Abstract
During translation termination, release factor protein catalyzes a hydrolytic reaction in the large subunit peptidyl transferase center to release the finished polypeptide chain. While the mechanism of catalysis of peptide release remains obscure, important contributing factors have been identified including conserved active site nucleotides and a GGQ tripeptide motif in the release factor. Here we describe pre-steady-state kinetic and nucleophile competition experiments to examine release factor contributions to the rate and specificity of peptide release. We find that while unacylated tRNA stimulates release in a non-discriminating manner, RF1 is very specific for water. Further analysis reveals that amino acid Q235 of the RF1 GGQ motif is critical for the observed specificity. These data lead to a model where release factors make two distinct contributions to catalysis – a relatively non-specific activation of the catalytic center and specific selection of water as a nucleophile facilitated by Q235.
Introduction
Termination of protein synthesis takes place when a stop codon on the mRNA positioned in the A site of the ribosome is recognized by a class 1 release factor protein to release the growing polypeptide chain. The release factor, like tRNA, is a bifunctional molecule that recognizes both the ribosomal decoding center and the peptidyl transferase center (PTC) to promote specific molecular events. In bacteria there are two class 1 release factors, RF1 and RF2, with overlapping specificity for the three stop codons, UAG, UGA and UAA. Upon stop codon recognition, the release factor catalyzes the hydrolysis of the peptidyl-tRNA ester bond in the active site of the large ribosomal subunit, releasing the completed polypeptide. While the release reaction is chemically similar to peptidyl transfer, it uses the weaker nucleophile, water, instead of the primary amine of the aminoacyl-tRNA. We are interested in understanding how these two very different bifunctional “adaptor” molecules, one composed of RNA and the other of protein, bind to and manipulate the same functional centers on the ribosome to achieve distinct functional outcomes.
Much recent biochemical and structural work focusing on the molecular mechanism of peptidyl transfer has concluded that the simpler peptidyl transfer (aminolysis) reaction depends (1) on a substrate-induced conformational rearrangement (referred to as induced fit) that positions the substrates for catalysis and (2) on a proton shuttle facilitated by the critically poised 2’OH of the peptidyl-tRNA substrate (reviewed in (Beringer and Rodnina, 2007b)). Evidence for the induced fit mechanism derives first from structural studies indicating that interaction between aminoacyl-tRNA and the ribosomal A loop triggers a series of conformational changes in the PT center that ultimately exposes the ester linkage of the P-site peptidyl-tRNA to nucleophilic attack (Schmeing et al., 2005b). Biochemical approaches have provided supporting evidence for the importance of these specific molecular interactions with the A loop for promoting catalysis in the active site (Beringer and Rodnina, 2007a; Brunelle et al., 2006; Kim and Green, 1999). Biochemical (Dorner et al., 2003), structural (Schmeing et al., 2005a) and computational (Das et al., 1999; Trobro and Aqvist, 2005) studies proposed the proton shuttle model for catalysis and the principal experimental evidence for the model derived from a study that evaluated the contribution of the 2’OH of the peptidyl-tRNA as worth at least 104-fold for catalysis (Weinger et al., 2004).
While mechanistic studies on the catalysis of peptidyl transfer have converged in recent years, relatively less progress in understanding the release reaction has been made, in large part because of the absence of high-resolution structural information on “release” complexes. Recent low resolution crystallographic and cryoEM structures of release complexes have confirmed biochemical and genetic studies suggesting that the universally conserved GGQ motif, found in both bacterial and eukaryotic release factors, is located within the peptidyl transferase center (Petry et al., 2005; Rawat et al., 2006; Rawat et al., 2003; Scarlett et al., 2003; Wilson et al., 2000). Such views suggest that the GGQ motif may be close enough to the catalytic reaction to contribute in some active and direct way to catalysis. It is also possible that interactions between the A loop and release factor simply promote conformational changes in the PTC, akin to those promoted by tRNA, to facilitate the catalytic reaction. These latter ideas are consistent with early experiments demonstrating that addition of unacylated tRNA to the ribosomal A site can stimulate peptide release (Caskey et al., 1971), as well as more recent studies indicating that this stimulation can amount to 300-fold under certain conditions (Zavialov et al., 2002). These data argue that RFs may function in large part by simply promoting conformational rearrangements in the PTC that open the peptidyl-tRNA substrate to nucleophilic attack.
The weaker nucleophilicity of water relative to the primary amine of an amino acid makes the release reaction more chemically challenging and thus more likely to depend on sophisticated enzyme mechanisms. Consistent with this apparent chemical challenge is the observation that mutation of the most proximal active site nucleotide residues (A2451, U2506, U2585, and A2602) underscores their importance for RF-mediated peptide release but not for the simpler peptidyl transfer reaction (Polacek et al., 2003; Youngman et al., 2004). The universally conserved GGQ tripeptide motif is known to be critical to in vivo function and to make substantial contributions to catalysis in vitro (Frolova et al., 1999; Mora et al., 2003; Song et al., 2000). In particular, the central glycine of the GGQ motif, when substituted with alanine (GAQ), has been shown to slow the rate constant for peptide release by four orders of magnitude (Zavialov et al., 2002). Mutation of the glutamine residue to alanine (GGA) or glutamate (GGE) had surprisingly modest effects on catalysis in different studies (4-fold effect on release under kcat/KM conditions (Mora et al., 2003) and 100-fold under kcat conditions (Mora et al., 2003; Seit-Nebi et al., 2001; Zavialov et al., 2002). In both bacteria and eukaryotes the glutamine of the GGQ motif is post-translationally methylated at the N5 position (Dincbas-Renqvist et al., 2000; Heurgue-Hamard et al., 2005). In bacteria, disruption of the methylase gene results in a slow growth phenotype in vivo but is not lethal (Nakahigashi et al., 2002). Moreover, in in vitro biochemical assays, loss of methylation at the glutamine residue has relatively modest (10-fold for RF2 and no effect for RF1) consequences for catalysis (Dincbas-Renqvist et al., 2000).
