Abstract
Aquaporin (AQP) water channels are abundant in the brain and spinal cord, where AQP1 and AQP4 are believed to play major roles in water metabolism and osmoregulation. Immunocytochemical analysis of the brain recently revealed that AQP4 has a highly polarized distribution, with marked expression in astrocyte end-feet that surround capillaries and form the glia limitans; however, the structural organization of AQP4 has remained unknown. In freeze-fracture replicas, astrocyte end-feet contain abundant square arrays of intramembrane particles that parallel the distribution of AQP4. To determine whether astrocyte and ependymocyte square arrays contain AQP4, we employed immunogold labeling of SDS-washed freeze-fracture replicas and stereoscopic confirmation of tissue binding. Antibodies to AQP4 directly labeled ≈33% of square arrays in astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Overall, 84% of labels were present beneath square arrays; 11% were beneath particle clusters that resembled square arrays that had been altered during fixation or cleaving; and 5% were beneath the much larger areas of glial plasma membrane that were devoid of square arrays. Based on this evidence that AQP4 is concentrated in glial square arrays, freeze-fracture methods may now provide biophysical insights regarding neuropathological states in which abnormal fluid shifts are accompanied by alterations in the aggregation state or the molecular architecture of square arrays.
Water transport is important in multiple physiological processes of the brain and spinal cord, including secretion and absorption of cerebrospinal fluid, movement of fluid across the blood–brain barrier, osmosensation, and regulation of renal water conservation. Precise control of cell volume is critical, because the brain is encased within the rigid cranium, and thus, even minor changes in water metabolism may result in fatal compressive cerebral edema. In other settings, brain swelling may produce neonatal hydrocephalus, failure of synaptic transmission, or altered neuronal excitability (1–3). Abnormal osmoregulation also may contribute to the pathogenesis of epilepsy, stroke, and trauma to the brain and spinal cord (4, 5).
Molecular pathways for the movement of water across biological membranes were unknown until the discovery of the aquaporin (AQP) family of membrane water channels (4, 6). Of these, two AQPs are expressed abundantly in the mammalian brain: AQP1 in the choroid plexus of the ventricles (where it apparently mediates secretion of cerebrospinal fluid; see refs. 4 and 7) and AQP4 in plasma membranes of ependymal cells and astrocytes, particularly in astrocyte end-feet that form the glia limitans and that surround capillaries (3). AQP4 is especially abundant in astrocytes and ependymocytes in osmosensory areas, including the supraoptic nucleus and subfornical organ (3). These sites suggest a role for AQP4 in water transport between the brain parenchyma, cerebrospinal fluid, and blood, as well as a role in osmoregulation.
Naturally occurring membranes enriched in AQP1 contain intramembrane particles (IMPs). Nevertheless, membrane reconstitution of AQP1 at high concentration yielded highly ordered two-dimensional crystalline lattices of AQP1, with a tetrameric assembly of subunits (8). The major intrinsic protein of lens MIP26 (or AQP0) occurs in lens fibers as tetragonal arrays (9). Similar “square arrays”** also are found in the renal collecting-duct principal cell, where multiple AQPs are expressed (12). These precedents suggest that some AQPs reside in native tissues as square arrays.
In contrast, early freeze-fracture studies of astrocyte end-feet revealed an abundance of naturally occurring square arrays of IMPs in the plasma membranes directly facing capillaries, as well as in the end-feet that form the glia limitans bordering the subarachnoid space (13–15). These areas correspond to the sites were AQP4 is expressed (3). The possibility that square arrays in the brain and spinal cord are formed from AQP4 was also suggested by the apparent absence of square arrays in brains from mice with AQP4 gene disruption (16). By using a new direct-labeling method for fracture-labeling IMPs in SDS-washed membranes (17), we sought to establish whether AQP4 proteins (i) are organized within square arrays in astrocytes and ependymocytes from rat brain and spinal cord, (ii) exist primarily as dispersed IMPs outside of square arrays, or (iii) occur in both clustered and dispersed IMP distributions.
MATERIALS AND METHODS
Antibodies and Immunogold Labels.
For correlation with data from previous reports (3), affinity-purified antibodies to rat AQP4 were generated against a synthetic peptide corresponding to amino acids 280–296 (LL182; see ref. 18), a domain predicted to lie in the middle of the 71-residue cytoplasmic C terminus (19). Monoclonal antibodies to gap-junction connexin-43 (Cx43) and polyclonal antibodies conjugated to 10- and 20-nm gold beads (goat anti-mouse and goat anti-rabbit IgG) were obtained from Chemicon.
Animals.
