Abstract
A small conserved open reading frame in the plastid genome, ycf9, encodes a putative membrane protein of 62 amino acids. To determine the function of this reading frame we have constructed a knockout allele for targeted disruption of ycf9. This allele was introduced into the tobacco plastid genome by biolistic transformation to replace the wild-type ycf9 allele. Homoplasmic ycf9 knockout plants displayed no phenotype under normal growth conditions. However, under low light conditions, their growth rate was significantly reduced as compared with the wild-type, due to a lowered efficiency of the light reaction of photosynthesis. We show that this phenotype is caused by the deficiency in a pigment–protein complex of the light-harvesting antenna of photosystem II and hence by a reduced efficiency of photon capture when light availability is limiting. Our results indicate that, in contrast to the current view, light-harvesting complexes do not only consist of the classical pigment-binding proteins, but may contain small structural subunits in addition. These subunits appear to be crucial architectural factors for the assembly and/or maintenance of stable light-harvesting complexes.
Keywords: chloroplast, LHCII, light-harvesting complex, photosynthesis, ycf9
Introduction
During the past decade, the plastid genomes from a number of higher and lower plant species have been completely sequenced. The picture that has emerged from these analyses is that plastid genomes show a remarkably conserved structure and coding capacity across large phylogenetic distances. The vast majority of plastid genome-encoded genes can be grouped into two classes: genetic system genes (rRNAs, tRNAs, ribosomal proteins, subunits of an Escherichia coli-like RNA polymerase) and photosynthesis genes (subunits of the photosystems I and II, the cytochrome b6f complex, the ATP synthase, large subunit of Rubisco). The few remaining genes have functions other than gene expression and photosynthesis and specify, for example, components of certain biosynthetic pathways (e.g., an acetyl-CoA-carboxylase subunit).
In addition to the >100 identified plastid genes, the chloroplast genomes of higher plants carry a number of open reading frames of unknown function designated ycf (hypothetical chloroplast reading frames; for an overview see Maier et al. 1995). These reading frames usually display strong interspecific conservation and therefore are assumed to be genuine genes. Recent methodological progress has opened new avenues to the functional characterization of plastid genome-encoded open reading frames. The successful development of transformation technologies for chloroplast genomes (Boynton et al. 1988; Svab et al. 1990; Svab and Maliga 1993) has facilitated reverse genetics approaches in plastids. The presence of an efficient homologous recombination system in chloroplasts allows the targeted inactivation of plastid-encoded reading frames by deletional or insertional mutagenesis (for review see Rochaix 1997; Bock et al. 1999). The resulting transplastomic plants can be purified to homoplasmy: all wild-type plastid genome copies are eliminated through continuous antibiotic selection and the plants eventually carry a homogeneous population of transformed plastid genomes. Such homoplasmic plants will be deficient for the gene product encoded by the open reading frame of interest and detailed analysis of their phenotype will allow conclusions about the function(s) of the reading frame in the wild-type. Reverse genetics experiments in the unicellular green alga Chlamydomonas reinhardtii (Monod et al. 1994; Takahashi et al. 1996) and, more recently, in the higher plant tobacco (Ruf et al. 1997; Burrows et al. 1998) have contributed substantially to the advancement of our understanding of both photosynthesis and chloroplast gene expression.
In this work, we have analyzed by reverse genetics a small conserved chloroplast open reading frame, ycf9. Generation of knockout tobacco plants revealed that ycf9 is a photosynthesis-related gene which, however, is not essential for photoautotrophic growth. We show that ycf9 encodes a novel structural component of a light-harvesting complex (LHC) of photosystem II (PSII). This component appears to be required for the stable integration of the pigment-binding LHC protein CP26 into the antenna of PSII.
Materials and Methods
Plant Material and Growth Conditions
Sterile tobacco plants (Nicotiana tabacum cv. Petit Havana) were grown on agar-solidified MS medium containing 30 g/liter sucrose (Murashige and Skoog 1962). Homoplasmic transplastomic lines were rooted and propagated on the same medium. For protein isolation and physiological measurements, wild-type and transformed plants were planted in pots and kept under greenhouse conditions. Plants grown under standard light (4,000–7,000 lux), high light (27,000 lux), or low light (100–300 lux) conditions were used for physiological measurements.
Construction of a Plastid Transformation Vector for Inactivation of ycf9
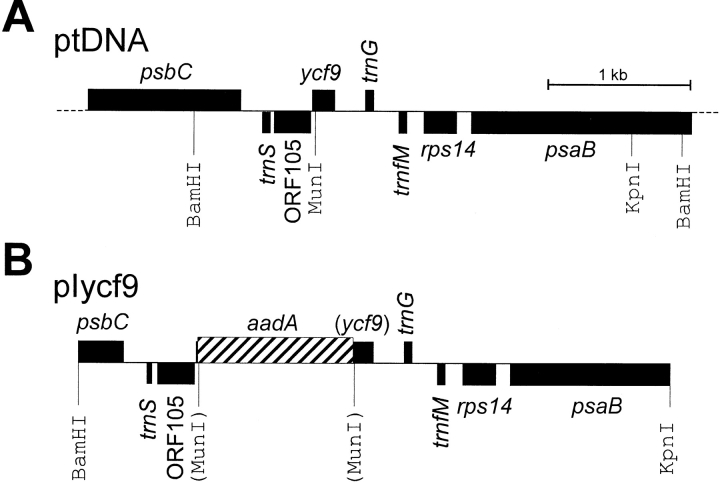
The region of the tobacco chloroplast genome containing the ycf9 reading frame was excised from a SalI ptDNA clone as a BamHI/KpnI fragment (Shinozaki et al. 1986; Fig. 1 A). The fragment was ligated into a similarly cut Bluescript vector (Stratagene). The ycf9 knockout allele was created by digestion with MunI, followed by a fill-in reaction of the recessed ends with Klenow DNA polymerase. A chimeric aadA gene conferring resistance to aminoglycoside antibiotics (Svab and Maliga 1993) was inserted into the blunted MunI site to disrupt ycf9 and to facilitate selection of chloroplast transformants. A plasmid clone carrying the aadA gene in the same orientation as ycf9 yielded transformation vector pIycf9 (Fig. 1 B).
Figure 1.
Construction of a plastid transformation vector for targeted disruption of the ycf9 open reading frame. A, Physical and restriction map of the region of the tobacco chloroplast genome containing ycf9. Genes above the line are transcribed from the left to the right, genes below the line are transcribed in the opposite direction. Restriction sites relevant for cloning and RFLP analysis are indicated. B, Map of the plastid DNA targeting fragment in the transformation vector pIycf9. The selectable marker gene, a chimeric aadA, was inserted downstream of the ycf9 initiation codon and is transcribed in the same direction as ycf9 in the wild type. Restriction sites destroyed during construction by ligation of heterologous ends are shown in parentheses.