Here we examine the effects of a large number of mutations in the GGQ region of the class I release factor in the context of a nucleophile partitioning analysis. These studies reveal two distinct components of release factor function, one dependent on a general activation of the catalytic center and the other dependent on specific selection of water by the highly conserved glutamine of the GGQ motif.
Results
RF1 mutagenesis and effects on peptide release activity
To address the role of RF1 active site amino acids on the rate of peptide release, we made a series of alanine mutations in and surrounding the invariant GGQ motif. We chose to target amino acids that are conserved among bacterial RFs, particularly those with interesting chemical properties that may contribute to catalysis (Fig 1A). The effect of these mutations on peptide release catalysis was determined using a pre-steady-state kinetic assay which follows the release of f-[35S]-Met from a ribosome complex carrying initiator f-[35S]-Met-tRNAfMet in the P site and a model mRNA carrying a Shine-Dalgarno sequence, an AUG start codon in the P site and a UAA stop codon in the A site. In other experiments, the use of more “termination like” complexes carrying a dipeptidyl-tRNA in the P site showed an overall decrease in the rate of release from the ribosome complex thus prompting us to use the simpler fMet release system (E.Y. and R.G., unpublished observations), though we note that other studies have documented suboptimal release behavior with such complexes (McCaughan et al., 1998). Complexes were formed in a buffer similar to one previously used for our extensive characterization of the peptidyl transfer reaction (Youngman et al., 2004). The peptide release reactions were carried out at 37°C. Reactions were started with addition of 5 μM RF1, far exceeding the known Kd of 47 pM for wild type RF1 binding to such ribosome complexes (Zavialov et al., 2002). Aliquots were removed and various time points were quenched in 25% formic acid. The reaction products were resolved by electrophoresis on cellulose TLC plates. The data were quantitated and fit to single-exponential curves to determine the rate constants for peptide release. For any mutants that exhibited rate defects, RF titrations were performed to ensure saturation conditions so that binding defects were not analyzed.
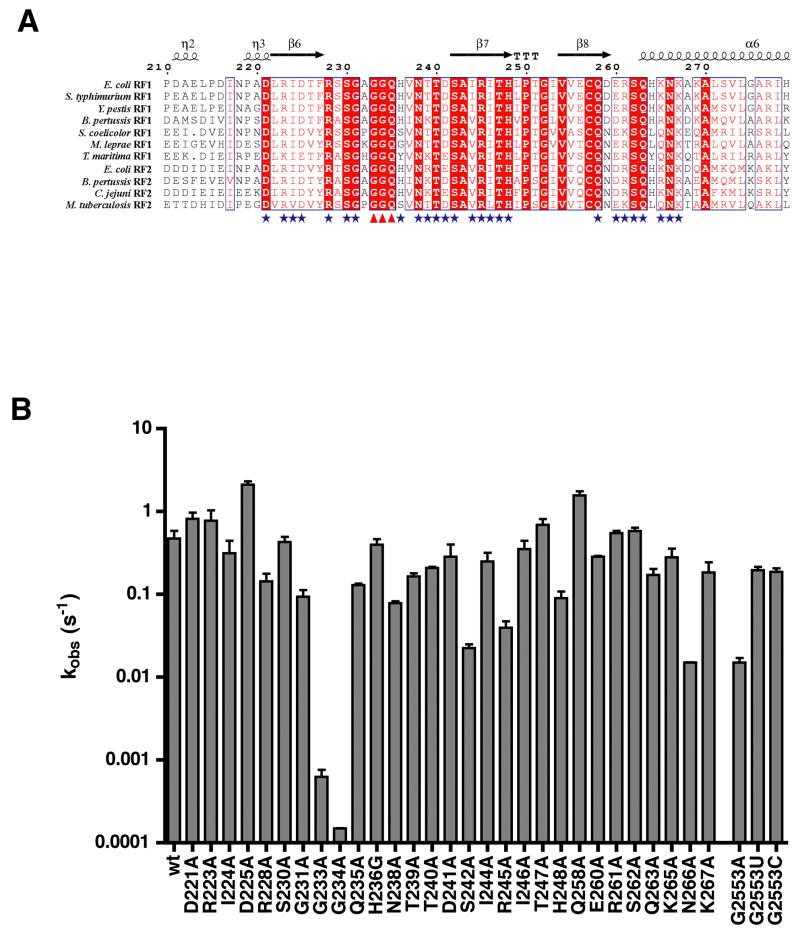
Figure 1. Mutations outside the RF GGQ (G233-Q235) motif have modest rate defects.
(A) Alignment of the GGQ domain of bacterial release factors RF1 and RF2 created with ESPript (http://espript.ibcp.fr/) showing conserved amino acids (boxed in red) and amino acids with conserved properties (in red lettering). Domain structural elements are shown above the alignment. Amino acids mutated in this study are indicated with a blue star and the mutated GGQ motif is indicated with red triangles.
(B) Log-scale graph showing the measured rate constants for hydrolysis (kobs (s−1)) for each RF1 and G2553 ribosome mutant. Rate constant for N266A was measured with 25 μM RF whereas all other measurements were carried out with 5 μM RF. Error bars represent the standard error in the measured rate constant from at least two experiments.