For light-microscopic and thin-section electron-microscopic immunocytochemistry, rats were perfused with 0.1% glutaraldehyde and 2% formaldehyde in 0.1 M cacodylate, pH 7.4. For freeze-fracture immunocytochemistry, adult Sprague–Dawley rats (two males and eight females) were fixed for 10 min by perfusion with 1% formaldehyde in Sorenson’s phosphate buffer (SPB), pH 7.4.
Immunoblotting.
Cerebellum, spinal cord, and cerebral cortex (including hippocampus) were homogenized (0.3 M sucrose/25 mM imidazole/1 mM EDTA, pH 7.2/8.5 μM leupeptin/1 mM phenylmethylsulfonyl fluoride). After centrifugation at 4,000 × g for 15 min at 4°C, the supernatant was centrifuged at 17,000 or 200,000 × g for 1 h. Pellets were solubilized in Laemmli sample buffer containing 2% SDS and run on 12% polyacrylamide minigels. Immunoblots were analyzed with affinity-purified anti-AQP4 (18) and visualized with the Enhanced Chemiluminescence System (Amersham).
Immunocytochemistry.
Fixed-tissue blocks of brain and spinal cord were infiltrated for 30 min with 2.3 M sucrose containing 2% formaldehyde, mounted on holders, and rapidly frozen in liquid nitrogen. For light-microscopic immunocytochemistry, frozen-tissue blocks were cryosectioned (0.8 μm; Ultracut S Cryoultramicrotome, Reichert), and sections were incubated with affinity-purified anti-AQP4 antibodies. Labeling with horseradish peroxidase-conjugated secondary antibody (P448 1:100; Dako) was visualized (i.e., amplified) with diaminobenzidine. For immunoelectron microscopy, frozen blocks were freeze-substituted and embedded in Lowicryl HM20 (Reichert). Ultrathin sections (40–60 nm) were incubated with affinity-purified anti-AQP4. Labeling was visualized with goat anti-rabbit IgG conjugated to 10-nm colloidal gold particles (Biocell Laboratories).
Freeze-Fracture Immunocytochemistry.
Vibratome brain slices were cut from the suprachiasmatic nucleus and hippocampus; spinal-cord slices were cut from the mid-thoracic level. Tissue slices were frozen ultrarapidly with a metal-mirror freezing device (20), freeze-fractured at −175 to −185°C in a JEOL RFD-9010C freeze-fracture device, shadowed with ≤1 nm of platinum/carbon, and replicated with 10 nm of pure carbon (21). Frozen replicated samples were bonded to a gold index grid by using 2% Lexan plastic (GE Plastics, Pittsfield, MA) that had been dissolved in dichloroethane, thawed, and photography-mapped by confocal microscopy (22). Tissue components other than membrane proteins adsorbed to the platinum/carbon replica were removed by washing the replicas in 2.5% SDS in 10 mM Tris⋅HCl, pH 8.6 for 18–24 h at 37–55°C with constant stirring (modified from the procedure described in ref. 17). Samples were rinsed three times in SPB and immersed for 30 min in “labeling-blocking buffer” (LBB), which consisted of 10% heat-inactivated goat serum and 0.5% teleost gelatin (Sigma) in 0.15 M SPB (23). LBB-protected replicas were placed in primary antibody solution (1:100 in LBB). For double-labeling experiments, species-specific primary antibodies were mixed in LBB, as were the species-specific immunogold secondary antibodies. After secondary labeling for 90 min, samples were rinsed five times (25 min total) in SPB, three times in distilled water, and air dried. In double-labeling experiments, each label served as a control for the other label, thereby allowing sources and rates of nonspecific and “false-negative” labeling to be identified.
After labeling, replicas were coated on the (former) tissue side with a second 10-nm carbon layer, which prevented the displacement of gold labels during the subsequent removal of the Lexan support film (22). Replicas were examined in a JEOL 2000 EX-II transmission electron microscope operated at 100 kV. Selected images are presented as stereoscopic images (included angle of 8°) to document that specific gold labels were present only on the cytoplasmic side of the replica.
Analysis of Immunogold Labeling.
After membrane splitting and replication with platinum/carbon, vigorous washing with SDS dissolves away most of the tissue. However, sufficient numbers of the replicated proteins remain adsorbed to the replica to permit direct immunocytochemical labeling and semiquantitative analysis (17, 24). This method is best suited for studying densely packed protein arrays consisting of identical epitopes.
We photographed 10 replicas from 10 labeling experiments, yielding 24 areas of astrocyte and ependymocyte plasma membrane totaling 24 μm2 (range: 0.1–3.5 μm2 per image). Gold labels were counted beneath (i) P-face square arrays; (ii) E-face square arrays; (iii) IMP clusters on astrocyte and ependymocyte membrane P faces; (iv) P faces that did not contain square arrays or IMP clusters; (v) surrounding areas of replicated extracellular space; and (vi) replicated plasma membranes of neurons, oligodendrocytes, and vascular endothelial cells. Analysis of the latter two areas allowed rates of nonspecific binding to be determined.