Chloroplast Transformation and Selection of Homoplasmic Transplastomic Tobacco Lines
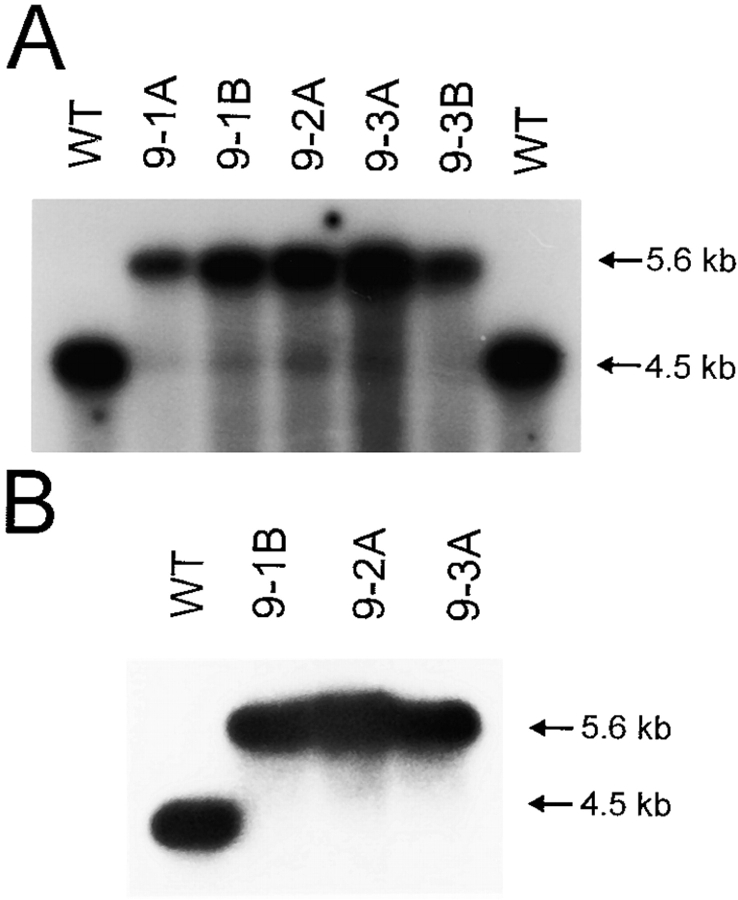
Young leaves from sterile tobacco plants were bombarded with plasmid pIycf9-coated tungsten particles using a biolistic gun (PDS1000He; BioRad). Primary spectinomycin-resistant lines were selected on RMOP regeneration medium containing 500 mg/liter spectinomycin dihydrochloride (Svab et al. 1990; Svab and Maliga 1993). Plastid transformants were identified by PCR amplification using the primer pair P10 (5′-AACCTCCTATAGACTAGGC-3′; complementary to the psbA 3′-untranslated region of the chimeric aadA gene) and P11 (5′-AGCGAAATGTAGTGCTTACG-3′; derived from the 3′ portion of the aadA coding region). Three independent transplastomic lines were subjected to four additional rounds of regeneration on RMOP/spectinomycin to obtain homoplasmic tissue. Homoplasmy was verified by restriction fragment length polymorphism (RFLP) analyses (see Fig. 3).
Figure 3.
RFLP analyses as test for homoplasmy of ycf9 knockout plants. A, Restriction analysis of total cellular DNA with BamHI. Three independently generated transplastomic lines (designated 9-1, 9-2, and 9-3; capital letters mark individually regenerated plants from one and the same line) and two wild-type controls (WT) are shown. Digestion with BamHI and hybridization to a radio-labeled ycf9-specific probe yields a 4.5-kb fragment for the wild-type genome and a 5.6-kb fragment for the transplastome (see Fig. 1). Note presence of a weak band of wild-type size in all transplastomic lines in addition to the 5.6-kb band characteristic of transformed plastid genomes. B, Restriction analysis of purified chloroplast DNA. Digestion with BamHI and hybridization to the same probe as in A detects only the 5.6-kb fragment diagnostic of the transplastome. Absence of the weak band of wild-type size detected in the total DNA samples from all transplastomic lines indicates homoplasmy and suggests the presence of promiscuous ycf9 copies DNA in an extraplastidic cellular compartment.
Isolation of Nucleic Acids and Hybridization Procedures
Total plant DNA was isolated by a minipreparation procedure (Doyle and Doyle 1990). For chloroplast DNA extraction, organelles were purified in Percoll gradients (Bock 1998). Restriction enzyme-digested DNA samples were separated on 1% agarose gels and blotted onto Hybond N nylon membranes (Amersham). For hybridization, α[32P]dATP-labeled probes were generated by random priming (Multiprime DNA labeling system; Amersham). A radiolabeled PCR product covering the entire coding region of ycf9 (generated with primer pair: Ycf9A, 5′-GCTGATAGAGGGATCAAAT-3′; Ycf9B, 5′-GGGTCATTTTGGTTTTGGG-3′) was used as probe for the RFLP analysis. Hybridizations were carried out at 65–68°C in Rapid Hybridization Buffer (Amersham).
Protein Isolation Procedures
Thylakoid proteins from wild-type and mutant tissue were isolated from total leaf material or isolated chloroplasts following published procedures (Machold et al. 1979). To analyze the association of the Ycf9 protein with the thylakoid membrane, purified thylakoids from wild-type and mutant plants (75 μg chlorophyll per 300 μl wash solution) were washed for 20 min at room temperature in 2 M NaCl, 2 M KJ, 0.1 M Na2CO3, 0.1 M NaOH, or 4 M urea. After centrifugation for 20 min at 40,000 g, pelleted thylakoids were washed once with a solution of 20 mM EDTA and 50 mM Hepes, pH 8.0. Washed thylakoids and supernatants were subsequently analyzed by PAGE.
Purification of LHC complexes and other protein complexes of the thylakoid membrane was carried out according to published protocols (Feng and McCarty 1990; Eshaghi et al. 1999).
SDS-PAGE and Western Blot Analyses
Isolated thylakoid or soluble proteins were separated on tricine-SDS polyacrylamide gels (Schägger and von Jagow 1987) and stained with Coomassie blue or silver, according to standard protocols (Coligan et al. 1995). For Western blot analyses, electrophoretically separated proteins were transferred to Hybond ECL nitrocellulose membranes (Amersham) using the Trans-Blot SD semidry transfer cell (BioRad) with a standard transfer buffer (182 mM glycine, 20 mM Tris, 20% methanol, 0.05% SDS). Immunoblot detection was performed using the enhanced chemoluminescence system (ECL; Amersham).
Mass Spectroscopy
Matrix-assisted laser desorption/ionization mass spectroscopy (MALDI) was performed with a Voyager STR DE mass spectrometer (PE Biosystems). LHCs purified from wild-type and ycf9 knockout plants were cocrystallized with dihydroxybenzoic acid and analyzed in the linear mode with an accelerating voltage of 20 kV. All measurements were externally calibrated for the low molecular weight range. Data were confirmed by several independent measurements.
Physiological Measurements
Determination of PSII activity was performed on dark-adapted leaves from wild-type and mutant plants as described previously (Ruf et al. 1997). The redox state of the reaction center chlorophyll P700 of photosystem I (PSI) was monitored by following the apparent changes in absorbance of dark-adapted leaves from wild-type and mutant plants at 830-nm wavelength (Schreiber et al. 1988) using a PAM fluorimeter with a modified emitter/detector unit. For rereduction of PSI, leaves were exposed to white light pulses of 4,000 μE/m2s.
