Summary
The authors recently reported that adoptive immunotherapy with autologous tumor-reactive tumor infiltrating lymphocytes (TILs) immediately following a conditioning nonmyeloablative chemotherapy regimen resulted in an enhanced clinical response rate in patients with metastatic melanoma. These observations led to the current studies, which are focused on a detailed analysis of the T-cell antigen reactivity as well as the in vivo persistence of T cells in melanoma patient 2098, who experienced a complete regression of all metastatic lesions in lungs and soft tissues following therapy. Screening of an autologous tumor cell cDNA library using transferred TILs resulted in the identification of novel mutated growth arrest-specific gene 7 (GAS7) and glyceral-dehyde-3-phosphate dehydrogenase (GAPDH) gene transcripts. Direct sequence analysis of the expressed T-cell receptor beta chain variable regions showed that the transferred TILs contained multiple T-cell clonotypes, at least six of which persisted in peripheral blood for a month or more following transfer. The persistent T cells recognized both the mutated GAS7 and GAPDH. These persistent tumor-reactive T-cell clones were detected in tumor cell samples obtained from the patient following adoptive cell transfer and appeared to be represented at higher levels in the tumor sample obtained 1 month following transfer than in the peripheral blood obtained at the same time. Overall, these results indicate that multiple tumor-reactive T cells can persist in the peripheral blood and at the tumor site for prolonged times following adoptive transfer and thus may be responsible for the complete tumor regression in this patient.
Keywords: melanoma, adoptive cell transfer, T-cell receptor beta chain variable regions, T-cell persistence, tumor antigens
A doptive transfer immunotherapy with populations of tumor infiltrating lymphocytes (TILs) with antitumor reactivity can mediate the regression of cancer in patients with metastatic melanoma.1–3 Factors that may influence the efficacy of these treatments include the survival and persistence of transferred lymphocytes in vivo, the ability of transferred lymphocytes to home to local lymph nodes or tumor sites, the maintenance of effector function in the transferred T cells, and the expression of tumor and HLA antigens by tumor cells.
Several observations indicate that the ability of tumor-reactive T cells to persist in vivo following adoptive transfer can have a significant impact on the response to therapy. In prior trials adoptive transfer of T-cell clones reactive with either MART-1 or gp100 peptides persisted at levels of 1% to 2% of the peripheral blood CD8+ T cells and could not be detected at all for 2 weeks after transfer. No objective clinical responses were observed.4 In a recent clinical trial employing the adoptive transfer of polyclonal populations of in vitro cultured autologous TILs to patients who received prior nonmyeloablative conditioning, however, 6 of the 13 patients treated with this regimen showed objective clinical responses. For two of the patients in this trial who showed nearly complete regression of multiple metastases, individual HLA-A2-restricted MART-1 reactive T-cell clones that expressed unrelated T-cell receptor beta chain variable region (TRBV) sequences underwent significant expansion following adoptive transfer. One represented over 50% of the T cells in the peripheral circulation of the two responders for periods greater than 4 months.1 The in vivo persistence of tumor-reactive T cells was also shown in another melanoma patient who exhibited nearly complete tumor regression after adoptive cell transfer following nonmyeloablative chemotherapy.5 These observations suggested that the persistence of adoptively transferred tumor-reactive T cells in peripheral blood was associated with tumor regression.
An additional factor that may play an important role in the efficacy of tumor treatments is the ability of transferred T cells to migrate to tumor sites; this can be influenced by several factors, including the response to chemokines as well as the expression of lymph node homing molecules such as CCR76 and CD62L.7 The accumulation of dense infiltrates of adoptively transferred tumor-reactive T cells that expressed dominant TRBV gene products was observed in tumor deposits resected from two patients between 2 and 3 weeks following adoptive transfer.1 The cell infiltration was associated with upregulation of HLA class I and class II expression on tumor cells, which may have been a consequence of the local release of IFN-γ following antigen stimulation of tumor-reactive T cells. In another report, adoptively transferred MART-1 reactive T-cell clones were tracked using soluble HLA-A2 peptide tetramers.4 Three days following T-cell infusion, the percentage of MART-tetramer positive in peripheral blood was estimated to be approximately 1%, whereas nearly 40% of the T cells that were isolated from a tumor nodule bound to this complex. It was also reported that Mart-1 tetramer positive T cells were detected at increased frequencies in the peripheral blood of one patient at a level that approached 2%, and radiologic observations carried out with T cells that were labeled with 111Indium, as well as through the use of 18F-fluorodeoxyglucose, indicated that T cells specifically accumulated in tumor deposits as early as 48 hours following adoptive transfer.8 Other studies have shown the apparent accumulation of T cells that express particular BV gene products9,10; however, the significance of these findings is not clear, as the ability of these cells to recognize tumor cells has not, in general, been directly examined.
The antigenic specificity of clonotypes present in the administered T-cell populations represents another factor that may influence clinical responses. The tumor antigens that have been identified to this point represent both widely expressed non-mutated antigens as well as mutated antigens that are generally expressed in only one or a small number of tumors.11–15 The results of some studies suggest that recognition of the non-mutated melanocyte differentiation antigen gp100 by HLA-A2-restricted TILs significantly correlated with clinical response to adoptive immunotherapy with TILs in HLA-A2-positive melanoma patients.16 In other cases, response to adoptive immunotherapy was associated with the transfer of dominant T-cell populations that recognize mutated antigens.5,17
In the current study, the antigen reactivity and persistence of T-cell clonotypes present in the TILs administrated to a patient who experienced a complete response following adoptive transfer were examined. The results show that the persistence of multiple tumor-reactive T cells is associated with tumor regression.
MATERIALS AND METHODS
Clinical Response Evaluation
Patient 2098 underwent computed axial tomography of the chest, abdomen, and pelvis prior to adoptive cell transfer and after the treatment. The sum of the longest diameters of all tumors (World Health Organization Response Evaluation Criteria in Solid Tumors) before and after therapy was calculated. A partial response was defined as a decrease of at least 50% (but < 100%) in the sum of the longest diameters of all evaluable metastases lasting at least 1 month with no new or enlarging tumors, and a complete response required the disappearance of all lesions with the appearance of no new lesions.
Patient Samples
Samples obtained from patient 2098 included TIL 2098 that were derived from the resected tumor by culturing in 6,000 IU/mL of IL-2,18 peripheral blood mononuclear cells (PBMCs), and tumor samples. PBMC samples were obtained prior to transfer and at different time points following adoptive transfer of autologous TILs. Tumor samples were obtained prior to transfer and at different time points following adoptive transfer of autologous TILs by fine-needle aspiration.