Wild-type RF1 catalyzed peptide release with a rate constant of 0.5 s−1, consistent with rates reported previously (Zavialov et al., 2002). Among the RF1 variants tested, G233A and G234A of the GGQ motif had by far the greatest defects in peptide release, with rate constants of 6.3 × 10−4 s−1 and 1.5 × 10−4 s−1, respectively (Fig 1B). These 800 to 3300-fold decreases in release rate are consistent with the documented importance of the GGQ glycine residues on peptide release by RF1 (Zavialov et al., 2002). Other mutations in the region of the GGQ motif, by contrast, had relatively little effect on the peptide release rate (ranging from 1- to 6-fold). Notably, mutant Q235A of the GGQ motif had only a 4-fold effect on the rate constant for release, consistent with one earlier report (Mora et al., 2003). Some intermediate defects of 13 to 33-fold were associated with mutations in two conserved amino acids in β-strand 7 (S242A 0.022 s−1 and R245A 0.040 s−1), and one mutation in α-helix 6 (N266A 0.015 s−1), indicating that these positions may be sites of interaction with the ribosome or may be important for the conformation of the RF protein. Interestingly, two of the mutants, D225A and Q258A, had release rate constants 3 to 4-fold faster than that of wild type.
A loop mutagenesis and effects on peptide release activity
Biochemical and structural studies recently provided evidence for a conformational rearrangement that activates the large subunit catalytic center for peptidyl transfer (Schmeing et al., 2005b). This rearrangement is thought to depend critically on the formation of specific pairing interactions between the CCA-end of the incoming aminoacyl-tRNA and the universally conserved nucleotide G2553 of the A loop of the 23S rRNA. It seems likely that the peptide release reaction promoted by the RFs might depend on similar rearrangements in the active site to activate the peptidyl-tRNA for hydrolysis. To investigate the possibility that interaction between the release factors and the A loop contributes to peptide release we determined the peptide release rates of ribosomes carrying mutations at G2553. Ribosome variants were co-expressed with wild type in E. coli from a plasmid-borne version of 23S rRNA and purified using our previously described affinity-purification procedure (Youngman and Green, 2005). The G2553A mutation had a 33-fold defect in the rate constant for peptide release, while the other two nucleotide substitutions, G2553C and G2553U, had near wild type rate constants of 0.2 s−1 (Fig 1B). These data are comparable to our earlier unpublished data describing similar defects associated with these mutations for the peptidyl transfer reaction (J.B. and R.G., unpublished observations).
Specificity of release activity assessed with nucleophile partitioning
We next performed a nucleophile partitioning experiment to test whether RF-mediated peptide release on the ribosome depends on specific interactions with water by the RF or the ribosome that serve to promote catalysis. By using exogenous nucleophiles that can effectively compete with H2O in the release reaction, one can determine the specificity of the reaction relative to the inherent nucleophilicity of the compounds in solution. For these partitioning experiments, we chose hydroxylamine, a small molecule known to be a potent nucleophile with a pKa in the physiological range. As above, ribosome complexes were prepared containing an initiator f-[35S]-Met-tRNAfMet in the P site and a model mRNA that places a UAA stop codon in the A site. A 5 M stock solution of hydroxylamine (HA) was adjusted to pH 7.5 with HCl, above the known pKa of 6.0 at 25°C (Robinson and Bower, 1961). We then identified conditions where HA effectively competes with water in solution (55 M) at pH 7.5 to release the peptidyl moiety from tRNA. Incubation of f-Met-tRNAfMet with HA yielded distinct products readily resolved by electrophoretic TLC (Fig 2A). We see a single species that represents the aminolysis product and two faster moving species that represent the hydrolysis product. The doublet nature of the hydrolysis product depends on the exact conditions of the electrophoretic TLC, consistent with multiple oxidation states of methionine compounded with salt front phenomena. While both the amine and the hydroxyl moieties of hydroxylamine could in principal function as nucleophiles to release the peptide from the tRNA, the mobility of the HA-mediated release product at pH 2.8 is consistent with it representing the aminolysis product. The use of the amine rather than the hydroxyl nucleophile is consistent with the documented nucleophilicity of this moiety due to the alpha effect of the neighboring non-bonded electrons (Edwards and Pearson, 1962; Jencks and Carriuolo, 1960).
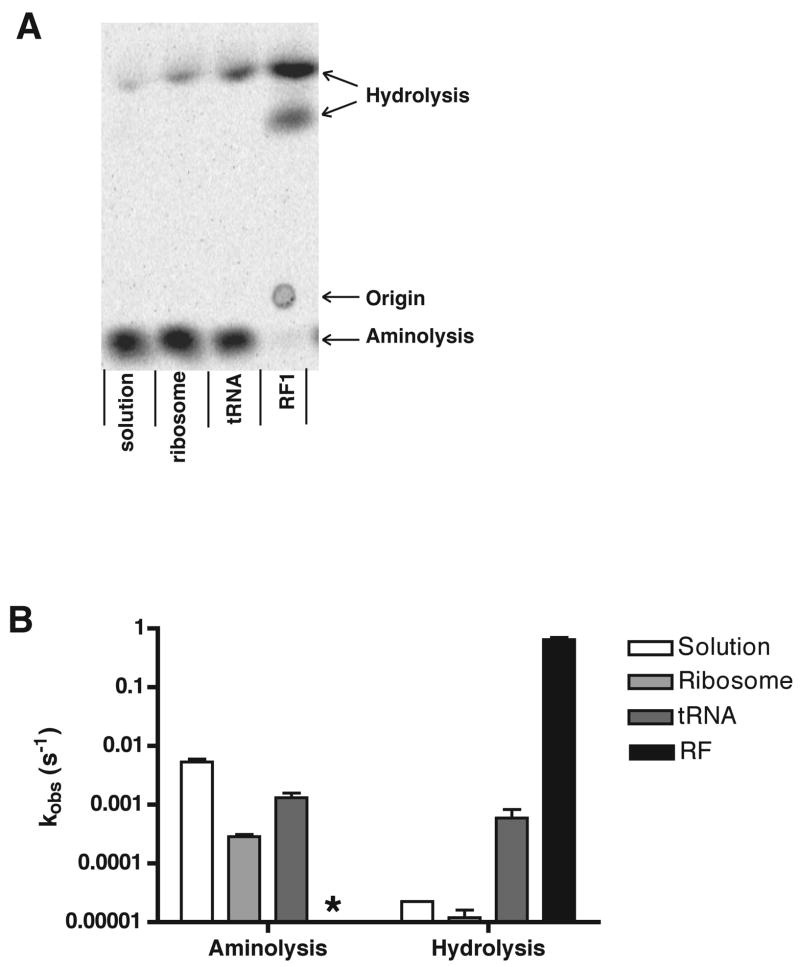
Figure 2. Peptide release catalyzed by RF1 exhibits high specificity for water.