Net oxygen evolution was measured at room temperature with a Clark electrode (Hansatech) in suspensions of isolated chloroplasts lysed in a hypotonic medium (Laasch 1987). Phenyl-1,4-benzoquinone (1 mM) was used as an electron acceptor. A halogen lamp (KL1500; Schott) served as a light source and was connected to the reaction vessel through fiber optics. Different light intensities were adjusted with neutral density filters.
Results
Targeted Inactivation of the Tobacco Chloroplast ycf9 Reading Frame
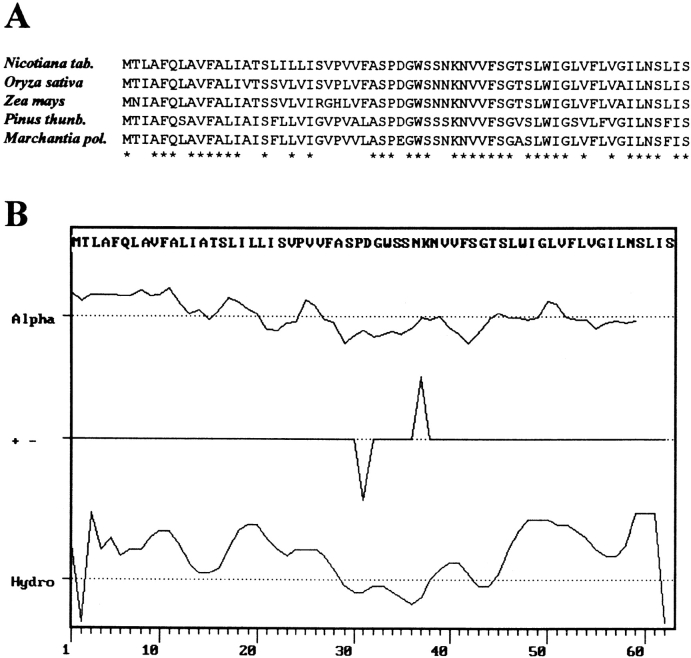
The chloroplast genomes of higher plants harbor in their large single-copy region a small conserved open reading frame designated ycf9 (Fig. 1 A). The putative gene product of ycf9 is a protein of 62 amino acids (Fig. 2 A) consisting of two potential membrane-spanning helices that are highly hydrophobic (Fig. 2 B). A hydrophilic domain separates the two hydrophobic stretches from each other suggesting that the Ycf9 protein spans twice either the thylakoid membrane or the inner membrane of the chloroplast envelope. Active transcription of ycf9 (Yao et al. 1989), together with the presence of strongly homologous reading frames in cyanobacteria (Kaneko et al. 1996) and algae (Evrard et al. 1990), suggest that ycf9 is a genuine gene the product of which participates in an evolutionarily conserved cellular function.
Figure 2.
Properties of the putative Ycf9 gene product. A, Alignment of the ycf9-derived amino acid sequences from four higher plant species and the liverwort Marchantia polymorpha. Amino acid positions identical in all species listed here are denoted by asterisks. B, Predicted α-helical domains, location of charged amino acid residues, and hydropathy blot of the tobacco Ycf9 protein. The protein has two potential membrane-spanning highly hydrophobic domains separated by a hydrophilic stretch. GenBank/EMBL/DDBJ accession numbers are: Z00044 (Nicotiana), X15901 (Oryza), X86563 (Zea), D17510 (Pinus), and X04465 (Marchantia).
To determine the function of ycf9, we have taken a reverse genetics approach and constructed a knockout allele for targeted disruption of the tobacco ycf9 (Fig. 1). The knockout allele was introduced into the tobacco plastid genome by particle bombardment-mediated chloroplast transformation (Svab et al. 1990; Svab and Maliga 1993). From 30 bombarded leaf samples, altogether seven chloroplast transformants were selected as verified by PCR and DNA gel blot analyses (Fig. 3). Three independent transplastomic lines (Nt-pIycf9-1, Nt-pIycf9-2, and Nt-pIycf9-3) were subjected to additional rounds of regeneration on spectinomycin-containing medium to obtain homoplasmic tissue.
Presence of Promiscuous ycf9 Copies and Homoplasmy of ycf9 Knockout Plants
According to our and others' experience (Maliga and Nixon 1998), homoplasmy of plastid transformants typically is achieved after subjecting the primary transformant to one to two additional rounds of regeneration on antibiotic-containing medium. We were surprised that in RFLP analyses performed after the fourth round of regeneration, all transformants carrying the ycf9 knockout allele showed, in addition to a strong band for the transplastomic fragment, a faint hybridization signal that corresponds in size to the restriction fragment from the wild-type genome (Fig. 3 A). However, we believed that the wild-type–like hybridization signal was not caused by true heteroplasmy of the chloroplast transformants, but rather by the presence of promiscuous plastid DNA in one of the other two genomes of the plant cell. It is well established that during evolution large fragments of chloroplast DNA integrated in the nuclear and mitochondrial genomes (Stern and Lonsdale 1982; Ayliffe and Timmis 1992; Nakazono and Hirai 1993; Ayliffe et al. 1998). Such promiscuous plastid DNA fragments can exhibit a remarkably high degree of sequence conservation at the DNA level (Ayliffe and Timmis 1992). If a promiscuous origin of the wild-type–like hybridization signals (Fig. 3 A) were indeed the case, these signals should be absent in Southern blot analyses using purified chloroplast DNA instead of total cellular DNA. Alternatively, if the transplastomic plants were still heteroplasmic, the wild-type signal should also persist upon analysis of isolated chloroplast DNA. When we repeated the RFLP analysis using the same restriction enzyme, but purified chloroplast DNA, no hybridization signal for the wild-type could be detected anymore (Fig. 3 B), strongly suggesting that our ycf9 knockout plants are homoplasmic and that promiscuous copies of the ycf9 region are present in tobacco cells.
Homoplasmy of ycf9 knockout plants was further confirmed by our subsequent biochemical analyses (see below) and by the lack of segregation of both phenotype and antibiotic resistance in the F1 generation.
Thylakoid Membranes from ycf9 Knockout Plants Lack a Small Protein
When transferred to the soil, ycf9 knockout plants grew photoautotrophically and displayed no obvious mutant phenotype under standard greenhouse conditions. This indicates that disruption of ycf9 does not abolish any essential function in plastid gene expression or cellular metabolism. Moreover, wild-type–like photosynthetic activity suggests that all of the major protein complexes of the thylakoid membrane (PSII, PSI, cytochrome b6f complex, and ATP synthase) are functional in ycf9 knockout plants. Intact photosynthetic electron transfer in ycf9 knockout plants was further confirmed by fluorescence measurements of PSII and PSI activities (data not shown).
