Abstract
C4 photosynthesis presents a sophisticated integration of two complementary cell types, mesophyll and bundle sheath cells. It relies on the differential expression of the genes encoding the component enzymes and transporters of this pathway. The entry enzyme of C4 photosynthesis, phosphoenolpyruvate carboxylase (PEPC), is found exclusively in mesophyll cells, and the expression of the corresponding gene is regulated at the transcriptional level. In the C4 dicot Flaveria trinervia, the mesophyll-specific expression of the C4 PEPC gene (ppcA) depends on a 41-bp segment in the distal promoter region referred to as MEM1 (for mesophyll expression module1). Here, we show that a MEM1 sequence found in the orthologous ppcA gene from the C3 species Flaveria pringlei is not able to direct mesophyll-specific gene expression. The two orthologous MEM1 sequences of F. pringlei and F. trinervia differ at two positions, a G-to-A exchange and the insertion of the tetranucleotide CACT. Changes at these two positions in the C3 MEM1 sequence were necessary and sufficient to create a mesophyll-specificity element during C4 evolution. The MEM1 of F. trinervia enhances mesophyll expression and concomitantly represses expression in bundle sheath cells and vascular bundles.
INTRODUCTION
The photosynthetic C4 cycle is a sophisticated add-on to the C3 photosynthetic pathway. It is characterized by an initial CO2 fixation step in the mesophyll cells by the oxygen-insensitive phosphoenolpyruvate carboxylase (PEPC), resulting in the C4 acids malate and/or aspartate. These C4 acids are subsequently transported to neighboring bundle sheath cells, where they are decarboxylated. The released CO2 is refixed by ribulose 1,5-bisphosphate carboxylase/oxygenase. Due to this prefixation of CO2 in the mesophyll cells, the photosynthetic C4 cycle acts as a pump that delivers high concentrations of CO2 to the site of ribulose 1,5-bisphosphate carboxylase/oxygenase in the bundle sheath cells. As a consequence of this CO2-concentrating mechanism, photorespiration in C4 plants is minimized and the net photosynthesis rate is increased (Hatch, 1987).
This division of labor between mesophyll and bundle sheath cells depends on differential gene expression (Nelson and Dengler, 1992). In NADP-malic enzyme–type C4 species, PEPC, NADP-malate dehydrogenase, and pyruvate orthophosphate dikinase are specifically expressed in mesophyll cells, whereas the decarboxylating enzymes, for instance NADP-dependent malic enzyme, and the secondary carboxylase ribulose 1,5-bisphosphate carboxylase/oxygenase, are expressed exclusively in bundle sheath cells. The cell-specific expression of these genes is regulated predominantly by transcription (Sheen, 1999); however, posttranscriptional control has been reported too (Patel et al., 2006).
The C4 pathway evolved independently >45 times during the evolution of angiosperms (Sage, 2004). The genes encoding the C4 isoforms of the C4 cycle enzymes originated from nonphotosynthetic progenitor genes that were already present in C3 ancestral species. To meet the special requirements of the C4 photosynthetic pathway, the expression program of the C3 progenitor genes had to be changed to a high and selective expression in the mesophyll or bundle sheath cells of the leaf, and the enzymes themselves had to be adapted to the metabolic and regulatory context of the C4 cycle (Bauwe and Chollet, 1986).
To gain insight into the evolution of C4 genes, we are using the entry enzyme of the C4 cycle, PEPC, as the model C4 enzyme/gene (Westhoff et al., 1997; Westhoff and Gowik, 2004) and the dicot genus Flaveria (Asteraceae) as the experimental system (Powell, 1978). This genus contains closely related C3, C4, and numerous C3-C4 intermediate species (Ku et al., 1991; McKown et al., 2005). These C3-C4 species differ quantitatively in the expression of C4 photosynthetic traits and are considered, at least partly, evolutionary intermediates (Monson and Moore, 1989).
The photosynthetic PEPCs of C4 Flaveria species are encoded by the ppcA gene class, whose orthologs are also found in C3 and C3-C4 intermediate species of this genus (Hermans and Westhoff, 1992). Analysis of ppcA promoter–β-glucuronidase (GUS) reporter gene fusions in the C4 plant F. bidentis revealed that the ppcA promoter of the C4 species F. trinervia directs a high expression of the reporter gene in the mesophyll cells. The orthologous ppcA promoter of the C3 plant F. pringlei, however, is weak and does not show any apparent cell or organ specificity (Stockhaus et al., 1997).
Detailed promoter reporter gene studies of the C4 ppcA gene in transgenic F. bidentis plants indicated that the proximal region (−1 to −570) and the distal region (−1566 to −2141) are sufficient for high mesophyll-specific expression of the GUS reporter gene (Gowik et al., 2004). While the proximal promoter region mediates very low basal promoter activity, the distal region confers a mesophyll expression component when fused to the ppcA promoter of F. pringlei. By dissection of the C4 distal region, a 41-bp module, named MEM1 (for mesophyll expression module1), was identified that together with the C4 proximal region is sufficient for mesophyll-specific reporter gene expression (Gowik et al., 2004).
MEM1 could be subdivided into two submodules, A and B, of 11 and 30 bp, respectively. The comparison of MEM1 sequences from C4 Flaveria species and of MEM1 homologs from C3 species of this genus revealed that the A submodules of the C4 and C4-like species have a guanine at their first nucleotide position, while an adenine is present in the A submodules of the C3 plants (Gowik et al., 2004). An additional difference is related to the tetranucleotide CACT, which is present in the B submodules of the C4 and C4-like plants but absent from the B submodules of the C3 plants (Gowik et al., 2004).
These data suggested that both nucleotide polymorphisms are involved in determining the mesophyll-specific expression of the C4 ppcA gene (Gowik et al., 2004). Hence, this investigation was initiated to identify at the nucleotide level the determinants for the mesophyll-specific expression of the C4 ppcA gene. By the analysis of MEM1 deletion– and substitution–reporter gene constructs in transgenic F. bidentis plants, it was found that both submodules of the MEM1 have to be present in the C4-specific state (i.e., the guanine in the A submodule and the tetranucleotide CACT in the B submodule of MEM1) in order to provide mesophyll-specific expression of the reporter gene. The C4 MEM1 behaves as an enhancer of mesophyll expression and, in addition, as a repressor of ppcA gene expression in the bundle sheath cells and the vascular bundles.
RESULTS
MEM1 Displays Transcriptional Enhancing and Repressor Activities
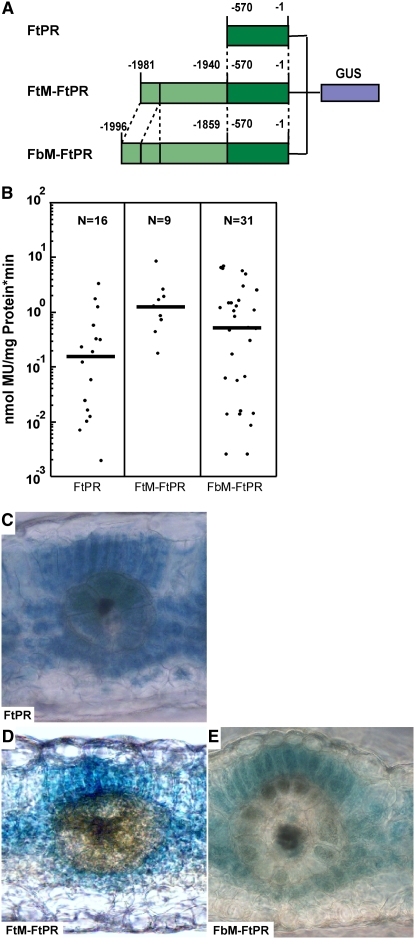
The proximal promoter region of the ppcA gene of F. trinervia (C4) (construct FtPR in Figure 1A) revealed only very weak activity in previous experiments, preventing an unequivocal in situ analysis (Gowik et al., 2004). To elucidate the expression specificity of this basal promoter, this experiment was repeated and F. bidentis was retransformed with this construct, and the histochemical activity of the GUS reporter gene was analyzed in the 16 obtained transgenic plants. The majority of the plants showed no GUS staining. In all five stainable plants, the GUS reporter gene was found to be expressed in mesophyll and bundle sheath cells but also in the vascular tissue (Figure 1C). Thus, the basal ppcA promoter directs no cell specificity.
