SUMMARY
The ability to sense and respond appropriately to environmental changes is a primary requirement of all living organisms. In response to phosphate limitation, Saccharomyces cerevisiae induces transcription of a set of genes involved in the regulation of phosphate acquisition from the ambient environment. A signal transduction pathway (the PHO pathway) mediates this response, with Pho85-Pho80 playing a vital role. Here we report the x-ray structure of Pho85-Pho80, the first for CDK-cyclin complex functioning in transcriptional regulation in response to environmental changes. The structure revealed a specific salt link between a Pho85 arginine and a Pho80 aspartate that makes phosphorylation of the Pho85 activation loop dispensable and that maintains a Pho80 loop conformation for possible substrate recognition. It further showed two new sites on the Pho80 cyclin for high affinity binding of the transcription factor substrate (Pho4) and the CDK inhibitor (Pho81) that are markedly distant to each other and the active site.
INTRODUCTION
Inorganic phosphate is an essential nutrient for all organisms, as it is required for the biosynthesis of diverse cellular components including nucleic acids, proteins, lipids, sugars and phospho-metabolites. The budding yeast Saccharomyces cerevisiae phosphate-responsive signaling system (known as the PHO pathway) senses and responds to changes in the concentration of inorganic phosphate in the medium [(Toh-e et al., 1973; Ueda et al., 1975); reviewed in (Carroll and O’Shea, 2002)]. Through this pathway, many PHO genes are repressed in high-phosphate conditions and induced in conditions of phosphate limitation. Central to the PHO pathway is a CDK-cyclin complex, Pho85-Pho80, whose activity is regulated in response to extracellular phosphate availability (Kaffman et al., 1994; Schneider et al., 1994; Toh-e et al., 1988). Pho81, a CDK inhibitor (CKI), binds to Pho85-Pho80 when cells are grown in both high- and no-phosphate conditions, but inhibits the kinase activity only during phosphate limitation (Schneider et al., 1994). The Pho85-Pho80-Pho81 complex regulates the location and activity of Pho4 (Kaffman et al., 1994), a transcription factor required for expression of phosphate-responsive genes, including PHO5, which encodes a secreted phosphatase. In high-phosphate medium, Pho85-Pho80 phosphorylates and inactivates Pho4. In medium devoid of phosphate, Pho81 inhibits Pho85-Pho80, leading to accumulation of unphosphorylated Pho4 in the nucleus and activation of PHO5 transcription.
Pho85, through its association with nine other Pho85 cyclins (called Pcls) (Measday et al., 1997), is one of the most versatile CDKs. Pcls target Pho85 to different substrates and thus other cellular functions (Carroll and O’Shea, 2002; Toh-e and Nishizawa, 2001), but the structural basis for substrate targeting is unclear. Of the diverse cellular functions regulated by Pho85, the PHO pathway is by far the best studied.
Despite significant similarity between Pho85 and the cell cycle CDKs, especially Cdc28/CDK2 (Toh-e et al., 1988), Pho85 possesses several prominent distinct features. Whereas phosphorylation of a conserved threonine or serine residue on the kinase subunit activation loop is required for full activation of CDK-cyclin complexes functioning in cell cycle [reviewed in (Morgan, 1996; Russo et al., 1996b)], it is dispensable for Pho85-Pho80 kinase function (Nishizawa et al., 1999). The residue at the +3 position of the consensus sequence (S/TPXK/R, where S/T is the phosphorylatable residue and X is any residue) of the substrates of most cell cycle CDK-cyclin complexes differs radically from that (SPXI/L) of the five phosphorylation sites on the Pho4 substrate of Pho85-Pho80 (O’Neill et al., 1996). Moreover, tight interaction between Pho80 and a site distal to the phosphorylation sites in Pho4 enhances catalytic efficiency by orders of magnitude and enables semi-processive phosphorylation (Byrne et al., 2004; Jeffery et al., 2001). The inhibitory domain of the Pho81 CKI differs from those of the two major types of mammalian CKIs, the INK4s and Cip/Kips, of cell cycle regulation (Huang et al., 2001). In addition, unlike CKIs of the cell cycle CDK-cyclin complexes which either target the kinase solely or both kinase and cyclin [reviewed in (Endicott et al., 1999)], Pho81 interacts with Pho85-Pho80 primarily through association with the Pho80 subunit (Schneider et al., 1994). Interestingly, Pho81 has the unusual property of forming a stable complex with Pho85-Pho80 under both high- and low-phosphate concentrations, but only inhibiting under low phosphate conditions (Schneider et al., 1994). Recently, it has been reported that kinase inhibition by the constitutively associated Pho81 requires myo-D-heptakisphosphate (IP7) (Lee et al., 2007). To gain new insights into the molecular nature of the distinct features of the Pho85 in conjunction with Pho80 cyclin, as well as the role of the complex in the PHO pathway, we carried out the determination of the crystal structure of the Pho85-Pho80 complex.
RESULTS AND DISCUSSION
Structure of the Pho85-Pho80 Complex
Crystals of the complex of full-length Pho85 (305 residues) and Pho80 (293 residues) were obtained by the hanging drop method and optimized by the addition of strontium to the precipitant and incubation at 10°C (see Supplemental Data). The structure, with two Pho85-Pho80 complexes in the asymmetric unit and an unusually high water content (~80%), was determined by single wavelength anomalous dispersion (SAD) and refined to 2.9 Å resolution (Table 1). The structure was used subsequently to determine, at an identical resolution, the structure of the isomorphous co-crystal of Pho85-Pho80 and the nonhydrolyzable ATP analog, ATP-γ-S (Table 1). The co-crystal structure is the focus of this report.
Table 1.