RNA Isolation
Total RNA was prepared from TIL 2098 sample, PBMC samples, and tumor samples using the RNeasy Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. Total RNA solutions were quantitated by spectrophotometry and the quality was verified by gel electrophoresis.
5′RACE Analysis of TRBV Gene Expression
The IMGT TRBV gene nomenclature was used in this research and is available at http://imgt.cines.fr/textes/IMG-Trepertoire/LocusGenes/nomenclatures/human/TRB/TRBV/Hu_TRBVnom.html. 5′RACE analysis of TRBV gene expression was carried out by the SMART RACE cDNA Amplification Kit (BD Biosciences Clontech, Palo Alto, CA) according to the manufacturer’s instructions. Briefly, 0.5 μg RNA from each sample was used in reverse transcription. 2.5 μL of diluted reverse transcription mixture was used in the PCR amplification of TRBV sequences in the presence of 3′TCRBCN primer. 3′TCRBCN primer anneals the constant region of the TCR β chain, and its sequence is 5′-CGA GGT AAA GCC ACA GTC T -3′. After cloning of TRBV PCR products using the pcDNA3.1/V5-His TOPO TA Expression Kit (Invitrogen, Carlsbad, CA), at least 96 clones were analyzed by an automated DNA sequencer (ABI PRISM 3100-Avant Genetic Analyze, Applied Biosystems, Foster City, CA). The sequence data were analyzed by comparison with known TRBV sequences using Vector NTI Suite 8 (InforMax, Frederick, MD).
RT-PCR of TRBV12-4 Gene Expression
Five microliters of diluted RT mixture from TIL 2098, PBMC1w, PBMC8w, and PBMC32w samples, which were the same samples for 5′RACE analysis, were used in the PCR amplification of TRBV12-4 sequences in the presence of 3′TCRBCN primer and TRBV12-4 specific primer. TRBV12-4 specific primer anneals the variable region of the TRBV12-4 β chain, and its sequence is 5′-AGG TGA CAG AGA TGG GAC AA-3′. PCR product was purified, cloned, and sequenced as for 5′RACE analysis. The sequences of the diversity region of TRBV12-4 were analyzed using Vector NTI Suite 8.
IFN-γ Release Assay
1 × 105 T cells were co-cultured with 1 × 105 tumor cells in a 96-well flat plate with 200 μL of T-cell assay medium (RPMI1640 supplemented with 2% fetal calf serum, 2 mM glutamine, 10 mM HEPES buffer, 0.55 mM DMSO, and 20 CU IL-2) in a humidified incubator at 37°C and 5% CO2 for 20 hours. The concentration of IFN-γ in the supernatant was determined by ELISA using a monoclonal antibody against human IFN-γ (Endogen, Woburn, MA).
FACS Analysis of CD107a Expression
1 × 106 TIL 2098 T cells were co-cultured with 1 × 106 allogeneic 888mel or autologous 2098mel tumor cells in a 6-well flat plate with 2 mL T-cell assay medium in a humidified incubator at 37°C and 5% CO2 for 20 hours. The cell samples were collected, washed with 5% fetal calf serum in 1× PBS, and stained with anti-CD107a FITC- or PE-conjugated antibody and anti-CD8 antibody (BD Biosciences, San Jose, CA) as well as TCR VB14 PE-conjugated or VB22 FITC-conjugated antibody (Beckman Coulter, Fullerton, CA). After staining, the expression of CD107a in TIL 2098 T cells was analyzed in a FACSCalibur (Becton Dickinson).
Tumor Antigen Identification
The SuperScript Plasmid System with Gateway Technology for cDNA Synthesis and Cloning Kit (Invitrogen, Carlsbad, CA) was used in the construction of an oligo(dT)-primed cDNA library according to the manufacturer’s protocol. The OrientExpress Random Primer cDNA Synthesis Kit (Novagen, Madison, WI) was used in the construction of a random primer-primed cDNA library according to the manufacturer’s protocol. Four micrograms of poly(A) RNA was purified from the total RNA sample of 2098mel tumor cells by using the PolyATract mRNA Isolation System Kit (Promega, Madison, WI) and was used in each library construction. The cDNA library was divided in pools of approximately 200 bacterial colonies per well, and 0.3 μg of DNA was transfected into 75,000 HEK293 cells that were stably transfected with the HLA-A2 gene construct in 96-well plates by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), and 18 hours after transfection, 1 × 105 TIL 2098 T cells were added to each well. Supernatants from each well were then harvested 24 hours later and tested for IFN-γ release by ELISA. cDNAs from the positive wells were further screened until the single cDNA construct per well was obtained. Individual single cDNAs isolated from positive wells were sequenced using the Big-Dye Terminator V 1.1 Kit (Applied Biosystems, Foster City, CA) and analyzed using an ABI Prism 3100-Avant Genetic Analyzer.
Peptides were synthesized using a solid-phase method based on standard F-moc chemistry on a multiple peptide synthesizer (Gilson, Worthington, OH). The purity of the peptides, as analyzed by mass spectrometry (Tufts Core Facility, Boston, MA), was estimated to be more than 90%. Lyophilized peptides were resuspended in DMSO and incubated at varying concentrations with 1 × 105 1978EBV-B cells in 200 μL T-cell assay medium at 37°C for 2 hours, washed with T-cell assay medium twice, and co-cultured with 1 × 105 TIL 2098 T cells in 200 μL T-cell medium in a humidified incubator at 37°C and 5% CO2 for 20 hours. After culture, IFN secretion was measured by ELISA.
T-Cell Cloning
T cells were cloned from the TIL 2098 sample by limiting dilution. Briefly, five cells per well were seeded in 96-well round-bottomed plates with 200 μL of X-Vivo 15 medium (Cambrex, Baltimore, MD) supplemented with 10% fetal calf serum, 10 mM HEPES buffer, 0.55 mM DMSO, 50 CU/mL IL-2, 3 μg/mL OKT3, and 5 × 104 irradiated allogeneic PBMCs. After 2 weeks of culture, T cells from growth-positive wells were transferred into new 96-well round-bottomed plates and tested for the ability to release IFN-γ in response to autologous 2098mel tumor cells. The TRBV gene products expressed by tumor reactive T-cell clones were identified using 5′RACE analysis of TRBV gene expression.
