a hemizygous xrn3 mutant, and a homozygous xrn4 mutant. Homozygous fry1 mutants overaccumulate MIR168a and MIR164b loops but not stem-loops or partial stem-loops. miR164 and miR168 accumulation in fry1 homozygous mutants is shown in (B). RNA extracted from MIR168a-overexpressing inflorescences was used to indicate the positions of MIR168a stem-loops and partial stem-loops, which are undetectable in wild-type inflorescences. The positions of 32P-labeled RNA oligonucleotides (nt) are noted. U6 hybridization is shown as a loading control. Normalized values of loop RNAs and miRNAs to U6 snRNA, with Col levels set at 1.0, are indicated.
(E) RNA gel blot analysis of MIR168a and MIR164b loop accumulation in control Col, xrn2, hypomorphic xrn3, xrn4, xrn2 xrn3, xrn2 xrn4, xrn3 xrn4, and fry1 mutant inflorescences. All mutations are homozygous. xrn3-containing mutants overaccumulate loops, although to a lesser extent than fry1 mutants. U6 hybridization is shown as a loading control. Normalized values of loop RNAs to U6 snRNA, with Col levels set at 1.0, are indicated.
Together, these data indicate that FRY1 functions in the degradation of nonfunctional end products of the miRNA pathway, such as MIRNA loops and miRNA target cleavage products, suggesting an impairment of exoribonuclease activity in fry1 mutants. This proposal is consistent with the impairment of exoribonuclease activity observed in yeast hal2 mutants (Dichtl et al., 1997). Indeed, FRY1/HOS2/SAL1 is one of six Arabidopsis orthologs of yeast Hal2 (Figure 1B), and there are several lines of evidence to suggest that FRY1 and Hal2 are functionally similar. First, both FRY1 and Hal2 convert 3′-phosphoadenosine 5′-phosphate (PAP), a toxic byproduct of sulfur assimilation (Saito, 2004), into 5′ AMP + Pi (Murguia et al., 1995; Quintero et al., 1996; Xiong et al., 2001). Furthermore, expression of Arabidopsis FRY1 complements PAP toxicity in yeast hal2 mutants, allowing these mutants to grow when sulfate is the sole sulfur source (Quintero et al., 1996). Finally, both Hal2 and FRY1 are sensitive to NaCl and lithium exposure (Quintero et al., 1996; Dichtl et al., 1997; Xiong et al., 2004). Increased PAP accumulation in hal2 yeast mutants suppresses the 5′→3′ exoribonucleases Rat1 and Xrn1 (Dichtl et al., 1997), and Arabidopsis has three orthologs of Rat1 (XRN2, XRN3, and XRN4) and no apparent ortholog of Xrn1 (Kastenmayer and Green, 2000). Therefore, it is possible that, as in hal2 mutants, fry1 mutants have impaired XRN activity. Arabidopsis XRN2 and XRN3 act in the nucleus, whereas Arabidopsis XRN4 acts in the cytoplasm (Kastenmayer and Green, 2000). The RNA targets of XRN2 and XRN3 are unknown, whereas XRN4 functions in the degradation of miRNA target cleavage products (Souret et al., 2004) and in the suppression of S-PTGS (Gazzani et al., 2004).
XRN2 and XRN3 Participate in MIRNA-Derived Loop Degradation, and Combined xrn2, xrn3, and xrn4 Mutations Phenocopy fry1
To test the hypothesis that XRN activities could be suppressed in fry1 mutants, we compared the developmental and molecular phenotypes of xrn2, xrn3, and xrn4 mutants with those of fry1 mutants. Homozygous complete loss-of-function xrn2-1, xrn2-2, xrn4-6, and xrn4-7 mutants did not exhibit the developmental defects of fry1 mutants (Figure 2; data not shown). Homozygous complete loss-of-function xrn3-1 and xrn3-2 mutants could not be recovered due to embryo-lethality (data not shown). However, one mutant line carrying an insertion within the promoter of the XRN3 gene (see Supplemental Figure 1 online) produced viable homozygous plants (Figure 2). The hypomorphic xrn3-3 mutant exhibited reduced XRN3 mRNA accumulation (see Supplemental Figure 1 online) and, although less developmentally impaired than fry1 mutants, the xrn3-3 mutant exhibited a similar set of developmental defects, including crinkly, rounded leaves and delayed flowering (Figure 2).