Data Collection and Refinement Statistics
| SeMet SADa | Nativeb | Native with bound ATP-γ-Sb | |
|---|---|---|---|
| Diffraction Data | |||
| Space group | P3121 | P3121 | P3121 |
| Cell dimensions | a=b=147.1 Å, c=212.4 Å | a=b=147.8 Å, c=212.8 Å | a=b=146.6 Å. c=212.8 Å |
| α=β=90°; γ=120° | α=β=90°; γ=120° | α=β=90°; γ=120° | |
| Wavelength | 0.97926 peak | 0.97948 | 1.0332 |
| Resolution (Å) | 30 – 3.0 | 15 – 2.9 | 12 – 2.9 |
| Total reflections | 32,066 | 52,156 | 53,149 |
| Rsymc,d (%) | 12.2 (53.5) | 9.6 (29.0) | 9.9 (24.8) |
| I/σI | 9.6 (2.3) | 12.2 (2.4) | 11.0 (2.4) |
| Completenessd (%) | 99.9 (99.9) | 88.4 (71.2) | 91.1 (75.6) |
| Redundancy d | 4.6 (4.6) | 3.2 (2.2) | 3.1 (2.0) |
| Refinement | |||
| Resolution (Å) | 15 – 2.9 | 12 – 2.9 | |
| Total reflections | 52,156 | 53,149 | |
| R-factore/R- free | 0.281/0.315 | 0.287/0.325 | |
| Rmsd | |||
| bond length | 0.008 | 0.007 | |
| bond angles | 1.318 | 1.320 | |
Data collected at beamline ID29, ESRF (1° oscillation) and processed with MOSFLM (Leslie, 1992).
Data collected at beamline ID19, APS (1° oscillation) and processed with HKL2000 (Otwinowski and Minor, 1997).
Rsym= ΣhΣi|Ii(h)−<I(h)>|/ΣhΣiI(h)).
Numbers in parenthesis are for outer shell data.
R-factor = Σ(|Fo|−k |Fc|)/Σ|Fo. R-free was calculated using a random 5% of the reflection data that was omitted in the refinement.
Pho85 has a typical CDK fold, consisting of two distinct lobes – a smaller N-terminal (or N) lobe composed mainly of a five-stranded antiparallel β sheet and the PSTAIRE helix and a much larger C-terminal (or C) lobe rich in a helices (Figure 1A). The structure is not unexpected given amino acid sequence identity as high as ~55% between Pho85 and CDK2, the mammalian cell cycle regulator (Morgan, 1997), and CDK5, an important signal transducer in neuronal development (Smith et al., 2001) and the closest known functional homolog of yeast Pho85 (Huang et al., 1999; Nishizawa et al., 1999). The ATP analog ATP-γ-S is bound in the cleft between the N and C lobes of Pho85 (Figures 1A) and held in place by nearly identical residues that bind the AMPPNP in the structure of the CDK2-cyclin A-peptide substrate-AMPPNP-Mg2+ complex (Brown et al., 1999). The cleft further harbors the catalytic center, which contains the conserved catalytic triad of residues in CDKs (Lys33, Glu51 and Asp145 in CDK2 and Lys36, Glu53 and Asp151 in Pho85).
Figure 1. Structure of Pho85-Pho80.
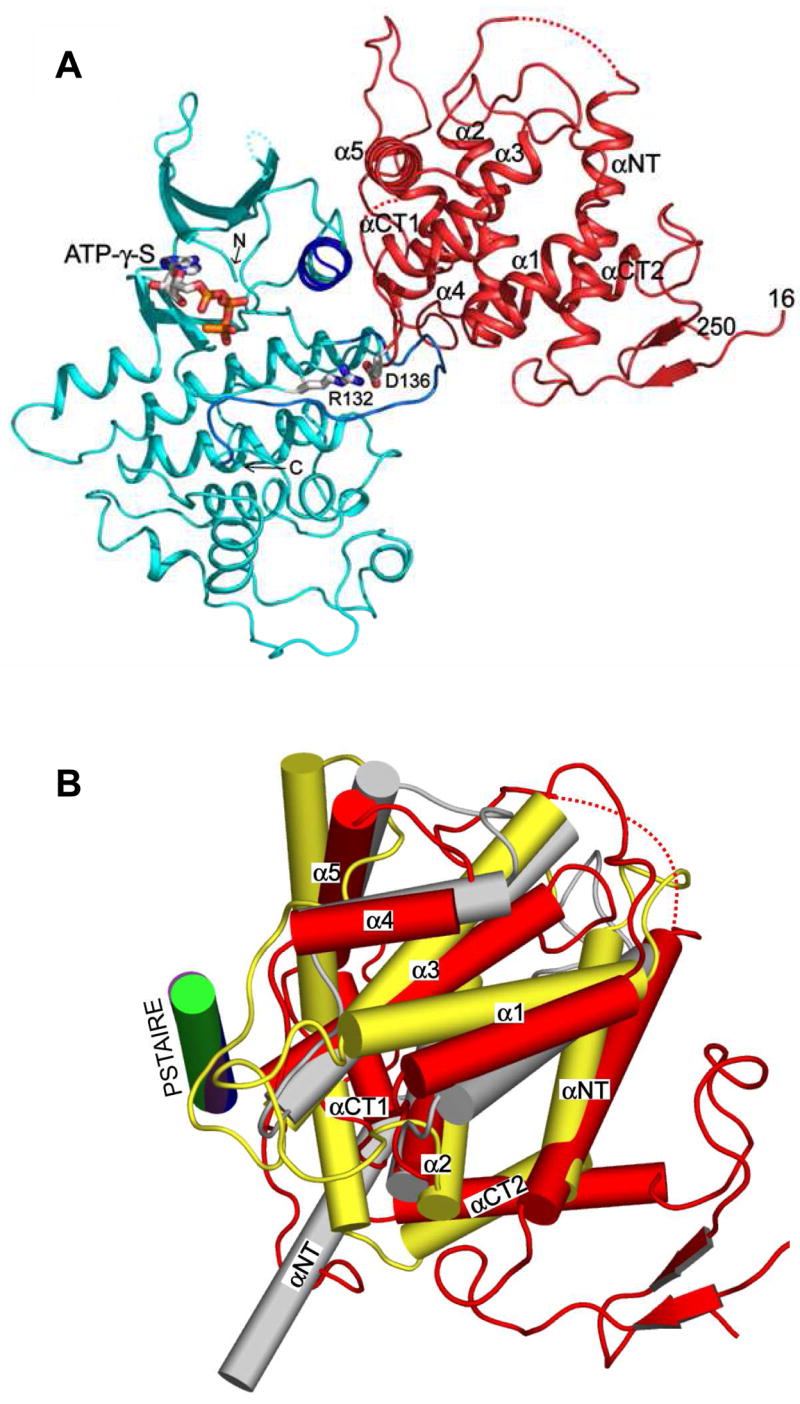
(A) Ribbon backbone trace of the crystal structure of the Pho85-Pho80 complex with the bound ATP analog, ATP-γ-S. Pho85 is shown in cyan, and its PSTAIRE helix and activation loop in blue and marine, respectively. Pho80 is depicted in red. The ATP-γ-S is represented in ball-and-stick model. The amino- and carboxy-terminal ends of Pho85 are marked by N and C, respectively, and those of Pho80 are denoted by residue numbers 16 and 250, respectively. The first 15 and last 43 residues in Pho80 are not visible (or disordered) in the electron density map, indicating their flexibilities. The long loops in Pho80 include the visible N-terminal 17- and C-terminal 22-residue loops, the 25-residue loop between αNT and α1, and the 33-residue loop between αCT1 and αCT2 helices (see Supplementary Figure S1). The two dotted lines in the Pho80 trace represent disordered segments (see Supplementary Figure S1). R132 on Pho85 and D136 on Pho80 form a salt link (details shown in Figure 2).