Gold beads were defined as associated with square arrays if they were located directly beneath or within 20 nm of the array (i.e., within a radius two antibody molecules in length). The density of gold labels (no. per μm2) was calculated for square arrays (alone) and for areas of astrocyte and ependymocyte plasma membranes devoid of square arrays. These values were compared as an estimate of the ratio of AQP4 in assembled vs. unassembled states.
RESULTS
Specificity of Anti-AQP4.
As reported (3), affinity-purified anti-AQP4 recognized 29- and 31-kDa bands, corresponding to the two alternative translation-initiation variants of AQP4 (Fig. 1A), as well as a higher molecular mass band corresponding to undissociated oligomeric AQP4. Immunocytochemical staining revealed abundant labeling of glial processes in thin cryosections of spinal cord (Fig. 1B), cerebellum, and hippocampus (3). Prominent labeling appeared in pericapillary glial processes (indicated by arrows in Fig. 1B) and glia limitans, but other astrocyte processes were labeled weakly or not at all.
Figure 1.
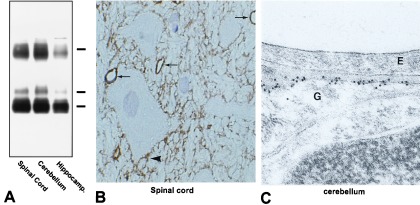
(A) Immunoblot of membrane fractions from spinal cord, cerebellum, and cerebral cortex (plus hippocampus). The blot was probed with affinity-purified anti-AQP4 and shows 29- and 31-kDa bands (splice variants) and a higher molecular mass band (undissociated-AQP4 oligomers). (B) Thin cryosection (0.9 μm) of spinal cord. Extensive AQP4-immunolabeling is observed on glial processes adjacent to capillaries (arrows) and adjacent to neurons (arrowhead). (×670.) (C) Ultrathin Lowicryl section of cerebellum. AQP4-labeling is associated with the glial process (G) on the side directly facing the capillary endothelium (E). (×72,000.)
In ultrathin Lowicryl sections of cerebellum (Fig. 1C) and spinal cord, immunoelectron microscopy revealed abundant labeling of AQP4 in glial processes adjacent to capillary endothelium, with negligible labeling of other glial membrane domains. Thus, AQP4 immunolabeling has a distinctly polarized distribution, primarily adjacent to capillary end-feet, the glia limitans, and ependymal-cell basolateral plasma membranes (3). Immunolabeling controls performed with nonimmune IgG or peptide-adsorbed anti-AQP4 were negative (not shown).
AQP4 Labeling in Freeze-Fracture Replicas of Ependymal Cells.
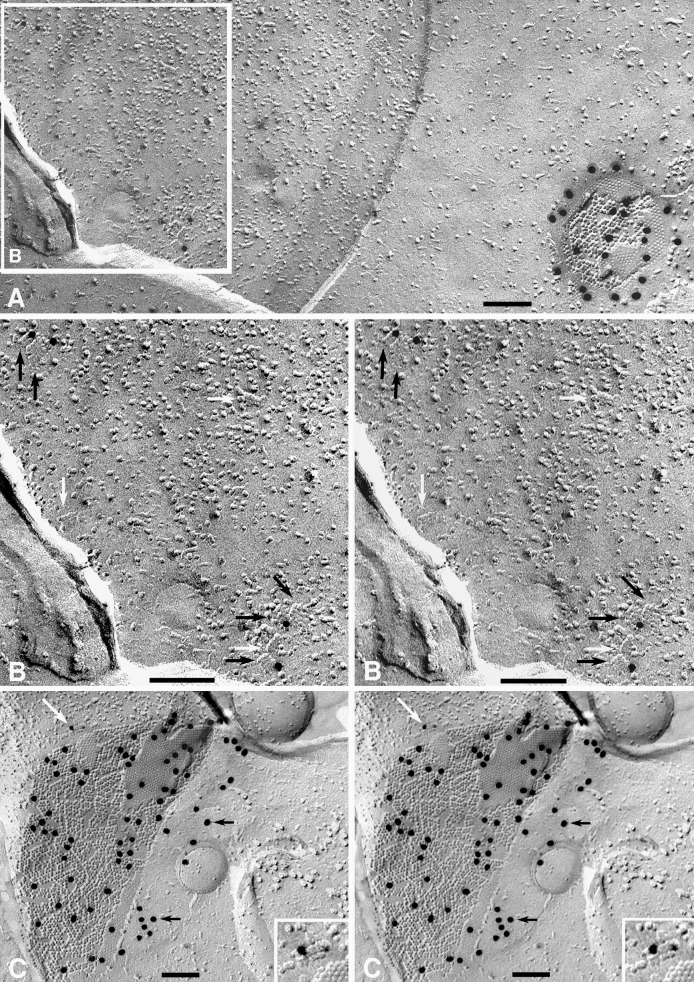
Samples from rat brain and spinal cord were labeled with anti-AQP4 or with anti-AQP4 plus anti-Cx43. At low magnification, conspicuous 20-nm gold particles revealed the presence of Cx43 in gap junctions of ependymal cells (Fig. 2A), confirming previous immunocytochemical studies (25). Discrete (nonclumped) anti-Cx43 gold labels were present at 100–300 per μm2 in gap junctions vs. 0.1–1 per μm2 in nonjunctional plasma membranes, thereby showing labeling with high specificity, moderate efficiency, and low background.