Excised MIRNA loops overaccumulated in hypomorphic xrn3-3 mutants, although to a level lower than in fry1 mutants (Figure 5E), suggesting that MIRNA loop degradation activity is more efficient in xrn3-3 mutants than in fry1 mutants, either because xrn3-3 is a hypomorphic mutant or because another XRN activity contributes to MIRNA loop degradation. XRN2 and XRN3 encode very similar nuclear proteins that could have partially redundant activities, and it is possible that the activities of both proteins are suppressed in fry1 mutants. Consistent with this possibility, xrn2-1 xrn3-3 double mutants accumulated excised MIRNA loops to a higher level than xrn3-3 single mutants (Figure 5E). In addition, the developmental defects of xrn2-1 xrn3-3 double mutants more closely resembled those of fry1 mutants than did xrn3-3 single mutants or xrn2-1 xrn4-6 and xrn3-3 xrn4-6 double mutants (Figure 2). However, xrn2-1 xrn3-3 double mutants were not phenotypically identical to fry1 mutants. This observation is likely because FRY1 has multiple functions and the developmental phenotypes of fry1 mutants likely result from the collective perturbation of several different pathways.
XRN2, XRN3, and XRN4 Act Individually as PTGS Suppressors
XRN4 previously was designated an endogenous suppressor of S-PTGS because xrn4 mutants showed an increased rate of S-PTGS, presumably due to the reduced degradation of aberrant transgene RNAs, which could trigger S-PTGS (Gazzani et al., 2004). Thus, if fry1 dampens XRN4 activity, the restoration of S-PTGS in two hypomorphic ago1 mutant backgrounds could be due to increased triggering of S-PTGS, which could compensate for reduced AGO1 slicer activity. Indeed, double mutants between the complete loss-of-function xrn4-1 mutant and ago1-27 or ago1-33 mutants triggered L1 S-PTGS as efficiently as L1 control plants (Figure 6A). However, if mutations in FRY1 do not completely inhibit XRN activities, as suggested by the fact that complete loss-of-function xrn3 mutants, but not complete loss-of-function fry1 mutants, are embryo-lethal, the coordinated dampening of the other two XRN activities by fry1 may be required to restore S-PTGS in ago1. To investigate this possibility, we generated double mutants between ago1-27 or ago1-33 and xrn2 and xrn3 mutants. Owing to 35S-mediated interference of L1 silencing, the xrn2-1 mutant was discarded and 35S-free xrn2-2 and xrn3-3 T-DNA mutants were used in our analysis. S-PTGS of L1 was restored in only 4% of ago1-27 xrn2-2 and was not restored in ago1-27 xrn3-3 mutants (Figure 6A). However, S-PTGS of L1 was restored in 15 and 20% of ago1-33 xrn2-2 and ago1-33 xrn3-3 mutants, respectively, suggesting that, like XRN4, XRN2 and XRN3 could act as an endogenous suppressor of S-PTGS, although to a lesser extent than XRN4. The restoration of PTGS in xrn3-3 ago1-33 but not in xrn3-3 ago1-27 confirmed that PTGS can be more easily restored in ago1-33 compared with ago1-27 (Figure 3).
Figure 6.
XRN2 and XRN3 Are Endogenous Suppressors of PTGS.
GUS protein activities contributed by the homozygous transgene loci L1 (A), Hc1 (B), and 6b4 (C) in control, ago1, xrn, and fry1 mutant leaves. Data are expressed as percentages of silenced plants in each genetic background. The number of individual plants analyzed is indicated above each bar.