RESULTS
Tumor Regression in a Patient Receiving Adoptive Immunotherapy
A detailed study was carried out in patient 2098 that showed a complete response to adoptive immunotherapy with autologous tumor reactive TILs in an attempt to better understand the factors involved with successful treatment. Substantial shrinkage of multiple metastatic tumors in the lung and soft tissues was observed 2 months following treatment (Fig. 1), and by 1 year after treatment all tumor had disappeared.
FIGURE 1.
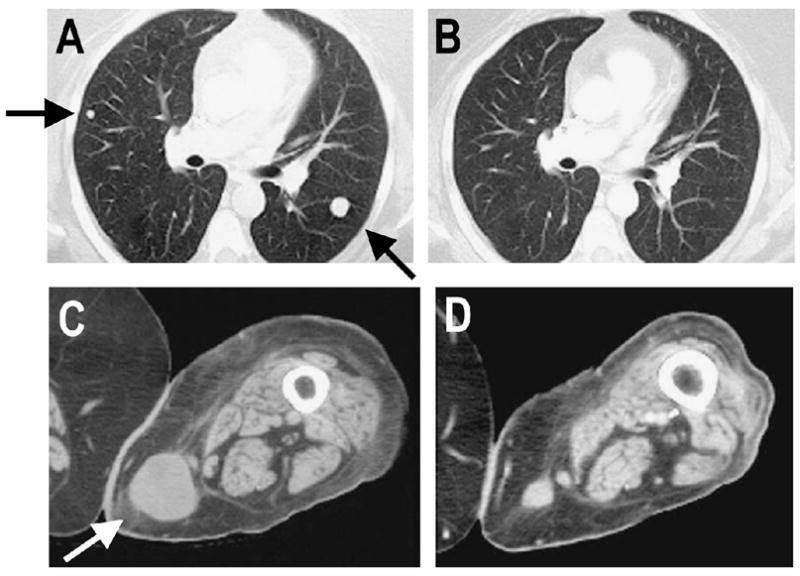
Clinical response in melanoma patient 2098 after adoptive cell transfer. Computed axial tomography scans showed the disease status prior to adoptive cell transfer (A) and 2 months after treatment (B) in the lung and prior to adoptive cell transfer (C) and 2 months after treatment (D) in the subcutaneous site. Arrows show sites of metastases.
Persistence of T-Cell Clones From TIL 2098 in Peripheral Blood Following Adoptive Cell Transfer
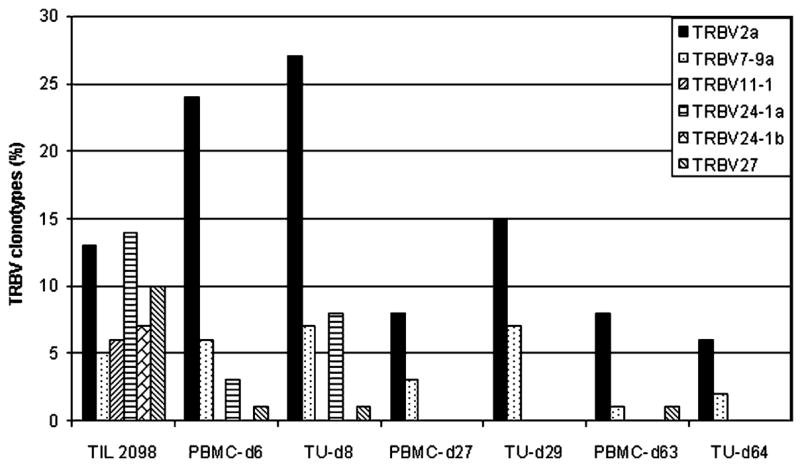
To obtain an unbiased sample of T-cell clonotypes present in TIL as well as peripheral blood lymphocytes (PBLs) obtained following adoptive transfer, the expressed TRBV genes were analyzed using a 5′RACE technique. Considerable heterogeneity was observed in TIL 2098, as 37 distinct clonotypes were identified following the sequencing of 96 cDNA clones amplified from this polyclonal T-cell population (Fig. 2). Six dominant clonotypes derived from TRBV2, 7-9, 11-1, 24-1, 24-1, and 27 germline gene families were identified that represented at least 5% of the sequences amplified from TIL 2098 mRNA. Eleven of the 37 T-cell clonotypes detected in TIL 2098 were also present at a level of at least 1% in PBMC samples that were obtained 6 days following adoptive transfer, whereas only five of the original TIL clonotypes were detected in a sample obtained 4 weeks following transfer.
FIGURE 2.
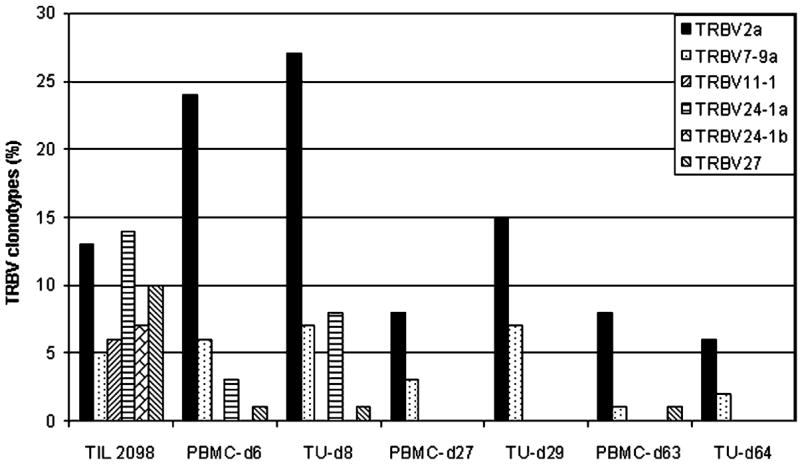
Diversity of TRBV sequences expressed in the in vitro expanded and transferred TILs from melanoma patient 2098 and T-cell persistence in the peripheral blood. TRBV sequences amplified from the resected tumor sample (2098Tu) used in TIL 2098 generation, TIL 2098, and PBMC-d6 and PBMC-d27 samples obtained at 6 days and 27 days respectively following transfer were aligned with each other as well as to germline TRBV sequences, and the percentage corresponding to each of the individual clonotypes was determined. Following TRBV family names, lowercase letters are used to distinguish unique sequences that are derived from the same TRBV family but that contain distinct CDR3 junctional region sequences. A minimum of 96 clones were sequenced from each sample.