(B) Relative positions of Pho80 (red), cyclin A (grey) and p25 (yellow) following superposition of their cognate kinase subunits as they occur in the structures of the Pho85-Pho80-ATP-γ-S complex, the phosphorylated CDK2-cyclin A complex (Brown et al., 1999) and CDK5-p25 (Tarricone et al., 2001). The orientation of Pho80 is identical to that shown in panel A. The depiction of the α-helices by cylinders emphasizes the differences between the relative orientations of the cyclins. p25 lacks the equivalent of the 2-turn α4 helices in Pho80 and cyclin A, and in its place is a turn which includes the P247 residue. The PSTAIRE (Pho85 and CDK2) or PSAALRE (p25) helices are shown in blue (Pho85), green (CDK2) and purple (CDK5). Although the PSTAIRE/PSSALRE helices are completely superimposable, the contacts they make with their cognate cyclins vary (e.g., see Figures S2 and S3). In the truncated cyclin A structure, only the first helix (αNT) and the first cyclin box are shown. The succeeding two helices (α1′ and α2′) in cyclin A (not shown) constitute the first two helices of the second cyclin box and adopt positions different from those of αCT1 and αCT2, respectively, in Pho80 or p25. The αNT helix of cyclin A makes significant interaction with the C-terminal lobe of CDK2 (Russo et al., 1996b). In contrast, the αNT helices of Pho80 and p25, which approximately adopt similar positions, do not participate in kinase binding as they lie on the opposite side of the cyclin structures to the kinase-cyclin interface or the location of αNT in cyclin A.
Pho80 has a single globular domain structure composed of eight helices, identified as αNT for the first helix, α1 to α5 for the core helix bundle and αCT1 and αCT2 for the last two helices that cap one end of the helix bundle and bring the C-terminal loop close to the N-terminal loop (Figures 1A and B). Pho80 contains several unusually long meandering loops, including those at both termini and between helices αNT and α1, and αCT1 and αCT2 (Figure 1A and Figure S1 in Supplemental Data). Furthermore, unlike the cyclins functioning in the cell cycle [e.g., the structure of the 259-residue fragment of vertebrate cyclin A bound to CDK2 (Jeffrey et al., 1995)], which contain tandem duplications of five-helix bundles (also known as cyclin boxes), the Pho80 structure shows only one cyclin box, which corresponds to the core helices α1 to α5 (Figures 1A and B). This structural result solidifies previous sequence comparison indicating one cyclin box for Pho80 and all other nine Pcls (Moffat et al., 2000) (see also Supplementary Figure S1). p25, the 147-residue fragment of the p35 activator of CDK5, is to our knowledge the only other cyclin whose crystal structure (Tarricone et al., 2001) shows a single cyclin box (minus the missing α4), as well as the presence of αNT helix and two additional C-terminal helices at locations quite close to those in Pho80 (Figure 1B). However, superposition of the Pho80 and p25 cyclin structures indicates only 84 overlapped α-carbons with rmsd of 2.0 Å, of which a significant number (59 α-carbons) are confined to α3, α4 and α5 helices that contact the region adjacent to the active site cleft of the cognate kinase subunits. As in the case of many other cyclins, Pho80 and p25, as well as the Pcls (Supplementary Figure S1), show no significant sequence similarity.
Superposition of the CDK subunits as they occur in the structure of the fully activated, phosphorylated CDK2-cyclin A complex (Brown et al., 1999) and those of the Pho85-Pho80-ATP-γ-S and CDK5-p25 (Tarricone et al., 2001) complexes, indicates no gross changes in the kinase conformation. Moreover, the superposition indicates that similar regions, mainly the PSTAIRE (Pho85 and CDK2)/PSSALRE (CDK5) helix and its preceding loop and the activation (or T-) loop, are engaged in major interactions with the regulatory partners. The superposition also clearly shows the differences in the relative dispositions of the equivalent helices and in the lengths and conformations of the inter-helix connecting loops in the cyclin subunits (Figure 1B). Consequently, the involvements of the helices and loops of the cyclins at the interface with their cognate kinases vary (for example, see Figures S2 and S3 in Supplemental Data). These variations, indicative of the plasticity of the CDK-cyclin interfaces, are underscored by comparing the three different complex structures in the following framework. In the Pho85-Pho80 complex, the cyclin helices (mostly the set of helices α3, α4 and α5) are involved in slightly fewer contacts (~45%) at the interface than the loop regions (~55%) located mainly between α3 and α4, α5 and αCT1 and αCT1 and αCT2. In contrast, in the CDK5-p25 complex, the cyclin makes significantly more contacts (~75%) via the same set of helices and α6 than via principally one loop that follows α3. The CDK2-cyclin A interface portrays the extreme case; the distribution is ~85% helices, contributed mostly by α3, α5 and αNT and ~15% loops, provided almost entirely by the one between α5 and α1′ helices, the first helix of the second cyclin box.
A New and Specific Mechanism for Circumventing Activation Loop Phosphorylation
Despite the difference in the juxtapositions of Pho80 and p25 with their kinase partners, the two complexes share one key functional feature that is distinct from the CDK-cyclin complexes involved in cell cycle regulation. Whereas phosphorylation of the T/S residue on the T-loop (e.g., T160 of CDK2) by the CDK activating enzyme is obligatory for full activation of the cell cycle CDK-cyclin complexes, it is not required for Pho85-Pho80 and CDK5-p35 despite the presence of an equivalent serine residue on their T-loops (S166 on Pho85 and S159 on CDK5) (Espinoza et al., 1998; Nishizawa et al., 1999; Poon et al., 1997). Phosphorylation of the T-loop T160 residue of CDK2-cyclin A causes a large conformational rearrangement that results in the opening of the substrate site and proper formation of the entire active site region [reviewed in (Pavletich, 1999; Russo et al., 1996b)]. Moreover, the presence of the phosphoryl adduct dictates the strong preference for the positively charged residue at the +3 position of the consensus sequence (SPXK/R) of substrates of cell cycle CDK-cyclin complexes. The crystal structure of phosphorylated CDK2-cyclin A in the presence of a peptide substrate containing the consensus sequence SPRK, AMPPNP and Mg2+, illustrates that the structural basis for this substrate specificity is a salt link formed between the phosphoryl adduct and the +3 positively charged residue (Brown et al., 1999). The dispensability of the T-loop phosphorylation in the Pho85-Pho80 complex and the strong preference for large aliphatic side chains (I/L) at the +3 position of the consensus sequence of the five phosphorylation sites on the Pho4 substrate indicate that Pho80 binding is sufficient for full activation of the kinase activity and that a different strategy is used for the recognition of the residue at the +3 position.