Figure 1.
Analysis of the ppcA GUS Reporter Gene Constructs FtPR, FtM-FtPR, and FbM-FtPR in Transgenic F. bidentis.
(A) Schematic presentation of the ppcA GUS chimerical genes. Nucleotide numbers refer to the translation initiation codon. The Ft MEM1 region is indicated by light green boxes, and the proximal region (PR) is indicated by dark green boxes. The state of the C3/C4-associated polymorphisms in the A submodule (G or A) and the B submodule (presence or absence of CACT) of MEM1 are indicated.
(B) GUS activities in leaves of transgenic F. bidentis plants. The median values (black bars) of the GUS activities are expressed in nanomoles of the reaction product 4-methylumbelliferone (MU) generated per milligram of protein per minute. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column.
(C) to (E) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with the FtPR (C), FtM-FtPR (D), or FbM-FtPR (E) construct. Incubation times were 25 h (C), 12 h (D), and 24 h (E).
The fusion of the MEM1 module of the C4 ppcA promoter of F. trinervia to the proximal promoter region (construct FtM-FtPR in Figure 1A) results in a statistically significant (P < 0.01) eightfold elevated promoter activity compared with the activity of the proximal region alone (Figure 1B), indicating that MEM1 of F. trinervia contains transcriptional enhancing activity. In situ analysis revealed clear mesophyll-specific expression, while no GUS activity could be detected in the bundle sheath cells and the vascular bundles (Figure 1D). Therefore, MEM1 of F. trinervia not only enhances expression in mesophyll cells but concomitantly represses expression in bundle sheath cells and vascular tissues.
Insertions between the A and B Submodules of MEM1 Do Not Affect Mesophyll Specificity
The MEM1 of F. trinervia is unique in that the A and B submodules are fused together with no intermediate sequence. By contrast, the A and B submodules of MEM1 of the C4, C4-like, and C3 species of Flaveria are separated by ∼90 to 100 bp of intervening sequences (Gowik et al., 2004). To investigate the effect of these spacer sequences on mesophyll expression specificity, MEM1 of the C4 plant F. bidentis was fused to the proximal region of the ppcA promoter of F. trinervia and the resulting promoter reporter gene construct FbM-FtPR (Figure 1A) was transformed into F. bidentis. Histochemical analysis of transgenic plants, carrying the chimerical gene, showed that FbM-FtPR directs GUS expression in mesophyll cells (Figure 1E) not distinguishable from that of the FtM-FtPR promoter. The expression strength of FbM-FtPR is approximately four times higher than that of the proximal promoter, reinforcing the notion that a C4 MEM1 adds a mesophyll-specific enhancing activity to the proximal promoter. We conclude from these experiments that the spacer between the A and B submodules in MEM1 of F. bidentis does not contain any cis-regulatory element of relevance for the mesophyll specificity of gene expression.
The C3-Type MEM1 of F. pringlei Does Not Confer Mesophyll-Specific Gene Expression
The comparison of MEM1 sequences from Flaveria species differing in the mode of photosynthesis and preliminary experiments described by Gowik et al. (2004) suggested that the single G/A nucleotide polymorphism in the A submodule and the insertion/deletion of the CACT tetranucleotide in the B submodule might be key determinants for mesophyll specificity. If so, a C3-type MEM1 element, when fused to the proximal promoter region of the C4 ppcA promoter, should not direct mesophyll-specific gene expression.
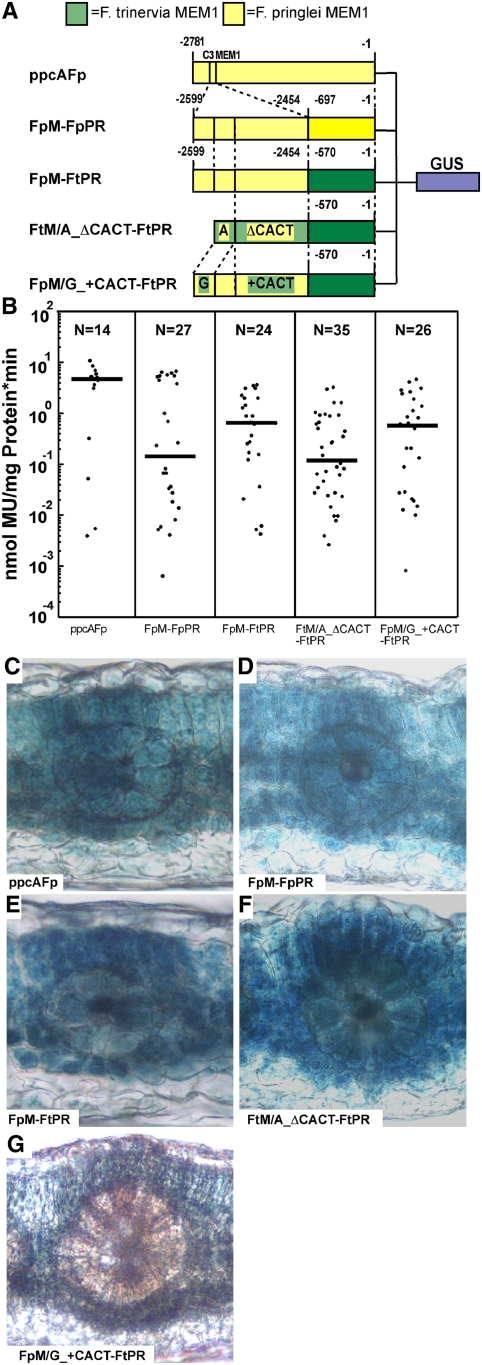
In order to examine the expression specificity of a C3-type MEM1, we used the orthologous ppcA promoter of the C3 species F. pringlei. The previously examined ppcA promoter construct of F. pringlei did not contain the complete MEM1 (Stockhaus et al., 1997). Hence, it was necessary to isolate a bona fide complete ppcA promoter of F. pringlei (i.e., containing the entire MEM1 element) by vectorette PCR. The obtained fragment of 2781 bp was fused to the GUS reporter gene (construct ppcAFp in Figure 2A), and the expression profile and strength of this promoter were analyzed in transgenic F. bidentis. Histochemical analysis of ppcAFp revealed expression in mesophyll cells and bundle sheath cells and in the vascular bundle (Figure 2C). It follows that this promoter does not show any apparent cell-specific expression. Its expression profile as well as its strength (Figure 2B) is indistinguishable from those of the previously analyzed ppcA promoter of F. pringlei, which was truncated by 198 bp (Stockhaus et al., 1997).
Figure 2.
Analysis of the ppcA GUS Reporter Gene Constructs ppcAFp, FpM-FpPR, FpM-FtPR, FtM/A_ΔCACT-FtPR, and FpM/G_+CACT-FtPR in Transgenic F. bidentis.
(A) Schematic presentation of the ppcA GUS chimerical genes. Nucleotide numbers refer to the translation initiation codon. The Ft MEM1 region is indicated by light green boxes, and the Fp MEM1 region is indicated by light yellow boxes. The Ft proximal region (PR) is indicated by dark green boxes, and the Fp PR is indicated by dark yellow boxes. The state of the C3/C4-associated polymorphisms in the A submodule (G or A) and the B submodule (presence or absence of CACT) of MEM1 are indicated.
(B) GUS activities in leaves of transgenic F. bidentis plants. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column. Median values (black bars) are shown. MU, 4-methylumbelliferone.