Since the computer analyses had predicted membrane localization of the putative Ycf9 gene product (Fig. 2), we suspected the Ycf9 protein to be present in thylakoid membranes. Therefore, we compared the protein patterns of thylakoid membranes from wild-type and ycf9 knockout plants. Indeed, in high-resolution polyacrylamide gels, we detected a single band among the low molecular weight thylakoid proteins that was completely absent from ycf9 knockout plants (Fig. 4 A) suggesting that this band could correspond to the Ycf9 gene product. The protein is not stained by Coomassie, but stains well with silver. Comparison of the thylakoid protein patterns of ycf9 knockout plants with those of tobacco mutants lacking PSI or the cytochrome b6f complex (Fig. 4 A) revealed that the putative Ycf9 protein is present also in the absence of PSI or cytochrome b6f. Presence of wild-type levels of all major protein complexes of the thylakoid membrane in ycf9 knockout plants was confirmed by Western blot analyses with antibodies against key proteins of PSII, PSI, the cytochrome b6f complex, and the chloroplast ATP synthase (data not shown).
Figure 4.
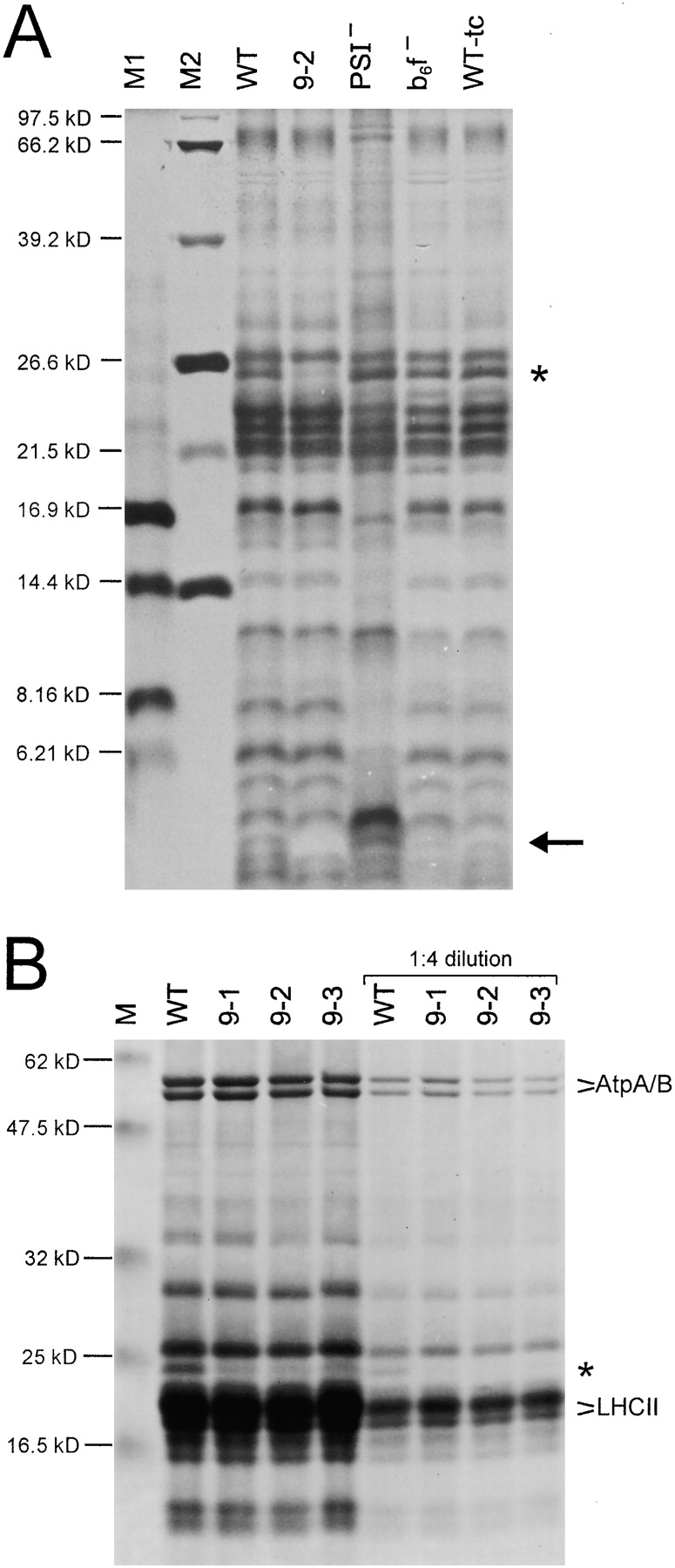
Analysis of thylakoid proteins from wild-type (WT) and ycf9 knockout plants. A, Silver staining of thylakoid proteins separated by electrophoresis in a 16.5% polyacrylamide (PAA) gel. For comparison, a mutant lacking PSI (PSI−; Ruf et al. 1997), a mutant lacking the cytochrome b6f complex (b6f−; Hager et al. 1999) and a wild-type in tissue culture (WT-tc) are also shown. The putative Ycf9 protein is marked by an arrow and is clearly present in all samples except for the thylakoids from the ycf9 knockout (9-2). Note that, as expected for a hydrophobic protein, the putative Ycf9 protein migrates faster than a protein of the calculated molecular weight of Ycf9 (6.58 kD). The asterisk marks a protein specifically reduced in ycf9 knockout plants (see part B). M1, M2: molecular weight markers. (B) Coomassie staining of thylakoid proteins from the wild-type and three independently generated ycf9 knockout plants (9-1, 9-2, and 9-3). Thylakoid proteins were electrophoresed in a 10% PAA gel. Two different protein concentrations were loaded. All ycf9 knockout plants clearly show specifically a drastic reduction of a thylakoid protein with an approximate molecular weight of 25 kD (asterisk). Selected prominent thylakoid proteins are marked (AtpA/AtpB, LHCII proteins). M, molecular weight marker.
The Putative Ycf9 Protein Is Tightly Associated with Thylakoid Membranes
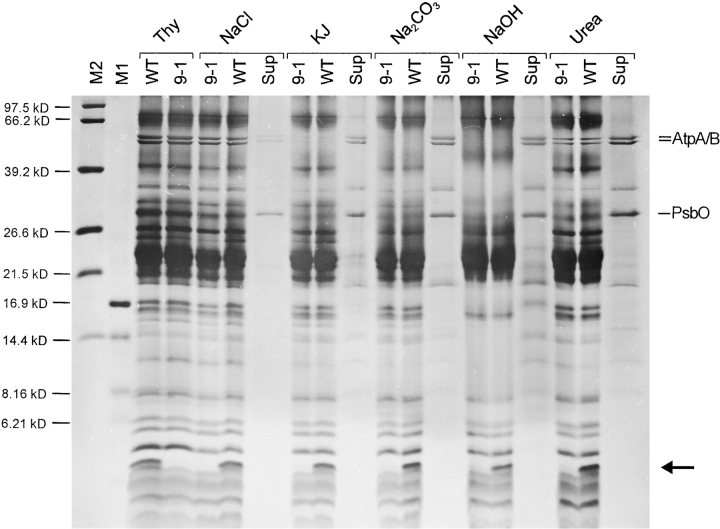
The hydrophobic properties of the potential Ycf9 protein (Fig. 2), as well as its coisolation with thylakoid membranes (Fig. 4 A), suggest that Ycf9 is a genuine thylakoid protein. To unambiguously prove the tight association of the Ycf9 gene product with the thylakoid membrane and to exclude possible isolation artifacts, we applied washes of purified thylakoids with chaotropic salts, alkali treatment of thylakoid membranes, and treatment with urea. All these conditions are known to remove proteins that are only loosely associated with membranes, as well as peripheral membrane proteins (e.g., Boudreau et al. 1997). Even upon the most stringent treatments (2 M KJ, 0.1 M Na2CO3, or 0.1 M NaOH) the potential Ycf9 protein remained exclusively in the membrane fraction, whereas peripheral thylakoid membrane proteins (e.g., the water-splitting complex of PSII and the CF1 part of the ATP synthase) were effectively released from the membranes (Fig. 5). These data confirm the tight association of the putative Ycf9 protein with photosynthetic membranes and suggest that Ycf9 is an integral thylakoid protein.