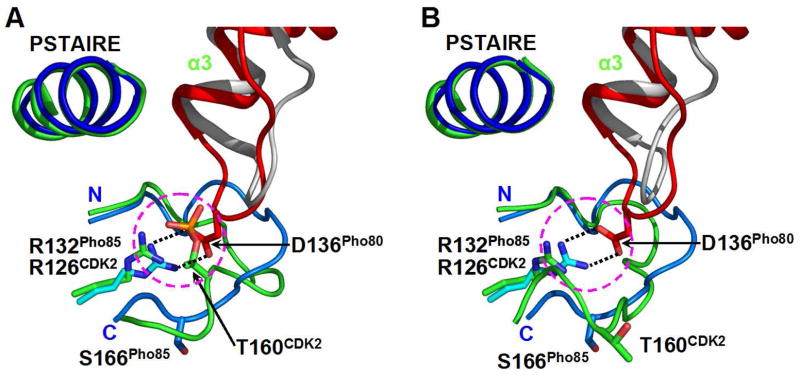
Our Pho85-Pho80 structure analysis has led to the discovery of a charge-coupling interaction between the kinase and the cyclin subunits that does away with activation loop phosphorylation. The interaction is between the guanidinium group of R132 on the C-lobe of Pho85 and the carboxylate group of D136 on the six-residue α3-α4 loop (named D-loop) of Pho80 (Figures 1A and 2). The charge-coupling interaction matches closely that occurring in the phosphorylated CDK2-cyclin A complex, between the guanidinium group of CDK2 R126, the equivalent of Pho85 R132, and the phosphoryl adduct to the T-loop T160 residue (Figure 2A). Thus, the R132–D136 salt link makes T-loop phosphorylation of Pho85 no longer necessary for full kinase activation. The fact that the R126 in the phosphorylated CDK2 structure remains in an identical position in the unphosphorylated kinase subunit (Figure 2B) indicates its major role in dictating the ultimate position of the phosphoryl group required for full kinase activation and substrate recognition. In contrast, if the T-loop S166 of Pho85 were phosphorylatable, the presence of Pho80 D136 would prevent the phosphoryl group from assuming a position similar to that of the phosphoryl group on T160 of CDK2. Support for the importance of the R132–D136 salt link is provided by the observation of loss of Pho80 function following very conservative mutation of D136 to N (Madden et al., 1990). Moreover, the complete conservation of Pho80 D136 in all of the other nine Pcls (See Figure S1 in Supplemental Data), highlights the arginine–aspartate salt link as a new feature of a large family of CDK-cyclin complexes essential in the regulation of diverse cellular functions.
Figure 2. A Salt Link Between Pho85 R132 and Pho80 D136 Which Makes T-loop Phosphorylation Dispensable.
(A) Relative positions of the loop between α3 and α4 in Pho80 (red) and cyclin A (grey) and the PSTAIRE helices and segments of the activation loops in Pho85 (residues 155 – 168) (marine) and phosphorylated (T160) CDK2 (residues 149 – 163) (green). The overlay is based on the superposition of the structures of the kinase subunits as they occur in the structures of the Pho85-Pho80-ATP-γ-S complex, and the phosphorylated CDK2-cyclin A complex (Brown et al., 1999). The α3-α4 loop in Pho80 deploys D136, hence the “D-loop” name. The activation loop T160 (with the phosphoryl adduct) of CDK2 corresponds to S166 in Pho85. Pho85 R132 and CDK2 R126 are superimposable. CDK5 R125 adopts almost identical position as those of Pho85 R132 and CDK2 R126 (data not shown). Pho80 D136 has no counterpart in cyclin A and p25. A more global view of the D-loop and the salt link is shown in Figure 1A.
(B) Similar superposition as shown in panel A, except that the CDK2 structure is the unphosphorylated form (Jeffrey et al., 1995). The position of R126 of CDK2 is essentially unchanged from that in the phosphorylated CDK2 (panel A). Note the significant change in the conformation of the segment of the T-loop of CDK2 following phosphorylation (compare with that in panel A).
Potential Involvement of the D-loop in Substrate Recognition
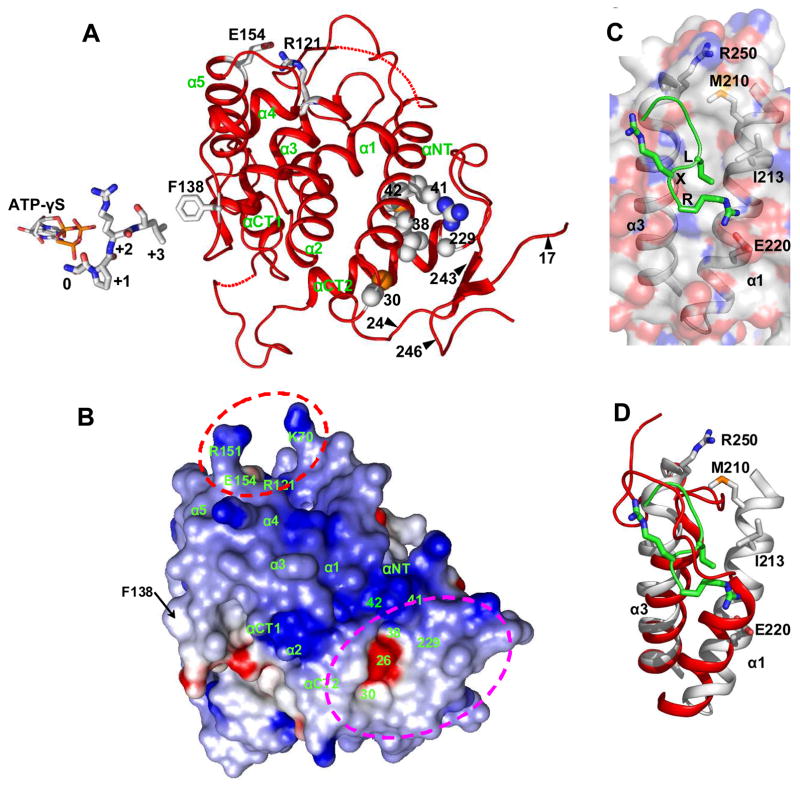
The CDK5-p25 complex crystal structure offered the first detailed view of a phosphorylation-independent, fully active CDK-cyclin complex (Tarricone et al., 2001). Moreover, a modeling study, followed by mutational analysis, has indicated the importance of the carboxylate side chain of E240, located close to the beginning of the loop that follows the α3 helix of p25, in the recognition of the positively charged residue at the +3 position of the substrate consensus sequence SPXK/R (Tarricone et al., 2001). Interestingly, F138, the sole hydrophobic residue on the D- (or α3-α4) loop in Pho80 (Figure 3A and Figure S1 in the Supplemental Data), is in a position to mimic the role of the p25 E240, in recognizing the I/L at the +3 position of the consensus sequence (SPXI/L) of the five phosphorylation sites on the Pho4 substrate of Pho85-Pho80 (O’Neill et al., 1996).