Analysis of the levels of the six dominant clonotypes derived from TIL 2098 indicated that there was a wide variation in the persistence of individual clonotypes (see Fig. 2). One of the clonotypes detected in the transferred TIL 2098 that expressed a rearranged product of the BV2 gene, designated 2098BV2a, represented 13% of the sequences derived from TIL 2098, increased to 23% of the sequences derived from PBMC obtained 1 week following transfer, and then decreased to 8% of the PBMC sequences 1 month following transfer. The 2098BV7-9a clonotype represented 5% of the TIL sequences and was maintained at a similar level for 1 week following transfer, and then declined to a level of 3% 1 month following transfer. Furthermore, the 2098BV2a and 2098BV7-9a clonotypes were detected in the PBMCs obtained at 8 months after treatment at levels of 6% and 1%, respectively, indicating that these two T-cell clones persisted for prolonged times. In contrast, the dominant 2098BV11-1, 24-1a, 24-1b, and 27 clonotypes showed a rapid decline and were either not detected or were present at relatively low levels 6 days as well as 1 month following transfer. None of the clonotypes detected in TIL 2098 were detected in PBMCs obtained from patient 2098 prior to adoptive cell transfer, indicating that the T-cell clones detected in peripheral blood after treatment were likely derived from the in vitro cultured TILs.
Analysis of the sample of uncultured tumor that was used to derive TIL 2098 revealed that only three of the clonotypes derived from TIL 2098 T cells—2098BV5-5, 7-9c, and 20-1a—could be detected in this sample (see Fig. 2). These did not represent the dominant clonotypes that were detected in TIL 2098, as these three clones represented only 1% of the sequences identified in the administered TILs, and thus the in vitro growth of TILs appeared to result in a dramatic skewing of the repertoire of T cells that were originally present in the uncultured tumor sample. These three clonotypes were also detected in the day-6 PBMC sample, but only one clonotype, 2098BV20-1a, was present in the later sample.
Persistence of T-Cell Clones in Tumor Sites Following Adoptive Cell Transfer
The frequency of dominant clonotypes that represent 5% or more of the sequences derived from TIL 2098 were then compared between PBMC samples obtained at approximately 1, 4, and 9 weeks following transfer and uncultured tumor samples obtained from a regressing tumor lesion that was biopsied at similar times following the T-cell transfer (Fig. 3). Three of the dominant clonotypes—2098BV2a, 7-9a, and 27—were detected at similar levels in PBMC and tumor samples obtained approximately 1 week following transfer. Approximately 4 weeks following transfer, when substantial tumor regression was taking place, the levels of the 2098BV2a and 7-9a clonotypes detected in tumor samples were somewhat higher than the levels detected in the PBMC sample; 9 weeks following transfer, the 2098BV2a, 7-9a, and 27 sequences were detected in PBMCs, while the 2098BV2a and 7-9a sequences were detected in the uncultured tumor sample.
FIGURE 3.
Comparison of T-cell persistence between the peripheral blood and tumor sites of melanoma patient 2098 after adoptive cell transfer. The TRBV sequences expressed in TIL 2098 as well as PBMC-6d, PBMC-27d, and PBMC-63d samples obtained at 6, 27, and 63 days following adoptive TIL 2098 transfer were compared with the TRBV sequences expressed in tumor samples obtained at 8, 29, and 64 days following adoptive TIL 2098 transfer, designated TU-d8, TU-d29, and TU-d64.
Tumor Reactivity of TIL 2098 T Cells
Initial analysis of the specificity of TIL 2098 T cells showed that allogeneic melanoma cell lines, including five melanoma cell lines that shared expression of HLA-A2 with autologous tumor cells, failed to stimulate significant cytokine release from TIL 2098 T cells (Fig. 4). The population of tumor reactive T cells present in TIL 2098 appeared to be predominantly HLA-A2 restricted, as greater than 80% of the response to autologous tumor cells could be blocked by prior incubation with an anti-HLA-A2 antibody, MA2.1, and allogeneic HLA-A2 positive transformed EBV-B cells failed to stimulate significant cytokine release from TIL 2098. After prior culture with IL-2, PBMCs obtained from patient 2098 8 weeks following adoptive cell transfer secreted high levels of IFN-γ in response to autologous tumor cells and only relatively low levels of cytokine in response to allogeneic melanomas, indicating that persistent T cells retained the ability to respond specifically to autologous tumor cells. PBMCs obtained prior to adoptive transfer failed to respond to autologous 2098mel (data not shown), which was consistent with the observation that none of the sequences detected in TILs were detected in peripheral blood at this time.
FIGURE 4.
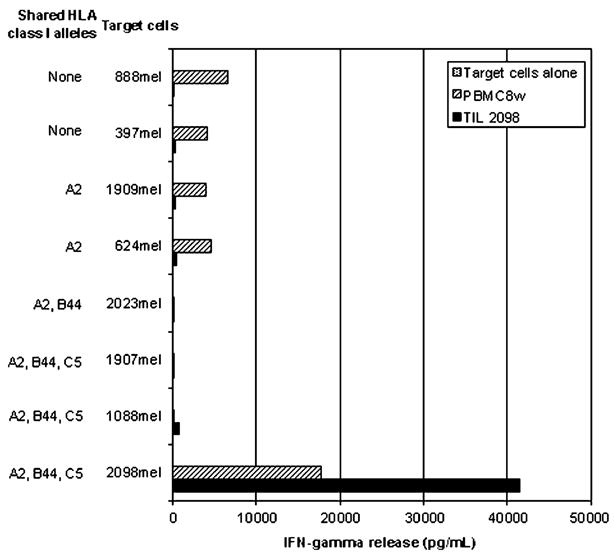
Tumor reactivity of TIL 2098. Tumor reactivity of TIL 2098 as well as a PBMC sample obtained at 8 weeks following adoptive cell transfer (PBMC8w) was evaluated in an IFN-γ release assay. PBMC8w was cultured in 500 CU/mL IL-2 for 16 hours before assay. The tumor reactivity of TIL 2098 and PBMC8w cells was tested with autologous 2098 tumor cells, allogeneic HLA-A2 positive 624mel, 1088mel, 1907mel, 1909mel, and 2023mel tumor cells, and HLA-A2 negative 397mel and 888mel tumor cells.
