Abstract
We describe a Drosophila gene, orbit, that encodes a conserved 165-kD microtubule-associated protein (MAP) with GTP binding motifs. Hypomorphic mutations in orbit lead to a maternal effect resulting in branched and bent mitotic spindles in the syncytial embryo. In the larval central nervous system, such mutants have an elevated mitotic index with some mitotic cells showing an increase in ploidy. Amorphic alleles show late lethality and greater frequencies of hyperploid mitotic cells. The presence of cells in the hypomorphic mutant in which the chromosomes can be arranged, either in a circular metaphase or an anaphase-like configuration on monopolar spindles, suggests that polyploidy arises through spindle and chromosome segregation defects rather than defects in cytokinesis. A role for the Orbit protein in regulating microtubule behavior in mitosis is suggested by its association with microtubules throughout the spindle at all mitotic stages, by its copurification with microtubules from embryonic extracts, and by the finding that the Orbit protein directly binds to MAP-free microtubules in a GTP-dependent manner.
Keywords: mitosis, microtubule-associated protein, Drosophila melanogaster, mitotic spindle, cen-, trosome
Introduction
The requirement for microtubules in the mitotic spindle is self-evident, and yet the role of nonneuronal microtubule-associated proteins (MAPs) in its function is poorly understood (for review, see Hyman and Karsenti 1996). Formation of the spindle requires that the interphase microtubule network be reorganized as a result of an increase in microtubule turnover at mitotic entry. This is possibly due to the dynamic instability of microtubules that oscillate between periods of growth and shrinkage and is due to an increase in the frequency of transitions from the polymerization to depolymerization phases, the frequency of catastrophe (for review, see Desai and Mitchison 1997). How the catastrophe rate increases at the onset of mitosis has been puzzling, as most known MAPs have the property of stabilizing microtubules. However, recent studies have identified proteins that promote microtubule catastrophe, such as op18 or stathmin, the microtubule-severing ATPase katanin, and the kin I family of kinesins (Belmont and Mitchison 1996; Hartman et al. 1998; Desai et al. 1999).
Studies on the mitotic roles of MAPs have concentrated upon the use of Xenopus as a model system as it offers advantages for experimentation in vitro. This work has identified several MAPs that localize to the mitotic spindle, including XMAP230, MAP4, XMAP215, and XMAP310 (Gard and Kirschner 1987; Andersen et al. 1994; Vasquez et al. 1994; Ookata et al. 1995; Andersen and Karsenti 1997; Charrasse et al. 1998) and offers the possibilities for direct studies of their effects upon microtubule dynamics.
We have chosen to search for mitotic regulators in Drosophila melanogaster, which offers the possibility of studying the effects of mitotic mutations within the intact cell. The characterization of maternal-effect mutants of Drosophila is a powerful route towards the identification of such genes. In many cases, maternal-effect mitotic defects reflect a specific requirement for the product of the affected gene for cell division throughout development. The proteins encoded by such genes may have either regulatory roles or be part of the structural components of the mitotic apparatus. The protein kinases encoded by the polo and aurora (aur) genes, for example, were first identified through hypomorphic mutations, which, when homozygous in the mother, result in gross mitotic defects within the embryo (Sunkel and Glover 1988; Glover et al. 1995). However, the functions of these kinases in cell division throughout development has been revealed through the study of series of mutant alleles that show developmental arrest at different stages. Sullivan et al. 1993 identified a number of maternal-effect mutations affecting the cytoskeletal organization of syncytial embryos. One of these, nuclear fallout (nuf), encodes a protein that concentrates at the centrosomes during prophase and is cytoplasmic during the rest of the nuclear cycle (Rothwell et al. 1998). However, none of the genes originally identified by maternal-effect mutations have yet been shown to encode a MAP, although the potential for identifying such mutants is evident. In fact, some alleles of abnormal spindle (asp), which encodes a protein associated with the polar regions of the mitotic spindle, exhibit a maternal effect on syncytial mitoses (Gonzalez et al. 1990; Saunders et al. 1997).
Alternative biochemical approaches to identify Drosophila MAPs also take advantage of the maternal dowry of proteins essential for the syncytial mitoses. One strategy has been to use libraries of mAbs to search for proteins that display dynamic patterns of localization during the mitotic cycle (Frasch et al. 1986). This set of antibodies was successful in identifying genes encoding a Drosophila homologue of the vertebrate regulator of chromatin condensation (RCC1; Frasch 1991), and a centrosomal antigen now known as CP190 (Whitfield et al. 1988). Another strategy was to purify molecules based upon their ability to bind actin (Miller et al. 1989) or microtubules (Kellogg et al. 1989), and then raise antibodies against individual proteins. Some 50 proteins were identified that would bind to microtubules and mAbs raised to 24 of them. One of the first to be cloned also proved to be CP190, which in turn was used as an affinity reagent to identify a second centrosomal associated antigen, CP60 (Kellogg and Alberts 1992). The function of the majority of these proteins still remains uncertain because of the lack of mutations or assays of their molecular function.
We have continued the direct genetic approach in a search for mutants that identify genes encoding MAPs, expecting that these would give rise to spindle defects in the syncytial mitoses, and also show defective cell divisions at other developmental stages. In this paper, we report the characterization of one such novel gene, orbit. We show that orbit encodes a novel 165-kD MAP and discuss possible functions for this protein suggested by the phenotypes of an allelic series of orbit mutants.
Materials and Methods
Immunofluorescent Staining of Embryos
Immunostaining of embryos from wild-type or mutant flies was carried out either as described by Gonzalez and Glover 1993 or by fixing dechorionated embryos with freshly prepared 4% paraformaldehyde in buffer B (45 mM KCl, 15 mM NaCl, 10 mM phosphate buffer, pH 6.8) at room temperature for 5 min and a further 25 min at 4°C. Microtubules were detected with the rat anti–α-tubulin antibody, YL1/2 (Sera-Lab) and either an FITC or Texas red-conjugated anti-rat IgG antibody (Jackson Laboratories). Centrosomes were revealed with the polyclonal rabbit serum, Rb188 (Whitfield et al. 1988) and Cy5-conjugated secondary anti-rabbit IgG antibody (Jackson Laboratories). Orbit was localized using an affinity-purified rabbit antibody against residues 1–632 (see below) diluted 1:100 in PBS containing 0.1% Triton X-100, and an FITC-conjugated anti-rabbit IgG antibody. DNA was stained with 1 μg/ml of propidium iodide. The preparations were observed using either an MRC600 or MRC1024 laser confocal microscope (BioRad). Images were processed and merged in pseudocolor using Photoshop version 5 (Adobe Systems).
Cytological Analysis of Larval Central Nervous System and Chromosome In Situ Hybridization
Wild-type and mutant larval CNSs were fixed and squashed in aceto-orcein, and whole-mount preparations of the larval CNS were carried out as previously described (Inoue and Glover 1998). For in situ hybridization, polytene chromosomes were prepared from salivary glands of late third instar larvae heterozygous for orbit and wild-type. Biotin-labeled probe was prepared from the P-lacW plasmid. Hybridization and signal detection were performed as described earlier (Deak et al. 1997). The specimens were examined using phase-contrast optics and hybridization signals were assigned to chromosome bands referring the revised salivary chromosome maps of Heino et al. 1994.
Reversion Analysis of orbit
Reversion analysis to test whether the P-lacW integrated at 78C is responsible for sterility and mitotic phenotypes of orbit were carried out by mating y w; orbit/TM3, Sb Δ2,3 dysgenic males with w; sr e Pr ca/TM6B, Hu Tb females. A total of 27 w − progenies were scored among 147 F1 flies with e + Hu or e + Pr markers. Each w − fly was individually backcrossed with y w; orbit/TM6C flies. From 27 independent crosses, females trans-heterozygous for each potential revertant allele without the w + marker over orbit were selected and tested for ability to produce viable progeny. Of the 27 w − derivatives of orbit, 22 turned out to be phenotypic revertants. To confirm the correlation of the phenotypic reversion of orbit with the loss of the P-lacW, we established homozygous lines of three phenotypic revertants, orbit 2, orbit 3, and orbit 4, and then examined genomic DNA organization by Southern hybridization using rescued fragments flanking both sides of the P-lacW element as probes. It was confirmed that all three revertants are imprecise and have internal deletions in the P-lacW sequence.
P Element Mediated Rescue
Genomic DNA fragments of 1.3-kb and 1.4-kb flanking the orbit insertion were isolated by plasmid rescue as previously described (Deak et al. 1997). These were used as probes to isolate several cosmid clones from the European Drosophila Genome Project cosmid library. The 14-kb BamHI fragment of cosmid 193F11 containing the orbit transcription unit was subcloned into a transformation vector pUAST (Brand and Perrimon 1993) by replacing a short BamHI fragment within the vector. The resulting plasmid was microinjected at 18°C into embryos derived from w; Δ2,3 females crossed with w; sr e Pr/TM6. Three w + transformants were recovered from 97 fertile G0 adults. It was confirmed that they each contained the intact transgene by genomic Southern hybridization. All transformed lines carried the transgene on the third chromosome, necessitating the generation of a recombined third chromosome carrying one transgene (at 56.5) and orbit. Females of the genotype w; ru h th st orbit/P[w + orbit +] sr e Pr were crossed to ru h th st cu sr e ca/TM3 males. Among the F1 progeny, seven males with recombined chromosomes carrying th and sr were selected and mated to w; orbit3 /TM6C females. Offspring having the recombined chromosome and the orbit 3 chromosome were examined by Southern hybridization for the presence of BamHI RFLPs characteristic of orbit, orbit + (on the transgene), and orbit 3. One of selected seven recombined chromosomes was a ru h th orbit P[w + orbit +] chromosome. Both reduced viability and sterility in orbit /orbit3 females and males were fully rescued in ru h th orbit P[w + orbit +]/orbit3 flies. An analogous procedure was followed to examine the ability of the transgene to rescue the lethality of orbit 3 /Df(3L)orbit2. Females of genotype w; orbit 3/P[w + orbit +] sr e Pr were crossed to w; ru h th st cu sr e ca/TM3 males. Flies carrying the transgene were first selected by the presence of the w + marker in the next generation. Individual lines were then established and flies examined by Southern hybridization for BamHI RFLPs to identify lines with a recombined chromosome carrying orbit 3 and P[w + orbit+]. Females of genotype w; orbit 3 P[w + orbit +] sr e Pr/TM6C were mated to w; Df(3L)orbit2/TM6C males. Among the F1 progeny, expected numbers of Sb + Pr flies were scored and those flies showed normal fertility in both sexes.