Figure 3. Three Functional Sites of the Pho80 Cyclin Subunit.
(A) Ribbon trace of Pho80. The regions of the three functional sites are identified as follows (see also text): The bound ATP-γ-S (stick representation) marking the location of the kinase active site region; the cluster of residues C30, L38, R41, M42 and G229 and segments of residues 17 to 24 and 243 to 246 forming a large cavity for possible tight interaction with the site on Pho4 distal to the phosphorylatable sites; and the salt link between R121 and E154 in a region for docking the inhibitor, Pho81. The peptide near the ATP-γ-S corresponds to a model of the the SPRL consensus sequence of the five Pho4 phosphorylations sites (O’Neill et al., 1996). Placement of the peptide was derived by superimposing the atomic coordinates of only the Pho85 and CDK2 subunits in the Pho85-Pho80-ATP-γ-S structure and the structure of the phosphorylated CDK2-cyclin A with a bound peptide containing the SPRK consensus sequence and AMPPNP-Mg2+ (Brown et al., 1999). Changing the lysine at the +3 position to leucine to conform to the Pho4 consensus sequence, SPXI/L, indicates that the leucine sidechain would be within van der Waals distance to the F138 located on the α3-α4 (or D-) loop in Pho80.
(B) Electrostatic surface potential surface of Pho80 contoured at −10kT (red) and +10kT (blue) calculated with GRASP (Nicholls et al., 1991). Pho80 is in identical orientation as in panel A. The region involved in binding of the CKI Pho81 is enclosed by red dashed lines. The region involved in docking of the site on Pho4 distal to the phosphorylation sites is enclosed in magenta dashed lines.
(C) Hydrophobic patch on cyclin A for docking the RXL motif of substrates or CDK inhibitors. The bound ligand, with backbone trace in green, shows only the RRL sequence (the equivalent of the RXL motif) of the substrate recruitment peptide RRLFGE of p107 (Brown et al., 1999). The residues M210, I213 and E220 on α1 and R250 on α3 define the specificity of the patch in a group of cell cycle cyclins for the RXL motif (Endicott et al., 1999).
(D) Identical to panel C, but with the surface of cyclin A removed for clarity, and the α1 and α3 helices and their preceding loops of Pho80 superimposed. The first two turns of α1 of cyclin A, which provides M210 and I213, are missing in the corresponding α1 of Pho80.
To assess the role of Pho80 F138 in substrate phosphorylation, we analyzed the effects of substitutions of this residue (Table 2). The Pho80 F138A mutation caused a 3.5- and 7.8-fold decrease in kcat/KM for phosphorylation of the natural substrate, Pho4, and the specific 24-residue peptide substrate (called SPVI), respectively. These results, together with an even greater (15.7-fold) reduction in the phosphorylation of Pho4 caused by the mutation F138E, suggest a major role for Pho80 F138 in attaining full kinase activity. The approximately three orders of magnitude greater catalytic efficiency of wild-type Pho85-Pho80 towards Pho4 compared to SPVI has been previously observed (Jeffery et al., 2001) and is attributed to the interaction between a site in Pho4 distal to the phosphorylation sites and a site in the Pho80 cyclin [(Byrne et al., 2004); discussed further below]. The 9.8 fold decrease in activity towards SPVA, a non-specific peptide substrate, is consistent with the strong preference for the large aliphatic sidechain at +3 position of the consensus sequence of the Pho4 phosphorylation sites. These observations suggest that F138 of Pho80 and position +3 of the substrate are important for efficient phosphorylation. If F138 interacts favorably with I in the +3 position of the substrate and the only effect of F138A is to prevent this interaction, we expect that the F138A mutation will decrease phosphorylation of the SPVI substrate, but that SPVA will be phosphorylated with efficiency comparable to the wild-type Pho80. Unexpectedly, the decrease in phosphorylation resulting from the F138A mutation is similar for both SPVI and SPVA substrates (14.9- vs 11.1-fold), suggesting that mutation of F138 may have additional effects on the structure of the kinase which cause a general reduction in catalytic efficiency.
Table 2.
Probing the Role of F138 in Pho80 in Substrate Recognition
| Substrate |
kcat/KM (M−1s−1)a |
||
|---|---|---|---|
| Pho85-Pho80 | Pho85-Pho80 F138A | Pho85-Pho80 F138E | |
| Pho4 | 6.6 ± 1.1 × 106 | 1.9 ± 0.5 × 106 | 4.2 ± 1.2 × 105 |
| SPVIb | 3353 ± 2650 | 431 ± 224 | — |
| SPVAc | 225 ± 78 | 39 ± 1 | — |
Kinase assays were done in triplicate. (See Supplementary Material).
SPVI is an identification of the peptide substrate used in the kinase assay. The sequence of the peptide substrate used is SAEGVVVASESPVIAPHGSTHARSY, which corresponds to the segment (residues 213 to 237) of the serine phosphorylation site 6 (or SP6) in Pho4 (Jeffery et al., 2001). The consensus sequence (SPVI) is in bold.