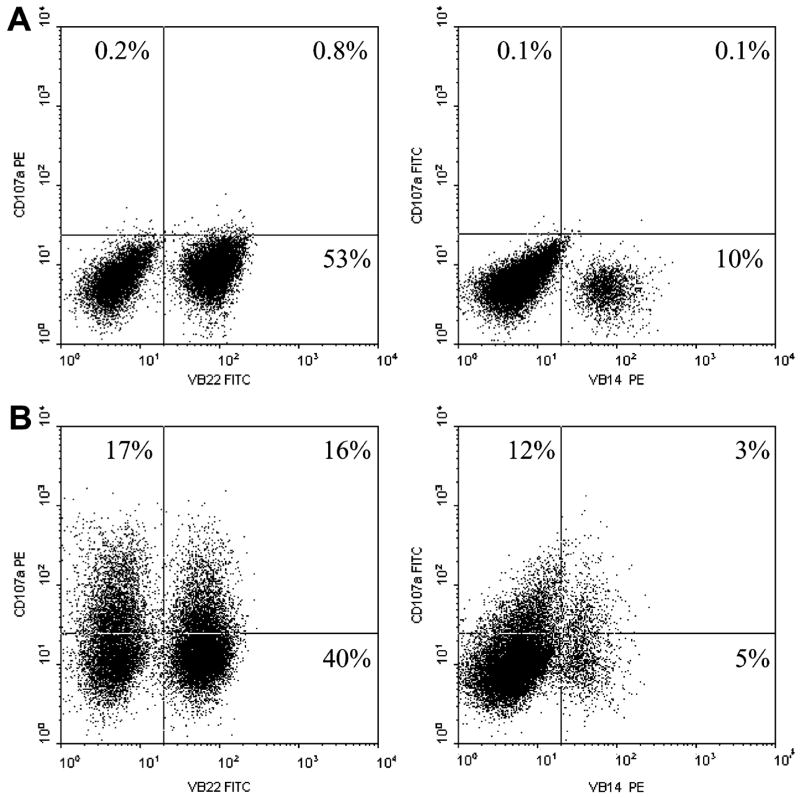
The dominant T-cell clonotypes present in TIL 2098 were then examined for their ability to recognize autologous tumor cells using a recently developed assay that measures cell surface expression of the CD107a (LAMP-1) protein, which is rapidly mobilized from intracellular cytotoxic granules following T-cell activation.19 The results of CD107a cell surface expression assay indicated that both the TRBV2 and TRBV27 clonotypes, measured using anti-VB22 and anti-VB14 antibodies respectively, responded specifically to autologous melanoma but not a non-HLA-A2 positive allogeneic melanoma (Fig. 5). These results indicated that a minimum of two of the dominant persistent T-cell clones derived from TIL 2098 were tumor reactive.
FIGURE 5.
Upregulation of CD107a cell surface expression in response to tumor stimulation in TIL 2098. 1 × 105 TIL 2098 T cells were co-cultured with 1 × 105 allogeneic 888mel tumor cells (A) or 1 × 105 autologous 2098mel tumor cells (B) for 16 hours. CD107a expression as well as TRBV2 and TRBV27 expression detected by corresponding TCR VB22 antibody and VB12 antibody respectively in TIL 2098 T cells was analyzed by FACS.
Identification of Antigens Recognized by TIL 2098 T Cells
In an attempt to further characterize the antigen reactivity of TIL 2098 T cells, pools of cDNAs derived from an autologous tumor cell cDNA library were then screened for their ability to stimulate IFN-γ release from the bulk TIL population following the transfection of HLA-A2 positive target cells. The sequence of one of the clones isolated from a positive pool represented a partial cDNA that corresponded to the growth arrest-specific gene 7 (GAS7). The single difference that was noted between the reported GAS7 sequence and the sequence isolated from the melanoma cDNA library, a C-to-T substitution within codon 225, resulted in a substitution of tyrosine for the histidine encoded within the normal gene (Table 1). A full-length glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was also identified following screening of the cDNA library with TIL 2098. This cDNA contained a G-to-A substitution in codon 175, resulting in a substitution of isoleucine for methionine at this position (see Table 1).
TABLE 1.
Antigenic Epitope Sequences and Frequency of GAS7 and GAPDH Wild-Type/Mutant Transcripts in Autologous 2098mel Tumor Cells
| Peptide Name | Peptide Sequence | Percentage of Wild-Type or Mutant Transcripts |
|---|---|---|
| GAS7-9merWt | LADEAEVHL | 92% |
| GAS7-9merMut | LADEAEVYL | 8% |
| GAS7-10merWt | SLADEAEVHL | |
| GAS7-10merMut | SLADEAEVYL | |
| GAPDH-9merWt | GIVEGLMIT | 88% |
| GAPDH-9merMut | GIVEGLITT | 12% |
| GAPDH-10merWt | GIVEGLMTTV | |
| GAPDH-10merMut | GIVEGLITTV |
Mutated amino acids are noted in bold and italic, and corresponding wild-type amino acids are noted in bold. The percentage of wild-type or mutant GAS7/GAPDH transcripts in 2098mel tumor cells was determined by sequencing 64 GAS7 or GAPDH constructs.
Both of the alterations that were identified from the 2098mel cDNA library represented mutations, as all of the GAS7 and GAPDH sequences that were amplified from normal T cells derived from patient 2098 corresponded to the known germline gene sequences at these positions. Sequences corresponding to the normal GAS7 and GAPDH gene products were also amplified from 2098mel, indicating that the tumor cells may also express a second copy of each of these genes. Mutated GAS7 products represented 8% of the transcripts that were amplified from the 2098mel cell line, and mutated GAPDH products represented 12% of the transcripts amplified from autologous tumor cells (see Table 1). These results indicated either that expression of these mutated and normal gene products was unbalanced in 2098mel, or a portion of the autologous 2098mel cells express only a normal copy of these genes and may lack expression of the mutated GAS7 and GAPDH gene products.
Several peptides that were encoded within the region encompassed by the point mutation in the GAS7 transcript and that corresponded to the HLA-A2 binding motif were then synthesized in an attempt to identify the T-cell epitopes encoded by this gene product (see Table 1). The TIL 2098 T cells recognized target cells pulsed with the mutated 9-mer peptide LADEAEVHL, corresponding to the GAS7 amino acids 218–226 (GAS7:218–226Mut) and the 10-mer peptide SLADEAEVHL (GAS7:217–226Mut) but failed to respond to the normal variant of either peptide (Fig. 6A). A comparison of the peptide titration, as well as the maximal level of cytokine release stimulated by the pulsed target cells, indicated that the 10-mer peptide was recognized more efficiently than the 9-mer.
FIGURE 6.