(C) to (G) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with ppcAFp (C), FpM-FpPR (D), FpM-FtPR (E), FtM/A_ΔCACT-FtPR (F), and FpM/G_+CACT-FtPR (G). Incubation times were 2 h (C), 3.5 h (D), 18 h (E), 17 h (F), and 3 h (G).
To assess the expression specificity of the C3-type MEM1 of F. pringlei in the context of the proximal region of the C4 ppcA promoter of F. trinervia, the corresponding construct FpM-FtPR (Figure 2A) was prepared and introduced into F. bidentis. For comparison, the C3-type MEM1 was also fused to its native proximal region (FpM-FpPR in Figure 2A). Both promoter constructs gave rise to the same expression pattern as ppcAFp (i.e., GUS expression was detected in the mesophyll and bundle sheath cells and in the vascular tissue) (Figures 2D and 2E). This suggests that the C3/C4-associated sequence polymorphisms in MEM1 are necessary for the mesophyll specificity of gene expression and that the proximal ppcA promoter segment does not interfere with the pattern of expression.
The FpM-FpPR promoter exhibits the same pattern of reporter gene expression as the bona fide complete ppcA promoter of F. pringlei (Figures 2C and 2D). However, the FpM-FpPR promoter is ∼26 times lower in expression strength than the complete promoter (Figure 2B). This indicates that the nucleotide sequences between MEM1 and the proximal part of the ppcA promoter of F. pringlei contain elements that enhance the overall expression of this C3-type ppcA promoter. This is similar to what was observed when the expression strength of the corresponding constructs of the C4 ppcA promoter were analyzed (Gowik et al., 2004).
Conversion of a C3 to a C4 MEM1 and Vice Versa Reveals That Two C3/C4-Associated Nucleotide Sequence Polymorphisms Are Sufficient for Mesophyll Expression
In order to clarify whether the two C3/C4-associated differences in MEM1 are the only determinants of mesophyll specificity in a C4-type MEM1, the A and B submodules of the C4 MEM1 of F. trinervia were changed into C3-type MEM1 submodules (FtM/A_ΔCACT-FtPR in Figure 2A) and the A and B submodules of the C3 MEM1 of F. pringlei were changed into C4-type MEM1 submodules (FpM/G_+CACT-FtPR in Figure 2A). The C3-type FtM/A_ΔCACT-FtPR promoter construct revealed the same expression pattern as the C3-type promoter constructs FpM-FpPR and FpM-FtPR (i.e., GUS staining was found in mesophyll and bundle sheath cells and in the vascular bundles) (Figures 2D to 2F). The C4-equivalent FpM/G_+CACT-FtPR promoter construct, however, directed mesophyll-specific expression behavior (Figure 2G).
It follows that the change of the A and B submodules of MEM1 from a C4 state into a C3 state results in the loss of expression specificity. On the other hand, the change of MEM1 from a C3 state into a C4 state leads to the acquisition of mesophyll specificity by repressing gene activity in the bundle sheath cells and the vascular bundles.
The A and B Submodules of MEM1 Are Both Required for Mesophyll-Specific Expression
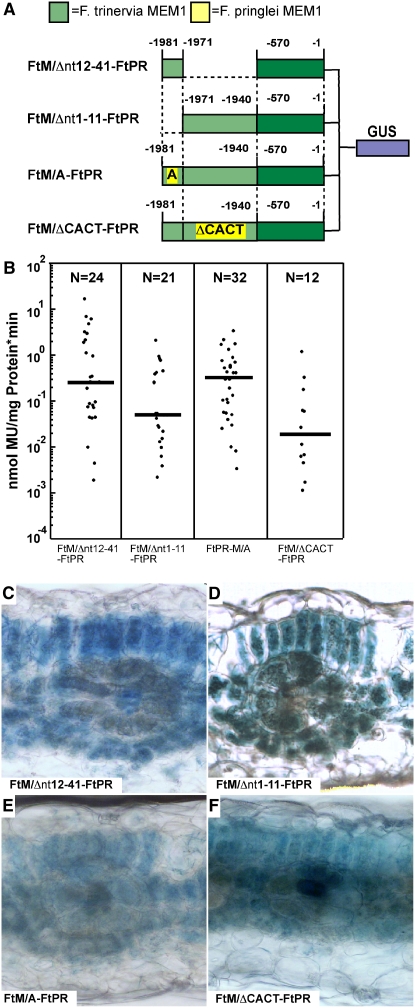
The two C4-associated polymorphisms correlate with mesophyll specificity. In order to find out whether both C3- and C4-correlated differences are necessary for mesophyll-specific gene expression, we followed two strategies. First, we divided the F. trinervia MEM1 into the 11-bp A submodule and the 30-bp B submodule and fused each submodule with the C4 proximal promoter region (constructs FtM/Δnt12-41-FtPR and FtM/Δnt1-11-FtPR in Figure 3A). Second, we converted independently the A and B submodules of the C4 MEM1 into C3-type submodules and combined each chimeric MEM1 module with the proximal part of the C4 ppcA promoter (constructs FtM/A-FtPR and FtM/ΔCACT-FtPR in Figure 3A) (Gowik et al., 2004).
Figure 3.
Analysis of the ppcA GUS Reporter Gene Constructs FtM-FtPR/Δnt12-41, FtM-FtPR/Δnt1-11, FtM/A-FtPR, and FtM/ΔCACT-FtPR in Transgenic F. bidentis.
(A) Schematic presentation of the ppcA GUS chimerical genes. Nucleotide numbers refer to the translation initiation codon. The Ft MEM1 region is indicated by light green boxes, and the Ft proximal region (PR) is indicated by dark green boxes. The state of the C3/C4-associated polymorphisms in the A submodule (G or A) and the B submodule (presence or absence of CACT) of MEM1 are indicated.
(B) GUS activities in leaves of transgenic F. bidentis plants. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column. Median values (black bars) are shown. MU, 4-methylumbelliferone.
(C) to (F) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with FtM/Δnt12-41-FtPR (C), FtM/Δnt1-11-FtPR (D), FtM/A-FtPR (E), and FtM/ΔCACT-FtPR (F). Incubation times were 21 h ([C], [D], and [F]) and 24 h (E). Note that some FtM/ΔCACT-FtPR plants were analyzed previously by Gowik et al. (2004). In the course of this study, they were reanalyzed by extending the staining period to 2 d in order to increase the sensitivity of detection. In addition, new plants were generated.
Deletion of one submodule (constructs FtM/Δnt12-41-FtPR and FtM/Δnt1-11-FtPR) caused a loss of mesophyll specificity (Figures 3C and 3D) (i.e., the great majority of the GUS-stainable plants expressed the reporter gene in mesophyll and bundle sheath cells as well in the vascular tissue). The same expression pattern (i.e., the loss of mesophyll specificity) was also observed when one submodule was in the C3 state and the other remained in the C4 state (Figures 3E and 3F).
We conclude that both C3- and C4-associated nucleotide sequence polymorphisms in MEM1 have to be in the C4 state for robust mesophyll-specific gene expression and that one C4-type submodule is not sufficient.
Evolutionary Origin of MEM1 in the Genus Flaveria
The experiments performed confirm the significance of both C3- and C4-associated nucleotide polymorphisms in MEM1 as being indispensable for mesophyll-specific ppcA gene expression in C4 Flaveria species. A previously performed comparative analysis of ppcA gene sequences of two C4 plants (F. trinervia and F. bidentis), two C4-like plants (F. palmerii and F. vaginata), and two C3 plants (F. cronquistii and F. pringlei) revealed MEM1-like sequences in the ppcA gene of all six Flaveria species (Gowik et al., 2004). A C4-specifc guanine at the first position in the A submodule and the tetranucleotide CACT in the B submodule is found in the MEM1 of the C4 and C4-like plants, whereas in the MEM1 of the two C3 plants, a C3-specific adenine is present and the tetranucleotide is absent (Gowik et al., 2004).