Orbit Antibody and Western Blot Analysis
A 2.0-kb EcoRI fragment of the cDNA clone pOrb1 was inserted in frame into the EcoRI site of the expression vector pGEX-2T (Pharmacia). The resulting plasmid expresses a fusion protein of a polypeptide corresponding to amino acids 1–632 and a stretch of 10 amino acids from the 5′ untranslated region of the cDNA with the COOH terminus of glutathione S-transferase (GST) protein. The recombinant Orbit protein was purified on a glutathione-Sepharose (Pharmacia) column. Antiserum was prepared by injecting rabbits as described (Harlow and Lane 1988). We affinity-purified antibodies specific to the Orbit protein from antiserum with GST-Orbit conjugated Sepharose after preabsorption with GST-conjugated Sepharose. For Western blot analysis, Canton-S females or orbit females were dissected and approximately equal volumes of ovaries were collected. Generally, ten larval wild-type brains or an approximately equal volume of brains from orbit 3 /Df(3L)orbit 2 third instar larvae were collected separately. Samples containing ∼20 μg of protein were electrophoresed and were transferred to a PVDF membrane (BioRad). To detect Orbit, the blots were incubated with the affinity-purified antibody diluted 1:1,500, followed by incubation with HRP-conjugated anti-rabbit IgG.
Microtubule Preparation and Overlay Assays
Microtubules were purified from 0–3-h-old Drosophila embryos essentially as described previously (Saunders et al. 1997). About 3 ml of embryos were homogenized with a Dounce homogenizer in 2 vol of ice-cold lysis buffer (0.1 M Pipes/NaOH, pH 6.6, 5 mM EGTA, 1 mM MgSO4, 0.9 M glycerol, 1 mM DTT, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). The microtubules were depolymerized by incubation on ice for 15 min, and the extract was then centrifuged at 16,000 g for 30 min at 4°C. The supernatant was recentrifuged at 135,000 g for 90 min at 4°C. Microtubules in this later supernatant were polymerized by addition of GTP to 1 mM and taxol to 20 μM and incubation at room temperature for 30 min. A 3-ml aliquot of the extract was layered on top of 3-ml 15% sucrose cushion prepared in lysis buffer. After centrifuging at 54,000 g for 30 min at 20°C using a swing out rotor, the microtubule pellet was resuspended in lysis buffer.
Microtubule overlay assays were performed as previously described (Saunders et al. 1997). 500 ng per lane of recombinant Asp (pAsp36), recombinant Orbit (see above), and BSA (Sigma Chemical Co.) were fractionated by 10% SDS-PAGE and blotted onto PVDF membranes (Millipore). The membranes were preincubated in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20) containing 5% low fat powdered milk for 1 h and then washed three times for 15 min in lysis buffer. The filters were then incubated for 30 min in lysis buffer containing either 1 mM GDP, 1 mM GTP, or 1 mM GTP-γ-S. MAP-free bovine brain tubulin (Molecular Probes) was polymerized at a concentration of 2 μg/ml in lysis buffer by addition of GTP to a final concentration of 1 mM and incubated at 37°C for 30 min. The nucleotide solutions were removed and the buffer containing polymerized microtubules added to the membranes for incubation for 1 h at 37°C with addition of taxol at a final concentration of 10 μM for the final 30 min. The blots were then washed three times with TBST and the bound tubulin detected using standard Western blot procedures using anti–β-tubulin antibodies (Boehringer Mannheim) at 2.5 μg/ml and the Super Signal detection system (Pierce).
Microtubule Binding Assays
Microtubules were polymerized with taxol in the absence of GTP as described in the previous section. Different concentrations of tubulin were used. Bacterially expressed Orbit was diluted to 200 ng/μl in microtubule lysis buffer (see previous section) and centrifuged for 30 min in a refrigerated Eppendorf centrifuge at top speed in order to remove any insoluble protein. This was mixed with the microtubule preparation (final volume of 20 μl) and incubated for 30 min at 37°C in the presence of GTP, GDP, or GTP-γ-S (1 mM each). Microtubules were sedimented by centrifugation for 30 min in an Eppendorf centrifuge and both the pellet and the supernatant were saved. The pellet was washed twice with 200 μl of lysis buffer and loaded, together with the supernatant on 10% polyacrylamide gels. The presence of microtubules after immunoblotting was assessed by staining the membranes with Ponceau S (Sigma Chemical Co.).
Results
orbit Identifies a Novel Locus Required for the Nuclear Division Cycles in Syncytial Embryos
We identified the original orbit mutation within a subset of a collection of P element–induced mutants (Deak et al. 1997) that showed maternal-effect lethality. Homozygous orbit mutants were fully viable, except that their development was delayed by a number of days in crowded cultures. However, homozygous orbit females laid fewer than 10% of the eggs of wild-type females, the numbers decreasing markedly as mutant females aged. Consistently, ovaries of orbit females showed degeneration and contained fewer egg chambers than wild-type. Approximately one third of the orbit-derived eggs possessed no nuclei, suggesting there had been defective premeiotic and/or meiotic divisions, and of the remainder, >90% showed perturbation of the uniform distribution of the nuclei. 10–20% of embryos derived from orbit females cellularized, at least partially, but <1% hatched. Homozygous males for orbit mutation were also sterile.
To better understand how the abnormal nuclear density might arise in orbit-derived embryos, we examined the distribution and organization of centrosomes and spindle microtubules during the syncytial nuclear division cycle. Whereas in wild-type embryos there is a regular distribution of mitotic spindles at metaphase (Fig. 1 A), in orbit-derived embryos at a similar stage there are regions devoid of nuclei that contain free centrosomes that nucleate asters of microtubules (Fig. 1 C). Moreover, additional centrosomes appeared to become incorporated into spindles to form multipolar structures (Fig. 1 C, arrowheads). Free centrosomes could also be observed in fields of anaphase figures from the mutant (Fig. 1 D) in which the spindles were frequently excessively curved, bent, and sometimes wavy (Fig. 1 D, arrowheads). This defect appeared accentuated at telophase (Fig. 1 E) where the midbodies could be disoriented so that they are aligned at 90° rather than 180° to each other (Fig. 1 E, arrowheads). When syncytial orbit-derived embryos were stained with Hoechst to reveal DNA, we frequently observed nuclei that were more brightly stained than their neighbors. These appear to contain more than a diploid amount of DNA, and could either be the outcome of failed nuclear separation or the fusion of nuclei (Fig. 1 F, large arrowheads). In addition, nuclei connected by thin chromatin bridges were occasionally seen suggesting failure of chromatid disjunction (Fig. 1 F, arrows).
Figure 1.
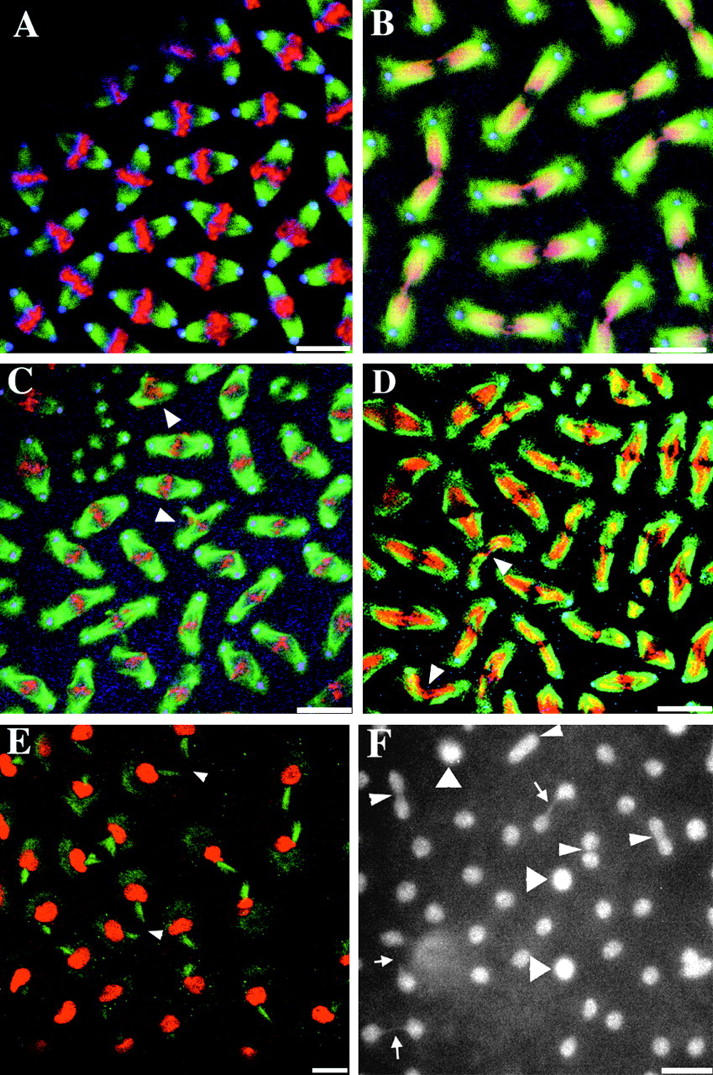
Mitotic defects in orbit-derived embryos. Wild-type embryos (A and B) and embryos derived from orbit females (C–E) were immunostained to visualize microtubules (green), centrosomes (blue), and simultaneously stained with propidium iodide for DNA (red). Normal metaphase (A) or anaphase (B) figures. C, A field of metaphase figures shows a region in the top left quadrant that has free centrosomes not associated with chromosomes. Arrowheads indicate tripolar metaphase figures. D, A field of anaphase figures showing examples of extremely curved spindle microtubule arrays (arrowheads). E, A field of telophase figures in which the spindle has become so bent as to position the midbody microtubules attached to sister nuclei at right angles (arrowheads). F, A field of late telophase figures stained with Hoechst. Large arrowheads indicate three nuclei that appear to contain more than a diploid complement of DNA because of their larger size and brighter staining. There are several pairs of sister nuclei that remain connected by chromatin bridges (arrows) and other pairs of sister nuclei in close association, as if they are either beginning to undertake telophase before having fully separated at anaphase, or that they are undergoing nuclear fusion (small arrowheads). Bars, 10 μm.