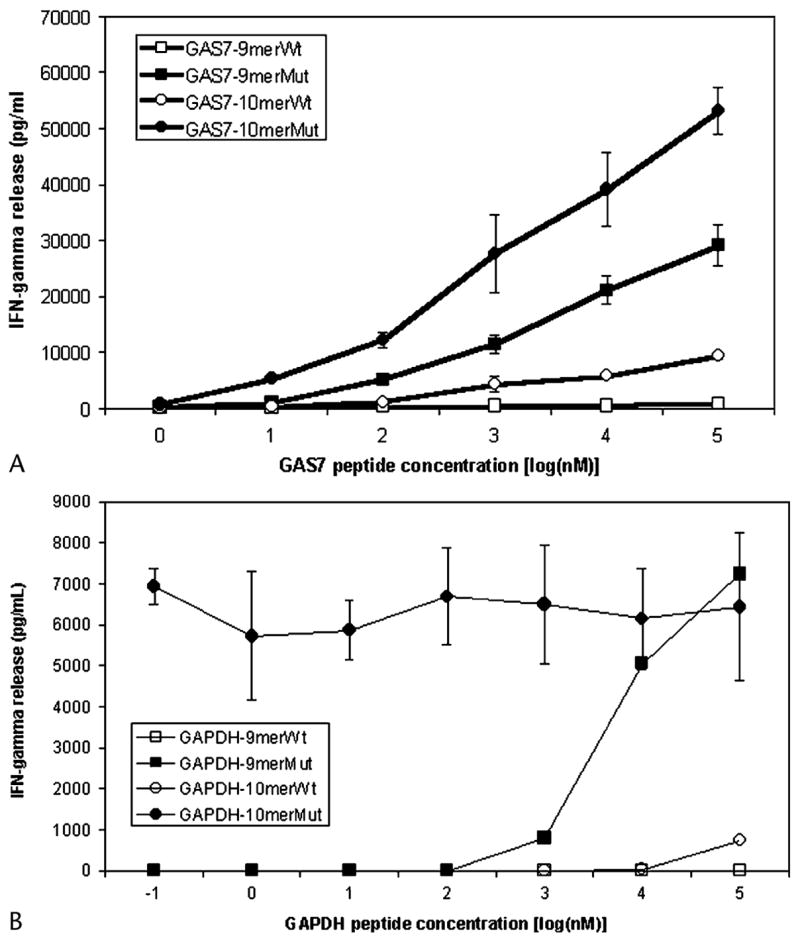
Titration of GAS7 and GAPDH antigenic epitopes recognized by TIL 2098. 1 × 105 1978EBV-B cells were pulsed with different concentrations of GAS7 (A) or GAPDH (B) peptides for 2 hours, washed with T-cell assay medium, and co-cultured with 1 × 105 TIL 2098 T cells for 16 hours. The concentration of IFN-γ in the culture medium released by T cells was measured by ELISA assay. The sequences of antigenic epitopes are shown in Table 1.
A similar approach was used to identify the GAPDH peptide epitope (see Table 1). The mutated GAPDH 10-mer peptide GIVEGLITTV (GAPDH:169–178Mut) stimulated the release of high levels of IFN-γ from TIL 2098 T cells (see Fig. 6B). An overlapping 9-mer peptide, GIVEGLITT, also stimulated high levels of IFN-γ production when target cells were pulsed with 1 μM concentrations or greater, but the 10-mer peptide stimulated high levels of cytokine release even at a concentration of 0.1 nM. Target cells incubated with another overlapping peptide, IVEGLITTV, also stimulated the release of high levels of IFN-γ, but a peptide titration assay indicated again that high levels of cytokine release were stimulated only when target cells were pulsed with a concentrations of 2 μM or greater (data not shown). These results indicated that epitopes derived from mutated GAS7 and GAPDH transcripts represented novel tumor antigens recognized by TIL 2098 T cells.
Antigen Reactivity of T-Cell Clonotypes
Multiple T-cell clones were then isolated from TIL 2098 T cells in an attempt to further characterize the specificity of the dominant T-cell clonotypes present within this TIL. The 2098BV7-9a and 27 clonotypes recognized the mutated GAS7 product (Fig. 7A). A T-cell clone that was not detected in the original TRBV sequence analysis carried out on TIL 2098 T cells and that expressed the TRBV12-4 gene product recognized the mutated GAPDH peptide. This clonotype appeared to be represented in TIL 2098 T cells at a level of approximately 0.1%, but it appeared to be present at a significantly higher frequency in a sample of PBMCs obtained 1 week following transfer, and was detected 8 weeks following transfer at levels that appeared to be lower than those observed in the transferred TILs (see Fig. 7B). The 2098BV2a clonotype failed to recognize either the mutated GAS7 or GAPDH gene products, although this T-cell clone strongly recognized autologous 2098mel. The screening of over 1.25 × 106 cDNA clones derived from either an oligo(dT)-primed or a random primer-primed cDNA library of 2098mel by TIL 2098 and 2098BV2a enriched T cells failed to result in identification of the tumor antigen recognized by the 2098BV2a T-cell clone. In addition, the 2098BV20-1a T-cell clone that was detected at a level of 1% in TIL 2098 as well as the uncultured tumor sample used to generate TIL 2098 persisted in the peripheral blood for 1 month after treatment (see Fig. 2). Results of an IFN-γ release assay indicated that the 2098BV20-1a T-cell clone did not recognize the autologous tumor cells (data not shown). Taken together, these results indicate that TIL 2098 predominantly or exclusively recognizes mutated gene products, and that the tumor regression observed in some patients following adoptive transfer can be mediated by T cells that recognize mutated gene products.
FIGURE 7.