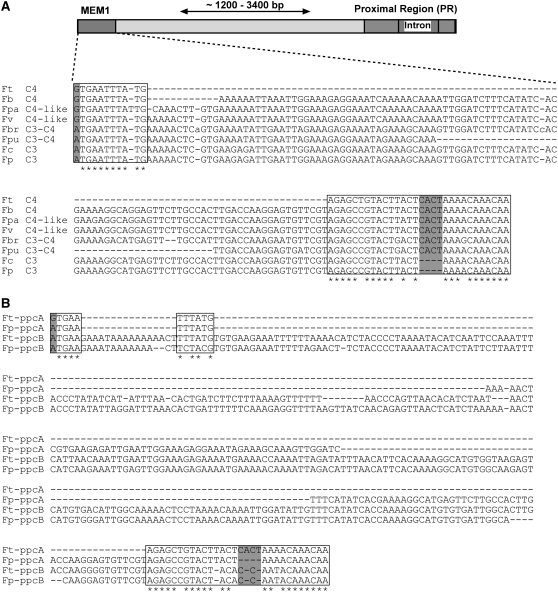
According to the recently published phylogeny of the genus Flaveria, which was based on both morphological and molecular characteristics, all C4 and C4-like species form a distinct clade (clade A), while the C3 species F. pringlei and F. cronquistii are basal (McKown et al., 2005). The C3-C4 intermediate species of the genus are contained within clade B (McKown et al., 2005). We were interested in whether a MEM1 sequence is also found in the ppcA genes of C3-C4 intermediate Flaveria species and which states of MEM1 submodules are present. We selected F. pubescens and F. brownii, the latter of which is considered to be a C4-like C3-C4 intermediate. ppcA promoter sequences for F. brownii and F. pubescens with a length of 4030 and 4596 bp, respectively, were isolated, and MEM1 sequences were identified in each of these ppcA promoters. In the F. brownii promoter, MEM1 was located at 3830 bp upstream of the translational start, while in the F. pubescens promoter, MEM1 was positioned at 4008 bp upstream of the ATG start codon. The MEM1 sequences of both Flaveria species revealed an intermediate character: they possess a C3-specifc adenine at the first position in the A submodules, whereas the B submodules are of a C4 type, due to the presence of the tetranucleotide CACT (Figure 4A).
Figure 4.
Comparisons of MEM1 and MEM1-Like Sequences.
(A) MEM1 sequences of the ppcA promoters from C4, C4-like, C3-C4 intermediate, and C3 Flaveria species. Ft, F. trinervia; Fb, F. bidentis; Fpa, F. palmerii; Fv, F. vaginata; Fbr, F. brownii; Fpu, F. pubescens; Fc, F. cronquistii; Fp, F. pringlei.
(B) Comparison of MEM1 sequences from the ppcA promoters of F. trinervia (Ft) and F. pringlei (Fp) with their MEM1-like counterparts from the ppcB genes. The MEM1 A and B submodules are highlighted by boxes. Asterisks label identical nucleotides in the A or B submodule of all promoters. Gray bars indicate the single nucleotide difference in the A submodule and the insertion/deletion of the CACT tetranucleotide in the B submodule. For a comparison of the whole promoters, see Supplemental Figure 1 online.
Phylogenetic analysis indicated that the current ppcA and ppcB genes arose from an ancestral ppcB-like gene by gene duplication (Bläsing et al., 2002). In order to elucidate the evolutionary origin of MEM1 further, we analyzed ppcB promoter sequences from the C4 plant F. trinervia and the C3 plant F. pringlei (Ernst and Westhoff, 1996). In both promoters, MEM1-like sequences could be identified, hereafter referred to as MEM1*. The MEM1* modules of the ppcB genes of F. trinervia and F. pringlei are quite similar. Both MEM1* submodules are in the C3 state, with an adenine at the first nucleotide position of the A submodules and no CACT motif in the B submodules. The A submodules of MEM1* of the ppcB genes are not contiguous; rather, they are interrupted by insertions of 13 and 15 bp, respectively (Figure 4B; see Supplemental Figure 1 online).
The Full-Length C4 ppcA Promoter of F. trinervia Is Active in the C3 Plant Arabidopsis thaliana but Does Not Show Cell Specificity of Expression in the Leaves
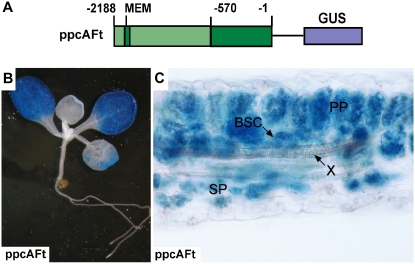
The data presented demonstrate that the evolutionary transition from C3 to C4 involved changes in the cis-regulatory modules of the C4 genes. However, they leave open whether this transition was also accompanied by trans-regulatory changes that modify the activity or expression of factors interacting with these cis-regulatory modules (Wray et al., 2003). Do C3 Flaveria species already contain the trans-regulatory system that could interpret the cis-regulatory logic of the C4 ppcA promoter correctly to be active only in mesophyll cells? Unfortunately, a transformation protocol for C3 Flaveria species does not exist; therefore, the expression specificities of C3- and C4-type MEM1 or of the corresponding full-size promoters could not be investigated in the homologous background. However, anatomical data clearly indicate that leaves of C3 dicots contain bundle sheath cells (Esau, 1977; Kinsman and Pyke, 1998), suggesting that the differentiation of the leaf chlorenchyma into mesophyll and bundle sheath cells is not restricted to C4 plants but is a common feature of many C3 angiosperms. Therefore, we wanted to know whether a heterologous C3 dicot with documented bundle sheath cells, the Brassicaceae species Arabidopsis (Kinsman and Pyke, 1998), correctly recognizes the cis-regulatory elements for mesophyll-specific gene expression of an asteracean C4 dicot. We introduced the full-size ppcA promoter of F. trinervia linked to the GUS reporter gene into Arabidopsis and analyzed its expression specificity in the leaf.
Transformation of Arabidopsis plants with the C4 ppcA promoter–GUS reporter gene resulted in a distinct GUS expression in the leaves. Several independent transgenic Arabidopsis lines were investigated, and most of them showed strong GUS staining in the leaves, while almost no GUS activity was histochemically detectable in stems or roots (Figure 5B). This indicates that the activity of this promoter in roots and stems is much lower than that in leaves.
Figure 5.
Analysis of the ppcA GUS Reporter Gene Construct FtppcA in Transgenic Arabidopsis.
(A) Schematic presentation of the ppcA GUS chimerical gene. Nucleotide numbers refer to the translation initiation codon. The Ft MEM1 region and the proximal region (PR) are indicated by dark green boxes.
(B) GUS staining of a transgenic Arabidopsis seedling. Incubation time was 2.5 h.
(C) Histochemical localization of GUS activity in a leaf section of a transgenic Arabidopsis plant transformed with FtppcA. Incubation time was 2.5 h. BSC, bundle sheath cells; PP, palisade parenchyma; SP, sponge parenchyma; X, xylem.
Histochemical analysis of GUS activity in the leaves showed that the promoter was active in palisade and spongy parenchyma, in the bundle sheath, and in the vascular bundles (Figure 5C). It follows that the C4 ppcA promoter exhibits no cell specificity in the leaves of the C3 plant Arabidopsis.
DISCUSSION
The acquisition of new functions for a preexisting gene, usually through changes in expression patterns and/or functional modifications of the encoded protein, is known as gene cooption and requires gene duplication events (Olson, 2006). Changes in the expression of a particular gene can result from alterations either in its cis-regulatory sequences or in the deployment and function of the transcription factors that control gene expression, or both (Love et al., 2007). Evolutionary biologists have collected convincing evidence that supports the view that changes in the spatiotemporal expression patterns of genes are a principal mechanism for the evolution of novelty, both in morphological and biochemical traits (Doebley and Lukens, 1998). The multiple independent origins of C4 photosynthesis in the angiosperms provide a good example for studying the evolution of novel morphological and biochemical traits. The evolution of C4 cycle enzymes is proposed to have required gene duplication with subsequent diversification through neofunctionalization (Monson, 2003).