(A) Electrophoretic TLC plate with endpoints of a nucleophile partitioning assay with peptidyl-tRNA; free in solution, bound to the P site of the ribosome, on the ribosome with an unacylated tRNA in the A site or on the ribosome with RF1 in the A site.
(B) Log-scale plot of observed rate constants of aminolysis and hydrolysis in a partitioning assay with 1 M hydroxylamine. Rate constants were determined by multiplying kobs by the endpoint of the individual aminolysis or hydrolysis reaction respectively. Data are presented as mean +/− standard error. The hydrolysis rate in solution had a very low endpoint, but its measurement could be verified by comparison to the hydrolysis rate in the absence of hydroxylamine. In the reaction with release factor, no aminolysis product was observed and thus the rate could not be determined (*).
Endpoints for release of f-[35S]-Met from the aminoacylated tRNA were measured in solution under a range of concentrations of HA – 100 mM HA yielded 79% aminolysis and 1 M HA yielded >99% aminolysis. As with any bifurcated reaction, the endpoint of the reaction is equivalent to k1/k1+k2 (where k1 will be kobs-HA and k2 will be kobs-water). Independent measurement of kobs-water for each complex (in the absence of added HA) allows for confirmation of kobs-water measured under conditions where the predominant product derives from HA-mediated aminolysis (thus making direct measurement of kobs-water challenging).
The partitioning assay was next used to evaluate the contributions made by the ribosome and the RF to the specificity of the release reaction. To do this, we first looked at the rates of hydrolysis and aminolysis when f-Met-tRNA was bound in the P site of the ribosome and either a stop or sense codon was found in the A site (but where no A-site ligand was bound). In both complexes, binding of peptidyl-tRNA to the ribosome (either stop or sense coding) modestly protects it from nucleophilic attack by ca. 3- to 20-fold. As in solution, at the chosen concentration of 1 M HA, the aminolysis reaction dominates, proceeding at 2.9 × 10−4 s−1, while the hydrolysis reaction proceeds at an observed rate of 1.2 × 10−5 s−1 (Fig 2B).
We next added a near-saturating concentration of unacylated tRNAPhe to a ribosome complex programmed with a UUU codon in the A site and examined the rates of aminolysis and hydrolysis. In this case, binding of the tRNA to the A site stimulates both the hydrolysis reaction by 50-fold and the aminolysis reaction by 5-fold, consistent with previous reports (Caskey et al., 1971; Zavialov et al., 2002). In a final experiment, RF1 was added to the UAA-programmed ribosome complexes and rates of aminolysis and hydrolysis were observed. In this case, hydrolysis of the peptidyl tRNA was dramatically stimulated (53000-fold) such that no aminolysis product is observed over background. These constraints place an upper limit of 50-fold on the stimulation of aminolysis by RF1 on the ribosome (based on a 2% upper limit for the aminolysis endpoint), though the value could be considerably lower. These data indicate that RF1 has at least a 1000-fold preference for water relative to HA. These results suggest that the interaction of RF with the ribosome complex makes specific contributions to catalysis with water that are distinct from a general activation of the catalytic center.
Molecular contributors to water specificity during RF-mediated release on the ribosome
The nucleophile partitioning experiments provide us with strong evidence that the release factor:ribosome complex exhibits a strong preference for water as the nucleophile in the release reaction. To identify potential contributors to this specificity, we asked whether mutation of any of the known components (ribosome, RF or peptidyl-tRNA) leads to an observable loss in specificity. The molecular signature of such a loss of specificity is predicted to be an increased level of aminolysis in the RF-catalyzed release reaction. We began by screening various mutants in RF1 and the ribosome active site to look for enhanced levels of aminolysis in the presence of 1 M HA. In a simple endpoint assay, several variants emerged exhibiting strikingly increased levels of aminolysis. All three alanine substitutions in the GGQ motif (Fig 3A left) yielded levels of aminolysis visible above background. We next evaluated several ribosome variants with mutations in the active site of the large subunit and in two cases, A2602C and U2585C, we observed a slight increase in the amount of aminolysis relative to wild type levels (Fig 3A right). The 2’OH of the peptidyl-tRNA substrate, which is known to be critical for peptidyl transfer (Weinger et al., 2004), was also tested for effects on release specificity. We tested its involvement by forming release complexes carrying a dipeptidyl tRNA in the P site (fMet-Lys-tRNALys) with either a normal ribose group at the A76 position or a deoxy substition. We then evaluated the effects of the deoxy substitution on hydrolysis and aminolysis and found that it did not affect the specificity of the reaction (data not shown).