Figure 5.
Tight association of the putative Ycf9 protein with the thylakoid membrane. Purified thylakoid membranes from the wild-type (WT) and a ycf9 knockout plant (9-1) were washed with 2 M NaCl, 2M KJ, 0.1 M Na2CO3, 0.1 M NaOH, or 4 M urea. For each wash, the patterns of the proteins remaining in the membranes (for the wild-type and the mutant) and the supernatant (for the wild-type) are shown. For comparison, untreated thylakoids (Thy) are also shown. Note that peripheral thylakoid proteins are most effectively dissolved from the membrane with KJ, Na2CO3, and NaOH. Selected marker proteins released from the thylakoids and found in the supernatants (Sup) are labeled: AtpA/B, CF1α/β subunits of the ATP synthase; PsbO, 33-kD manganese-stabilizing protein of the oxygen-evolving complex of PSII. The putative Ycf9 protein is marked with an arrow and clearly remains membrane-associated upon all treatments. M1 and M2, molecular weight markers.
Lack of Ycf9 Gene Product Is Associated with Reduced Amounts of the LHC Protein CP26
When we analyzed the thylakoid protein patterns of wild-type and ycf9 knockout plants, we noticed that the absence of Ycf9 gene product is accompanied by a significant reduction in the amount of a single thylakoid protein of ∼25 kD (Fig. 4 B). As it seemed reasonable to us to assume that the function of the Ycf9 gene product is related to this 25-kD protein, we were interested in identifying the association of this protein with a specific protein complex in the thylakoid membrane. Therefore, we fractionated thylakoid membranes and isolated all major protein complexes. All fractions were subsequently assayed for the presence of the 25-kD protein that could be identified easily, making use of the quantitative difference between the wild-type and the ycf9 knockout (Fig. 4 B). In these analyses, the 25-kD protein was detected exclusively in purified LHCs of PSII (Fig. 6 A), but not in any of the other purified thylakoidal protein complexes (PSI, PSII, or ATP synthase; data not shown). The isolated LHCs showed the typical pattern of pigment-binding LHC proteins (e.g., Boekema et al. 1999) and readily identified the 25-kD protein as being the LHC protein CP26 (Fig. 6 A). The molecular identity of the 25-kD protein was confirmed by Western blotting with antibodies against selected subunits of the LHCs of PSI and PSII (Fig. 6 B and data not shown).
Figure 6.
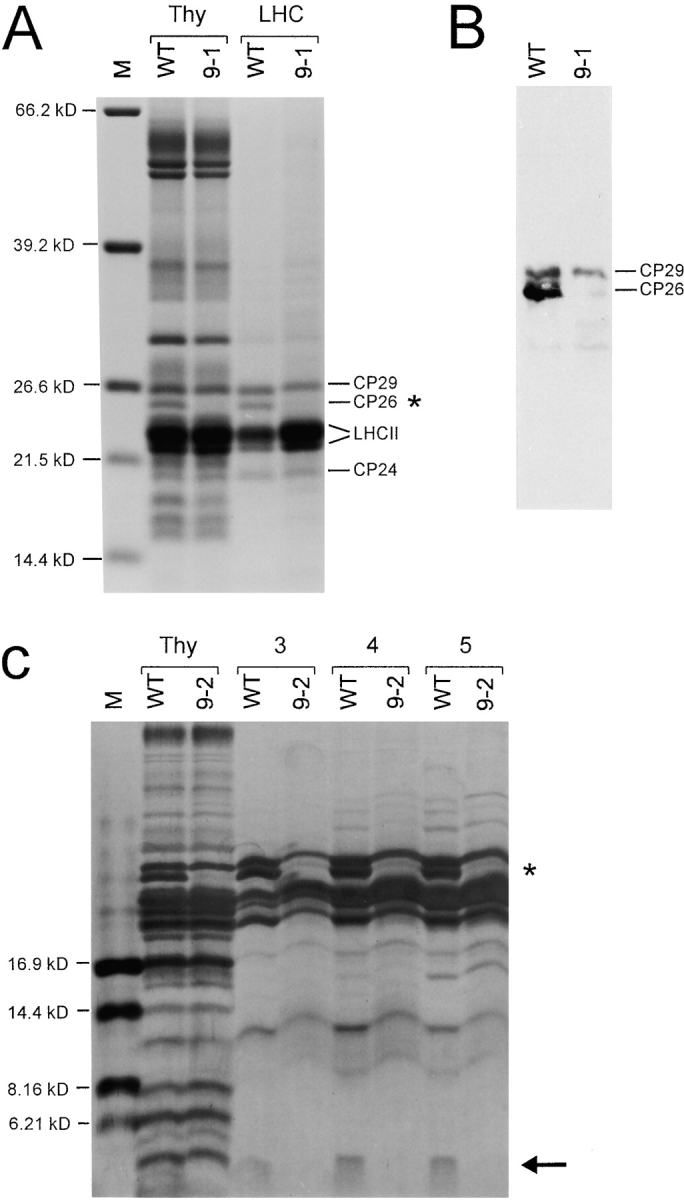
Analysis of purified PSII LHCs from wild-type and ycf9 knockout plants. A, Identification of the 25-kD protein. Thy, Thylakoid proteins; LHC, purified LHCs. LHC proteins were separated in a 10% PAA gel and subsequently stained with Coomassie blue. The known LHC proteins (LHCII proteins, CP24, CP26, and CP29) are indicated (e.g., Boekema et al. 1999). Comparison of the LHC protein patterns of the wild-type (WT) and the ycf9 knockout (9-1) clearly identifies the 25-kD protein as CP26 (asterisk). B, Confirmation of the identity of the 25-kD protein by probing blotted thylakoid proteins of the wild-type (WT) and the ycf9 knockout (9-1) with an anti-CP26 antibody. Owing to the homology of all pigment-binding LHC proteins, the antibody inevitably shows some cross-reactivity with CP29 (see Di Paolo et al. 1990; Sigrist and Staehelin 1992) and, to a lesser extent, with some smaller LHC proteins. C, Detection of the putative Ycf9 protein in isolated LHCs. The three major LHC-containing fractions (marked 3, 4, and 5) of the sucrose gradient (Eshaghi et al. 1999) are shown. LHC proteins were electrophoresed in a 16.5% PAA gel and stained with silver. The LHC proteins migrate slightly slower than the corresponding proteins in the thylakoid preparation due to the high sucrose content in the gradient fractions. Note presence of the protein in the wild-type (arrow), but complete absence from LHCs of the ycf9 knockout plant (9-2). The identity of the missing protein with the Ycf9 gene product was further confirmed by mass spectroscopy (see text for details). CP26 is marked by an asterisk. M, molecular weight markers.