Molecular Mapping of orbit
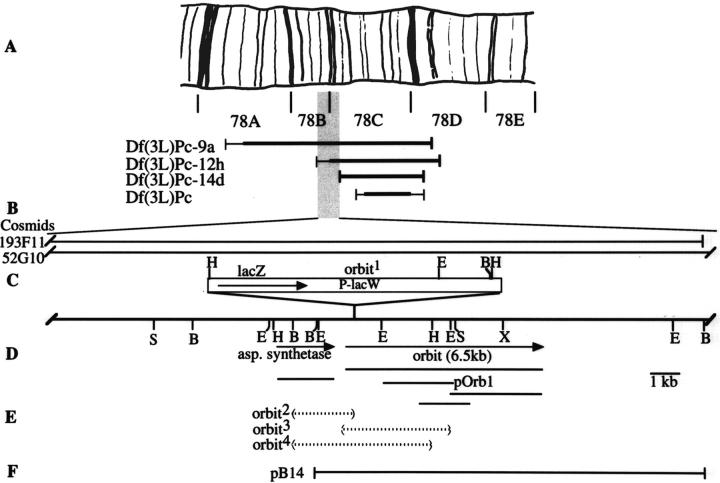
Sterility of the orbit homozygotes of both sexes could be reverted under dysgenic conditions suggesting that the P element is responsible for the mutation. In situ hybridization to polytene chromosomes showed a single P element at 78C. Consistently, the locus maps by recombination to 46.6 between st and cu. Deficiency mapping (Fig. 2 A) placed the orbit locus in the cytological interval 78B3-C1 to 78C2, defined by the proximal breakpoints of the chromosome deficiencies Df(3L)Pc-12h and Df(3L)Pc-14d. One male sterile, sa (White-Cooper et al. 1998), and four lethal mutations, l(3)neo29, l(3)07615, l(3)78Cb, and l(3)78Ca that map in the 78C region each complemented orbit, suggesting orbit is a previously unidentified gene. Hemizygotes for orbit showed reduced viability and exhibited the abnormal adult external morphologies typical of several cell cycle mutants, including slightly roughened eyes, thin and short bristles, and less frequently, notched wings. This indicates the hypomorphic nature of orbit and suggests a requirement for orbit gene function for the proliferation of imaginal tissues.
Figure 2.
Cytological and physical maps of the orbit region. A, Polytene chromosome map of division 78. The cytological extents of the four deficiencies are shown as thick lines below the map. Thin lines indicate uncertainties in the deficiency breakpoints. The orbit mutation was uncovered by Df(3L)Pc-9a and Df(3L)Pc-12h, but not by Df(3L)Pc-14d or Df(3L)Pc. This places orbit within the chromosome interval 78B3-C1 to 78C2. B, Horizontal lines represent the genomic regions contained by two cosmid clones, 52G10 and 193F11. C, Restriction enzyme map of the orbit region. Cleavage sites are: B, BamHI; E, EcoRI; H, HindIII; S, SalI. D, Extents of the orbit and asparagine synthetase transcription units in which the arrowhead indicates the 3′ end of the transcript. The cDNA clones for the two transcription units were isolated from 0–8- or 0–22-h embryonic cDNA libraries, or from a testis cDNA library. E, The extent of the genomic deletions associated with the three lethal alleles is represented as dotted lines between parentheses. The deletion associated with orbit 2 extends from within the 0.9-kb BamHI fragment in the aspartate synthetase gene to within the P element. The deletion associated with orbit 4 also extends from within the 0.9-kb BamHI fragment to a position within the 2.0-kb EcoRI–HindIII fragment of the orbit gene. The 3-kb deletion resulting in the orbit 3 mutation extends from the 5′ regulatory region to within the 2.0-kb EcoRI–HindIII fragment of the orbit gene without affecting the function of the asparagine synthetase gene. F, The 14-kb BamHI fragment from cosmid 193F11 was used for germ-line transformation.
In addition to wild-type revertants, mobilization of the P-lacW element also generated three imprecise excisions resulting in lethal mutations that we initially named orbit 2, orbit 3, and orbit 4. Most homozygotes of orbit 3 and orbit 4 or trans-heterozygotes between these two alleles died during the third instar larval stage. Hemizygotes for orbit 2 over Df(3L)Pc-9a survive until third instar larvae or to early pupae, although orbit 2 homozygotes died at an early larval stage due to an another spontaneous lethal mutation not included in the Df(3L)Pc-9a interval. Some of the trans-heterozygotes between orbit 2 and orbit 3 or between orbit 2 and orbit 4 survived until early pupal stage.
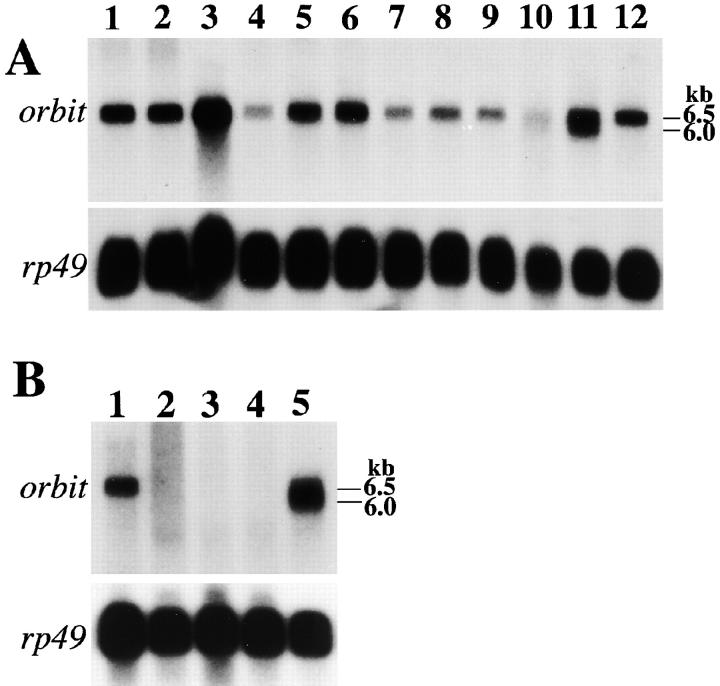
The molecular nature of these imprecise excisions became apparent after cloning DNA flanking the P-lacW insertion at 78C (Fig. 2 D). Northern blot analysis using the 6-kb HindIII genomic fragment spanning the P element insertion site as a probe revealed two transcription units: one of 6.5-kb that encodes a protein with a novel open reading frame, and a second of 2.2-kb that encodes a Drosophila homologue of the asparagine synthetase. The P element responsible for the orbit mutation was inserted 503 bp upstream of the first ATG of the 6.5-kb transcription unit. The 6.5-kb transcript, present at all developmental stages, and a 6.0-kb variant found in males, could barely be detected in orbit homozygotes (Fig. 3). Southern blot analysis suggested that the imprecise excision in orbit 3 had created a 3-kb deletion extending from the 5′ regulatory region into the coding region of the 6.5-kb transcription unit. The deletions in orbit 2 and orbit 4 extended from within the 6.5-kb transcription unit into the adjacent asparagine synthetase gene (Fig. 2 E) leading us to rename these mutations Df(3L)orbit2 and Df(3L)orbit4, respectively. To confirm that the 6.5-kb transcription unit was orbit, we carried out a germline transformation experiment with a 14-kb BamHI fragment of genomic DNA carrying the entire 6.5-kb transcription unit and the 3′ third of the asparagine synthetase gene (see Materials and Methods). Transformants carrying this transgene rescued the female sterility and the reduced viability in orbit/orbit 3 and Df(3L)orbit2/orbit 3 transheterozygotes.
Figure 3.
