Abstract
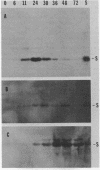
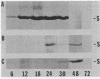
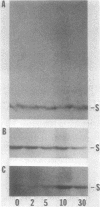
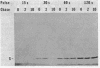
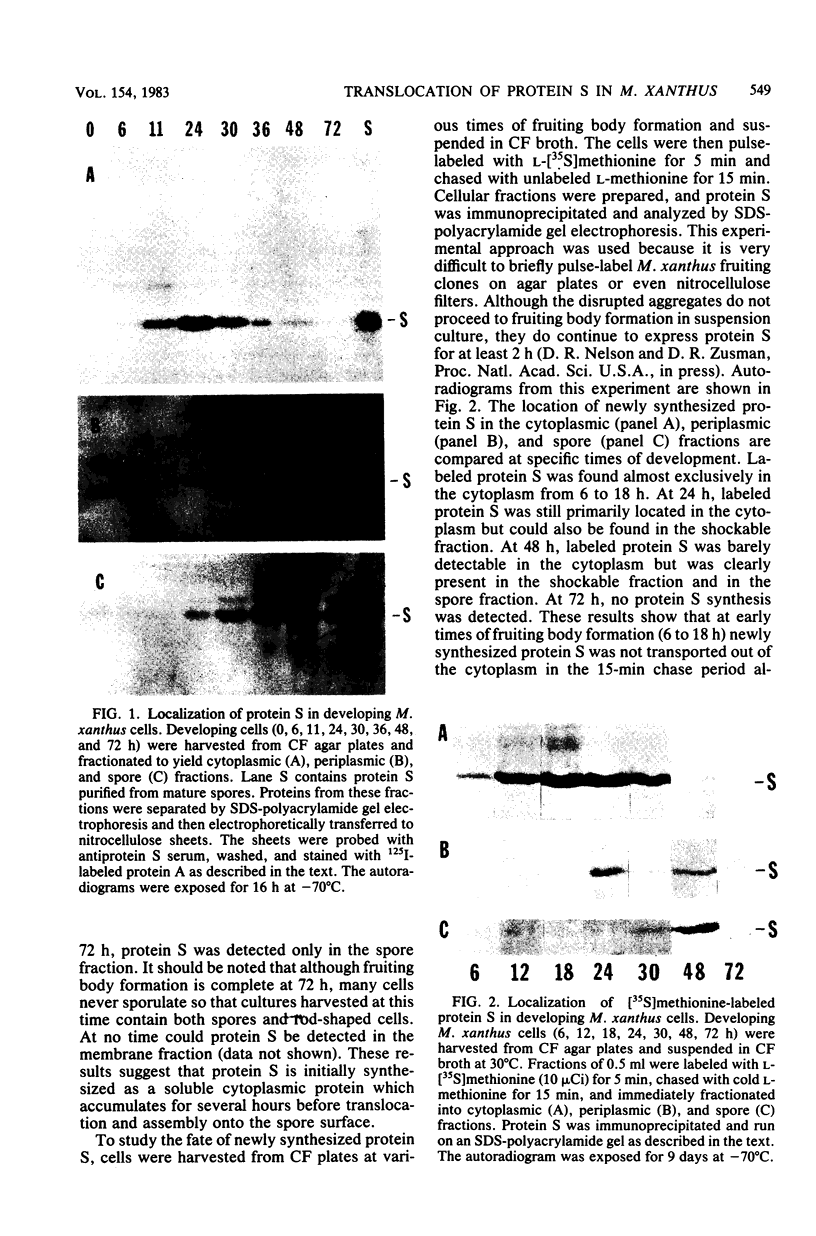
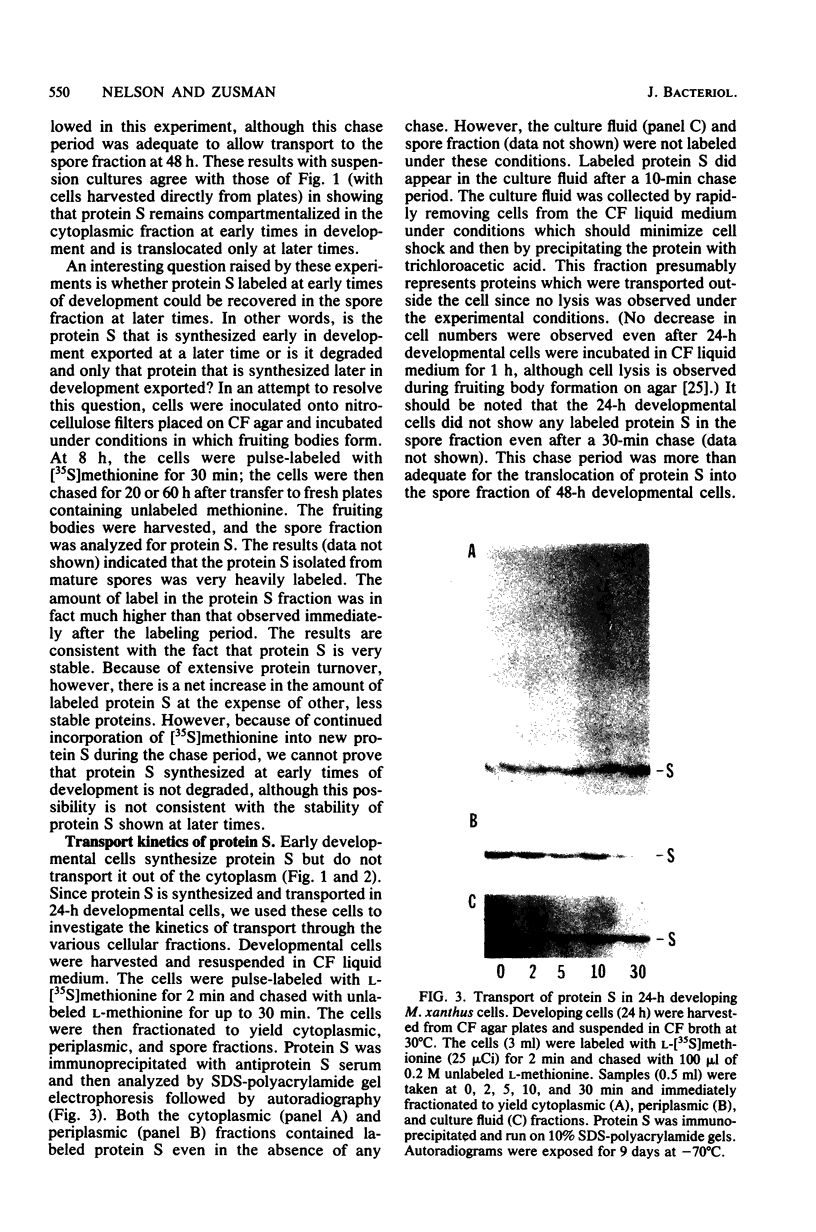
Protein S, the most abundant soluble protein synthesized by Myxococcus xanthus FB during early fruiting body formation, accumulates in the soluble fraction of developing cells, reaching a peak at about 24 h; at late stages of fruiting body formation, protein S is found on the surface of spores (M. Inouye et al. Proc. Natl. Acad. Sci. U.S.A. 76:209-213, 1979). In this study, the transport and localization of protein S were investigated. Cells were fractionated to give osmotic shock, membrane, cytoplasmic, and spore fractions. The various fractions were then analyzed for protein S. Protein S was first detected in the cytoplasmic fraction at about 3 to 6 h of development. However, transport of protein S through the cytoplasmic membrane was not observed until 15 to 18 h of development. Thus, protein S is unusual among translocated proteins in that it accumulates as a soluble cytoplasmic protein before translocation. Biosynthesis of protein S ceased after 48 h; by 72 h, protein S was only found on the surface of spores. Pulse-chase experiments were performed to determine the transport kinetics of protein S. The results showed that in 24-h developing cells, the transport of protein S across the cytoplasmic membrane was rapid, occurring in less than 2 min. However, transport across the outer membrane was slow, requiring 10 to 15 min. Pulses of 15 s with [35S]methionine failed to reveal any short-lived precursor form in immunoprecipitated material separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Isoelectric focusing also failed to detect any precursor form of protein S. Thus, protein S appears to be translocated in the absence of a cleaved signal peptide.