To investigate further the contribution of XRN3 and XRN2 during silencing, we examined the effects of 35S-free xrn2-2, xrn3-3, and xrn4-1 mutations during silencing of the 35S-GUS Hc1 locus, which spontaneously triggers PTGS at a frequency of 20% in a wild-type background (Elmayan et al., 1998), and the 35S-GUS 6b4 locus, which does not spontaneously trigger PTGS (Beclin et al., 2002). The frequencies of Hc1-PTGS and 6b4-PTGS were elevated to 100 and 55%, respectively, in xrn4-1 mutants (Figures 6B and 6C), confirming the role of XRN4 as an endogenous suppressor of PTGS. In addition, the frequencies of Hc1-PTGS were elevated to 47 and 94% and the frequencies of 6b4-PTGS were elevated to 5 and 12% in xrn2-2 and xrn3-3, respectively (Figures 6B and 6C), indicating that, like XRN4, XRN2 and XRN3 act as endogenous suppressors of S-PTGS, although to a lesser extent. Therefore, the efficient restoration of L1 S-PTGS in ago1-27 fry1-4 and ago1-33 fry1-5 likely results from the simultaneous suppression of XRN2, XRN3, and XRN4 by fry1.
xrn4 and fry1 Mutants Are Hyperresistant to CMV Infection
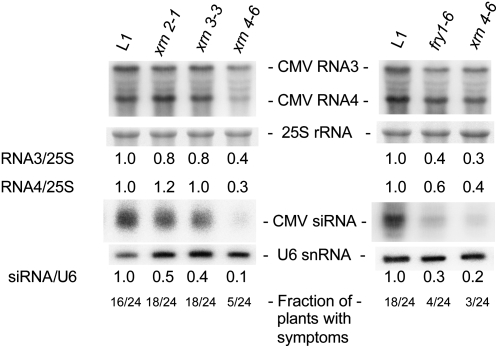
Our results show that, along with XRN4 (Gazzani et al., 2004), XRN2 and XRN3 act as endogenous S-PTGS suppressors and that FRY1 likely suppresses PTGS by negatively regulating these XRNs. In addition to transgenes, viruses are targeted by PTGS, and mutations in PTGS genes (AGO1, DCL4, HEN1, RDR6, SDE3, SDE5, and SGS3) lead to virus hypersusceptibility in plants (Mourrain et al., 2000; Dalmay et al., 2001; Morel et al., 2002; Boutet et al., 2003; Bouche et al., 2006; Hernandez-Pinzon et al., 2007). To examine the effects of xrn and fry mutations on CMV, a classic RNA virus PTGS model in plants, we infected xrn2, xrn3, xrn4, and fry1 mutants with CMV. Twenty-one days after infection, there was no obvious effect on CMV RNA accumulation in xrn2 and xrn3 mutants, although there was a slight decrease in CMV siRNA accumulation (Figure 7). This subtle effect likely reflects the different localizations of CMV (cytoplasm) versus XRN2 and XRN3 (nucleus). By contrast, both xrn4 and fry1 mutants had reduced CMV RNA accumulation, reduced CMV-derived siRNA accumulation, and fewer plants showed virus symptoms compared with control plants (Figure 7), indicating that both mutants were hyperresistant to CMV infection. These results are consistent with those we observed for transgene silencing and indicate that XRN4 and FRY1 suppress virus-induced PTGS.
Figure 7.
xrn4 and fry1 Mutants Have Reduced CMV Accumulation.
RNA gel blot analyses of CMV RNA3 and RNA4 and CMV-derived siRNA accumulation in control L1 and xrn2, xrn3, xrn4, and fry1 mutant 29-d-old rosettes. Both fry1 and xrn4 mutants accumulate less CMV RNA and thus less CMV-derived siRNAs than control L1 plants. 25S rRNA hybridization is shown as a control for mRNA gel blots. Normalized values of CMV RNA3 and RNA4 to 25S rRNA, with L1 levels set at 1.0, are indicated. U6 hybridization is shown as a loading control for siRNA gel blots. Normalized values of CMV siRNAs to U6 snRNA, with L1 levels set at 1.0, are indicated. The number of symptomatic plants out of 24 CMV-infected plants is indicated at the bottom of each lane.