Northern analysis of orbit expression. A, Developmental expression of orbit in wild-type animals. Lane 1, 0–2-h embryos; lane 2, 2–4-h embryos; lane 3, 4–8-h embryos; lane 4, 8–12-h embryos; lane 5, 12–16-h embryos; lane 6, 16–22-h embryos; lane 7, first instar larvae; lane 8, second instar larvae; lane 9, third instar larvae; lane 10, pupae; lane 11, adult males; lane 12, adult females. 20 μg of total RNA was loaded on each lane. A 2-kb EcoRI fragment from an orbit cDNA clone was used as a probe. The same filter was then rehybridized with rp49 (ribosomal protein 49) cDNA as a loading control. The signal in lane 4 of this particular blot is anomalous, apparently due to a combination of underloading and a transfer problem. B, Expression of orbit in adult flies homozygous for orbit. Lane 1, wild-type adult females; lane 2 and 3, orbit/orbit adult females; lane 4, orbit/orbit adult males; lane 5, wild-type adult males. 20 μg of total RNA was loaded on each lane. A loading control as in A is shown at the bottom.
Mutations at orbit Locus Lead to an Accumulation of Polyploid Cells with Hypercondensed Chromosomes in the Larval Central Nervous System
The rapidity of the nuclear division cycles in syncytial embryos, and the absence of certain checkpoint controls make it difficult to observe primary defects in cell cycle mutants. The availability of additional orbit alleles showing late larval lethality therefore prompted us to investigate the effects of these mutations on progression of mitosis in dividing somatic cells. We found mitotic defects in cells of the larval CNS not only in the amorphic alleles generated by P element mobilization, but also in the original orbit mutant. In contrast to wild-type cells (Fig. 4A and Fig. B; Table ) where hyperploid cells are never seen, ∼6% of total metaphase cells in squashed preparations of the larval CNS from the orbit homozygotes contained more than a diploid complement of chromosomes. The overall mitotic index in the larval CNS was almost three times higher than wild-type (Table ), and moreover, the proportion of diploid cells in metaphase to anaphase was two times higher in orbit than in wild-type, indicative of a delay in the passage though this mitotic transition. The majority of the polyploid figures and 18% of the diploid figures in orbit homozygotes contained hypercondensed chromosomes, suggesting that these cells had been delayed in mitosis for a significant period of time (Fig. 4C and Fig. G; Table ). In orbit, the level of hyperploidy of most cells did not exceed 8N. Such hyperploidy could arise by a failure of either chromosome segregation or cytokinesis (Gatti and Baker 1989). However, the metaphase arrest or delay would seem to indicate the former possibility as being more likely. An early mitotic defect is also suggested by our observation of a low frequency of circular mitotic figures (Fig. 4 D) similar to those seen in mgr (merry-go-round) and aur mutants (Gonzalez et al. 1988; Glover et al. 1995). These have all the major chromosomes arranged in a circle with their centromeres inward and arms oriented toward the periphery and the fourth chromosomes at the center, an arrangement that is extremely rare in squashed preparations of wild-type brains. We could also observe monopolar anaphase-like figures similar to the one presented in Fig. 4 E, in which chromatids appear as if pulled towards a single pole.
Figure 4.
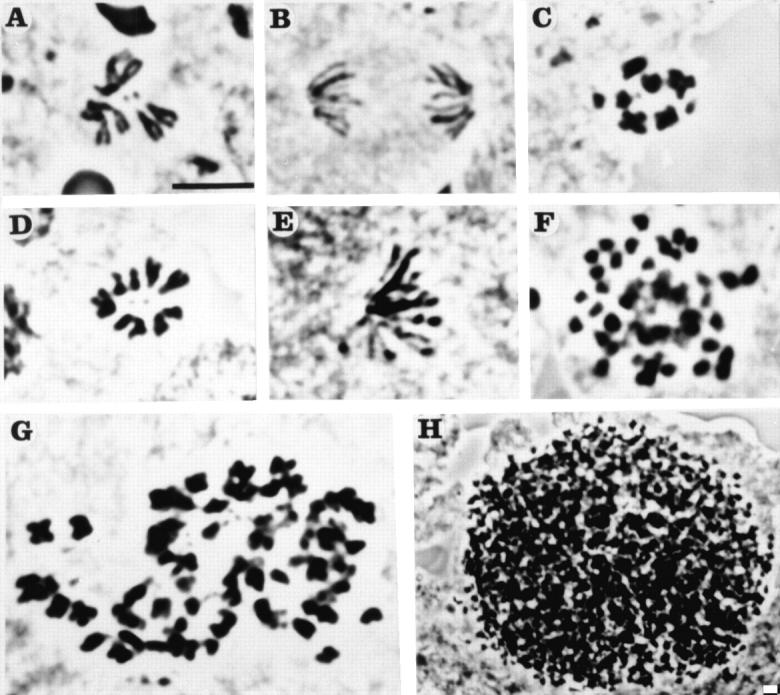
Cytological phenotypes in squashed preparations of larval CNS cells from third instar larvae homozygous for orbit or from third instar larvae trans-heterozygous for orbit 3 and Df(3L)orbit2. A diploid wild-type metaphase (A) or anaphase (B) figure. C, A diploid metaphase figure with hypercondensed chromosomes from orbit larva. D, A circular mitotic figure from orbit homozygotes. Note that for a cell to be scored as having a circular figure, all arms of the major chromosomes were oriented to the periphery and the small dot-like fourth chromosomes were in the center of the circle. E, A monopolar anaphase-like figure in which all chromatids appear to be pulled toward a single pole. F, A polyploid mitotic figure from the genotype orbit 3 /Df(3L)orbit2, in which the chromosomes are highly condensed. G, A polyploid mitotic figure containing hypercondensed chromosomes from an orbit larva. H, A view at lower magnification of an extremely hyperploid mitotic figure from an orbit 3 /Df(3L)orbit2 larva. More than 100 dot-like chromosomes were contained in this single mitotic cell. Bar, 10 μm.
Table 1.
Mitotic Phenotypes of Larval CNS Cells in orbit1
| Genotype | Fields scored | Polyploidy figures | Hypercondensed figures | CMFs | Metaphase to anaphase ratio | Mitotic index |
|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||||
| +/+ | 501 | 0 (0) | 0 (0) | 0 (0) | 3:1 | 0.9 |
| orbit1/orbit1 | 341 | 44 (6) | 118 (18) | 21 (3) | 6:1 | 2.4 |
| orbit1/Df(3L)Pc-9a | 488 | 120 (13) | 248 (32) | 38 (5) | 10:1 | 2.0 |
| orbit1/orbit2 | 375 | 165 (16) | 345 (40) | 25 (3) | 11:1 | 3.0 |
| orbit1/orbit3 | 444 | 128 (10) | 448 (34) | 38 (3) | 20:1 | 3.4 |
| orbit1/orbit4 | 188 | 75 (15) | 150 (4) | 15 (4) | 21:1 | 2.8 |
In orbit/Df(3L)Pc-9a hemizygotes, the proportion of polyploid cells, the proportion of diploid metaphase figures containing hyper-condensed chromosomes, and the ratio of metaphase to anaphase figures all increased (Table ), confirming the hypomorphic nature of this allele. Heterozygotes of orbit and each of the three mutations, Df(3L)orbit2, orbit3, and Df(3L)orbit4, showed comparable phenotypes to orbit/Df(3L)Pc-9a, consistent with the amorphic nature of these three lethal alleles resulting from sequence deletions. The proportion of the circular metaphase figures in hemizygotes for orbit or trans-heterozygotes between orbit and the amorphic alleles did not increase significantly compared with that in orbit homozygotes. We examined the loss of orbit function in orbit 3/Df(3L)Pc-9a or orbit 3/Df(3L)orbit2. The third instar larvae of these mutants were lacking imaginal discs and had a small larval CNS characteristic of many cell division cycle mutants. They exhibited an extremely high proportion of polyploid cells of between 80 to 90% of the total metaphase figures (Table ; Fig. 4). The extent of polyploidy was also increased in these mutants with >30% possessing greater than an 8N chromosome complement. The mitotic figure presented in Fig. 4 H shows a typically polyploid cell from a orbit 3/Df(3L)orbit 2 trans-heterozygote that has >100 extremely hypercondensed chromosomes. In contrast to the hypomorphic mutant orbit, we were unable to detect either circular metaphase or anaphase figures within these amorphic mutant brains. It appears that these highly polyploid cells are the result of multiple cell cycles in which chromosome segregation has failed.
Table 2.
Polyploid Mitotic Figures in Lethal orbit Alleles
| Genotype | Fields scored | Polyploid cells | >8N polyploid | Hypercondensed diploid cells | Anaphase figures | Mitotic index |
|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | (%) | |||
| orbit2/Df(3L)Pc-9a | 324 | 59 (85) | 29 (50) | 100 | 0 | 0.21 |
| orbit2/orbit3 | 467 | 87 (81) | 46 (43) | 100 | 0 | 0.23 |
| orbit2/orbit4 | 555 | 79 (91) | 47 (46) | 100 | 0 | 0.16 |
| orbit3/Df(3L)Pc-9a | 98 | 130 (84) | 42 (32) | 100 | 0 | 1.6 |
There appears to be no additional cell cycle defect resulting from the loss of the asparagine synthetase gene in addition to orbit since the mitotic phenotypes of Df(3L)Pc-9a/Df(3L)orbit2 and Df(3L)orbit2/Df(3L)orbit4 are identical to the orbit 3 amorphic mutant.