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F., Nikaido K. Two-dimensional gel electrophoresis of membrane proteins. Biochemistry. 1976 Feb 10;15(3):616–623. doi: 10.1021/bi00648a026. [DOI] [PubMed] [Google Scholar]
- Bassford P. J., Jr, Silhavy T. J., Beckwith J. R. Use of gene fusion to study secretion of maltose-binding protein into Escherichia coli periplasm. J Bacteriol. 1979 Jul;139(1):19–31. doi: 10.1128/jb.139.1.19-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975 Dec;67(3):852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev Biol. 1979 Feb;68(2):579–591. doi: 10.1016/0012-1606(79)90228-8. [DOI] [PubMed] [Google Scholar]
- Ito K., Bassford P. J., Jr, Beckwith J. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell. 1981 Jun;24(3):707–717. doi: 10.1016/0092-8674(81)90097-0. [DOI] [PubMed] [Google Scholar]
- Kaiser D., Manoil C., Dworkin M. Myxobacteria: cell interactions, genetics, and development. Annu Rev Microbiol. 1979;33:595–639. doi: 10.1146/annurev.mi.33.100179.003115. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lingappa V. R., Lingappa J. R., Blobel G. Chicken ovalbumin contains an internal signal sequence. Nature. 1979 Sep 13;281(5727):117–121. doi: 10.1038/281117a0. [DOI] [PubMed] [Google Scholar]
- Manoil C., Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J Bacteriol. 1980 Jan;141(1):305–315. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek R. L., Walsh K. A., Palmiter R. D. The signal sequence of ovalbumin is located near the NH2 terminus. J Biol Chem. 1982 Oct 25;257(20):12245–12251. [PubMed] [Google Scholar]