The Ycf9 Protein Copurifies with PSII LHCs
To establish a clear link between the absence of Ycf9 gene product and the reduction of the LHC protein CP26, we set out to test whether the putative Ycf9 protein is also part of the LHCs of PSII. As Ycf9 stains well with silver (Fig. 4 A and 5), we separated purified thylakoidal protein complexes from the wild-type and the ycf9 knockout mutant on high-percentage polyacrylamide gels that were subsequently subjected to silver staining. Like CP26, the putative Ycf9 protein was detected exclusively in purified LHC (Fig. 6 C and data not shown). No difference between the wild-type and the ycf9 knockout plants was detectable with either Coomassie or silver staining in any of the other protein complexes of the thylakoid membrane (not shown).
To unambiguously demonstrate that the single small thylakoid protein missing in the knockout plants is indeed the Ycf9 protein, we comparatively analyzed purified LHCs from the wild-type and the mutant by MALDI-TOF mass spectroscopy, a technique suitable to determine molecular masses with extraordinarily high accuracy. These analyses detected a single low molecular weight protein present in wild-type LHCs, but absent from mutant LHCs. The molecular mass of this protein was determined to be 6,581 D, which exactly corresponds to the theoretical molecular mass of the Ycf9 protein (including the NH2-terminal formyl group of the chloroplast initiator amino acid N-formyl methionine). This confirms that the single protein present in the wild-type, but missing from thylakoid preparations, as well as LHC preparations of ycf9 knockout plants (Fig. 4 A, 5, and 6 C), is indeed the Ycf9 gene product.
In sum, presence of the Ycf9 and CP26 proteins in one and the same complex confirms that ycf9 knockout plants have a specific defect in the light-harvesting antenna of PSII and, moreover, suggests that the reduced amounts of CP26 in the knockout plants are directly correlated with the absence of the Ycf9 protein.
Our finding that the ycf9 gene product functions in photosynthesis is consistent with the absence of the ycf9 reading frame from the plastid genome of the nonphotosynthetic holoparasitic plant Epifagus virginiana, which has lost practically all plastid-encoded photosynthesis-related genes during evolution (Wolfe et al. 1992).
The Ycf9 Protein Is Likely to Act as Stability Factor for CP26
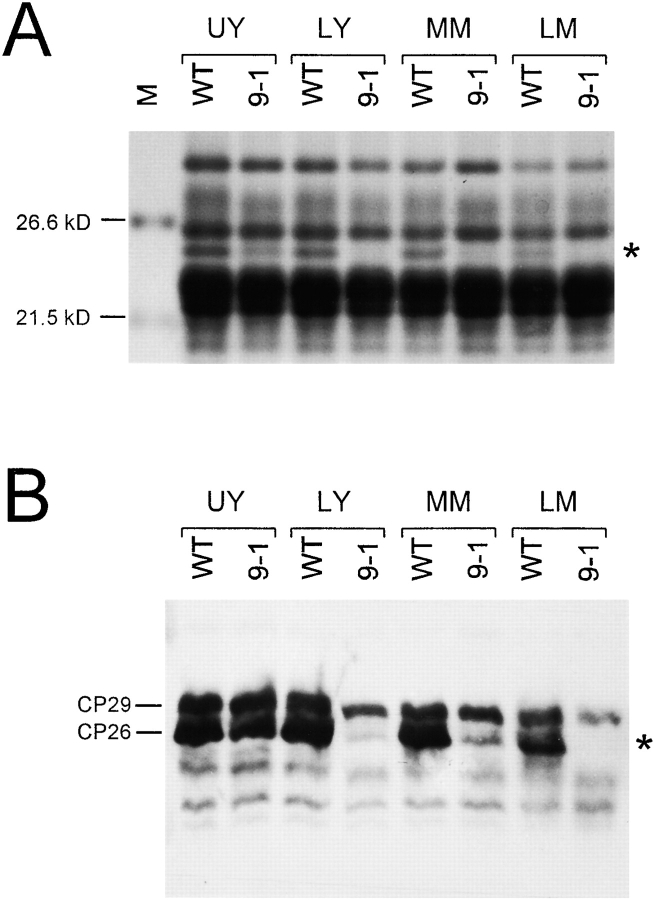
Several possible reasons can be envisaged for the reduced amounts of CP26 in the absence of Ycf9 protein. Reasonable hypotheses would be, for example, that Ycf9 assists in the incorporation of CP26 into the PSII antenna or, alternatively, that Ycf9 is required for the stable association of CP26 with PSII. To assess the amount of CP26 present in the ycf9 knockout plants quantitatively and to determine possible changes during plant development, we isolated thylakoid proteins from leaves at different stages of expansion, as well as leaves from plants of different age. Interestingly, very young leaves of ycf9 knockout plants were found to accumulate small, but significant amounts of CP26, whereas CP26 was virtually completely absent from old leaf material (Fig. 7). This indicates that, early in development, when photosystems and LHCs are assembled and hence CP26 synthesis is high, the absence of Ycf9 protein has less dramatic effects than later in development, when the synthesis of CP26 proteins is lower. This may suggest that the Ycf9 protein enhances the stability of the CP26 complex and that, when Ycf9 is missing, CP26 is more readily lost from the LHC or is more sensitive to degradation. Upon analyzing both stromal protein fractions and purified thylakoids or LHCs, neither accumulation of significant amounts of free CP26 in the stroma nor presence of any CP26 degradation products was detected in ycf9 knockout plants (Fig. 7 B and data not shown). This is consistent with the idea that CP26 is only stable when incorporated in the thylakoid membrane, whereas unassembled or disassembled CP26 is degraded rapidly.
Figure 7.
Analysis of the amounts of CP26 in thylakoids isolated from leaves at different stages. UY, Upper leaves (harvested from the top of young plants of ∼30-cm height); LY, lower leaves from the same plants; MM, fully expanded leaves (harvested from the middle parts of mature plants at the time of flowering); LM, older leaves (harvested from the lower parts of the same plants). A, Coomassie staining of isolated thylakoid proteins separated in a 10% PAA gel. Whereas CP26 is clearly detectable in very young leaf material of the ycf9 knockout plant (9-1), it is practically undetectable in older leaves. WT, Wild-type; M, molecular weight marker. B, Western blot analysis using an anti-CP26 antibody. The analysis confirms virtually complete lack of CP26 in mature leaf tissue of ycf9 knockout plants. The CP29 protein cross-reacting with this antibody (Di Paolo et al. 1990; Fig. 6 B) is also indicated.
ycf9 Knockout Plants Have a Reduced Photosynthetic Capacity
CP26 is part of the light-harvesting antenna of PSII and forms a subcomplex also referred to as LHCIIc (for review see, e.g., Wollman et al. 1999). CP26 builds one of the three so-called minor LHCs. Together with the two other minor complexes, CP24 (LHCIId) and CP29 (LHCIIa), and the trimeric major light-harvesting complex LHCII (LHCIIb), CP26 forms the chlorophyll a/b-containing antenna of PSII. Having established that the knockout of the ycf9 reading frame results in a deficiency in the LHCs of PSII, we were interested in the phenotypic consequences of this defect. This was of particular interest, since data on specific functions of any of the subcomplexes of the PSII antenna are largely lacking (Flachmann 1997).