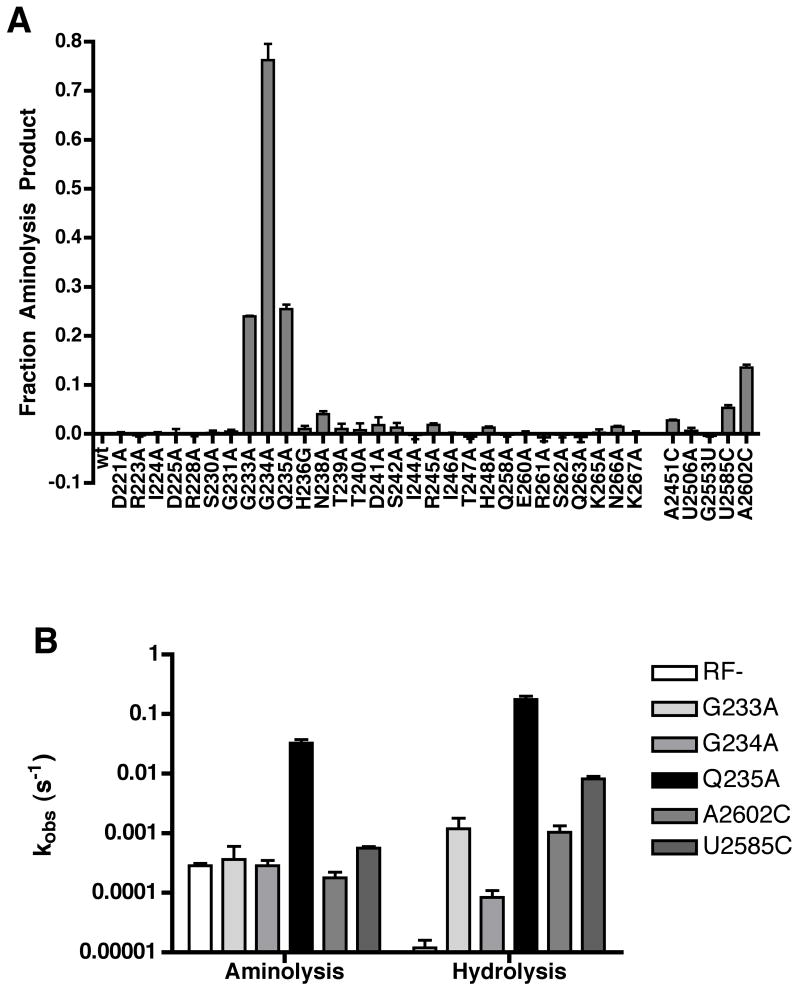
Figure 3. Mutation of Q235 in RF1 is associated with loss of specificity for water.
(A) Nucleophile partitioning assay endpoints with RF1 and ribosome active site mutants. Incubation times varied for the different mutants to ensure that the endpoint of the reaction was measured (slow variants needed longer time points). The endpoint was defined as the fraction of aminolysis product to total release product and was normalized to zero for background aminolysis in the wild type reaction. Data are presented as the mean +/− standard error of at least two measurements.
(B) Observed rate constants of aminolysis and hydrolysis for RF1 and ribosome mutants that exhibit increased aminolysis endpoints in a nucleophile partitioning assay presented on a log-scale plot. Rate constants were calculated by multiplying kobs by the endpoint of the individual aminolysis or hydrolysis reaction respectively. Each rate constant was measured at least twice and is presented as the mean +/− standard error.
We noticed that a number of variants identified in the endpoint assay in Fig 3A were known to be deficient in their rate of catalysis (G233A, G234A, A2602C and U2585C) (Fig 1) (Mora et al., 2003; Polacek et al., 2003; Youngman et al., 2004; Zavialov et al., 2002). We further realized that with such slow variants, the background rate of aminolysis could contribute to the observed aminolysis. In this case, such variants would be red herrings in a search for specificity defects. This possibility was evaluated by looking at the actual rates of aminolysis and hydrolysis for each of the variants displaying increased levels of aminolysis in the endpoint assay. Interestingly only the Q235A variant displayed an increase in the observed rate of the aminolysis reaction (115-fold) whereas all other variants simply displayed defects in the rate of hydrolysis relative to the wild type (Fig 3B). These data together identify Q235 as uniquely associated with the specificity of the release reaction for water.
Q235 side chain size is a critical determinant of RF1 specificity
To further explore the role of Q235 in peptide release specificity we prepared a series of mutations at position Q235 of RF1 and tested each variant for overall activity and for specificity defects in the partitioning assay. Two of the RF1 mutants, Q235N and Q235D, had drastically reduced release activity with defects of 7500 to 9500-fold, comparable to the defects of the GAQ variant above. Most of the variants, however, had more modest defects in overall catalysis ranging from 2 to 75-fold (Fig 4A). As for specificity, there appears to be a correlation between shortening of the side-chain of the amino acid at this position and a loss of specificity for water. Beginning with the serine substitution (GGS), we begin to see a visible increase in the amount of aminolysis product produced in an endpoint assay, and this product becomes even more abundant with the GGA and GGG variants (Fig 4B). As above, measurement of the observed rates of aminolysis and hydrolysis are critical for evaluating actual changes in specificity (as opposed to simple losses in activity) of the various mutants. The rate analysis presented in Fig 4C indicates that three of the ten Q variants (Q235S, Q235A and Q235G) result in losses in specificity for water in the RF-mediated release reaction. These three variants are relatively active overall (4 to 30-fold down) and substantially accelerate the aminolysis reaction (15 to 170-fold) (Table 1). It is important to note that the greatest losses of specificity are not associated with the least active variants per se. For example, the Q235L and Q235T variants are compromised in their release activity, but do not yield elevated levels of aminolysis, suggesting that the aminolysis reaction is similarly diminished. The Q235N and Q235D variants, which are very compromised in their hydrolysis activity, are also slow in the aminolysis reaction compared to the minus RF control (by about 25-fold).