In this study, we investigated the evolution of a cis-regulatory module, MEM1, that is necessary and sufficient for mesophyll-specific gene expression. We found that changes at two positions in the 41-bp module (i.e., an A-to-G conversion in the A submodule and the addition of the tetranucleotide CACT in the B submodule) convert an element with no obvious function into a mesophyll-specificity module.
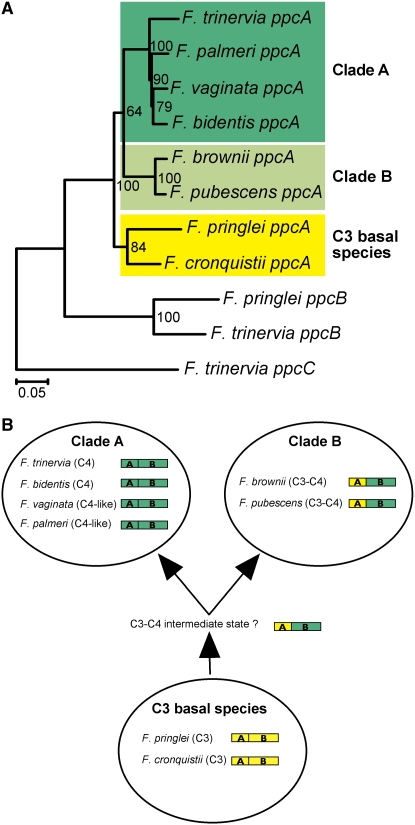
MEM1 sequences were identified in the promoters of ppcA genes from eight different C4, C4-like, C3-C4 intermediate, and C3 Flaveria species (Figure 4A). A phylogenetic tree based on the sequences of these eight promoters is in agreement with the phylogeny of the genus Flaveria as proposed by McKown et al. (2005) (Figure 6A). The ppcA promoter tree indicates that the ppcA genes of clade A and clade B evolved separately from the ancestral ppcA genes, which resemble the ppcA genes of the present basal C3 Flaveria species. This supports the conclusion of McKown and coworkers (2005) that C4 traits evolved independently in Flaveria clades A and B.
Figure 6.
MEM1 Evolution in the Genus Flaveria.
(A) Phylogenetic tree of the ppcA, ppcB, and ppcC PEPC promoter sequences of Flaveria. The sequence alignment was constructed with Dialign (Morgenstern et al., 1998) and corrected by hand. The tree was generated by the distance method as implemented in PAUP 4.0b10 (Swofford, 2002), where uninformative characteristics were excluded. Bootstrap analysis was performed with 1000 replications, and the obtained bootstrap values are indicated.
(B) Model of MEM1 evolution in the genus Flaveria. The model relies on the phylogeny of the genus Flaveria, which is based on morphological and molecular data sets (McKown et al., 2005).
Comparison of the MEM1 sequences from the eight Flaveria species exhibiting different modes of photosynthesis (Figure 4A) suggests a scenario for how a ubiquitously expressed C3-type PEPC gene was converted into a cell-specific C4-type PEPC gene (Figure 6B). All C4 and C4-like species of clade A possess MEM1 sequences with both submodules in the C4 state, while the two basal C3 species have C3-type submodules. The MEM1 modules of the C4-like and the C3-C4 intermediate species of clade B are intermediate, with a C3-type submodule A and a C4-type submodule B. Based on the clear phylogenetic separation of the two clades, a logical hypothesis is that the last common ancestor of clades A and B should already have had a C4-type submodule B and that the insertion of the tetranucleotide CACT in the B submodule of MEM1 occurred before the formation of the A and B clades. A further evolutionary step, an A-to-G exchange in the A submodule of MEM1, led to the formation of a mesophyll-specificity element in the A clade.
How can these changes in a cis-regulatory cell-specificity element of gene expression be reconciled with the evolutionary pathway of C4 traits in Flaveria? Sage (2004) proposed a framework model of how C4 photosynthesis evolves. Since this photosynthetic pathway evolved independently even within some angiosperm families, Sage concluded that the evolution of C4 photosynthesis required genetic and anatomical preconditions. At the genome level, large numbers of duplicated genes should exist that would allow the modification of the duplicated copies without risking deleterious effects caused by the loss of the original gene functions. At the level of leaf anatomy, the distance between mesophyll and bundle sheath cells should have declined, either by a reduction in the interveinal distance or by the enhancement of the bundle sheath layer.
In Flaveria, the duplication of a pre-ppcB–like progenitor ppc gene led to the ppcA gene class that is present in all Flaveria species and served as the evolutionary substrate for the creation of the C4 isoform PEPC gene (Westhoff and Gowik, 2004). The present ppcB genes contain a MEM1-like sequence (Figure 4B); therefore, an ancestral MEM1-like module was already present in the pre-ppcB genes. This MEM1-like module was further modified in the progenitor C3 species of Flaveria, and a MEM1 module evolved that required only two relatively small changes in order to become an expression module for mesophyll specificity. A C3-type MEM1 module is found in the basal C3 Flaveria species F. pringlei and F. cronquistii, which are typical C3 plants based on their biochemical and physiological characteristics (Edwards and Ku, 1987). The two species lack pronounced C4 anatomical and vein-patterning characteristics, indicating that structural preconditioning is limited (McKown and Dengler, 2007). Instead, the basal C3 species exhibit preconditioning at the cis-regulatory level.
The derived C3 species F. robusta, by contrast, is already preconditioned anatomically. It shows a higher vein density compared with F. pringlei and F. cronquistii (McKown and Dengler, 2007), but its biochemical and physiological features are still clearly C3 (Edwards and Ku, 1987). It would be interesting to know whether the anatomical preconditioning is accompanied by further advances in MEM1 evolution and whether the CACT tetranucleotide has already been inserted into the B submodule of MEM1.
Clade A of Flaveria is solidly C4 or C4-like, with the exception of F. ramosissima, which is the only true C3-C4 intermediate of this clade and represents its basal species (McKown and Dengler, 2007). According to physiological data, the ppcA PEPC should not be expressed mesophyll-specifically in F. ramosissima (Ku et al., 1991); therefore, one would expect a MEM1 with submodule A in the C3 state.
All species of clade B are C3-C4 intermediates except for F. brownii, which has been classified as a C4-like C3-C4 intermediate (Cheng et al., 1988). F. pubescens is much less advanced than F. brownii in progression toward C4 photosynthesis; therefore, these two species may be regarded as representative for the degree of C3-C4 intermediacy found in clade B of Flaveria (Edwards and Ku, 1987). We infer from this that MEM1 did not change during further evolution of the B clade species (i.e., its A submodule remains in the C3 state). The presence of a hybrid MEM1 module in all Flaveria species of the B clade suggests that their ppcA PEPC should not be expressed in a mesophyll-specific manner. This is in accordance with previous reports (Reed and Chollet, 1985). Even in the C4-like species F. brownii, PEPC activity is not strictly compartmentalized and is found in bundle sheath cells (Cheng et al., 1989). This reinforces the notion that a MEM1 with both submodules in the C4 state is imperative for a mesophyll-specific transcription of the ppcA PEPC genes. It suggests furthermore that MEM1 may be the only, or at least the dominant, cis-regulatory module for the mesophyll specificity of ppcA PEPC gene expression in Flaveria.