Monopolar and Multipolar Spindles in orbit Cells
We extended our observations of the orbit mutant phenotype in whole-mount preparations of the larval CNS by examining mitotic spindles and centrosomes as revealed by immunostaining. In contrast to the bipolar spindles organized by two fully separated centrosomes observed in all wild-type cells (Fig. 5 A), ∼10% of mitotic cells observed in the larval CNS of the hypomorphic mutant orbit contained a polyploid set of chromosomes associated with spindles that were frequently multipolar (Fig. 5 B). Consistent with the quantitation of orcein-stained figures in squashed preparations, some 3–5% of mitotic cells contained monopolar mitotic spindles. In some cases, these had a hemispindle-like structure in which chromosomes appeared to be pulled towards a single pole (Fig. 5 C). In others, condensed chromosomes were arranged around a single centrosome on the same plane (Fig. 5 D). These figures appear to correspond to the circular mitotic figures presented in squashed preparations of larval CNS (Fig. 5 D). The finding of monopolar spindles, together with cells that have a reduced number of centrosomal bodies relative to their chromosome complement, suggests that a primary defect in orbit mutants might be a failure of spindle pole separation that ultimately leads to polyploidy.
Figure 5.

Mitotic figures in whole-mount preparations of larval CNS cells from orbit homozygotes. A–C, Chromosomes were stained with propidium iodide (left column and red in the merged images presented in the right-most column). Simultaneously, the centrosomes were also visualized in the same channel of the microscope by immunostaining using Rb188, followed by Texas red-conjugated anti-rabbit IgG. Microtubules were immunostained as described in Materials and Methods (middle column and green in the merged images of the right-hand column). A, A normal bipolar wild-type metaphase spindle organized by fully separated centrosomes at opposite poles. B, A highly polyploid cell containing at least three foci of microtubule organizing activity (arrowheads). The weaker staining pole is actually on a lower confocal plane than the other two. C, A cell that contains a monopolar mitotic spindle in an anaphase-like configuration. The chromatids appear to be pulled toward the single pole (arrows). In D, chromosomes are stained with propidium iodide (red in the left column and the merged image at the extreme right) and centrosomes with Rb188 (green in middle column and the merged image). This cell contains a circular monopolar mitotic figure in which a single centrosome is situated in the center of chromosomes on the same confocal plane. Bars, 10 μm.
The orbit Gene Product Belongs to a Novel Family of Proteins Containing Putative GTP Binding Sites in a Highly Basic Domain
The sequence of orbit cDNAs revealed that the gene encodes a protein of 1,492 amino-acids (Fig. 6). The first ATG consistent with Drosophila translation initiation consensus (Cavener and Cavener 1993) is found at position 769 of the 5,959 nucleotide cDNA sequence. A poly(A) addition signal AATAAA lies at position 5,892. The novel protein contains a centrally located highly basic region (pI = 11.0) of 472 amino acids, flanked on both sides by short stretches of acidic residues. Within the basic domain are two consensus sites for phosphorylation by P34cdc2, and two putative GTP-binding motifs. The motif GGGTGTG (residues 544–550) closely resembles the glycine-rich peptide which interacts with the guanine or phosphate groups of the bound GTP in β-tubulin and in the Escherichia coli FtsZ protein (Nogales et al. 1998). The sequence NKLD (residues 400–403) corresponds to the NKXD (X for any amino acid residues) consensus motif which can interact with the purine base of the bound nucleotide in the GTPase superfamily (Burns and Farrell 1996). A BLAST search with the Orbit protein sequence revealed the presence of four closely related proteins from other organisms: two identified by the human putative open reading frame, KIAA0622 and KIAA0627 (Ishikawa et al. 1998), and two, R107.6 and ZC84.3, predicted from the C. elegans genome sequencing project (Wilson et al. 1994). The homologies fall into two regions, HR1 lying between residues 290 and 1,068, and HR2 between residues 1,093 and 1,271. As the regions of homology between the five proteins lie in register, it is likely that they are the signature of a family of related proteins. Moreover, the basic domain contained in the HR1 is a common feature of all five proteins, and the consensus sequences for cdc2 phosphorylation are found within or in the vicinity of this basic region in all except ZC84.3. The NKXD motifs are also conserved in the two human homologues. Five of the most strikingly conserved motifs shared by these proteins are presented in Fig. 7 B. Basic domains are a characteristic of microtubule-binding proteins and in this context, it is of interest that one of the conserved motifs of HR1 (residues 326–350) shows considerable similarity to the sequence involved in the binding of human MAP4 to microtubules (Olson et al. 1995). Furthermore, another conserved sequence within the basic domain (residues 479–506) has similarity to a motif in Stu1, a MAP identified from budding yeast (Pasqualone and Huffaker 1994).
Figure 6.

The amino acid sequence of the Orbit protein. A basic domain with a predicted pI of 11.0 flanked by short acidic regions of pI 4.1 and 4.3, respectively, are shown in boxes. The sequence shows extensive homology with a human putative proteins, KIAA0622 and KIAA0627, and related proteins from C. elegans, R107.6 and ZC84.3, which are underlined. Consensus sites for phosphorylation by p34cdc2 are shaded. The possible GTP-binding motif, GGGTGTG, identical to the GTP binding site in the FtsZ protein of E. coli, and NKLD, which matches a consensus motif for GTP binding in the GTPase family, are indicated by double underlining. These sequence data are available from GenBank/EMBL/DDBJ under accession number AB031048.
Figure 7.
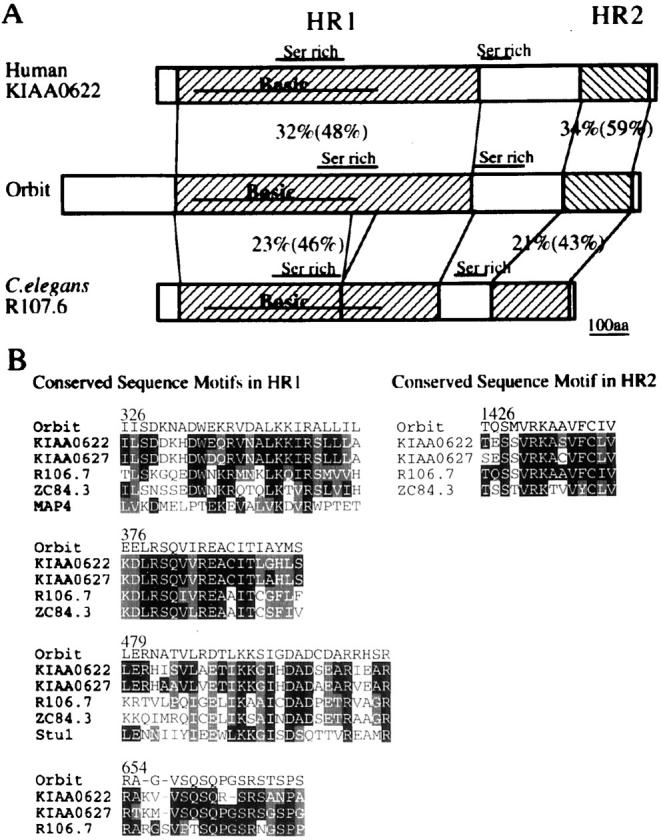
Conserved domains of the Orbit protein, and its human and C. elegans counterparts. A, Schematic representation of Orbit, KIAA0622 from human, and R107.6 from C. elegans. A segment of Orbit from residue 290–1068, which we designate HR1, shows 32% identity (48% similarity) and 23% identity (46% similarity) of amino acid sequence to a corresponding region of KIAA622 and R107.6, respectively. A second segment of Orbit from residue 1291–1471, which we designate as HR2, shows 34% identity (59% similarity) and 21% identity (43% similarity) to corresponding regions of KIAA0622 and R107.6, respectively. The basic domain and a serine-rich domain characteristics of HR1 in Orbit are found in conserved positions in the related proteins. A second serine-rich domain flanks HR1 in each case. B, Alignment of highly conserved sequence motifs within the HR1 and HR2 domains. One of these conserved sequence motifs (residue 326–350) shows a considerable similarity to a stretch of amino acid sequence within human MAP4 involved in a microtubule-binding. Another of these conserved sequence motifs (residue 479–506) has similarity to a sequence within the MAP Stu1p from budding yeast. Identical residues are in solid boxes and conservative amino acid changes are in light shaded boxes.
Orbit Associates with Microtubules in a GTP-dependent Manner
The presence of motifs within a basic domain that have similarity to MAP domains suggested that the Orbit protein might itself bind microtubules and that this could explain its requirement in mitosis. To investigate potential interactions of Orbit with microtubules, we first raised a polyclonal antibody against a fusion protein between GST and a polypeptide corresponding to amino acid residues 1–632. The affinity-purified antibody recognizes a polypeptide of 160–170 kD in immunoblots of extracts from ovaries or third instar larval brains (Fig. 8 A), which is greatly reduced in ovaries of homozygous orbit females, and barely detectable in the third instar larval brains from the transheterozygotes between the two amorphic alleles orbit 2 and orbit 3. Note that increased amounts of tissue extract were loaded from the mutant brains and ovaries to normalize total protein loaded. We conclude, therefore, that this band corresponds to the Orbit protein. The molecular weight of Orbit estimated from its electrophoretic mobility is in good agreement with the molecular weight of 165,420 D, calculated from the amino acid sequence. We then purified microtubules from Drosophila embryo extracts by taxol-induced polymerization, followed by centrifugation, and salt washing of the pellet. We found that the Orbit protein copurified with β-tubulin in this preparation and so, by this criterion, is a novel MAP (Fig. 8 B).
Figure 8.