DISCUSSION
In this study, we identify the exoribonucleases XRN2 and XRN3 and FRY1 as PTGS suppressors. Previously, the exoribonuclease XRN4 was shown to suppress PTGS, likely through the degradation of RdRp templates. In this way, mutations in XRN4 lead to the buildup of aberrant, uncapped RNAs, which can trigger PTGS of previously expressed messages by presumably serving as RdRp templates (Gazzani et al., 2004). XRN4 also was shown to degrade miRNA cleavage products (Souret et al., 2004), which lack a 5′ cap structure. While XRN4 is cytoplasmic, XRN2 and XRN3 are nuclear proteins (Kastenmayer and Green, 2000). Our analyses of PTGS efficiency in the hypomorphic xrn3-3 mutant and the null xrn2-2 mutant (Figures 6B and 6C) suggest that XRN2 and XRN3 contribute to the degradation of aberrant transgene RNAs in the nucleus, complementary to the degradation of aberrant transgene RNAs by XRN4 in the cytoplasm (Figure 8). However, although mutations in all three XRNs independently trigger PTGS of a sense transgene, the endogenous RNA targets of the XRNs are different: XRN4 degrades cytoplasmic, uncapped messages like miRNA target cleavage products (Gazzani et al., 2004; Souret et al., 2004), whereas XRN2 and XRN3 degrade MIRNA-derived loops excised during miRNA maturation in the nucleus (Figure 8). The different localization of XRN4 (cytoplasm) versus XRN2 and XRN3 (nucleus) likely explains why xrn4 but not xrn2 or xrn3 mutants are hyperresistant to infection by the cytoplasmic virus CMV (Figure 7).
Figure 8.
Model for FRY1, XRN2, XRN3, and XRN4 Activity during PTGS.
Shown at left is a simplified model for miRNA biogenesis and action. Normal miRNA maturation requires DCL1, SERRATE, HYL1, and HEN1 and liberates MIRNA loops, miRNA*, and mature miRNAs. miRNAs are exported from the nucleus by HASTY and incorporate into an AGO1-containing complex to direct the cleavage of partially complementary RNAs. MIRNA loops are substrates of both the nuclear XRN2 and XRN3, while 3′ target cleavage products are substrates of the cytoplasmic XRN4. Shown at right is a simplified model for transgene- and virus-induced silencing. Aberrant RNA deriving from transgene transcription and virus replication is converted to double-stranded RNA and processed into siRNAs by RDR6 and SGS3, and DCL2 and DCL4, respectively. siRNAs incorporate into an AGO1-containing complex to direct the cleavage of complementary RNAs. XRN2, XRN3, and XRN4 are endogenous suppressors of PTGS, and we propose that the transgene- and virus-derived aberrant RNAs that trigger PTGS are substrates of these XRNs. At center, FRY1 converts 3′-phosphoadenosine 5′-phosphate (PAP), a toxic byproduct of sulfate assimilation, into 5′ AMP + Pi. Our data, together with those from yeast, predict that XRN levels are regulated through a FRY1-dependent system that keeps PAP, an inhibitor of yeast XRN activity, levels in check. We propose that XRN activities are repressed in fry1 mutant plants, leading to the overaccumulation of aberrant RNAs that trigger PTGS.
Neither XRN2 nor XRN4 is critical for proper plant development. However, the embryo-lethality of null xrn3 alleles and the developmental defects exhibited by hypomorphic xrn3-3 mutants suggest an essential role of XRN3. xrn2 xrn3 double mutants exhibit stronger developmental defects and accumulate higher levels of MIRNA loops than hypomorphic xrn3-3 mutants, indicating that these two exoribonucleases have partially redundant roles during development, which likely reflects their partially redundant molecular activities. Because it is unlikely that the overaccumulation of MIRNA loops causes embryo-lethality and developmental defects, other targets of XRN3 and the precise reasons for embryo-lethality in xrn3 mutants remain to be identified.