SPVA, identical to SP6 peptide substrate except that the I residue at the +3 position of the consensus sequence is replaced by A.
The close similarity in the mechanism of substrate recognition by Pho85-Pho80 and CDK5-p25, being governed by F138 in Pho80 and E240 in p25, is in accord with the finding that the mammalian CDK5 is the closest functional homology of the yeast Pho85. The mechanism is consistent with the findings that Pho85 can bind to p35/p25 and phosphorylate CDK5 substrate or, vice versa, that CDK5 can bind to some of the Pho80/Pcls and phosphorylate Pho85 substrates (Huang et al., 1999; Nishizawa et al., 1999).
Whether the similar role of residues F138 of Pho80 and E240 of p25, located in a similar loop region, in recognizing the residue at the +3 position residue of the phosphorylation sites of the substrates also applies to the complexes of Pho85 with the other Pcls remains uncertain. Interestingly, the substrates that have been identified for several of the other complexes have different residues at the +3 positions (Supplementary Table S1). However, sequence alignment of Pho80 and Pcls indicates that the F138 residue varies between the other Pcls, but not in a manner that would be complementary to several of the different +3 residues of the substrates (Supplementary Figure S1 and Table S1).
A Unique Site on Pho80 for Tight Binding of Pho4 Substrate
The structure of Pho85-Pho80 has further provided insights into two other major roles of the regulatory subunit in the PHO signaling system owing to its tight interactions with a site on Pho4 distal to the phosphorylation sites and with the CDK inhibitor, Pho81. In addition to its potential role in interacting with the substrate in the active site region, Pho80 plays an important role in substrate selection through binding with the Pho4 distal site, thereby enhancing the catalytic efficiency by orders of magnitude (e.g., see Table 2) and enabling semi-processive phosphorylation of multiple serine residues (Byrne et al., 2004; Jeffery et al., 2001). The Pho4 distal site apparently comprises of two long segments (1–30 and 156–200) (Byrne et al., 2004; Jayaraman et al., 1994). To identify the site on Pho80 for tight interaction with the Pho4 distal site, we have examined the wealth of data from mutagenesis studies of Pho80, focusing initially on those single amino acid changes that produce stable protein mutants but unable to phosphorylate and inactivate Pho4 and mapping them onto the crystal structure. Point mutations of M42 to I, V or L in Pho80 were found to suppress constitutively-active alleles of PHO4, implicating this region, located in the αNT helix (Figure 3A), in Pho4 interaction (Okada and Toh-e, 1992). Interestingly, the three M42 mutations are able to suppress the constitutive phenotype of Pho4 L171 and 163–171 deletions (Okada and Toh-e, 1992), which are located in one of the two segments of the Pho4 sequence that comprise the distal, high-affinity site. Nine single amino acid mis-sense mutations of Pho80, excluding the initiator M1 residue, have also shown to cause the failure to repress PHO5 transcription at high phosphate levels (Madden et al., 1990). Four of these mutations cluster adjacent to each other in the Pho80 structure: C30Y, L38F and R41Q, which reside on the αNT helix and its preceding loop, and G229D, which reside close to the C-terminal end of αCT2 (Figure 3A). These four residues, together with M42, form a solvent-exposed, extended surface (Fig. 3B) remote from the active center or the Pho85-Pho80 interface. The other five mutations involving residues 130, 136, 148, 149 and 172 do not belong to another cluster or occupy positions near the active center, with the exception of the D-loop D136 (Figure 1A; discussed below). The extended cluster makes up a significant portion of an oblong shallow cavity punctuated by a small central hole (Figure 3B) that is further bounded by the combined regions of the ordered N- and C-terminal loops (Figure 3A). Support for the participation of the two terminal loops in Pho4 binding is provided by the finding (Madden et al., 1990) that deletion of the first 16 or the last 47 (247–293) residue segments (which interestingly are completely invisible in the electron density (Figure 1A)), caused no diminution of the Pho80 function, but further deletion of residues 17–24 or 243–246 (which are far from the Pho85-Pho80 interface) resulted in nonfunctional Pho80. The cavity and its close vicinity exhibit three different surfaces: an intense positive electrostatic surface contributed mainly by portions of helices αNT and αCT2, a negative surface associated with a single carboxylate confined on the central hole and a hydrophobic surface located primarily on the N- and C-terminal loop regions (Figure 3B). The role of hydrophobic interaction is consistent with the observation that five of the seven single amino acid substitutions in the two segments that cause Pho4 to be constitutively active are hydrophobic residues (Byrne et al., 2004). Hydrophobic interaction, combined with electrostatic interactions, may be sufficient to keep Pho4 in place in order to achieve multiple phosphorylations in a semi-processive manner.
To test the hypothesis that the residues C30, L38F, R41, and G229 are part of a distal binding site for Pho4, we generated the four Pho80 mutants known to be defective in phosphorylation of Pho4 in vivo (C30Y, L38F, R41Q, and G229D) and assayed their enzymatic activity. Each of Pho80 mutants was co-expressed and purified in a stable 1:1 complex with Pho85 (Supplementary Material). Consistent with proposed role of the residues in distal Pho4 binding, each of the mutations caused a 2.7 to 5.7 fold increase in KM values (Table 3). As reported previously for Pho4 mutants defective in interaction with Pho80-Pho85 (Byrne et al., 2004), this decrease in KM suggests that these residues may play a role in increasing the local concentration of Pho4. Surprisingly, the alterations in the putative distal site also caused a 1.8 to 8.1 fold decrease in kcat. This finding has two possible explanations: either the mutations introduced have caused a subtle change in the Pho80 structure, perturbing the Pho85-Pho80 kinase active center; or the mutations are not only involved in increasing the local concentration of Pho4, but play some other undetermined role in the mechanism of Pho4 phosphorylation.
Table 3.
Role of Distal Site Cluster on Pho80 in Pho4 Substrate Recognition
| Pho80 | Pho4a | SPVIa,b | ||
|---|---|---|---|---|
| KM (nM) | kcat (s−1) | kcat/KM (M−1s−1) | kcat/KM (M−1s−1) | |
| Native | 349.2 ± 56.1 | 11.4 ± 0.5 | 6.62 ± 1.12 × 106 | 3353 ± 2650 |
|
| ||||
| C30Y | 960.2 ± 233.8 | 2.4 ± 0.2 | 4.95 ± 1.64 × 105 | 2167 ± 166 |
| L38F | 1263.0 ± 119.5 | 6.5 ± 0.3 | 1.05 ± 0.194 × 106 | 1663 ± 859 |
| R41Q | 1999.4 ± 174.9 | 1.4 ± 0.1 | 1.42 ± 0.052 × 105 | 1856 ± 594 |
| G229D | 1562.7 ± 80.8 | 6.0 ± 0.1 | 7.66 ± 0.357 × 105 | 1525 ± 415 |
Kinase activity assays were performed in triplicate.