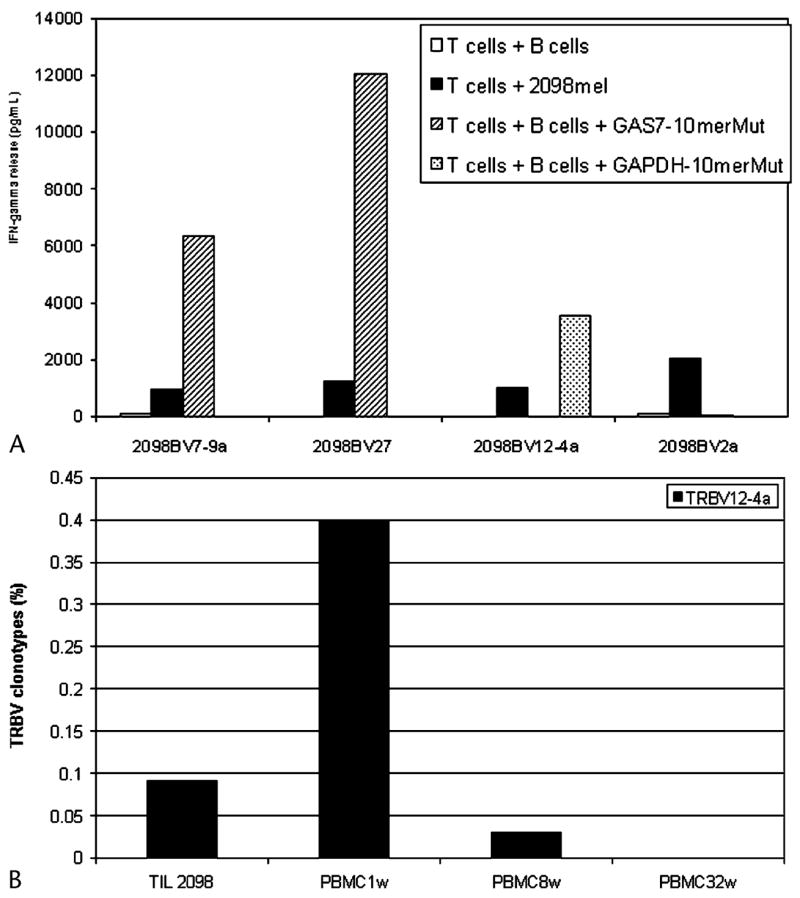
Determination of tumor antigen reactive T-cell clones and persistence of 2098BV12-4a T-cell clone in melanoma patient 2098. A, 1 × 105 cloned 2098BV2a, 2098BV7-9a, 2098BV12-4a, or 2098BV27 T cells were co-cultured for 16 hours with 1 × 105 2098mel tumor cells or 1978EBV-B cells pulsed with no peptide, 1 μM GAS7-10merMut, or 1 μM GAPDH-10merMut for 2 hours. The concentration of IFN-γ in the culture medium released by T cells was measured by ELISA assay. B, TRBV12-4 sequences were amplified from TIL 2098 as well as PBMC1w, PBMC8w, and PBMC32w samples obtained at 1, 8, and 32 weeks respectively following adoptive TIL 2098 transfer by RT-PCR using specific TRBV12-4 gene family primers. The percentage of TRBV12-4a sequences was normalized by FACS analysis in the total populations.
DISCUSSION
Several recent studies have begun to explore the role of T-cell persistence in clinical responses to the adoptive transfer of tumor reactive T cells. In one study, T cells generated by stimulation of PBMCs from HLA-A2-positive patients with the MART-1 peptide were adoptively transferred into patients and the survival of T cells in peripheral blood was tracked by staining using a MART-1 tetramer.8 Significant levels of tetramer positive cells could be detected within the first 3 days following transfer, when they generally ranged between 0.5% and 2% of peripheral blood CD8+ T cells, but by 2 weeks following transfer these levels had generally declined to background levels of less than 0.1%. In a trial carried out to evaluate responses to the adoptive transfer of highly tumor reactive T cell clones, no objective clinical responses were observed and cloned T cells could not generally be detected in peripheral blood beyond 2 days following transfer using a sensitive PCR technique.20 In a subsequent study, patients who received tumor reactive T-cell clones were treated with a prior nonmyeloablative chemotherapy regimen in an attempt to enhance the engraftment of adoptively transferred cells, but once again clinical responses were not observed.1 Preliminary analysis of samples obtained from patients in this trial, however, indicated that engraftment of the T-cell clones was not enhanced by the prior chemotherapy (M. Dudley, unpublished data). In a study of 13 patients who received an adoptive transfer of bulk populations of TILs following nonmyeloablative chemotherapy, two of the patients who showed nearly complete tumor regression were found to possess highly persistent T-cell clones that recognized the MART-1 tumor antigen.1 Further analysis of a patient analyzed in this trial who showed nearly complete tumor regression, patient 1913, revealed that a T-cell clone present in the adoptively transferred TILs that recognized a mutated HLA class I molecule expressed on autologous tumor cells persisted at levels of 14% to 25% for a period of 5 months following transfer.5 The present study represents an attempt to extend these findings by carrying out an extensive analysis of the antigen specificity and levels of T-cell persistence of individual clonotypes represented in a TIL that was associated with a complete response to adoptive immunotherapy. In this study, seven T-cell clonotypes—2098BV2a, 7-9a, 12-4a, 20-1a, 20-1b, 27, and 29-1c—derived from the infused TIL 2098 T cells persisted in peripheral blood for a period of 1 month or greater (see Figs. 2, 3, and 7B), and two of them persisted for greater than 8 months. The data suggest that T-cell persistence in vivo may be prerequisite to clinical responses after adoptive cell transfer therapy.
The results presented in several studies provide direct evidence that expanded populations of tumor reactive T cells may be present in tumors. In the study employing the adoptive transfer of TILs to patients receiving nonmyeloablative chemotherapy, immunohistochemical staining revealed that a dense infiltrate of T cells expressing the TRBV gene product expressed by dominant T-cell clonotypes present in peripheral blood was present in the regressing tumor lesions of a patient with a near-complete response.1 In another study employing the adoptive transfer of MART-1 reactive T-cell clones, MART-1 tetramer positive cells represented nearly 40% of the CD8+ T cells present within a tumor deposit 3 days following transfer, although no objective responses were seen.4 In the present study, four of the persistent T-cell clones that were detected in peripheral blood were also detected at similar levels in a regressing tumor lesion approximately 1 week following transfer. Two persistent clones were also detected 4 weeks following transfer at somewhat higher levels in a tumor sample than in a sample of peripheral blood drawn at the same time, and were present in the tumor sample at 9 weeks after treatment (see Fig. 3). Although varying degrees of contamination with blood could occur during collection of the fresh tumor samples, dense T-cell infiltrates were observed by immunohistochemical staining of the tumor lesions taken at the same time as the samples were taken for TRBV analysis (data not shown). The results indicate that the transferred T lymphocytes traffic to tumor sites and thus may play a role in tumor regression following adoptive cell transfer.
Mutated products of the GAS7 and GAPDH genes represented two of the antigens recognized by TIL 2098 T cells. Growth arrest-specific (gas) genes are expressed preferentially in cultured cells that enter a quiescent state following serum deprivation or growth to confluence,21 and a GAS7 translocation was involved in transforming multi-potent hematopoietic progenitors and inducing mixed lineage leukemias.22,23 The GAS7 gene product had also been identified as a molecule that was upregulated in cultured normal cells that had undergone cell cycle arrest following serum starvation.24 Thus, the mutated GAS7 gene isolated from 2098mel may have played a role in the development of this tumor. The GAPDH is a multifunctional protein.25 In addition to its integral role in glycolysis, converting glyceraldehyde-3-phosphate into 1,3-bisphosphoglycerate, GAPDH has been shown to have diverse biologic functions, including the signaling of apoptosis. GAPDH also participates in membrane, cytoplasmic, and nuclear functions for endocytosis, mRNA regulation, tRNA export, DNA replication, and DNA repair. Recently, GAPDH was identified as an endothelium-associated antigen by modified serologic analyses of recombinant cDNA expression libraries.26 It seems that GAPDH may play a role in tumor development in cancer patients. The data provide the evidence that mutated GAS7 and GAPDH tumor antigens may be involved in both tumor development and T-cell recognition.