Our forward genetic screen identified FRY1 as a suppressor of transgene-induced silencing in plants. We also showed that FRY1 is a suppressor of virus-induced silencing. FRY1 is a dual-function 3′(2′),5′-bisphosphate nucleotidase/inositol polyphosphate 1-phosphatase orthologous to Hal2 and CysQ in yeast and Escherichia coli, respectively (Neuwald et al., 1992; Glaser et al., 1993). fry1 mutants were isolated previously in altered stress response genetic screens, which implicated FRY1 in abscisic acid signaling and stress responses to cold, drought, and salinity (Quintero et al., 1996; Xiong et al., 2001, 2004). Whether the PTGS-suppressing activity of FRY1 is necessary for its previously reported functions remains to be determined. Our data suggest that fry1 mutations restore PTGS through the coordinated downregulation of XRN2, XRN3, and XRN4 activities (Figure 8). Rather than an absolute on/off switch model, our data are consistent with a fine-tuning modulation of XRN activities by FRY1. Indeed, null xrn3 alleles were embryo-lethal, whereas null fry1 alleles exhibited developmental defects but were viable, similar to the partial loss-of-function xrn3-3 allele, suggesting that fry1 partially inhibits XRN activities. In addition, whereas fry1 was able to restore L1 S-PTGS in the hypomorphic ago1-27 and ago1-33 mutants, it was unable to restore L1 S-PTGS in null ago1 mutants (data not shown), indicating that even in the presence of additional silencing triggers, none of the other nine AGO proteins could functionally substitute for AGO1. The reduced XRN activities in fry1 mutants likely lead to an increased accumulation of aberrant transgene RNA, the trigger of S-PTGS, to a level that could compensate for the reduced slicer activity in hypomorphic ago1-27 and ago1-33 mutants (Figure 8). Consistent with the hypothesis that fry1 reinforces S-PTGS initiation, fry1 was unable to restore L1 S-PTGS in hen1-4, rdr6 (sgs2-1), or sgs3-1 null alleles (data not shown), indicating that fry1-mediated reinforcement of the S-PTGS pathway requires the integrity of the pathway. Future analyses of the five other suppressor mutants recovered in this screen likely also will reveal novel silencing suppressors.
METHODS
Plant Materials
Arabidopsis thaliana 35S-GUS L1 and Hc1 lines and ago1-27, ago1-33, hen1-4, rdr6 (sgs2-1), sgs3-1, and xrn4-1 mutants have been described (Elmayan et al., 1998; Mourrain et al., 2000; Morel et al., 2002; Boutet et al., 2003; Gazzani et al., 2004; Sorin et al., 2005). fry1-6 (SALK_020882), xrn2-1 (SALK_041148), xrn2-2 (SAIL_781E02), xrn3-1 (SALK_129657), xrn3-2 (SAIL_762H09), xrn3-3 (SAIL_1172C07), xrn4-6 (SALK_014209), and xrn4-7 (SALK_125718) were recovered in the SAIL and SALK sequence-indexed T-DNA insertion collections (Sessions et al., 2002; Alonso et al., 2003). All plants were grown in standard long-day conditions (16 h of light, 8 h of dark) at 22°C. The list of primers used for genotyping T-DNA insertion lines is presented in Supplemental Table 1 online.
Mutant Screen, Genetic Analyses, and Map-Based Cloning
Ethyl methanesulfonate mutagenesis, mutant screen, and genetic complementation tests were performed as described before (Elmayan et al., 1998; Mourrain et al., 2000). For map-based cloning, suppressor mutants (in Col) were crossed to wild-type Landsberg erecta and the mutations were mapped in the GUS-silenced ago1-phenotypic F2 recombinant lines using Col/Landsberg erecta polymorphic markers. fry1-4 and fry1-5 were mapped between markers CER455203 and CER457067. T-DNA insertion lines disrupting genes located within this 240-kb interval were examined for developmental defects. Line fry1-6 (SALK_020882) disrupting FRY1 (At5g63980) exhibited molecular and developmental defects similar to fry1-4 and fry1-5.