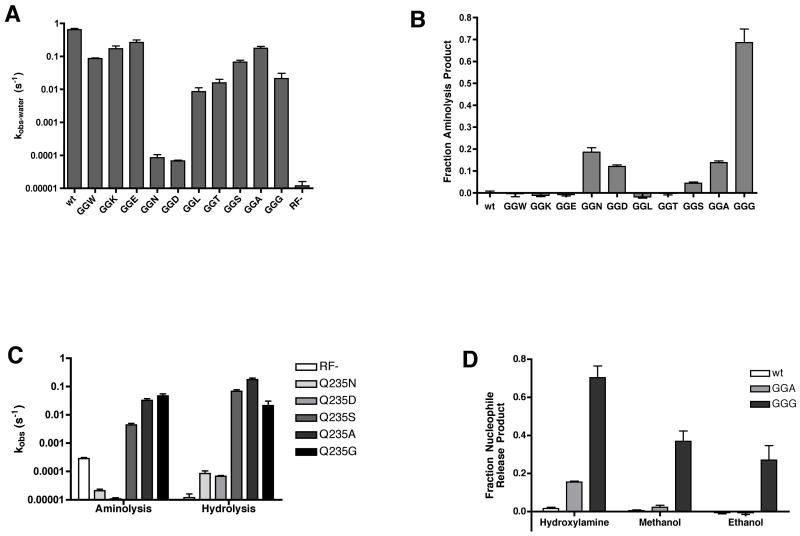
Figure 4. Mutations at Q235 reveal a correlation between side-chain size and water specificity.
(A) Hydrolysis rates of RF1 Q235 mutants presented on a log-scale plot. For mutants where detectable aminolysis product was observed (GGN, GGD, GGS, GGA, and GGG) the hydrolysis rate constant (kobs-water) was calculated from kobs × hydrolysis endpoint. For all other mutants, kobs = kobs-water. Rate constants were measured at least twice and are presented as mean +/− standard error.
(B) Endpoints of aminolysis reactions with a series of RF1 Q235 mutations. Endpoints were calculated as the fraction of aminolysis product to total release product, normalized to the wild type background aminolysis, and are presented as the mean +/− standard error of at least two measurements.
(C) Log-scale plot of rate constants for aminolysis and hydrolysis of RF1 Q235 mutants that exhibit increased aminolysis endpoints. The rate constants for aminolysis and hydrolysis are calculated from their respective endpoints multiplied by kobs. The mean +/− standard error of at least two measurements are presented.
(D) Aminolysis or alcoholysis endpoints of wild type, Q235A and Q235G RF1 measured in the presence of 1 M hydroxylamine, 10% methanol or 10% ethanol. Endpoints are measured as the fraction of aminolysis or alcoholysis product to total release product and presented as the mean +/− standard error of at least three measurements.
Table 1.
Rate constants for hydrolysis and aminolysis derived from nucleophile partitioning experiments
| Kobs-HA (s−1) | Fold increase (HA) | Kobs-water (s−1) | Fold increase (water) | Fold RF defect | |
|---|---|---|---|---|---|
| Solution | 5.3×10−3 ± 0.7×10−3 | 19 | 2.3×10−5 | 2 | |
| Ribosome | 2.9×10−4 ± 0.2×10−4 | 1.2×10−5 ± 0.4×10−5 | |||
| tRNA | 1.3×10−3 ± 0.3×10−3 | 5 | 5.9×10−4 ± 2.3×10−4 | 50 | |
| RF1 | cnd | 0.65 ± 0.06 | 53000 | ||
|
| |||||
|
Ribosome Mutants
| |||||
| A2602C | 1.8×10−4 ± 0.4×10−4 | 1 | 1.0×10−3 ± 0.3×10−3 | 85 | 620 |
| U2585C | 5.6×10−4 ± 0.3×10−4 | 2 | 8.2×10−3 ± 0.9×10−3 | 670 | 79 |
|
| |||||
|
RF1 Mutants
| |||||
| G233A | 3.6×10−4 ± 2.4×10−4 | 1 | 1.2×10−3 ± 0.6×10−3 | 97 | 540 |
| G234A | 2.9×10−4 ± 0.6×10−4 | 1 | 8.4×10−5 ± 2.6×10−5 | 7 | 7700 |
| Q235W | cnd | 0.086 ± 0.003 | 7000 | 7 | |
| Q235K | cnd | 0.17 ± 0.04 | 14000 | 4 | |
| Q235E | cnd | 0.27 ± 0.05 | 22000 | 2 | |
| Q235N | 2.1×10−5 ± 0.3×10−5 | −14 | 8.5×10−5 ± 2.0×10−5 | 7 | 7600 |
| Q235D | 1.1×10−5 ± 0.1×10−5 | −26 | 6.8×10−5 ± 0.4×10−5 | 6 | 9500 |
| Q235L | cnd | 0.0086 ± 0.0026 | 700 | 75 | |
| Q235T | cnd | 0.016 ± 0.004 | 1300 | 41 | |
| Q235S | 4.4×10−3 ± 0.6×10−3 | 15 | 0.068 ± 0.009 | 5500 | 10 |
| Q235A | 3.3×10−2 ± 0.5×10−2 | 113 | 0.18 ± 0.02 | 14000 | 4 |
| Q235G | 4.7×10−2 ± 0.8×10−2 | 165 | 0.021 ± 0.009 | 1700 | 30 |
cnd = could not determine
To extend these analyses, we asked whether the most permissive variants, Q235A and Q235G, are also permissive with respect to other nucleophiles. The results shown in Fig 4D indicate that the Q235G variant is the most permissive, allowing for substantial amounts of methanolysis and ethanolysis in the presence of 10% methanol or ethanol in solution. Other nucleophiles that were tested either resulted in overall inactivation of the ribosome or release factors (hydrazine, ethanolamine), or had a pKa too far from the physiological range to effectively compete with water at pH 7.5 (methylamine, ethylamine, ammonium).