SPVI represents the peptide substrate which corresponds to the segment (residues 213 to 237) of the serine phosphorylation site 6 (or SP6) in Pho4 (Jeffery et al., 2001). See Table 2, footnote a.
To determine if the distal site mutations somehow caused alteration of the active site, we assayed the kinase activity of the cyclin mutants towards the SPVI peptide substrate, which lacks a site for high affinity interaction with Pho80. If the active site of the kinase is perturbed, then we expect to see a similar decrease in kinase activity with the SPVI peptide as we saw for full-length Pho4. If, however, the loss in kinase activity is due solely to the lack of interaction between Pho4 and the putative distal site, then the Pho80 mutants should phosphorylate SPVI peptide in a manner similar to wild-type Pho80-Pho85. In each case, kcat/KM of the distal site mutants for the peptide substrate SPVI changed by no more than 2.2 fold (Table 3; compared to 6.3- to 46-fold change in kcat/KM for full-length Pho4). While this change in activity does not rule out a slight perturbation of the active site, it suggests that the primary effect of these mutations is to disrupt distal interactions with Pho4. The structural and biochemical data presented here suggests that this region is a site for distal, high-affinity Pho4 interaction.
Many other CDK-cyclin substrates are believed to also contain distal binding sites, and an RXL motif has been identified for recruiting substrates, as well as the Cip/Kip type CDK inhibitors, to the cell cycle cyclins [(Adams et al., 1996; Schulman et al., 1998); reviewed in (Endicott et al., 1999)]. Structure analysis has revealed a small hydrophobic patch on cyclin A for interacting with the RXL motif (Brown et al., 1999; Russo et al., 1996a) (Figure 3C). The hydrophobic patch, which is conserved among the A, B, D and E cyclins (Endicott et al., 1999), is located on the opposite side of the cyclin structure to the catalytic center and composed mostly of the N-terminal regions of helices α1 and α3 (Figures 3C). Although RXL motifs are present in the Pho4 distal binding site and the 80-residue segment of the Pho81 CDK inhibitor that is sufficient for kinase inhibition, they are not required for substrate and inhibitor binding to Pho85-Pho80 [(Huang et al., 2001) and Supplementary Data]. A clear explanation for this finding has emerged from the structure of Pho85-Pho80. Neither the hydrophobic patch on cyclin A nor the set of residues (M210, I213 and E220 on α1 and R250 on α3) that defines the specificity of the patch for the RXL motif (Endicott et al., 1999) (Figure 3C) is present in Pho80 (Figure 3D). In fact, the area in question in Pho80 bears no resemblance to that harboring the hydrophobic patch on cyclin A. As shown in the superimposed structures in Figure 3D, the area in Pho80 is overlaid mostly by segments of the loops preceding α1 and α3 of cyclin A and, moreover, the first two turns of the α1 helix in cyclin A, which provide M210 and I213, are nonexistent in Pho80. Interestingly, the hydrophobic patch on the cell cycle cyclins (Figure 3C) appears to be much smaller than the region in Pho80 for tight binding of Pho4 (Figures 3A and B). A larger region in Pho80 likely also reflects the need to accommodate and bind tightly a more extensive Pho4 distal site formed from the folding of two long discontinuous polypeptide segments
Location of a Site on Pho80 for Tight Binding of Pho81 CKI
The activity of Pho85-Pho80 is further regulated by a CDK inhibitor, Pho81 (Ogawa et al., 1995; Schneider et al., 1994). Pho81 is thus far the only well-established CKI of the Pho85-Pho80/Pcl family. It has the unusual property of forming a stable complex with Pho85-Pho80 under both high- and low-phosphate concentrations, but only inhibiting under low phosphate conditions (Schneider et al., 1994). Functional studies have demonstrated that Pho81 exerts its inhibitory activity by tightly binding mainly to Pho80 (Huang et al., 2001; Schneider et al., 1994) and identified a segment of 80 residues that is necessary and sufficient for CKI function (Huang et al., 2001). Genetic screen studies have also indicated that the region of R121 and E154 in Pho80 are involved in binding Pho81 (Schneider et al., 1994). R121 and E154 reside near the N-termini of helices α3 and α5, respectively, and form a salt link at the base of a solvent-exposed U-shaped surface, which is ~35 Å to the catalytic center (Figures 3A and B). The U-shaped surface and its close vicinity deploy positively charged residues, including K70 near the C-terminus of the αNT-α1 loop and R151 at the N-terminus of α5, which form the two sides of the U surface (Figs. 3A and B). Binding of the inhibitory segment may be accomplished partly by docking onto the U-shaped surface and, concomitantly, by making complementary electrostatic interactions with the nearby positively charged surface. Pho81 binding may also involve hydrophobic interaction since hydrophobic residues constitute half of the 80-residue primary sequence of the inhibitory domain of Pho81.
Recently, Lee et al. demonstrated that myo-D-heptakisphosphate (IP7) regulates Pho81 inhibition of Pho85-Pho80 in vivo and vitro (Lee et al., 2007). The positively charged nature of the putative Pho81 binding site and surrounding area (Figure 3B) could reflect a role in binding IP7.