Figure 2.
Replica of ependymal-cell plasma membrane in section of suprachiasmatic nucleus after double-labeling with anti-Cx43/20-nm gold beads and anti-AQP4/10 nm-gold beads. (A) The single gap junction seen in this image has 22 large gold beads beneath or within 20 nm of P face IMPs and E face pits. No other 20-nm gold beads are present. Calibration bars in all freeze-fracture images represent 0.1 μm. (×85,000.) (B) Inscribed area from Fig. 2A that contains four anti-AQP4/10-nm gold particles beneath five square arrays (black arrows). Three additional square arrays are unlabeled (white arrows). The high angle of shadowing in this area resulted in reduced specimen contrast. (×130,000.) (C) AQP4/10-nm gold-labeled square array adjacent to Cx43/20-nm gold-labeled gap junction between ependymal cells in spinal cord. The white arrow points to AQP4/10-nm gold-labeled square array (shown at higher magnification in the inset image). Black arrows point to cryptic labeling of an unreplicated portion of the gap junction (see text). (×70,000; Inset ×135,000.)
In double-labeled ependymocytes, anti-AQP4/10-nm gold labels were restricted almost exclusively to the relatively rare square arrays (Fig. 2A, inscribed area; shown in a higher-magnification stereoscopic image in Fig. 2B). Stereoscopic images showed that >99.5% of labels were attached beneath the replica (Fig. 2C). Gold-labeled square arrays adjacent to Cx43-labeled gap junctions (Fig. 2C) confirmed the differential specificity of anti-Cx43 labels for ependymocyte and astrocyte gap junctions vs. anti-AQP4 labels for square arrays. Moreover, minimal nonspecific labeling was observed, and no interaction of species-specific primary and secondary antibody labels was detected.
“Cryptic” labeling of undissolved remnants of gap junctions was seen at the margins of oblique- and crossfractured gap junctions (indicated by black arrows in Fig. 2C). Cryptic labeling extended up to 0.3 μm beyond the visible-margin portion of the gap junction and projected beneath the intervening replicated cytoplasm. (The samples were air dried, causing the labels to collapse against the replica.) In other labeled replica samples prepared by glutaraldehyde/osmium fixation and staining with uranyl acetate/lead citrate and examined as “whole mounts,” the labeled membrane debris was found to contain partially dissolved gap junctions. Thus, incompletely dissolved membrane fragments account for the cryptic labeling seen in unstained replicas (J.E.R. and T.Y., unpublished work). These data suggest that at the boundary layer between replica and tissue, unfractured and unreplicated membrane fragments are not always dissolved completely. The undissolved fragments may remain attached to and continuous with the replicated portions of gap junctions and other structures, thereby providing the basis for “cryptic” labeling in unstained replicas.
AQP4 Labeling of Astrocyte End-Feet.