While the C4-type MEM1 acts as an expression module for mesophyll specificity, the function of its C3 counterpart is not known. The incomplete C3-type MEM1 in the original ppcA promoter of F. pringlei resulted in the same expression pattern obtained for the complete promoter, suggesting that the C3 MEM1 is dispensable (Stockhaus et al., 1997) (Figure 2C). However, MEM1 is conserved in the two C3 species F. pringlei and F. cronquistii (Figure 4A), indicating that this sequence element is functional. Furthermore, MEM1-like (MEM1*) sequences are also found in the nonphotosynthetic ppcB PEPC genes of F. pringlei and F. trinervia (Ernst and Westhoff, 1996) (Figure 4B). While the complete ppcA and ppcB promoters of F. pringlei and F. trinervia share 19 to 22% identical nucleotides, 75 to 82% identical nucleotides are found if only the MEM1 and MEM1* sequences of these promoters are compared. This conservation of MEM1 and MEM1* sequences suggests a function of these elements also in the promoters of the ppcB and the nonphotosynthetic ppcA genes, even if this function remains obscure.
The full-size C4 ppcA promoter of F. trinervia does not show any cell specificity in the leaves of the heterologous C3 dicot Arabidopsis (i.e., the promoter is active in all leaf parenchyma cells, including the bundle sheath, and the vascular bundles). When this promoter was analyzed in tobacco (Nicotiana tabacum), a member of the Solanaceae, the promoter was found to be active in the palisade cells but not in the spongy parenchyma cells. No expression was observed in the vascular bundles (Stockhaus et al., 1994). Thus, in both heterologous C3 backgrounds, the mesophyll specificity of expression is not maintained but the expression patterns are different. One may conclude that the trans-regulatory systems operating in the leaf cells of Arabidopsis and tobacco differ from that of F. bidentis and that this difference causes the nonspecific expression of the C4 ppcA promoter. However, the multiple origin of C4 photosynthesis in the angiosperms could involve multiple independent selections of cis-regulatory modules for cell-specific gene expression; therefore, the mesophyll-specificity module MEM1 could be specific for Flaveria. Indeed, the Solanaceae does not contain any C4 species, nor does the Brassicaceae (Sage, 2004)—if ones treats the Brassicaceae and the Cleomaceae, the latter of which contains C4 species (Sage, 2004), as separate families (Hall et al., 2002).
Even within the Poaceae, the expression of C4 genes from the panicoid C4 grasses Zea mays and Panicum miliaceum (Matsuoka et al., 1994; Nomura et al., 2005b) and the chloridoid C4 grass Zoysia japonica (Nomura et al., 2005a) in the C3 grass Oryza sativa did not generally result in the maintenance of cell specificity. While some C4 gene promoters maintain their cell specificity of expression, others do not. Together, these data on the heterologous expression of C4 genes in C3 plants indicate that species- or genus-specific cis- and trans-regulatory systems evolved that may differ from gene to gene.
The C4 MEM1 is a cis-regulatory module with a dual function. It represses the expression of the linked gene in bundle sheath cells and the vascular tissue and concomitantly enhances transcription in the mesophyll cells. Interestingly, this silencing function of C4 MEM1 acts in both the bundle sheath cells and the vascular tissue, suggesting that with respect to the expression of the ppcA gene, both tissues are coregulated. In agreement with this, there is histological evidence that at least in C4 grasses of the NADP-malic enzyme subtype and in C4 Cyperaceae species, bundle sheath cells and vascular tissue are of common origin and derived from the procambrium, while mesophyll cells originate from the ground meristem (Dengler et al., 1985; Soros and Dengler, 2001).
Cis-regulatory enhancers and silencers function similarly. They are recognized by DNA binding proteins that influence transcription initiation either by contacting the transcription machinery directly via protein–protein interaction or by altering the chromatin structure by recruiting histone-modifying enzymes or chromatin-remodeling complexes (Blackwood and Kadonaga, 1998; Gaston and Jayaraman, 2003). Repressor proteins function in diverse ways, for instance by competing with transcriptional activator proteins for a common cis-regulatory element or by direct interaction with those proteins (Maldonado et al., 1999).
How transcriptional enhancement and repression can be explained mechanistically in the case of MEM1 is unclear at present. The two polymorphic sites are 25 bp apart in F. trinervia but are separated by 122 bp in F. bidentis. Since both MEM1 modules direct the same expression specificity, the distance of the two polymorphic sites is not relevant. One may infer that trans-regulatory factors bind separately to the A and B submodules of MEM1. Preliminary analysis with the yeast one-hybrid system revealed that trans-regulatory factors of the basic leucine zipper (bZIP) protein family interact strongly with the C4-type MEM1, while interaction with the C3-type MEM1 is relatively weak (Akyildiz, 2007). The binding sites of these bZIP-type proteins within MEM1 have not been determined, and their in vivo relevance for the control of expression of the ppcA gene in C4 Flaveria species has not yet been investigated. It is also not known whether other trans-regulatory factors may interact with these bZIP proteins and are required for the functioning of MEM1 (Després et al., 2000).
The data presented here show that small changes in nucleotide sequence were sufficient to create a novel mode of gene expression. Since such small changes are likely to occur in plant genomes quite easily, it is conceivable that the compartmentalized gene expression in C4 plants arose many times independently during the evolution of angiosperms. The molecular anatomy and evolution of a cis-regulatory module for cell-specific gene expression have been elucidated at the nucleotide level in this study. It will be interesting to know whether MEM1 represents a universal cis-regulatory module for mesophyll-specific gene expression in Flaveria. The C4 carbonic anhydrase of Flaveria is a good choice to answer this question (Burnell and Hatch, 1988). It may be even more interesting to investigate how mesophyll-specific gene expression was achieved in other families of the angiosperms that evolved C4 species. The genomes of the Brassicaceae are intensively studied at present; therefore, the genus Cleome, with its C4 and C3 species, might be a good model system for a comparative analysis at the genome level (Brown et al., 2005).
METHODS
DNA manipulations were performed according to Sambrook and Russell (2001). All DNA fragments created by PCR were confirmed by DNA sequencing. Plasmid pBluescript II SK+ [pBIISK(+); Stratagene] was used for standard cloning in Escherichia coli (Sambrook and Russell, 2001).
Construction of a Complete Promoter of Flaveria pringlei (ppcAFp)
Inspection of the previously used ppcA promoter of F. pringlei (named ppcA-L-Fp; nucleotides −1 to −2583) (Stockhaus et al., 1994) revealed that the A submodule of MEM1 was lacking. For the generation of a complete ppcA promoter of F. pringlei (nucleotides −1 to −2781), PCR was performed with the oligonucleotide primers Fp-5′HindIII and Fp-3′HpaI (Table 1) and genomic DNA of F. pringlei as template. The resulting DNA fragment was subcloned into pBIISK(+). After digestion with HindIII/HpaI, the released fragment was inserted into the HindIII/HpaI-digested vector ppcA-L-Fp (Stockhaus et al., 1994).
Table 1.
Oligonucleotides Used for the Generation of MEM1 Reporter Gene Vectors
| Promoter–GUS-Reporter Gene Vectors | Oligonucleotides/Primers | Sequences (5′ to 3′) |
|---|---|---|
| FtM/Δnt11-FtPR | FtDEΔAB5′HindIII | AGCTTAGAGCTGTACTTACTCACTAAAACAAACAAT |
| FtDEΔAB3′XbaI | CTAGATTGTTTGTTTTAGTGAGTAAGTACAGCTCTA | |
| FtM/Δnt12-41-FtPR | FtDEAΔB5′HindIII | AGCTTGTGAATTTATGT |
| FtDEAΔB3′XbaI | CTAGACATAAATTCACA | |
| FtM/A-FtPR | FtDEaB5′HindIII | AGCTTATGAATTTATGAGAGCTGTACTTACTCACTAAAACAAACAAT |
| FtDEaB3′XbaI | CTAGATTGTTTGTTTTAGTGAGTAAGTACAGCTCTCATAAATTCATA | |
| FtM/ΔCACT-FtPR | FtDEAb5′HindIII | AGCTTGTGAATTTATGAGAGCTGTACTTACTAAAACAAACAAT |
| FtDEAb3′XbaI | CTAGATTGTTTGTTTTAGTAAGTACAGCTCTCATAAATTCACA | |
| FtM/A_ΔCACT-FtPR | FtDEab5′HindIII | AGCTTATGAATTTATGAGAGCTGTACTTACTAAAACAAACAAT |
| FtDEab3′XbaI | CTAGATTGTTTGTTTTAGTAAGTACAGCTCTCATAAATTCATA | |
| FbM-FtPR | FbDEAB5′HindIII | GGGAAGCTTGTGAATTTATGAAAAAATTAAATTGGAAAGAGG |
| FbDEAB3′XbaI | GGGTCTAGATTGTTTGTTTTAGTGAGTAAGTACGGCTCTACGAACAC | |
| ppcAFp | Fp5′HindIII | GGGAAGCTTTTTCTTTTGTATTTGTTATTGTTTACG |
| Fp3′HpaI | GGGGTTAACGCCTCTATGTACAGAGAATACC | |
| FpM-FpPR FpM-FtPR | FpDEab5′HindIII | GGGAAGCTTATGAATTTATGAAAAACTCGTG |
| FpDEab3′XbaI | GGGTCTAGATTGTTTGTTTTAGTAAGTACG | |
| FpM/G_+CACT-FtPR | FpDEAB5′HindIII | GGGAAGCTTGTGAATTTATGAAAAACTCGTGAAGAG |
| FbDEAB3′XbaI | GGGTCTAGATTGTTTGTTTTAGTGAGTAAGTACGGCTCTACGAACAC |
Restriction sites of HindIII, XbaI, and HpaI are given in boldface letters.