Phylogenetic Analysis
Full-length sequences were aligned (see Supplemental Dataset 1 online) using ClustalW (Thompson et al., 1994). Midpoint rooted phylogenetic trees were generated in MEGA4 (Tamura et al., 2007) by the neighbor-joining method (Saitou and Nei, 1987) using the Poisson correction method (Zuckerkandl and Pauling, 1965). All positions containing gaps and missing data were eliminated from the dataset with the default assumption that the substitution patterns among lineages and the substitution rates among sites were homogeneous. There were 253 positions in the final dataset. The optimal tree is shown in Figure 1B. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985).
Molecular Analyses and Virus Infections
DNA sequencing, RNA gel blot analyses, and GUS fluorimetric assays were performed as described before (Boutet et al., 2003; Vaucheret et al., 2006), except for the GUS siRNA gel blot analysis, which was performed as described by Pall et al. (2007) using the Tris-borate–based Sequagel system (National Diagnostics). All RNA gel blots were of total RNA, except for the GUS mRNA gel blot and the GUS siRNA gel blot, which were of LiCl-fractionated high molecular mass RNA and low molecular mass RNA, respectively. Riboprobes for AGO1 mRNA, ARF mRNAs, and GUS siRNAs, random-primed DNA probes for GUS mRNA, and oligonucleotide probes for miRNA detection were as described previously (Boutet et al., 2003; Mallory et al., 2005; Vaucheret et al., 2006). CMV infections were as described (Mourrain et al., 2000; Boutet et al., 2003). Briefly, plants were grown in standard long-day conditions (16 h of light, 8 h of dark) at 22°C in vitro for 7 d and transplanted to soil, and 24 plants of each genotype were inoculated with CMV on day 8. Whole rosettes were collected from symptomatic plants and pooled at 21 d after inoculation and subjected to RNA analysis. Probes for CMV RNAs and siRNAs were as described (Bouche et al., 2006).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: FRY1, At5g63980; XRN2, At5g42540; XRN3, At1g75660; XRN4, At1g54490; AGO1, At1g48410.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. XRN3 Partial Nucleotide Sequence and mRNA Accumulation in xrn3-3 Hypomorphic Mutants.
Supplemental Table 1. List of Primers Used for Genotyping T-DNA Insertion Lines.
Supplemental Dataset 1. Text File of Amino Acid Alignments Used to Generate the Phylogenetic Tree Presented in Figure 1.
Supplementary Material
Acknowledgments
We thank Bruno Letarnec for plant care and Taline Elmayan for comments on the manuscript. Seeds from the SAIL and SALK sequence-indexed T-DNA insertion lines were provided by the European Arabidopsis Stock Center. This work was partly supported by a grant from the Agence Nationale pour la Recherche (Grant ANR-06-POGM-007-02) to H.V.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Hervé Vaucheret (herve.vaucheret@versailles.inra.fr) and Allison C. Mallory (amallory@versailles.inra.fr).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ambros, V., and Chen, X. (2007). The regulation of genes and genomes by small RNAs. Development 134 1635–1641. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Marathe, R., Ge, X., Herr, J.M., Jr., Mau, C., Mallory, A., Pruss, G., Bowman, L., and Vance, V.B. (2000). A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290 142–144. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., and Baulcombe, D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N., Tsai, C.H., Lie, M., Havecker, E., and Baulcombe, D.C. (September 12, 2007). The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. http://dx.doi.org/10.1016/j.cub.2007.08.039. [DOI] [PubMed]
- Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12 684–688. [DOI] [PubMed] [Google Scholar]
- Bortolamiol, D., Pazhouhandeh, M., Marrocco, K., Genschik, P., and Ziegler-Graff, V. (2007). The polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. [DOI] [PubMed]
- Bouche, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet, S., Vazquez, F., Liu, J., Béclin, C., Fagard, M., Gratias, A., Morel, J.B., Crété, P., Chen, X., and Vaucheret, H. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. (2002). The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16 2733–2742. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Horsefield, R., Braunstein, T.H., and Baulcombe, D.C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, B., Stevens, A., and Tollervey, D. (1997). Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16 7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Glaser, H.U., Thomas, D., Gaxiola, R., Montrichard, F., Surdin-Kerjan, Y., and Serrano, R. (1993). Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 12 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., and Jacobsen, S.E. (2007). Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Hernandez-Pinzon, I., Yelina, N.E., Schwach, F., Studholme, D.J., Baulcombe, D., and Dalmay, T. (2007). SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 50 140–148. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Molnar, A., Jones, A., and Baulcombe, D.C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 103 14994–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., Fahlgren, N., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., and Carrington, J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer, J.P., and Green, P.J. (2000). Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA 97 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, H., and Taira, K. (2005). Transcriptional gene silencing by short interfering RNAs. Curr. Opin. Mol. Ther. 7 125–131. [PubMed] [Google Scholar]
- Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E.J., Carrington, J.C., Liu, Y.P., Dolja, V.V., Calvino, L.F., Lopez-Moya, J.J., and Burgyan, J. (2006). Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Tej, S.S., Luo, S., Haudenschild, C.D., Meyers, B.C., and Green, P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Bartel, D.P., and Bartel, B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., and Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38 (suppl.): S31–S36. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., and Birchler, J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6 24–35. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Béclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Murguia, J.R., Belles, J.M., and Serrano, R. (1995). A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267 232–234. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., Krishnan, B.R., Brikun, I., Kulakauskas, S., Suziedelis, K., Tomcsanyi, T., Leyh, T.S., and Berg, D.E. (1992). CysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 174 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall, G.S., Codony-Servat, C., Byrne, J., Ritchie, L., and Hamilton, A. (2007). Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35 e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh, M., Dieterle, M., Marrocco, K., Lechner, E., Berry, B., Brault, V., Hemmer, O., Kretsch, T., Richards, K.E., Genschik, P., and Ziegler-Graff, V. (2006). F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA 103 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, L., and Meister, G. (2007). Argonaute proteins: Mediators of RNA silencing. Mol. Cell 26 611–623. [DOI] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Quintero, F.J., Garciadeblas, B., and Rodriguez-Navarro, A. (1996). The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, B.M., Pruss, G.J., and Vance, V.B. (2004). Plant viral suppressors of RNA silencing. Virus Res. 102 97–108. [DOI] [PubMed] [Google Scholar]
- Saito, K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sarmiento, C., Nigul, L., Kazantseva, J., Buschmann, M., and Truve, E. (2006). AtRLI2 is an endogenous suppressor of RNA silencing. Plant Mol. Biol. 61 153–163. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin, C., Bussell, J.D., Camus, I., Ljung, K., Kowalczyk, M., Geiss, G., McKhann, H., Garcion, C., Vaucheret, H., Sandberg, G., and Bellini, C. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17 1343–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret, F.F., Kastenmayer, J.P., and Green, P.J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15 173–183. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Mallory, A.C., and Bartel, D.P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. (2001). RNA silencing as a plant immune system against viruses. Trends Genet. 17 449–459. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M. (2005). The role of the RNAi machinery in heterochromatin formation. Cell 122 13–16. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411 834–842. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Lee, B., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Lee, H., Huang, R., and Zhu, J.K. (2004). A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J. 40 536–545. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., and Haley, B. (2005). Ribo-gnome: The big world of small RNAs. Science 309 1519–1524. [DOI] [PubMed] [Google Scholar]
- Zaratiegui, M., Irvine, D.V., and Martienssen, R.A. (2007). Noncoding RNAs and gene silencing. Cell 128 763–776. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yuan, Y.R., Pei, Y., Lin, S.S., Tuschl, T., Patel, D.J., and Chua, N.H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl, E., and Pauling, L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, V. Bryson and H.J. Vogel, eds (New York: Academic Press), pp. 97–166.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.