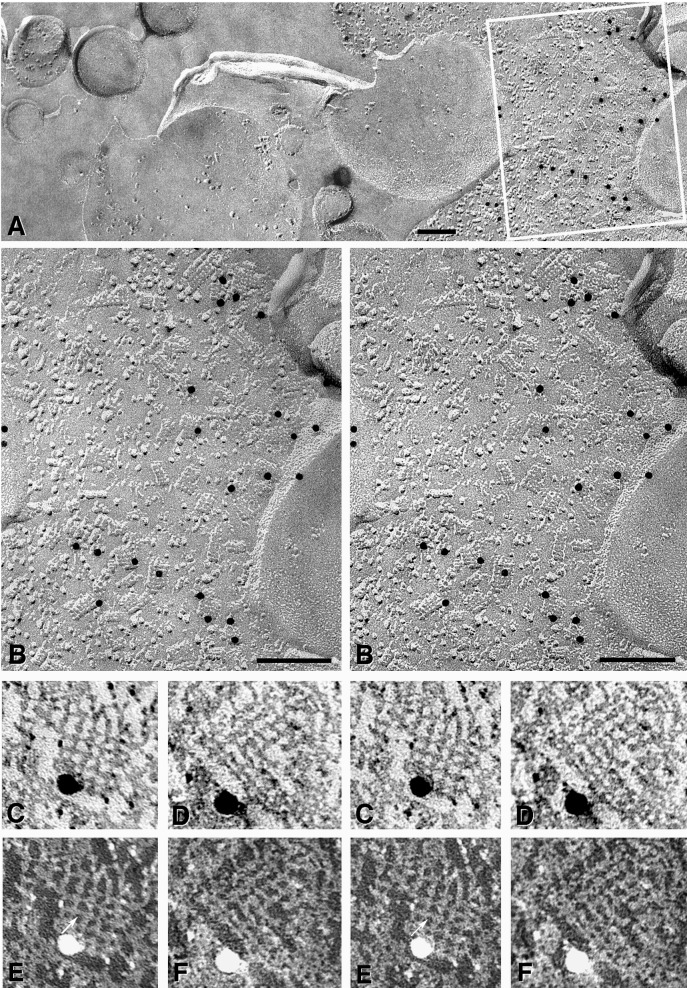
Square arrays are most abundant in astrocyte end-feet surrounding capillaries and in the glia limitans, where they may reach densities of 300–500 per μm2 (11). In samples of the glia limitans that had been single-labeled with anti-AQP4, 10-nm gold beads were present beneath astrocyte plasma membrane P faces (Fig. 3). All other areas of the replica (including extracellular space and nonastrocyte membranes) were essentially devoid of gold labels (left side of Fig. 3A), thereby showing a low level of nonspecific labeling.
Figure 3.
High-specificity anti-AQP4/10-nm gold labeling of square arrays in astrocyte end-feet. (A) Gold labels are confined almost exclusively to the astrocyte end-feet. No gold beads are seen on other membranes or attached to the replicated extracellular matrix (on the left side of the image). (×70,000.) (B) Stereoscopic image of the area inscribed in A reveals 17 gold particles directly beneath square arrays. (×135,000.) (C–F) High-magnification stereoscopic images of labeled AQP4 square arrays (C and D printed with white shadows; E and F printed with black shadows). In images printed with black shadows, 2-nm crossbridges (indicated by an arrow in E) are resolved linking the subunit IMPs (see text and ref. 21). (×450,000.)
In 24 μm2 of astrocyte plus ependymocyte plasma membrane, 183 gold beads were bound to 558 square arrays (i.e., ≈33% of square arrays were labeled). For example, of ≈50 square arrays in the area included in Fig. 3B, 17 (or 34%) were labeled; two gold beads were associated with clusters of IMPs that were not clearly identifiable as square arrays (see ref. 26), and four gold beads at the edge of the replicated portion of the astrocyte plasma membrane were attributed to cryptic labeling (see above). Labels were not found to be associated with the E face images of square arrays. Thus, AQP4-immunogold labeling was exclusively associated with P face replicas of square arrays. Based on an average area of 1270 nm2 per square array (or 1/800 μm2; see ref. 11 for similar estimates of maximum square array density), labeling density for square arrays was calculated to be 272 gold beads per μm2 of IMP lattice, equivalent to that observed for immunogold labeling of connexons within gap junctions.
Overall, gold labels on astrocyte and ependymocyte plasma membrane were present at 9.1 per μm2 of astrocyte and ependymocyte plasma membranes. Of these, 7.6 per μm2 (or 84%) were directly associated with square arrays, and 1.0 per μm2 (11%) were associated with square-array-sized clusters of distinctive elongated IMPs. (These may represent square arrays that had been altered by circulatory arrest during fixation; ref. 26.) The remaining 0.5 gold label per μm2 (5.5%) was observed in areas of plasma membrane that did not contain square arrays or IMP clusters. Nonspecific labeling of plasma membranes of other cell types, as well as on replicas of the extracellular space, was very low (<0.06 per μm2; area analyzed = 62.3 μm2). From these data, we calculated a “signal-to-noise” ratio (23) of ≈4000:1 (based on signal of 272 gold beads per μm2 for square arrays vs. noise of 0.06 per μm2).
At high magnification (Fig. 3 C–F), 2-nm crossbridges were seen to link the constituent IMPs of square arrays. (Fig. 3 C and D are printed with conventional “white shadows.” Fig. 3 E and F are printed with black shadows, which better reveal molecular detail. See ref. 21 for additional details of square-array substructure.) The identity of the 2-nm crossbridges linking IMPs within the square arrays is currently unknown, but they represent intramembrane domains of the square-array proteins, possibly including the B and E loops of AQP4 (4, 27). The crossbridges may contribute to the formation of the square lattice of IMPs, to the structural integrity of the arrays, or to possible stretch sensitivity of AQP4 water channels as established for ion channels (28).