Construction of the Fusion of MEM1 of F. pringlei with the Proximal Region of the ppcA Promoter of F. pringlei (FpM-FpPR)
For the generation of this construct, PCR was performed with the oligonucleotide primers FpDEab5′HindIII and FpDEab3′XbaI (Table 1) and genomic DNA of F. pringlei as template. The resulting DNA fragment was subcloned into pBIISK(+). After digestion with HindIII/XbaI, the resulting fragment was inserted into the HindIII/XbaI-digested vector ppcA-PRFp (Stockhaus et al., 1994).
Fusions of MEM1 Variants with the C4 Proximal Region of the ppcA Promoter of F. trinervia
The following promoter–GUS reporter gene fusions are based on the construct ppcA-PRFt-DR(+)Ft (Gowik et al., 2004). The constructs with native MEM1 of F. trinervia (ppcA-PRFt-DRabFt) and the MEM1 version lacking the CACT tetranucleotide in the B submodule (ppcA-PRFt-DRabFt-Δcact) have been described (Gowik et al., 2004). For reasons of nomenclature, they were renamed FtM-FtPR and FtM/ΔCACT-FtPR, respectively. For the generation of the two deletion constructs FtM/Δnt1-11-FtPR and FtM/Δnt12-4-FtPR, and for the two substitution constructs FtM/A-FtPR and FtM/A_ΔCACT-FtPR, the respective oligonucleotides were synthesized (Table 1) and annealed. The annealed oligonucleotides were digested with HindIII/XbaI and inserted into ppcA-PRFt-DR(+)Ft to replace the DR fragment.
In order to fuse MEM1 of F. bidentis (−1859 to −1996), MEM1 of F. pringlei (−2454 to −2538), and the C4-converted MEM1 of F. pringlei to the proximal region of the ppcA promoter of F. trinervia, PCR was performed using the primers depicted in Table 1 and genomic DNA of F. bidentis and F. pringlei as templates. The resulting DNA fragments were digested with HindIII/XbaI and subcloned into pBIISK(+). After an additional digestion with HindIII/XbaI, the released MEM1 fragments were inserted into ppcA-PRFt-DR(+)Ft to replace the DR fragment. The constructs were named FbM-FtPR, FpM-FtPR, and FpM/G_+CACT-FtPR, respectively.
Isolation of 5′ Flanking Sequences from the ppcA PEPC Genes of F. brownii and F. pubescens
The 5′ flanking regions of the ppcA genes of F. brownii and F. pubescens were isolated from total DNA by vectorette PCR (Siebert et al., 1995) with the Universal Genome Walker kit (Clontech) as described by Gowik et al. (2004).
Generation of Transgenic F. bidentis and Arabidopsis thaliana
The promoter–GUS reporter gene constructs were introduced into the Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991) via electroporation. F. bidentis and Arabidopsis were transformed as described (Chitty et al., 1994; Clough and Bent, 1998). Integration of the chimerical genes into the genome was confirmed by PCR.
Measurement of GUS Activity and Histochemical Analysis
T0 plants of F. bidentis were used for quantitative and histochemical analysis of the GUS reporter gene. For the analysis, mature F. bidentis plants, grown in the greenhouse up to 40 to 50 cm and before flower initiation, were used (Stockhaus et al., 1997). Arabidopsis T1 plants were grown either in tissue culture or on soil in the greenhouse. Histochemical analysis was performed as described (Stockhaus et al., 1997). Incubation times for GUS staining were varied according to the GUS activities of the individual plants. GUS activities were measured quantitatively as described (Jefferson, 1987). The average values of the data are expressed by medians, and Mann-Whitney U test statistics, as implemented in the software package Kaleidagraph version 3.6 for Mac OS X (Synergy Software; www.synergy.com), were used to test whether two data series differed from each other.
Phylogenetic Analysis
For phylogenetic analysis, sequences were aligned with Dialign (Morgenstern et al., 1998) and adjusted manually. The tree was generated by the distance method as implemented in PAUP 4.0b10 (Swofford, 2002), where uninformative characteristics were excluded. Bootstrap analysis was performed with 1000 replications.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers EF522173 (5′ upstream region of the ppcA gene of F. brownii) and EF522174 (5′ upstream region of the ppcA gene of F. pubescens).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Nucleotide Sequence Alignment of the 5′ Upstream Regions of the ppcA genes of F. trinervia, F. bidentis, F. brownii, F. pubescens, F. cronquistii, and F. pringlei and the ppcB Genes of F. trinervia and F. pringlei.
Supplemental Data Set 1. Text File of the Nucleotide Sequence Alignment Shown in Supplemental Figure 1 online.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 590 at Heinrich Heine University. We thank our gardeners for the careful cultivation of the plants in the greenhouse. We also thank K. Ernst and K. Meierhoff for carefully reading the manuscript and Christian Thalmann for assistance with the GUS staining of Arabidopsis seedlings.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter Westhoff (west@uni-duesseldorf.de).
Online version contains Web-only data.
References
- Akyildiz, M. (2007). Identification of cis- and trans-Regulatory Factors Controlling the Expression of the C4 Phosphoenolpyruvate Carboxylase Gene of the C4 Dicot Flaveria trinervia. PhD dissertation (Düsseldorf, Germany: Heinrich-Heine Universität).
- Bauwe, H., and Chollet, R. (1986). Kinetic properties of phosphoenolpyruvate carboxylase from C3, C4, and C3-C4 intermediate species of Flaveria (Asteraceae). Plant Physiol. 82 695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood, E.M., and Kadonaga, J.T. (1998). Going the distance: A current view of enhancer action. Science 281 60–63. [DOI] [PubMed] [Google Scholar]
- Bläsing, O.E., Ernst, K., Streubel, M., Westhoff, P., and Svensson, P. (2002). The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia—Implications for the evolution of C4 photosynthesis. Planta 215 448–456. [DOI] [PubMed] [Google Scholar]
- Brown, N.J., Parsley, K., and Hibberd, J.M. (2005). The future of C4 research—maize, Flaveria or Cleome? Trends Plant Sci. 10 215–221. [DOI] [PubMed] [Google Scholar]
- Burnell, J.N., and Hatch, M.D. (1988). Low bundle sheath carbonic anhydrase is apparently essential for effective C4 pathway operation. Plant Physiol. 86 1252–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.H., Moore, B.D., Edwards, G.E., and Ku, M.S.B. (1988). Photosynthesis in Flaveria brownii, a C4-like species. Leaf anatomy, characteristics of CO2 exchange, compartmentation of photosynthetic enzymes, and metabolism of 14CO2. Plant Physiol. 87 867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.H., Moore, B.D., Wu, J., Edwards, G.E., and Ku, M.S.B. (1989). Photosynthetic plasticity in Flaveria brownii. Growth irradiance and the expression of C4 photosynthesis. Plant Physiol. 89 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitty, J.A., Furbank, R.T., Marshall, J.S., Chen, Z., and Taylor, W.C. (1994). Genetic transformation of the C4 plant, Flaveria bidentis. Plant J. 6 949–956. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dengler, N.G., Dengler, R.E., and Hattersley, P.W. (1985). Differing ontogenetic origins of PCR (“Kranz”) sheaths in leaf blades of C4 grasses (Poaceae). Am. J. Bot. 72 284–302. [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290. [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, G.E., and Ku, M.S.B. (1987). Biochemistry of C3-C4 intermediates. In The Biochemistry of Plants, Vol. 10, M.D. Hatch and N.K. Boardman, eds (New York: Academic Press), pp. 275–325.