A, Levels of Orbit protein are greatly reduced in orbit mutants. Equivalent amounts of total protein extract corresponding to three wild-type ovaries (lane 1); ten orbit/orbit ovaries (lane 2); ten wild-type larval brains (lane 3); and 20 orbit 3 /Df(3L)orbit2 larval brains (lane 4), were blotted and detected with an affinity-purified Orbit antibody. The antibody recognizes a major band of 165 kD that is reduced in amount in extracts from orbit ovaries and is barely detectable in extracts of orbit larval brains. B, Orbit copurifies with microtubules. Western blot of the fractions obtained during the purification of microtubules from wild-type embryos (see Materials and Methods). The blot was probed with anti-Orbit (diluted 1:1,500) and antitubulin (diluted 1:4) antibodies. Lane 1, 20 μg of crude embryonic protein extract; lane 2, 20 μg of protein from the supernatant fraction after the centrifugation through sucrose; lane 3, 20 μg of proteins removed from the microtubule pellet by a 250 mM NaCl wash; and lane 4, 10 μg of the final microtubule fraction. C and D, Orbit binds to microtubules in the presence of GTP, but not its nonhydrolyzable analogue GTP-γ-S. Recombinant Asp (lane 1), Orbit protein (lane 2), and BSA (lane 3) were transferred to PVDF membranes, incubated with the indicated nucleotides, and subsequently polymerized microtubules. Binding of microtubules was assessed using anti-β-tubulin (see Materials and Methods). E, Orbit binds to microtubules in solution in the presence of GTP (I), but not GDP (II) or GTP-γ-S (III). Soluble Orbit protein was incubated with different concentrations of microtubules (as indicated on the figure) in the presence of GTP (I), GDP (II) or GTP-γ-S (III). Polymerized microtubules were recovered by centrifugation. Presence of Orbit in the microtubule pellet and the supernatant was assessed by immunoblot using anti-Orbit antibody.
We assessed the direct binding of Orbit to tubulin in microtubule overlay assays using phosphocellulose purified MAP-free tubulin (Fig. 8C and Fig. D). Recombinant Orbit protein containing the putative tubulin binding domain and the two GTP binding motifs were transferred to PVDF membranes (Materials and Methods) which were preincubated with GDP, GTP, or its nonhydrolyzable analogue GTP-γ-S. Recombinant Asp protein was used as a positive control for microtubule binding and BSA as a negative control. The filters were then incubated with polymerized microtubules and binding detected using antitubulin antibodies. We found that this segment of Asp protein would bind microtubules irrespective of the preincubation treatment. In contrast, Orbit would bind microtubules only when first incubated with GTP, but not with GDP (not shown) or GTP-γ-S (Fig. 8C and Fig. D).
To confirm the results obtained by the microtubule overlay assays, we have also assessed whereas Orbit would bind microtubules in a GTP-dependent way when in solution. Microtubules were polymerized with taxol in the absence of GTP and then incubated with soluble Orbit protein in the presence of GTP, GTP, or GTP-γ-S (Fig. 8 E, I, II, and III, respectively). Binding to microtubules was detected by Western blots after sedimentation of the tubulin polymers by centrifugation. In the presence of GTP, Orbit was found exclusively in the microtubule pellet, whereas the protein was in the supernatant when either GDP or GTP-γ-S were used. This was independent of the microtubule concentration. We conclude that to bind microtubules, Orbit must bind GTP.
To determine whether the Orbit protein is a component of the mitotic spindle, we performed immunostaining of syncytial blastoderm embryos using the affinity-purified antibody described above and compared the staining pattern with distribution of tubulin (Fig. 9). As syncytial embryos enter mitosis at prophase, Orbit protein accumulates distinctly at the periphery of nuclei in the polar regions showing extensive colocalization with tubulin as the spindle forms (Fig. 9 A). Throughout metaphase to anaphase, Orbit colocalizes with microtubules throughout the entire region of the mitotic spindle and its asters (Fig. 9B and Fig. C). The microtubule association remains with the midbody (Fig. 9 D), and some residual midbody staining appears to remain in interphase (Fig. 9 E).
Figure 9.
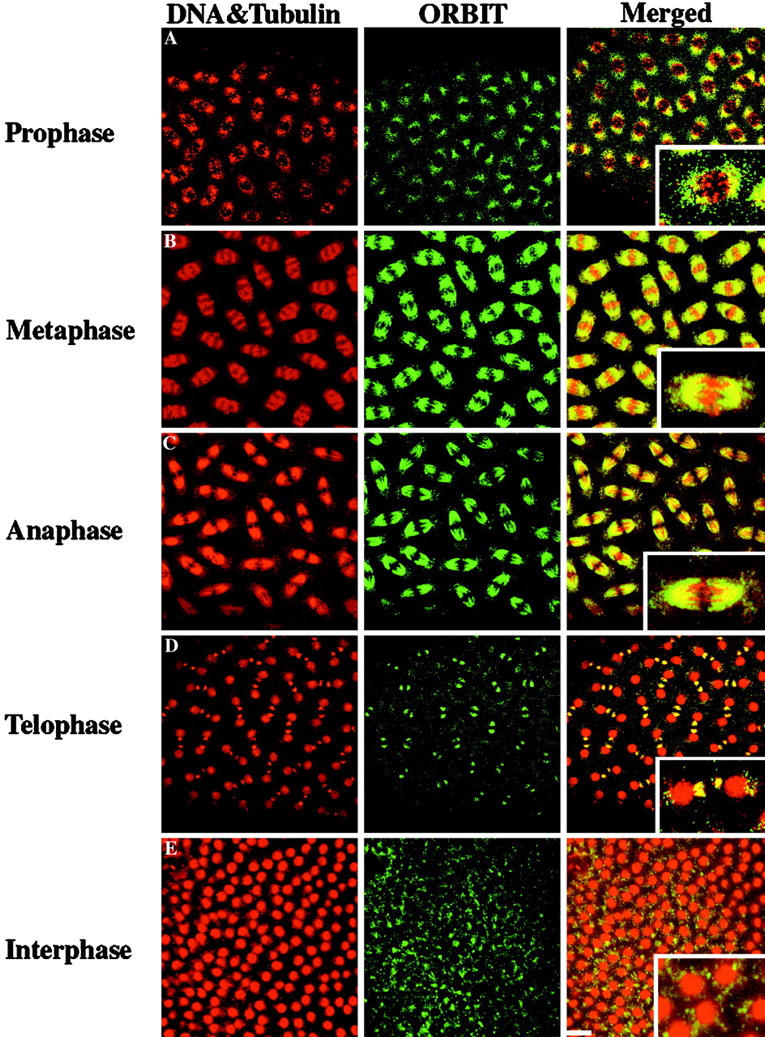
Immunolocalization of Orbit protein during the nuclear division cycle in syncytial blastoderm embryos. Wild-type syncytial embryos were simultaneously stained with propidium iodide to reveal DNA; antitubulin antibody, YL1/2; and Texas red-conjugated goat anti–rat IgG to reveal microtubules in the same channel of the microscope (red in the left and merged images on the extreme right). Staining of Orbit using the primary anti-Orbit rabbit antibody and FITC-conjugated goat anti–rabbit IgG is shown in the middle column and merged images in green. The mitotic phases are: A, prophase; B, metaphase; C, anaphase; and D, telophase. E, An interphase figure. Bar, 10 μm.
Discussion
Spindle Defects in orbit Embryos
The mutant phenotype of orbit is suggestive of a role for the wild-type gene in the functioning of the mitotic spindle consistent with the gene product being a novel MAP. This discovery helps overcome the difficulty in interpreting mitotic phenotypes in syncytial embryos derived from homozygous mutant females. Such difficulties arise since syncytial embryos lack certain checkpoints and so aspects of mitotic cycles can continue even though other steps are blocked. This is reflected by the finding of free centrosomes in orbit-derived embryos that appear to be undergoing autonomous duplication cycles, as seen in many other mitotic mutants. Maternal-effect mutations leading to mitotic defects are often hypomorphic, and have some residual function that allow the homozygous mothers to survive to adulthood partly assisted by a supply of wild-type gene product from the heterozygous grandmother. orbit is no exception to this rule, and indeed it proved possible to make amorphic alleles that show larval lethality by remobilization of the P element responsible for the original mutation. Nevertheless, the characteristic spindle defects of two types seen in orbit-derived embryos reflect the specific effect of diminution of the levels of Orbit protein. The branched spindles could either be an immediate consequence of failure in centrosome duplication or separation, or they could arise by capture of a free centrosome by an otherwise bipolar spindle. In either case, these defects, together with the high proportion of the spindles with wavy or bent arrays of microtubules, indicate a role for the Orbit protein in regulating function of spindle microtubules. Branched spindle defects are also seen in aur-derived embryos thought to be defective in aspects of centrosome separation (Glover et al. 1995), and as with orbit, are often associated with the generation of what appear to be tetraploid nuclei in the syncytial blastoderm. Such nuclei could arise either as a consequence of the failure of chromosome segregation, or a refusion of sister chromatids or sister nuclei after segregation. The finding of wavy and bent spindle microtubules, however, is not seen in aur-derived embryos and resembles more the maternal-effect phenotype described for certain alleles of asp (Gonzalez et al. 1990). Taken together, the different aspects of the maternal-effect phenotype suggest a primary defect in spindle microtubule function leading to failure of chromosome segregation.