Discussion
The mutational analysis of release factor function presented here provides new insight into the mechanism of peptide release on the ribosome. In a search for catalytically important amino acid residues on RF1, we found little evidence that elements outside of the broadly conserved GGQ motif contributed substantially to catalysis under kcat conditions. As previously reported, mutations of the glycines (G233 and G234) in the GGQ motif resulted in dramatic losses in catalytic activity -- 800-fold for the AGQ variant and 3300-fold for the GAQ variant -- while mutation of the universally conserved Q235 of RF1 had surprisingly modest (4-fold) effects. Our extended analysis of the region did identify several other residues having intermediate effects (10 to 30-fold) on the rate of peptide release. Two of these variants (S242A and R245A) are found in β strand 7 which closely approaches the GGQ motif while a third (N266A) very closely approaches S242 in the X-ray structure, suggesting that interaction between these two regions might be critical to release factor function.
We note that several variants (D225A and Q258A) had release activity that was slightly faster (3 to 4-fold) than wild type. The identification of fast variants with mutations in a region somewhat remote from the active site argues that the assay is at least partially rate limited by something other than chemistry, but not binding as reactions were carried out at saturation. It has been speculated that conformational changes could be rate limiting for release given that the conformation of isolated RF1 observed by X-ray crystallography is strikingly different than in its extended ribosome-bound state (Petry et al., 2005; Rawat et al., 2006; Shin et al., 2004; Vestergaard et al., 2005; Vestergaard et al., 2001; Zoldak et al., 2007). Recent molecular dynamics studies have argued that conserved histidine residues in the RF might be critical in mediating such proposed conformational rearrangements (Ma and Nussinov, 2004). Interestingly, one of the two fast RF1 variants, Q258A, found in this study is located in the interface of the two flexible release factor domains. Further experiments will be required to define such potential intermediates in the RF functional cycle.
The identification of a very small region of the RF protein with chemically inert amino acid moieties (glycine) as the critical component for catalysis substantially limits the number of mechanisms that must be considered. It seems likely that the critical glycines are important in allowing a tight turn of the loop of the release factor, thus facilitating precise binding of this region in the catalytic center. The large effects on catalysis resulting from the substitution of even an alanine at these positions are consistent with recent molecular dynamics simulations arguing for critical packing interactions facilitated by the extremely tight turn in the protein backbone in this region (Trobro and Aqvist, 2007). Disruption of the active site by the glycine substitutions could in turn also perturb rRNA elements known to be critical (Polacek et al., 2003; Youngman et al., 2004) or the peptidyl-tRNA substrate (and its catalytically essential 2’OH, J. Brunelle and R.G. unpublished data) in such a way as to diminish catalysis.
If release factors interact with the PTC to facilitate productive reorganization of the catalytic center, it might be predicted that they would trigger the same rearrangements as tRNA via interactions with the A loop. This idea was tested by looking at the effects of nucleotide substitutions at G2553 of the A loop on release factor mediated catalysis. The effects of these mutations were relatively modest, but in retrospect are generally consistent with the defects observed in the related peptidyl transfer reaction (J.B., E.Y, R.G. unpublished data). It is possible that nucleotide substitutions are so similar in their overall properties as to minimize potential effects on function, thus explaining the absence of strong effects on either peptidyl transfer or release. It is also possible that the RF utilizes distinct “trigger” points from tRNA to facilitate productive conformational rearrangements that we have not probed in this study. Further biochemical studies will be needed to elucidate a role, if any, for the A loop in peptide release.
We used hydroxylamine as a competitor nucleophile to try to decipher molecular contributors to the RF-mediated release reaction. We found that release of the peptide from the ribosome-bound tRNA could be promoted by nucleophiles other than water with essentially no selectivity for water both in the very slow background reaction (where the A site is empty) as well as in the slightly faster tRNA-promoted release reaction (Caskey et al., 1971; Zavialov et al., 2002). The non-specificity of these RF-independent events are easily reconciled with a number of early studies showing the peptidyl transferase center to be quite non-specific during the peptidyl transfer reaction (Lieberman and Dahlberg, 1995) and with recent structural and biochemical studies arguing that an induced fit mechanism can explain much of catalysis in this active site (Schmeing et al., 2005b). These data might also be anticipated based on the knowledge that this ribosomal active site must catalyze two distinct reactions during translation, the nucleophile specificity being determined by the incoming A-site substrate.
It is striking that the release reaction promoted by RF1 binding in the A site is highly specific for water. In the presence of 1 M hydroxylamine, we could detect no aminolysis product. This result allows us to provide a lower limit estimate that the RF is at least 1000-fold specific for water relative to HA, and is potentially even more specific. This observation suggests that the release factor interacts with the ribosomal active site in such a way as to exclude other nucleophiles from the reaction, possibly through steric clash with nucleophiles larger than water, or through specific contacts with the water molecule to promote catalysis.