CONCLUSION
Since the Pho85-Pho80 complex structure is to our knowledge the first for a member of the superfamily of Pho85-Pho80/Pcl CDK-cyclin complexes, it sets the stage for future comparison with structures of other complexes that regulate other cellular functions. The structure has shed new light on the remarkable multiple key functional roles that Pho80 plays in the PHO pathway by harboring sites for conferring substrate specificity, for tight interaction with the distal site of the Pho4 substrate and for high affinity binding of the novel inhibitory domain of the CKI Pho81. The binding sites for the Pho4 and Pho81 have heretofore not been seen in other CDK-cyclin complexes, and moreover, neither site contains the equivalent of the hydrophobic patch on cell cycle cyclins for interacting with the RXL motif of substrates or inhibitors. Pho80, in partnership with Pho85, further plays an important role in a prototypic mechanism for doing away with T-loop phosphorylation for full activation of the CDK-cyclin complex. By deploying both D136 and F138, the α3-α4 loop or D-loop of the Pho80 cyclin box has emerged as playing important dual roles in the activity of Pho85-Pho80. The charge coupling of Pho80 D136 with the Pho85 R132 makes activation loop phosphorylation no longer required for full kinase activation of the CDK-cyclin complex. The Pho80 F138 is potentially involved in the recognition of the I/L residue at position +3 of the consensus sequence of the five phosphorylatable sites on the Pho4 substrate. Further studies based on the heterodimer structure are required to further deepen our understanding of Pho85-Pho80 function, including the mode of binding of a substrate which would include the distal site, the strategy by which phosphorylation of multiple sites in Pho4 occurs in a semi-processive manner and the mechanism by which the Pho81 CKI, while remaining bound to Pho85-Pho80, modulates kinase activity as a function of phosphate concentration. Finally, structural studies of other Pho85-Pcl complexes are also necessary to unravel the mechanisms by which these complexes recognize different substrates.
EXPERIMENTAL PROCEDURE
Baterial Expression and Purification of Pho85-Pho80
Co-expression of His6-Pho85 and Pho80 in E. coli essentially follows the published procedure (Jeffery et al., 2001). The methods used in the purification of Pho85-Pho80 are described in the Supplemental Data.
Crystallization, Diffraction Data Collection and Structure Determination
Details regarding crystallization, diffraction data collection and structure determination by single wavelength anomalous dispersion technique can be found in the Supplemental Data. Statistics of the data collection and refinement are shown in Table 1.
Probing the Roles of the F138 Residue Close to the Active site and the Distal site on Pho80 in Substrate Recognition
Details regarding site-directed mutagenesis and kinase activity assay to assess the role of the Pho80 F138 and the potential site on Pho80 for interacting with the site on the Pho4 substrate distal to the phosphorylation sites are described in the Supplemental Data.
Supplementary Material
Acknowledgments
We thank Dr. Stephan Ginell and staff members for their assistance in data collection at beamline ID19, APS, which is supported by the Office of Research, U.S. Department of Energy. The work was supported by NIH grants GM051377 to E.K.O and GM068826 to F.A.Q, and by the Howard Hughes Medical Institute to E.K.O. and the Welch Foundation (Q-0581) to F.A.Q.,
Footnotes
Protein Data Bank ID Codes
Two sets of atomic coordinates have been deposited with ID codes 2PK9 and 2PMI.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nature Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- Byrne M, Miller N, Springer M, O’Shea EK. A distal, high-affinity binding site on the cyclin-CDK substrate Pho4 is important for its phosphorylation and regulation. J Mol Biol. 2004;335:57–70. doi: 10.1016/j.jmb.2003.10.035. [DOI] [PubMed] [Google Scholar]
- Carroll AS, O’Shea EK. Pho85 and signaling environmental conditions. Trends Biochem Sci. 2002;27:87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- Endicott JA, Noble ME, Tucker JA. Cyclin-dependent kinases: inhibition and substrate recognition. Curr Opin Struct Biol. 1999;9:738–744. doi: 10.1016/s0959-440x(99)00038-x. [DOI] [PubMed] [Google Scholar]
- Espinoza FH, Farrell A, Nourse JL, Chamberlin HM, Gileadi O, Morgan DO. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Patrick G, Moffat J, Tsai LH, Andrews B. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc Natl Acad Sci USA. 1999;96:14445–14450. doi: 10.1073/pnas.96.25.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Jeffery DA, Anthony MD, O’Shea EK. Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol Cell Biol. 2001;21:6695–6705. doi: 10.1128/MCB.21.19.6695-6705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman PS, Hirst K, Goding CR. The activation domain of a basic helix-loop-helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO J. 1994;13:2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery DA, Springer M, King DS, O’Shea EK. Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80-Pho85 is semi-processive with site preference. J Mol Biol. 2001;306:997–1010. doi: 10.1006/jmbi.2000.4417. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massaguâe J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O’Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EACBM Newsl Protein Crystallogr. 1992;26 [Google Scholar]
- Madden SL, Johnson DL, Bergman LW. Molecular and expression analysis of the negative regulators involved in the transcriptional regulation of acid phosphatase production in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5950–5957. doi: 10.1128/mcb.10.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Huang D, Andrews B. Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog Cell Cycle Res. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–772. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Suzuki K, Fujino M, Oguchi T, Toh-e A. The Pho85 kinase, a member of the yeast cyclin-dependent kinase (Cdk) family, has a regulation mechanism different from Cdks functioning throughout the cell cycle. Genes Cells. 1999;4:627–642. doi: 10.1046/j.1365-2443.1999.00290.x. [DOI] [PubMed] [Google Scholar]
- Ogawa N, Noguchi K, Sawai H, Yamashita Y, Yompakdee C, Oshima Y. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Toh-e A. A novel mutation occurring in the PHO80 gene suppresses the PHO4c mutations of Saccharomyces cerevisiae. Curr Genet. 1992;21:95–99. doi: 10.1007/BF00318466. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- Poon RY, Lew J, Hunter T. Identification of functional domains in the neuronal Cdk5 activator protein. J Biol Chem. 1997;272:5703–5708. doi: 10.1074/jbc.272.9.5703. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996a;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nature struct biol. 1996b;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- Schneider KR, Smith RL, O’Shea EK. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Greer PL, Tsai LH. Cdk5 on the brain. Cell Growth Differ. 2001;12:277–283. [PubMed] [Google Scholar]
- Tarricone C, Dhavan R, Peng J, Areces LB, Tsai LH, Musacchio A. Structure and regulation of the CDK5-p25(nck5a) complex. Mol Cell. 2001;8:657–669. doi: 10.1016/s1097-2765(01)00343-4. [DOI] [PubMed] [Google Scholar]
- Toh-e A, Nishizawa M. Structure and function of cyclin-dependent Pho85 kinase of Saccharomyces cerevisiae. J Gen Appl Microbiol. 2001;47:107–117. doi: 10.2323/jgam.47.107. [DOI] [PubMed] [Google Scholar]
- Toh-e A, Tanaka K, Uesono Y, Wickner RB. PHO85, a negative regulator of the PHO system, is a homologue of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- Toh-e A, Ueda Y, Kakimoto SI, Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973;113:727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Toh-e A, Oshima Y. Isolation and characterization of recessive, constitutive mutations for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975;122:911–922. doi: 10.1128/jb.122.3.911-922.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.