DISCUSSION
We have demonstrated direct immunogold labeling of AQP4 in square arrays in SDS-washed grid-mapped fracture replicas of astrocyte and ependymocyte plasma membranes from brain and spinal cord of mature rats. Labeling paralleled the distribution of AQP4 proteins in astrocyte plasma membranes as demonstrated by thin-section immunogold labeling (3). Within astroglial end-feet, 85–95% of AQP4 labeling was present beneath square arrays. Nevertheless, because the remaining surface area of astroglial cells is much larger than the portion present in end-feet, a significant portion of AQP4 could exist in those areas but at such a low concentration that it would not be detectable by our methods.
AQP4 antibodies label approximately the same proportion of IMPs in square arrays as connexin antibodies label IMPs in gap junctions (1–3% of IMPs; 100–300 labels per μm2 in both cases). These proportions support the hypothesis that AQP4 is the major component of square arrays. Nevertheless, our data do not preclude the possibility that accessory proteins may be present within the arrays, or that several classes of proteins may reside within an IMP array. For example, multiple connexins reside within individual gap junctions (29), and occludin and ZO-1 reside within liver tight-junction strands (17, 24). Thus, precedent establishes that multiple protein isoforms, and even multiple proteins, may be present in structurally homogenous IMP arrays. Square arrays may be no exception.
Historical Perspectives Regarding Square Arrays.
Square arrays have been observed in freeze-fracture replicas for more than 30 years (reviewed in ref. 11). Although substantial circumstantial evidence could have implicated AQP4 as a component in astrocyte square arrays (3, 12), it was known that AQPs in addition to AQP4 are present in other membranes that are enriched in square arrays. For example, AQP0 (or MIP26) is expressed only in lens-fiber cell membranes, where its presence in square arrays was documented by fracture-labeling (9). Moreover, because AQP0 is the major intrinsic protein of lens (constituting more than 50% of membrane protein, with much of the remainder consisting of connexins), lens square arrays necessarily consist predominantly of a single protein, AQP0. Likewise, the presence of the homolog AQPcic coincides with the distribution of square arrays in the water-channel-enriched filter chambers of sap-sucking insects (30). Moreover, square arrays appeared in cultured cell lines transfected with Aqp4 (31) but were absent in tissues from mice with disruption of the AQP4 gene (16). In contrast, at least one AQP, AQP1, does not occur naturally in square arrays in the choroid plexus or in erythrocytes, but AQP1 does form square lattices in artificial membrane bilayers (8).
Perivascular end-feet of rat-brain astrocytes (32) were known to be particularly enriched in square arrays (500–600 square arrays per μm2; ref. 11), whereas nonadjacent membrane domains had markedly reduced densities of square arrays (33). This distribution precisely reflected the areas of plasma membrane where AQP4 was identified by immunogold electron microscopy (3). However, strict correlation was not possible because the astrocyte end-feet also contained high concentrations of potassium “leakage” channels (34). Nevertheless, in rat brains, the developmental expression of square arrays (14) was remarkably similar to the ontogeny of AQP4 (19), and the presence of square arrays in paravitreous and perivascular end-feet of retinal Müller cells (35) was found to coincide exactly with the sites where AQP4 was shown by immunogold labeling of retina (36). In other tissues (tracheal epithelium, ref. 37; fast-twitch skeletal muscle fiber, refs. 38 and 39; and renal collecting duct, ref. 40), the presence of square arrays has been correlated with immunohistochemically identified AQP4 (12, 41, 42) or with areas of AQP4 expression (18). These observations heightened our suspicion that AQP4 is present within square arrays in brain and spinal cord. However, in none of these cases was direct labeling of square arrays demonstrated.
Potential Relevance to Central Nervous System Trauma and Disease.
Recognition that square arrays in astrocyte and ependymocyte plasma membranes are composed of AQP4 provides insight into the functions of these structures. Interestingly, the prediction that square arrays in renal collecting-duct cells are involved in cellular water transport predated the discovery of AQPs (40); however, the hypothesis appears to be correct. Also interesting are suggestions that square arrays may participate in potassium-channel activation (43) or potassium-buffering during neuronal activation (32, 34). This may reflect parallel distributions of unrelated proteins. Alternatively, coordination of the activities of separate AQP4 and potassium-leak channels may facilitate “potassium-siphoning” at astrocyte end-feet (44, 45) during osmoregulatory activity that follows both synaptic transmission and repetitive nerve firing.
AQP4 square-array-mediated water transport also may be of significance in several pathological conditions affecting the central nervous system. Square arrays are reported to be reduced or disrupted in astrocytes after circulatory arrest (ref. 26; compare ref. 11), and the inability to dissipate cerebral edema may reflect a reduction in levels of AQP4 or an altered assembly or distribution of the protein. Observations that reactive astrocytes exhibit reduced polarity (33) suggest that altered expression of AQP4 may contribute to recovery from neural injury. These observations warrant further investigation; applying fracture-labeling methods to tissues from these clinical disorders and others, such as epilepsy and hydrocephalus, may help to define alterations in the distribution of AQP4 square arrays during disruptions of water metabolism within the central nervous system.