- Morrison C. E., Zusman D. R. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J Bacteriol. 1979 Dec;140(3):1036–1042. doi: 10.1128/jb.140.3.1036-1042.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Cumsky M. G., Zusman D. R. Localization of myxobacterial hemagglutinin in the periplasmic space and on the cell surface of Myxococcus xanthus during developmental aggregation. J Biol Chem. 1981 Dec 10;256(23):12589–12595. [PubMed] [Google Scholar]
- Osborn M. J., Wu H. C. Proteins of the outer membrane of gram-negative bacteria. Annu Rev Microbiol. 1980;34:369–422. doi: 10.1146/annurev.mi.34.100180.002101. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Gagnon J., Walsh K. A. Ovalbumin: a secreted protein without a transient hydrophobic leader sequence. Proc Natl Acad Sci U S A. 1978 Jan;75(1):94–98. doi: 10.1073/pnas.75.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Synthesis of exported proteins by membrane-bound polysomes from Escherichia coli. Eur J Biochem. 1977 May 2;75(1):43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. I. Bacterial flagella. Annu Rev Microbiol. 1977;31:397–419. doi: 10.1146/annurev.mi.31.100177.002145. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. The assembly of proteins into biological membranes: The membrane trigger hypothesis. Annu Rev Biochem. 1979;48:23–45. doi: 10.1146/annurev.bi.48.070179.000323. [DOI] [PubMed] [Google Scholar]
- Wireman J. W., Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1977 Feb;129(2):798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]