Origins of Polyploidy in the orbit CNS
Defective spindle microtubule function is also evident in the developing larval central nervous system of orbit mutants. A high frequency of cells in a metaphase-like state suggests that the spindle integrity checkpoint has been activated to delay progression through mitosis. The high degree of chromosome condensation provides further evidence that the cells have been arrested at this point for some time, during which there has been continued activity of p34cdc2. There are two characteristic features of the arrested cells in the orbit mutant; a low frequency of monopolar mitotic structures and also polyploid cells. The proportion of polyploid cells increases when the orbit mutation is hemizygous, indicative of its hypomorphic nature. In the amorphic mutant combinations, monopolar figures are no longer seen and virtually all cells become polyploid and at much greater levels. Polyploid cells could arise either through a defect in chromosome segregation followed by exit from mitosis, and subsequent reentry into the next mitotic cycle, or alternatively, there can be a failure of cytokinesis. The findings of a high mitotic index with very few anaphases, and the presence of monopolar figures, strongly suggests to us that the polyploidy arises as a consequence of spindle defects leading to a failure of chromosome segregation. Of course, this would not preclude some function for the Orbit protein in the late mitotic spindle, the correct structure of which is essential for cytokinesis to take place (Williams et al. 1995; Adams et al. 1998). However, the low incidence of anaphase figures in orbit mutants suggests that mitotic events rarely proceed to this stage.
The high levels of polyploidy attained in cells of the amorphic orbit mutants indicate that they have gone through repeated cell cycles without division, and preclude analysis of the primary mutant defect. The hypomorphic orbit mutant on the other hand, allows us to glimpse those aspects of mitosis that are most sensitive to diminished Orbit function. The observation of a low frequency of monopolar spindle structures suggests that Orbit assists in promoting the correct separation of centrosomes to form a bipolar spindle structure. However, there would seem to be other requirements for the Orbit protein in the spindle since bipolar spindles do form, which then appear to undergo spindle checkpoint arrest at metaphase. Indeed, the centrosome separation defect may be secondary to a spindle microtubule function. In this respect, orbit mutants differ from mgr or aur, which appear to have a more direct role in centrosome separation. Not only is the frequency of monopolar mitotic figures lower in orbit than in mgr or aur, but also monopolar structures are not seen in amorphic orbit mutants, whereas they increase in frequency in amorphic aur mutants. This suggests a direct role for the Aurora protein kinase in centrosome separation, such that in its absence the mitotic cycle is definitively arrested at this point. In contrast, the decrease in mitotic index and accompanying increase in levels of polyploidy in amorphic orbit mutants is indicative of cells continually delayed and repeatedly leaking through the spindle integrity checkpoint. The structure of the monopolar figures also differs between these mutants. In mgr and aur, the chromosomes are invariably arranged in a circle in a metaphase-like state as if under tension with their centromeres pulled towards, but always at some distance from the center of, the circle and the chromosome arms pulled out towards the periphery. Similar figures are seen in orbit, but in addition, there are anaphase-like figures in which the centromeres appear to have been pulled into the immediate vicinity of a single pole. These cytological phenotypes more closely resemble those in the mutants for the kinesin-like protein, KLP61F, first thought to be required for centrosome separation at prophase (Heck et al. 1993), but then shown by antibody injection experiments to be required for maintenance of spindle bipolarity (Sharp et al. 1999a). The Orbit protein appears to be localized throughout the mitotic spindle like the KLP61F protein (Barton et al. 1995), although at the EM level it is apparent that KLP61F is not uniformly distributed (Sharp et al. 1999b).
Mitotic Functions of Microtubule-associated Proteins
Orbit protein is associated with all spindle and astral microtubules at all stages of the mitotic cycle, and microtubules from embryo cytoplasm copurify with Orbit protein attached to them. The lower ratio of Orbit:tubulin in the microtubule pellet fraction, compared with a crude extract could suggest that not all of the Orbit protein is bound to microtubules. Alternatively, the affinity of Orbit for the taxol polymerized microtubules used in our experiments could be lower than for naturally polymerized microtubules, a possibility currently under investigation. The primary sequence of the Orbit protein reveals it to be a basic protein, a characteristic of MAPs. Moreover, within these highly basic regions are motifs that strongly resemble sequences present in the vertebrate and yeast MAPs, MAP4 and Stu1p. Aizawa and colleagues 1990 described three distinctive features in the microtubule-binding domains of MAP2, tau, and MAP4. Polypeptides comprising these different domains cause microtubules assembled in vitro to adopt to take different shapes (Tokuraku et al. 1999). Similarly, microtubules assembled in the presence of the individual neuronal MAPs, MAP1A, MAP1B, and MAP2 were also shown to adopt a variety of shapes from “short and straight” to “long and bendy” (Pedrotti et al. 1996). Thus, the bending of spindle shape seen in orbit embryos may be indicative of requirement for the Orbit protein to confer a certain shape to the spindle microtubules.
Many of the first MAPs to be characterized were obtained from preparations of tubulin from mammalian brain, and are likely to have their primary function in the neuronal cytoskeleton. Nevertheless, it is now appreciated that some of these proteins are expressed in other tissues in which there is cell proliferation. Phosphorylation of the Xenopus homologue of MAP4 by both p34cdc2 and mitogen-activated-protein kinases appears important for its microtubule-binding and stabilizing properties and for chromosome movement during anaphase A (Shiina and Tsukita 1999). Similarly, Stathmin/Op18 is a protein that interacts with tubulin to inhibit microtubule polymerization. Overexpression of its nonphosphorylatable forms prevents mitotic spindle assembly in tissue culture cells (Belmont and Mitchison 1996; Marklund et al. 1996), whereas its phosphorylated form stimulates microtubule growth around chromosomes (Andersen and Karsenti 1997). In this context, the conserved p34cdc2 sites in Orbit may well play a role in regulating its function. Moreover, the abundance of serine residues within two regions of the protein may be indicative of sites for phosphorylation by other mitotic kinases, such as Polo or Aurora, whose consensus sites are not yet known.
Other MAPs can act through destabilizing the polymers, for example the Kin I kinesins (Desai et al. 1999), and others, such as katanin, can actually sever the microtubules (Hartman et al. 1998). Three Xenopus MAPs that each localize to the spindle have different effects on microtubule dynamics: XMAP215 promotes microtubule growth, XMAP230 decreases the catastrophe frequency, and XMAP310 increases the rescue frequency (Vasquez et al. 1994; Andersen and Karsenti 1997). It is of considerable future interest to determine whether the Orbit protein has any effect upon these parameters of microtubule dynamics.
At present the essential mitotic function of Orbit remains highly speculative, but our study has revealed a fascinating property of this novel protein; that it binds microtubules in a GTP-dependent manner. It is surprising that Orbit will bind to microtubules in the presence of GTP, but not in the presence of the nonhydrolyzable GTP-γ-S form since it is generally assumed that these nucleotides have similar structures. However, the vinca-alkaloid self-association of tubulin and microtubule assembly is sensitive to the precise modification of guanine nucleotide analogues and the salt concentration suggestive of an allosteric interaction (Vulevic and Correia 1997; Vulevic et al. 1997). The GTP binding site of Orbit has similarities to that of tubulin and so may also be very sensitive to modifications of the nucleotide. Another possibility is that Orbit is itself a GTPase and that its association with microtubules requires GTP hydrolysis. It is well established that tubulin and its prokaryotic counterpart, the FstZ protein, are GTPases (Mukherjee and Lutkenhaus 1998; Lu et al. 1998; reviewed by Nogales et al. 1999; Salimnia et al. 2000). The hydrolysis of GTP complexed to αβ-tubulin dimers at the plus ends of microtubules leads to their increased curvature and destabilization of the tubulin lattice (Muller-Reichert et al. 1998). Microtubule destabilizing agents, such as colchicine and nocodazole, are known to promote GTPase activity, whereas taxol binding to the inner surface of the microtubule counteracts the effects of GTP hydrolysis. It is of considerable interest to know whether Orbit has GTPase activity either intrinsically or in association with tubulin. Irrespective of whether Orbit can hydrolyze GTP, the finding of a new GTP binding MAP raises possible new complexities for the role of this nucleotide in regulating microtubule behavior.
It would be of interest to determine whether there is any interaction between Orbit and the Awd protein (Abnormal wing discs) a microtubule-associated NDP kinase that converts GDP to GTP (Biggs et al. 1990). awd mutants display hypercondensed chromosomes typical of those seen in colchicine-treated cells, suggesting activity of this enzyme is required for microtubule polymerization. Given our present findings, however, it is also likely that the Awd protein can influence other aspects of microtubule behavior. The availability of hypomorphic orbit mutants now raises the future possibility of using genetic screens to search for mutations that either enhance or suppress the orbit phenotype. Such mutations could identify genes encoding proteins that interact with or regulate Orbit protein function in the mitotic spindle.
Acknowledgments
We would like to thank Fumiko Hirose for supplying staged total RNA and Maria Deak for technical assistance at initial stages of the project.
This work was supported by a program grant from the Cancer Research Campaign (CRC), and by a Grant-in-Aid for Scientific Research (A) on Priority Areas from the Ministry of Education, Science and Culture of Japan. Project grant support was provided by the Medical Research Council and the Association for International Cancer Research. The CRC Cell Cycle Genetics Group is also a member of a TMR Network of the EU.
Footnotes
Michina Shiraki's present address is Graduate School of Biological Sciences, Nara Institute of Science and Technology, Takayama-cho, lkoma, Nara 630-0101, Japan.
Abbreviations used in this paper: asp, abnormal spindle; aur, aurora; GST, glutathione S-transferase; MAP, microtubule-associated protein; mgr, merry-go-round.