- Ernst, K., and Westhoff, P. (1996). The phosphoenolpyruvate carboxylase (ppc) gene family of Flaveria trinervia (C4) and F. pringlei (C3): Molecular characterization and expression analysis of the ppcB and ppcC genes. Plant Mol. Biol. 34 427–443. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1977). Anatomy of Seed Plants. (New York: John Wiley & Sons).
- Gaston, K., and Jayaraman, P.S. (2003). Transcriptional repression in eukaryotes: Repressors and repression mechanisms. Cell. Mol. Life Sci. 60 721–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik, U., Burscheidt, J., Akyildiz, M., Schlue, U., Koczor, M., Streubel, M., and Westhoff, P. (2004). Cis-regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.C., Sytsma, K.J., and Iltis, H.H. (2002). Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Am. J. Bot. 89 1826–1842. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D. (1987). C4 photosynthesis: A unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895 81–106. [Google Scholar]
- Hermans, J., and Westhoff, P. (1992). Homologous genes for the C4 isoform of phosphoenolpyruvate carboxylase in a C3- and a C4-Flaveria species. Mol. Gen. Genet. 234 275–284. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Kinsman, E.A., and Pyke, K.A. (1998). Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125 1815–1822. [DOI] [PubMed] [Google Scholar]
- Ku, M.S.B., Wu, J., Dai, Z., Scott, R.A., Chu, C., and Edwards, G.E. (1991). Photosynthetic and photorespiratory characteristics of Flaveria species. Plant Physiol. 96 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo, G.R., Stein, P.A., and Ludwig, R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9 963–967. [DOI] [PubMed] [Google Scholar]
- Love, A.C., Andrews, M.E., and Raff, R.A. (2007). Gene expression patterns in a novel animal appendage: The sea urchin. Evol. Dev. 9 51–68. [DOI] [PubMed] [Google Scholar]
- Maldonado, E., Hampsey, M., and Reinberg, D. (1999). Repression: Targeting the heart of the matter. Cell 99 455–458. [DOI] [PubMed] [Google Scholar]
- Matsuoka, M., Kyozuka, J., Shimamoto, K., and Kano-Murakami, Y. (1994). The promoters of two carboxylases in a C4 plant (maize) direct cell-specific, light-regulated expression in a C3 plant (rice). Plant J. 6 311–319. [DOI] [PubMed] [Google Scholar]
- McKown, A.D., and Dengler, N.G. (2007). Key innovations in the evolution of Kranz anatomy and C-4 vein pattern in Flaveria (Asteraceae). Am. J. Bot. 94 382–399. [DOI] [PubMed] [Google Scholar]
- McKown, A.D., Moncalvo, J.M., and Dengler, N.G. (2005). Phylogeny of Flaveria (Asteraceae) and of C4 photosynthesis evolution. Am. J. Bot. 92 1911–1928. [DOI] [PubMed] [Google Scholar]
- Monson, R.K. (2003). Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int. J. Plant Sci. 164 (suppl.): S43–S54. [Google Scholar]
- Monson, R.K., and Moore, B.D. (1989). On the significance of C3-C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant Cell Environ. 12 689–699. [Google Scholar]
- Morgenstern, B., Frech, K., Dress, A., and Werner, T. (1998). DIALIGN: Finding local similarities by multiple sequence alignment. Bioinformatics 14 290–294. [DOI] [PubMed] [Google Scholar]
- Nelson, T., and Dengler, N.G. (1992). Photosynthetic tissue differentiation in C4 plants. Int. J. Plant Sci. 153 (suppl.): S93–S105. [Google Scholar]
- Nomura, M., Higuchi, T., Ishida, Y., Ohta, S., Komari, T., Imaizumi, N., Miyao-Tokutomi, M., Matsuoka, M., and Tajima, S. (2005. a). Differential expression pattern of C4 bundle sheath expression genes in rice, a C3 plant. Plant Cell Physiol. 46 754–761. [DOI] [PubMed] [Google Scholar]
- Nomura, M., Higuchi, T., Katayama, K., Taniguchi, M., Miyao-Tokutomi, M., Matsuoka, M., and Tajima, S. (2005. b). The promoter for C4-type mitochondrial aspartate aminotransferase does not direct bundle sheath-specific expression in transgenic rice plants. Plant Cell Physiol. 46 743–753. [DOI] [PubMed] [Google Scholar]
- Olson, E.N. (2006). Gene regulatory networks in the evolution and development of the heart. Science 313 1922–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, M., Siegel, A.J., and Berry, J.O. (2006). Untranslated regions of FbRbcS1 mRNA mediate bundle sheath cell-specific gene expression in leaves of a C4 plant. J. Biol. Chem. 281 25485–25491. [DOI] [PubMed] [Google Scholar]
- Powell, A.M. (1978). Systematics of Flaveria (Flaveriinae-Asteraceae). Ann. Mo. Bot. Gard. 65 590–636. [Google Scholar]
- Reed, J.E., and Chollet, R. (1985). Immunofluorescent localization of phosphoenolpyruvate carboxylase and ribulose 1,5-bisphosphate carboxylase/oxygenase proteins in leaves of C3, C4 and C3-C4 intermediate Flaveria species. Planta 165 439–445. [DOI] [PubMed] [Google Scholar]
- Sage, R.F. (2004). The evolution of C4 photosynthesis. New Phytol. 161 341–370. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning. A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sheen, J. (1999). C4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 187–217. [DOI] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros, C.L., and Dengler, N.G. (2001). Ontogenetic derivation and cell differentiation in photosynthetic tissues of C3 and C4 Cyperaceae. Am. J. Bot. 88 992–1005. [PubMed] [Google Scholar]
- Stockhaus, J., Poetsch, W., Steinmüller, K., and Westhoff, P. (1994). Evolution of the C4 phosphoenolpyruvate carboxylase promoter of the C4 dicot Flaveria trinervia: An expression analysis in the C3 plant tobacco. Mol. Gen. Genet. 245 286–293. [DOI] [PubMed] [Google Scholar]
- Stockhaus, J., Schlue, U., Koczor, M., Chitty, J.A., Taylor, W.C., and Westhoff, P. (1997). The promoter of the gene encoding the C4 form of phosphoenolpyruvate carboxylase directs mesophyll specific expression in transgenic C4 Flaveria spp. Plant Cell 9 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0. (Sunderland, MA: Sinauer Associates).
- Westhoff, P., and Gowik, U. (2004). Evolution of C4 phosphoenolpyruvate carboxylase—genes and proteins: A case study with the genus Flaveria. Ann. Bot. (Lond.) 93 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff, P., Svensson, P., Ernst, K., Bläsing, O., Burscheidt, J., and Stockhaus, J. (1997). Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria. Aust. J. Plant Physiol. 24 429–436. [Google Scholar]
- Wray, G.A., Hahn, M.W., Abouheif, E., Balhoff, J.P., Pizer, M., Rockman, M.V., and Romano, L.A. (2003). The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20 1377–1419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.