Acknowledgments
We thank Kim Davidson, Gitte Christensen, and Annette Blak Rasmussen for expert technical assistance. This work was supported by National Institutes of Health Grants RO1 NS-31027 (to J.E.R.) and R01 HL48268 and EY11239 (to P.A.), and by grants from the Karen Elise Jensen Foundation and the Novo Nordisk Foundation (to S.N.).
ABBREVIATIONS
- AQP
Aquaporin
- Cx43
Connexin-43
- IMP
intramembrane particle
Footnotes
These distinctive IMP arrays were originally called “rectilinear arrays” (10), but subsequently have been called “square arrays,” “rectangular arrays,” “orthogonal arrays,” “orthogonal assemblies,” “assemblies,” “orthogonally arranged particles” (or “OAPs”), and “tetragonally arranged particles” (reviewed in ref. 11). Because these arrays are composed of equilateral rectangles, we employ the term “square arrays,” which more accurately describes the organization of their subunits.
References
- 1. Johnson A K, Loewy A D. In: Central Regulation of Autonomic Functions. Loewy A D, Spyer K M, editors. Vol. 14. London: Oxford Univ. Press; 1990. pp. 247–267. [Google Scholar]
- 2.Jefferys J G. Physiol Rev. 1995;75:689–723. doi: 10.1152/physrev.1995.75.4.689. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen S, Nagelhus E A, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen O P. J Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King L S, Agre P. Annu Rev Physiol. 1996;58:619–648. doi: 10.1146/annurev.ph.58.030196.003155. [DOI] [PubMed] [Google Scholar]
- 5.Schweitzer J S, Dudek F E. In: Contemporary Neuropharmacology: Pathogenesis, Neurotransmitters and Drug Action—Epilepsy. Gross R A, editor. 1998. inpress. [Google Scholar]
- 6.Knepper M A. Proc Natl Acad Sci USA. 1994;91:6255–6258. doi: 10.1073/pnas.91.14.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen S, Smith B L, Christensen E I, Agre P. Proc Natl Acad Sci USA. 1993;90:7275–7279. doi: 10.1073/pnas.90.15.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walz T, Tittmann P, Fuchs K H, Müller D J, Smith B L, Agre P, Gross H, Engel A. J Mol Biol. 1996;264:907–918. doi: 10.1006/jmbi.1996.0686. [DOI] [PubMed] [Google Scholar]
- 9.Zampighi G A, Hall J E, Ehring G R, Simon S A. J Cell Biol. 1989;108:2255–2275. doi: 10.1083/jcb.108.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreutziger G O. Proc Annu Meet Electron Microsc Soc Am. 1968;26:234–235. (abstr.). [Google Scholar]
- 11.Wolburg H. J Hirnforsch. 1995;36:239–258. [PubMed] [Google Scholar]
- 12.Verbavatz J-M, Van Hoek A N, Ma T, Sabolic I, Valenti G, Ellisman M H, Ausiello D A, Verkman A S, Brown D. J Cell Sci. 1994;107:1083–1094. doi: 10.1242/jcs.107.4.1083. [DOI] [PubMed] [Google Scholar]
- 13.Landis D M D, Reese T S. J Cell Biol. 1974;60:316–320. doi: 10.1083/jcb.60.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landis D M D. J Exp Biol. 1981;95:35–48. doi: 10.1242/jeb.95.1.35. [DOI] [PubMed] [Google Scholar]
- 15.Hatton J D, Ellisman M H. J Neurocytol. 1982;11:335–349. doi: 10.1007/BF01258250. [DOI] [PubMed] [Google Scholar]
- 16.Verbavatz J M, Ma T, Govin R, Verkman A S. J Cell Sci. 1997;110:2855–2860. doi: 10.1242/jcs.110.22.2855. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto K. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 18.Terris J, Ecelbarger C A, Marples D, Knepper M A, Nielsen S. Am J Physiol. 1995;269:F775–F785. doi: 10.1152/ajprenal.1995.269.6.F775. [DOI] [PubMed] [Google Scholar]
- 19.Jung J S, Bhat R V, Preston G M, Baraban J M, Agre P. Proc Natl Acad Sci USA. 1994;91:13052–13056. doi: 10.1073/pnas.91.26.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips T E, Boyne A F. J Electron Microsc Technol. 1984;1:9–29. [Google Scholar]