As mentioned above, ycf9 knockout plants displayed no obvious mutant phenotype under standard growth condition indicating that, in spite of the lowered amounts of CP26, the plants are still capable of capturing photons quite efficiently. We also did not observe significant differences to the wild-type when plants were grown under light stress conditions (27,000 lux), suggesting that the reduction in the CP26 LHC does not affect appreciably the photoprotective function suggested for the PSII antenna. However, when plants were kept under low light conditions (100–300 lux) we noticed reduced growth of the ycf9 knockout plants as compared with the wild-type control (Fig. 8) suggesting that the phenotype manifests when light harvesting becomes the limiting growth parameter. To confirm that this phenotype was caused by a reduced efficiency of light harvesting, we measured photosynthetic oxygen evolution of chloroplasts isolated from wild-type and mutant plants. These analyses revealed that the ycf9 knockout plants produce significantly less oxygen than the wild-type (Table ). This effect was particularly pronounced when oxygen evolution was measured under low light conditions (Table ), indicating that photosynthetic water splitting is less efficient in the mutant, most probably as the result of inefficient light harvesting.
Figure 8.
Phenotype of ycf9 knockout plants under low light conditions. Two typical mutant plants (Nt-pIycf9-1 and Nt-pIycf9-2) and two wild-type plants (WT) grown under identical conditions are shown. Plants were raised from seeds and kept at an average light intensity of 200 lux. Note significantly reduced growth of the ycf9 knockout plants as compared with the wild-type controls.
Table 1.
Photosynthetic Oxygen Evolution of Chloroplasts from Wild-type and ycf9 Knockout Plants
| Photon flux density | 14 μE | 55 μE | 192 μE | 1,150 μE | 1,530 μE |
|---|---|---|---|---|---|
| Wild-type | 2.91 | 13.6 | 40.8 | 116.6 | 127.5 |
| Nt-pIycf9 | 1.05 | 7.4 | 28.7 | 96.0 | 118.1 |
| Nt-pIycf9 (% of wild-type) | 36% | 54% | 70% | 82% | 93% |
Chloroplasts were isolated from young plants adapted to low light conditions. Oxygen evolution values are given in μmol oxygen per hour and mg chlorophyll.
Discussion
LHCs act as antennae for the absorption of light energy in photosynthetic membranes. They are pigment-rich protein complexes that trap photons and transfer them rapidly to the reaction centers in the two photosystems. In addition to their light-harvesting function, LHCs may be involved in photoprotection and proton channeling in PSII.
The largest portion of the photosynthetic pigments present in both prokaryotic and eukaryotic photoautotrophic organisms is found associated with the LHCs. Pigment molecules are bound by LHC proteins either noncovalently (carotenoids and chlorophylls) or covalently via thioester bonds (phycobilins). The presence of many different pigment types (e.g., chlorophyll a, chlorophyll b, violaxanthin, and β-carotene) ensures optimal utilization of the sunlight by the photosynthetic apparatus, in that a wide range of spectral qualities can be absorbed.
Sequence alignments have clearly demonstrated that all pigment-binding LHC proteins exhibit significant similarity to each other. Moreover, all classical LHC proteins show a characteristic topology (three α-helical transmembrane domains with stromal-exposed NH2 termini), strongly suggesting that they are evolutionarily related and that all LHC genes arose from a common ancestor. The LHC genes form a multigene family in all higher plant nuclear genomes with at least 30 members in Arabidopsis (Jansson 1999).
The light-harvesting complex of PSII is the most abundant pigment–protein complex in plants. It consists of four subcomplexes: the trimeric major LHC and three minor complexes, CP24, CP26, and CP29. A pool of the trimeric LHC plays a crucial role in the light energy distribution between the two photosystems. This regulation of light harvesting occurs through reversible redox-controlled LHCII protein phosphorylation resulting in lateral migration of LHCII away from PSII (state transition; for review see Allen and Nilsson 1994; Gal et al. 1994).
As all pigment-binding LHC proteins are nuclear-encoded, they are synthesized as precursor proteins in the cytosol and subsequently imported into the chloroplast compartment. Although in vitro experiments have provided some information about the assembly of LHCs inside the chloroplast (including the targeting of LHC proteins to the thylakoid membrane, pigment attachment to apo-LHC proteins, supercomplex formation with photosystem cores), many steps in antenna biogenesis and turnover are still elusive (for review see Wollman et al. 1999).
In the course of this work, we have identified a first LHC protein lacking the conserved pigment-binding motifs. This protein is encoded by the chloroplast ycf9 reading frame. As it is rather small (6.58 kD) and obviously lacks pigment-binding domains, the Ycf9 protein is unlikely to be directly involved in light harvesting. Instead, it appears to be a structural component in the absence of which the CP26 complex is unstable and tends to be lost from the PSII antenna. In accordance with the nomenclature conventions for plastid-encoded genes, we propose to rename the ycf9 reading frame lhbA (LHC of PSII associated factor 1).
Upon transfer to low light conditions, plants undergo a low irradiance-induced increase in PSII antenna size. As efficient light harvesting becomes particularly important when light availability is limiting, it is not surprising that our ycf9 knockout plants displayed a phenotype of mild growth retardation only under low light conditions (Fig. 8). This suggests that the Ycf9/LhbA protein (and hence CP26), while being of limited importance under optimal growth conditions, confer a selective advantage by contributing to maximum light harvesting under unfavorable environmental conditions.
As the lack of Ycf9/LhbA leads to destabilization of CP26, the ycf9 knockout plants provide an excellent experimental system to study the function of one of the minor LHCs, the CP26 complex. Detailed electron transfer studies by fluorescence measurements have indicated that, in the absence of CP26, energy transfer from the major PSII LHC (LHCII) to the PSII reaction center is less efficient in ycf9 knockout plants, confirming the phenotype of the mutant plants and suggesting that the CP26 complex is involved in the transmission of energy absorbed by the LHCII trimer to the PSII reaction center (Biehler, K., S. Ruf, and R. Bock, manuscript in preparation).
The LhbA/Ycf9 protein is unlikely to be the only structural protein of the light-harvesting antenna. In fact, in all our LHC preparations we detected several other small proteins by silver staining (Fig. 6 C and data not shown). This may suggest that LHCs contain, in addition to the classical pigment-binding proteins, a second class of LHC proteins that serve important functions in the assembly and/or maintenance of stable antenna complexes. The purification and characterization of potential other members of this new class of LHC proteins is currently underway.
Acknowledgments
We are grateful to Drs. Roberto Bassi (Università di Verona); Andrew L. Staehelin (University of Colorado at Boulder); and Michael Hippler, Christoph Beck, and Michael Schroda (Universität Freiburg) for generously providing antisera. We are indebted to Drs. Pal Maliga and Zora Svab (Rutgers University, NJ) for ptDNA clones and the chimeric aadA gene. Excellent technical assistance by Mrs Marita Hermann and photographic work by Mrs Monika Messerschmid is gratefully acknowledged.