Mutated GAS7 and GAPDH were recognized by the infused TIL 2098 (see Fig. 6), suggesting that they played a role in mediating tumor regression in patient 2098. It has been reported that T-cell clones reactive with the mutated HLA-A11 gene product and the mutated p14ARF product may have played a role in the nearly complete tumor regression in patient 1913 that was observed following adoptive cell transfer.5 Therefore, T cells that recognize mutated gene products may be especially effective in mediating tumor regression after adoptive cell transfer.
In summary, patient 2098 had a complete tumor regression after adoptive transfer of autologous anti-tumor TILs. Four T-cell clones with tumor-specific activities persisted in this patient after treatment. The 2098BV7-9a and 27 T-cell clones recognized mutated GAS7 and the 2098BV12-4a T-cell clone recognized mutated GAPDH, whereas the 2098BV2a T-cell clone recognized autologous tumor cells, but its antigen was not identified by all our efforts (see Fig. 7A). The unidentified antigen appeared to be limited in its expression to autologous tumor cells, and therefore it likely represents a mutant antigen. The results showed that multiple tumor reactive persistent T-cell clones might be responsible for tumor regression in patient 2098 after adoptive cell transfer. These results also provide clues to understand the mechanism of tumor regression in cancer patients and to improve the efficacy of adoptive cell transfer immunotherapy.
References
- 1.Dudley ME, Wunderlich JR, Robbins PF, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Packard BS, Aebersold PM, et al. Use of tumor infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. Preliminary report. N Engl J Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Yannelli JR, Yang JC, et al. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J Natl Cancer Inst. 1994;86:1159–1166. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 4.Yee C, Thompson JA, Byrd D, et al. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci USA. 2002;99:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, El-Gamil M, Dudley ME, et al. T cells associated with tumor regression recognize frameshifted products of the CDKN2A tumor suppressor gene locus and a mutated HLA class I gene product. J Immunol. 2004;172:6057–6064. doi: 10.4049/jimmunol.172.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller G, Lipp M. Shaping up adaptive immunity: the impact of CCR7 and CXCR5 on lymphocyte trafficking. Microcirculation. 2003;10:325–334. doi: 10.1038/sj.mn.7800197. [DOI] [PubMed] [Google Scholar]
- 7.Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 8.Meidenbauer N, Marienhagen J, Laumer M, et al. Survival and tumor localization of adoptively transferred Melan-A-specific T cells in melanoma patients. J Immunol. 2003;170:2161–2169. doi: 10.4049/jimmunol.170.4.2161. [DOI] [PubMed] [Google Scholar]
- 9.Pisarra P, Mortarini R, Salvi S, et al. High frequency of T cell clonal expansions in primary human melanoma. Involvement of a dominant clonotype in autologous tumor recognition. Cancer Immunol Immunother. 1999;48:39–46. doi: 10.1007/s002620050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willhauck M, Scheibenbogen C, Pawlita M, et al. Restricted T-cell receptor repertoire in melanoma metastases regressing after cytokine therapy. Cancer Res. 2003;63:3483–3485. [PubMed] [Google Scholar]
- 11.Paschen A, Eichmuller S, Schadendorf D. Identification of tumor antigens and T-cell epitopes, and its clinical application. Cancer Immunol Immunother. 2004;53:196–203. doi: 10.1007/s00262-003-0479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renkvist N, Castelli C, Robbins PF, et al. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins PF, Kawakami Y. Human tumor antigens recognized by T cells. Curr Opin Immunol. 1996;8:628–636. doi: 10.1016/s0952-7915(96)80078-1. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg SA. Progress in the development of immunotherapy for the treatment of patients with cancer. J Intern Med. 2001;250:462–475. doi: 10.1046/j.1365-2796.2001.00911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang RF, Rosenberg SA. Human tumor antigens for cancer vaccine development. Immunol Rev. 1999;170:85–100. doi: 10.1111/j.1600-065x.1999.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami Y, Dang N, Wang X, et al. Recognition of shared melanoma antigens in association with major HLA-A alleles by tumor infiltrating T lymphocytes from 123 patients with melanoma. J Immunother. 2000;23:17–27. doi: 10.1097/00002371-200001000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Robbins PF, El-Gamil M, Li YF, et al. A mutated b-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley ME, Wunderlich J, Shelton TE, et al. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332–342. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio V, Stuge TB, Singh N, et al. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- 20.Dudley ME, Wunderlich J, Nishimura MI, et al. Adoptive transfer of cloned melanoma-reactive T lymphocytes for the treatment of patients with metastatic melanoma. J Immunother. 2001;24:363–373. doi: 10.1097/00002371-200107000-00012. [DOI] [PubMed] [Google Scholar]
- 21.Ju YT, Chang AC, She BR, et al. gas7: a gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc Natl Acad Sci USA. 1998;95:11423–11428. doi: 10.1073/pnas.95.19.11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megonigal MD, Cheung NK, Rappaport EF, et al. Detection of leukemia-associated MLL-GAS7 translocation early during chemotherapy with DNA topoisomerase II inhibitors. Proc Natl Acad Sci USA. 2000;97:2814–2819. doi: 10.1073/pnas.050397097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.So CW, Karsunky H, Passegue E, et al. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–171. doi: 10.1016/s1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 24.She BR, Liou GG, Lin-Chao S. Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp Cell Res. 2002;273:34–44. doi: 10.1006/excr.2001.5435. [DOI] [PubMed] [Google Scholar]
- 25.Brown VM, Krynetski EY, Krynetskaia NF, et al. A novel CRM1-mediated nuclear export signal governs nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase following genotoxic stress. J Biol Chem. 2004;279:5984–5992. doi: 10.1074/jbc.M307071200. [DOI] [PubMed] [Google Scholar]
- 26.Zhong X, Ran YL, Lou JN, et al. Construction of human liver cancer vascular endothelium cDNA expression library and screening of the endothelium-associated antigen genes. World J Gastroenterol. 2004;10:1402–1408. doi: 10.3748/wjg.v10.i10.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]