- 21.Rash J E, Duffy H S, Dudek F E, Bilhartz B L, Whalen L R, Yasumura T. J Comp Neurol. 1997;388:265–292. doi: 10.1002/(sici)1096-9861(19971117)388:2<265::aid-cne6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Rash J E, Dillman R K, Morita M, Whalen L R, Guthrie P B, Fay-Guthrie D, Wheeler D W. In: Rapid Freezing, Freeze Fracture, and Deep Etching. Severs N J, Shotton D M, editors. Vol. 1. New York: Wiley-Liss; 1995. pp. 127–150. [Google Scholar]
- 23.Rash J E, Johnson T J A, Dinchuk J E, Duch D S, Levinson S R. In: Freeze-Fracture Studies of Membranes. Hui S W, editor. Vol. 3. Boca Raton, FL: CRC Press; 1990. pp. 41–59. [Google Scholar]
- 24.Fujimoto K. Histochem Cell Biol. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
- 25.Nagy J I, Ochalski P A Y, Li J, Hertzberg E L. Neuroscience. 1997;78:533–548. doi: 10.1016/s0306-4522(96)00584-2. [DOI] [PubMed] [Google Scholar]
- 26.Landis D M D, Reese T S. J Cell Biol. 1981;88:660–663. doi: 10.1083/jcb.88.3.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung J S, Preston G M, Smith B L, Guggino W B, Agre P. J Biol Chem. 1994;269:14648–14654. [PubMed] [Google Scholar]
- 28.Oliet S H, Bourque C W. Nature (London) 1993;364:341–343. doi: 10.1038/364341a0. [DOI] [PubMed] [Google Scholar]
- 29.Sosinsky G. Proc Natl Acad Sci USA. 1995;92:9210–9214. doi: 10.1073/pnas.92.20.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beuron F, Le Caherec F, Guillam M T, Cavalier A, Garret A, Tassan J P, Delamarche C, Schultz P, Mallouh B, Rolland J P, Hubert J F, Gouranton J, Thomas D. J Biol Chem. 1995;270:17414–17422. doi: 10.1074/jbc.270.29.17414. [DOI] [PubMed] [Google Scholar]
- 31.Yang B, Brown D, Verkman A S. J Biol Chem. 1996;271:4577–4580. [PubMed] [Google Scholar]
- 32.Risau W, Wolburg H. Trends Neurosci. 1990;13:174–178. doi: 10.1016/0166-2236(90)90043-a. [DOI] [PubMed] [Google Scholar]
- 33.Anders J J, Brightman M W. J Neurocytol. 1979;8:777–795. doi: 10.1007/BF01206676. [DOI] [PubMed] [Google Scholar]
- 34.Hatton J D, Ellisman M H. Cell Tissue Res. 1981;215:309–323. doi: 10.1007/BF00239117. [DOI] [PubMed] [Google Scholar]
- 35.Wolburg H, Berg K. Neurosci Lett. 1987;82:273–277. doi: 10.1016/0304-3940(87)90268-0. [DOI] [PubMed] [Google Scholar]
- 36.Nagelhus E A, Veruki M L, Torp R, Haug F-M, Laake J H, Nielsen S, Agre P, Ottersen O P. J Neurosci. 1998;18:2506–2519. doi: 10.1523/JNEUROSCI.18-07-02506.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue S, Hogg J C. J Ultrastruc Res. 1977;61:89–99. doi: 10.1016/s0022-5320(77)90008-9. [DOI] [PubMed] [Google Scholar]
- 38.Ellisman M H, Rash J E, Staehelin L A, Porter K R. J Cell Biol. 1976;68:752–774. doi: 10.1083/jcb.68.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rash J E, Ellisman M H. J Cell Biol. 1974;63:567–586. doi: 10.1083/jcb.63.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura T, Nagano T. J Electron Microsc Technol. 1985;34:364–372. [PubMed] [Google Scholar]
- 41.Frigeri A, Gropper M A, Umenishi F, Kawashima M, Brown D, Verkman A S. J Cell Sci. 1995;108:2933–3002. doi: 10.1242/jcs.108.9.2993. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen S, King L S, Christensen V M, Agre P. Am J Physiol. 1998;273:C1549–C1561. doi: 10.1152/ajpcell.1997.273.5.C1549. [DOI] [PubMed] [Google Scholar]
- 43.Gotow T, Hashimoto P H. Glia. 1989;2:273–285. doi: 10.1002/glia.440020409. [DOI] [PubMed] [Google Scholar]
- 44.Orkand R K, Nicholls J G, Kuffler S W. J Neurophysiol. 1966;29:788–806. doi: 10.1152/jn.1966.29.4.788. [DOI] [PubMed] [Google Scholar]
- 45.Newman E A. Science. 1986;233:453–454. doi: 10.1126/science.3726539. [DOI] [PMC free article] [PubMed] [Google Scholar]