This research was supported by a grant from the Deutsche Forschungsgemeinschaft (BO 1482/1-3) to R. Bock.
Footnotes
Abbreviations used in this paper: LHC, light-harvesting complex; LHCII, LHC of photosystem II; PSI, photosystem I; PSII, photosystem II; ptDNA, plastid DNA; RFLP, restriction fragment length polymorphism.
References
- Allen J.F., Nilsson A. Redox signalling and the structural basis of regulation of photosynthesis by protein phosphorylation. Ann. Rev. Genet. 1994;28:863–868. [Google Scholar]
- Ayliffe M.A., Timmis J.N. Tobacco nuclear DNA contains long tracts of homology to chloroplast DNA. Theor. Appl. Genet. 1992;85:229–238. doi: 10.1007/BF00222864. [DOI] [PubMed] [Google Scholar]
- Ayliffe M.A., Scott N.S., Timmis J.N. Analysis of plastid DNA-like sequences within the nuclear genomes of higher plants. Mol. Biol. Evol. 1998;15:738–745. doi: 10.1093/oxfordjournals.molbev.a025977. [DOI] [PubMed] [Google Scholar]
- Bock R. Analysis of RNA editing in plastids. Methods. 1998;15:75–83. doi: 10.1006/meth.1998.0607. [DOI] [PubMed] [Google Scholar]
- Bock R., Drescher A., Ruf S. Reverse genetics in higher plant plastids. In: Wagner E., Normann J., Greppin H., Hackstein J.H.P., Herrmann R.G., Kowallik K.V., Schenk H.E.A., Seckbach J., editors. From Symbiosis to Eukaryotism. Endocytobiology VII. Geneva University Press; Geneva: 1999. pp. 427–437. [Google Scholar]
- Boekema E.J., van Roon H., Calkoen F., Bassi R., Dekker J.P. Multiple types of association of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Biochemistry. 1999;38:2233–2239. doi: 10.1021/bi9827161. [DOI] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.-D. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6095–6104. doi: 10.1093/emboj/16.20.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., Gillham N.W., Harris E.H., Hosler J.P., Johnson A.M., Jones A.R., Randolph-Anderson B.L., Robertson D., Klein T.M., Shark K.B., Sanford J.C. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Burrows P.A., Sazanov L.A., Svab Z., Maliga P., Nixon P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan J.E., Dunn B.M., Ploegh H.L., Speicher D.W., Wingfield P.T. Current Protocols in Protein Science. John Wiley & Sons, Inc; New York: 1995. [Google Scholar]
- Di Paolo M.L., Dal Belin Peruffo A., Bassi R. Immunological studies on chlorophyll-a/b proteins and their distribution in thylakoid membrane domains. Planta. 1990;181:275–286. doi: 10.1007/BF00195877. [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Eshaghi S., Andersson B., Barber J. Isolation of a highly active PSII-LHCII supercomplex from thylakoid membranes by a direct method. FEBS Lett. 1999;446:23–26. doi: 10.1016/s0014-5793(99)00149-0. [DOI] [PubMed] [Google Scholar]
- Evrard J.-L., Weil J.-H., Kuntz M. An ORF potentially encoding a 6.5 kDa hydrophobic protein in chloroplasts is also present in the cyanellar genome of Cyanophora paradoxa . Plant Mol. Biol. 1990;15:779–781. doi: 10.1007/BF00016127. [DOI] [PubMed] [Google Scholar]
- Feng Y., McCarty R.E. Purification and reconstitution of active chloroplast F0 . J. Biol. Chem. 1990;265:5104–5109. [PubMed] [Google Scholar]
- Flachmann R. Composition of photosystem II antenna in light-harvesting complex II antisense tobacco plants at varying irradiances. Plant Physiol. 1997;113:787–794. doi: 10.1104/pp.113.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A., Zer H., Ohad I. Redox-controlled thylakoid protein phosphorylation. News and views. Ann. Rev. Genet. 1994;28:869–885. [Google Scholar]
- Hager M., Biehler K., Illerhaus J., Ruf S., Bock R. Targeted inactivation of the smallest plastid genome-encoded open reading frame reveals a novel and essential subunit of the cytochrome b6f complex. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:5834–5842. doi: 10.1093/emboj/18.21.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Laasch H. Non-photochemical quenching of chlorophyll a fluorescence in isolated chloroplasts under conditions of stressed photosynthesis. Planta. 1987;171:220–226. doi: 10.1007/BF00391097. [DOI] [PubMed] [Google Scholar]
- Machold O., Simpson D.J., Moller B.L. Chlorophyll-proteins of thylakoids from wild-type and mutants of barley (Hordeum vulgare L.) Carlsberg Res. Com. 1979;44:235–254. [Google Scholar]
- Maier R.M., Neckermann K., Igloi G.L., Kössel H. Complete sequence of the maize chloroplast genomegene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- Maliga P., Nixon P.J. Judging the homoplastomic state of plastid transformants. Trends Plant Sci. 1998;3:376–377. [Google Scholar]
- Monod C., Takahashi Y., Goldschmidt-Clermont M., Rochaix J.-D. The chloroplast ycf8 open reading frame encodes a photosystem II polypeptide which maintains photosynthetic activity under adverse growth conditions. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:2747–2754. doi: 10.1002/j.1460-2075.1994.tb06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962;15:493–497. [Google Scholar]
- Nakazono M., Hirai A. Identification of the entire set of transferred chloroplast DNA sequences in the mitochondrial genome of rice. Mol. Gen. Genet. 1993;236:341–346. doi: 10.1007/BF00277131. [DOI] [PubMed] [Google Scholar]
- Rochaix J.-D. Chloroplast reverse geneticsnew insights into the function of plastid genes. Trends Plant Sci. 1997;2:419–425. [Google Scholar]
- Ruf S., Kössel H., Bock R. Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 1997;139:95–102. doi: 10.1083/jcb.139.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schreiber U., Klughammer C., Neubauer C. Measuring P700 absorbance changes around 830 nm with a new type of pulse modulation system. Z. Naturforsch. 1988;43c:686–698. [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genomeits gene organization and expression. EMBO (Eur. Mol. Biol. Organ.) J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist M., Staehelin A.L. Identification of type 1 and type 2 light-harvesting chlorophyll a/b-binding proteins using monospecific antibodies. Biochim. Biophys. Acta. 1992;1098:191–200. doi: 10.1016/s0005-2728(05)80336-6. [DOI] [PubMed] [Google Scholar]
- Stern D.B., Lonsdale D.M. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature. 1982;299:698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- Svab Z., Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z., Hajdukiewicz P., Maliga P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Rahire M., Breyton C., Popot J.-L., Joliot P., Rochaix J.-D. The chloroplast ycf7 (petL) open reading frame of Chlamydomonas reinhardtii encodes a small functionally important subunit of the cytochrome b6f complex. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:3498–3506. [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Morden C.W., Palmer J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.-A., Minai L., Nechushtai R. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- Yao W.B., Meng B.Y., Tanaka M., Sugiura M. An additional promoter within the protein-coding region of the psbD-psbC gene cluster in tobacco chloroplast DNA. Nucleic Acids Res. 1989;17:9583–9591. doi: 10.1093/nar/17.23.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]