References
- Adams R.R., Tavares A.A., Salzberg A., Bellen H.J., Glover D.M. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H., Emori Y., Murofushi H., Kawasaki H., Sakai H., Suzuki K. Molecular cloning of a ubiquitously distributed microtubule-associated protein with Mr 190,000. J. Biol Chem. 1990;265:13849–13855. [PubMed] [Google Scholar]
- Andersen S.S., Karsenti E. XMAP310a Xenopus rescue-promoting factor localized to the mitotic spindle. J. Cell Biol. 1997;139:975–983. doi: 10.1083/jcb.139.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S.S., Buendia B., Dominguez J.E., Sawyer A., Karsenti E. Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopus laevis eggs and dividing cells. J. Cell Biol. 1994;127:1289–1299. doi: 10.1083/jcb.127.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N.R., Pereira A.J., Goldstein L.S. Motor activity and mitotic spindle localization of the Drosophila . Mol. Biol. Cell. 1995;6:1563–1574. doi: 10.1091/mbc.6.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Mitchison T.J. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Biggs J., Hersperger E., Steeg P.S., Liotta L.A., Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor-metastasis codes for a nucleoside diphosphate kinase. Cell. 1990;63:933–940. doi: 10.1016/0092-8674(90)90496-2. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burns R.G., Farrell K.W. Getting to the heart of β-tubulin. Trends Cell Biol. 1996;6:297–303. doi: 10.1016/0962-8924(96)10024-6. [DOI] [PubMed] [Google Scholar]
- Cavener D.R., Cavener B.A. Translation start sites and mRNA leaders. In: Maroni G., editor. An Atlas of Drosophila Genes. Oxford University Press; New York: 1993. pp. 359–377. [Google Scholar]
- Charrasse S., Schroeder M., Gauthier-Rouviere C., Ango F., Cassimeris L., Gard D.L., Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 1998;111:1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Deak P., Omar M.M., Saunders R.D., Pal M., Komonyi O., Szidonya J., Maroy P., Zhang Y., Ashburner M., Benos P. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogastercorrelation of physical and cytogenetic maps in chromosomal region 86E–87F. Genetics. 1997;147:1697–1722. doi: 10.1093/genetics/147.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Frasch M. The maternally expressed Drosophila gene encoding the chromatin-binding protein BJ1 is a homolog of the vertebrate gene regulator of chromatin condensation, RCC1 . EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:1225–1236. doi: 10.1002/j.1460-2075.1991.tb08064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Glover D.M., Saumweber H. Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J. Cell Sci. 1986;82:155–172. doi: 10.1242/jcs.82.1.155. [DOI] [PubMed] [Google Scholar]
- Gard D.L., Kirschner M.W. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Baker B.S. Genes controlling essential cell-cycle function in Drosophila melanogaster . Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Glover D.M., Leibowitz M.H., McLean D.A., Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Glover D.M. Techniques for studying mitosis in Drosophila . In: Brook R., Fantes P., editors. The Cell CycleA Practical Approach. IRL Press; Oxford: 1993. pp. 163–168. [Google Scholar]
- Gonzalez C., Casal J., Ripoll P. Functional monopolar spindles caused by mutation in mgr, a cell division gene of Drosophila melanogaster . J. Cell Sci. 1988;89:39–47. doi: 10.1242/jcs.89.1.39. [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Saunders R.D.C., Casal J., Molina I., Carmena M., Ripoll P., Glover D.M. Mutations at the asp locus of Drosophila leads to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J. Cell Sci. 1990;96:605–616. doi: 10.1242/jcs.96.4.605. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane E. AntibodiesA Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor Press, NY: 1988. [Google Scholar]
- Hartman J.J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R.D., McNally F.J. Katanin, a microtubule-severing protein, is a novel ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- Heck M.M., Pereira A., Pesavento P., Yannoni Y., Spradling A.C., Goldstein L.S. The kinesin-like protein KLP61F is essential for mitosis in Drosophila . J. Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino T.I., Saura A.O., Sorsa V. Salivary gland chromosome maps. Drosophila Infomation Service. 1994;1994:619–738. [Google Scholar]
- Hyman A.A., Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Inoue Y.H., Glover D.M. Involvement of the rolled/MAP kinase gene in Drosophila mitosisinteraction between genes for the MAP kinase cascade and abnormal spindle . Mol. Gen. Genet. 1998;258:334–341. doi: 10.1007/s004380050739. [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Nagase T., Suyama M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. DNA Res. 1998;5:169–176. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- Kellogg D.R., Alberts B.M. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol. Biol. Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R., Field C.M., Alberts B.M. Identification of microtubule-associated proteins in the centrosome, spindle, and kinetochore of the early Drosophila embryo. J. Cell Biol. 1989;109:2977–2991. doi: 10.1083/jcb.109.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Stricker J., Erickson H.P. FtsZ from Escherichia coli, Azobacter vinelandii, and Thermotoga maritimaquantitation, GTP hydrolysis and assembly. Cell Motil. Cytoskel. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Marklund U., Larsson N., Gradin H.M., Brattsand G., Gullberg M. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5290–5298. [PMC free article] [PubMed] [Google Scholar]
- Miller K.G., Field C.M., Alberts B.M. Actin-binding proteins from Drosophila embryosa complex network of interacting proteins detected by F-actin affinity chromatography. J. Cell Biol. 1989;109:2963–2975. doi: 10.1083/jcb.109.6.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:242–269. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T., Chretien D., Severin F., Hyman A.A. Structural changes at microtubule ends accompanying GTP hydrolysisinformation from a slowly hydrolyzable analogue of GTP, gunylyl(alpha,beta)methylenediphosphate. Proc. Natl. Acad. Sci. USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Downing K.H., Amos L.A., Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- Nogales E., Whittaker M., Milligan R.A., Downing K.H. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Olson K.R., McIntosh J.R., Olmsted J.B. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J. Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K., Hisanaga S., Bulinski J.C., Murofushi H., Aizawa H., Itoh T.J., Hotani H., Okumura E., Tachibana K., Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J. Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D., Huffaker T.C. STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti B., Ulloa L., Avila J., Islam K. Characterization of microtubule-associated protein MAP1Bphosphorylation state, light chains, and binding to microtubules. Biochemistry. 1996;35:3016–3023. doi: 10.1021/bi951314f. [DOI] [PubMed] [Google Scholar]
- Rothwell W.F., Fogarty P., Field C.M., Sullivan W. Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organization. Development. 1998;125:1295–1303. doi: 10.1242/dev.125.7.1295. [DOI] [PubMed] [Google Scholar]
- Salimnia H., Radia A., Bernatchez S., Beveridge T.J., Dillon J.R. Characterization of the ftsZ cell division gene of Neisseria gonorrhoeaeexpression in Escherichia coli and N. gonorrhoeae . Arch. Microbiol. 2000;173:10–20. doi: 10.1007/s002030050002. [DOI] [PubMed] [Google Scholar]
- Saunders R.D.C., Avides M.C., Howard T., Gonzalez C., Glover D.M. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., McDonald K.L., Brown H.M., Matthies H.J., Walczak C., Vale R.D., Mitchison T.J., Scholey J.M. Visualization of the bipolar kinesin, KLP61F, on microtubule bundles within spindles of Drosophila early embryos J. Cell Biol. 144 1999. 125 138a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Yu K.R., Sisson J.C., Sullivan W., Scholey J. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos Nature Cell Biol. 1 1999. 51 54b [DOI] [PubMed] [Google Scholar]
- Shiina N., Tsukita S. Mutations at phosphorylation sites of Xenopus microtubule-associated protein 4 affect its microtubule-binding ability and chromosome movement during mitosis. Mol. Biol. Cell. 1999;10:597–608. doi: 10.1091/mbc.10.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W., Fogarty P., Theurkauf W. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 1993;118:1245–1254. doi: 10.1242/dev.118.4.1245. [DOI] [PubMed] [Google Scholar]
- Sunkel C.E., Glover D.M. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Tokuraku K., Katsuki M., Nakagawa H., Kotani S. A new model for microtubule-associated protein (MAP)-induced microtubule assembly. The Pro-rich region of MAP4 promotes nucleation of microtubule assembly in vitro . Eur. J. Biochem. 1999;259:158–166. doi: 10.1046/j.1432-1327.1999.00019.x. [DOI] [PubMed] [Google Scholar]
- Vasquez R.J., Gard D.L., Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulevic B., Correia J.J. Thermodynamic and structural analysis of microtubule assemblythe role of GTP hydrolysis. Biochemistry. 1997;36:1357–1375. doi: 10.1016/S0006-3495(97)78782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulevic B., Lober S., Correia J.J. Role of guanine nucleotides in the vinblastin-induced self-association of tubulineffects off guanosine alpha, beta-methylenetrisphosphate and guanosine alpha, beta-methylenediphosphate. Biochemistry. 1997;36:12828–12835. doi: 10.1021/bi971120v. [DOI] [PubMed] [Google Scholar]
- White-Cooper H., Schafer M.A., Alphey L.S., Fuller M.T. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila . Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- Whitfield W.G., Millar S.E., Saumweber H., Frasch M., Glover D.M. Cloning of a gene encoding an antigen associated with the centrosome in Drosophila . J. Cell Sci. 1988;89:467–480. doi: 10.1242/jcs.89.4.467. [DOI] [PubMed] [Google Scholar]
- Williams B.C., Riedy M.F., Williams E.V., Gatti M., Goldberg M.L. The Drosophila kinesin-like protein KLP3A is a midbody component. J. Cell Biol. 1995;129:709–723. doi: 10.1083/jcb.129.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R., Ainscough R., Anderson K., Baynes C., Berks M., Bonfield J., Burton J., Connell M., Copsey T., Cooper J. 2.2Mb of contiguous nucleotide sequence from chromosome III of C. elegans . Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]