Our analysis of various mutant RFs using the nucleophile partitioning analysis has provided us with substantial insight into the apparent specificity of the RF-mediated release reaction for water. Several substitutions at Q235 yielded RF variants that have a clear loss in specificity for water. The serine, alanine and glycine substitutions had relatively modest effects on the rates of hydrolysis but resulted in substantial stimulation of the observed rate of aminolysis. The GGG variant was of particular interest since the relative ratio of aminolysis to hydrolysis products that it generated was very similar to that generated by deacylated tRNA binding to the A site of the ribosome. While the rates of release promoted by the two different A-site substrates are quite different (the GGG variant is 35-fold more stimulatory than tRNA), they both “open up” the active site of the ribosome to nucleophiles in a similar way that is largely non-discriminatory and simply reflects the relative nucleophilicity of water and hydroxylamine under the chosen conditions. This variant thus provides the clearest evidence for the idea that release factors make two distinct contributions to catalysis. The first contribution of the RF is to simply turn on the active site, potentially in a fashion related to the induced fit rearrangement stimulated by tRNA binding. We note that in the absence of high-resolution structural data it is impossible to determine whether the two activated states are equivalent or whether distinct mechanisms of activation are used by tRNAs and RFs. Based on the GGG RF variant, we estimate the contribution of general activation to be worth at least 1750-fold (comparing the GGG rate of hydrolysis to the minus RF rate on the ribosome). The fact that this rate enhancement is larger than that observed simply with peptidyl-tRNA free in solution is explained by the fact that ribosome:RF complex likely optimally constrains the ester linkage for attack. The second contribution of the RF is to the specificity of the reaction, helping to exclude larger nucleophiles by specifically positioning water in an ideal position for catalysis. The GGG, GGA and GGS variants thus behave essentially as loss of function mutants for the specificity component of catalysis, while maintaining the activation component of the RF.
We are particularly intrigued by the Q235N and Q235D RF1 variants which are substantially deficient in catalysis of both the hydrolysis and the aminolysis reactions. We note that the Q235E variant which simply substitutes an oxygen moiety for the amine functionality of the glutamine (to create the acid functionality) is quite an active variant, similar to previous reports (Mora et al., 2003; Seit-Nebi et al., 2001), suggesting that the carbonyl moiety might be most critical in mediating interactions with water for catalysis. Such a role for the carbonyl function was initially proposed in X-ray studies of the isolated eRF1 (Song et al., 2000) as well as in more recent molecular modeling studies (Trobro and Aqvist, 2007). It is possible that the strong negative effects of the N and D substitutions derive from the fact that these variants are very similar in structure to the Q and the E variants, but are a single carbon shorter. Perhaps their side chains hydrogen-bond to and orient water, like their Q and E cousins, but the oriented water in these cases is too remote from the labile ester bond to be involved in catalysis, but is close enough to prevent hydroxylamine from entering the site at all. Interestingly, the relatively large side chains W, K and L have modest effects on the hydrolysis rate and do not appear to result in a loss of specificity in the release reaction since we see no aminolysis product. Perhaps these large side-chains, which are similar in size to the natural methylated glutamine at this position, are able to maintain a reasonably tight pocket for water despite the lack of the putative orienting side-chain moiety. This specific role for the RF in providing a tight pocket for orienting water may also help to explain the previously observed defects in release activity associated with ribosome active site mutations (Polacek et al., 2003; Youngman et al., 2004). Subtle changes in the heterocyclic base in the active site could have substantial effects on the tightness of the pocket, thus making the untethered water substrate (as opposed to the tethered amino acid on aminoacyl-tRNA) sub-optimally positioned for catalysis.
Our results help to rationalize the observed universal conservation of the GGQ motif in evolutionarily unrelated and strikingly different structural contexts in bacterial and eukaryotic RFs and the fact that mutations at these three positions are not tolerated in vivo (Mora et al., 2003). The glycines of the GGQ motif are critical for efficient catalysis, resulting in major catalytic defects when mutated. The glutamine of the GGQ motif is critical in facilitating the water specific reaction as manifested in our partitioning experiments. And, while Q235 can be substituted in vitro with relatively minor effects on overall catalytic efficiency, such effects may well be too detrimental in vivo. We further note that Q235 may make more substantial contributions to catalysis than observed in our assay given that a rate-limiting step other than chemistry may be being followed. Our results reveal two distinct roles for the RF in catalyzing peptide release, a general activation of the catalytic center and a contribution to the specificity for water, thus accounting for much of the documented rate enhancement accomplished by release factors on the ribosome (Fig. 5). Such an understanding of RF function on the ribosome helps draw mechanistic connections to the likely more evolutionarily ancient function of this active site in catalyzing tRNA-activated peptidyl transfer.
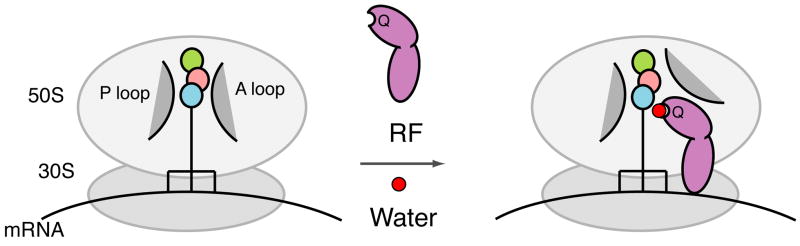
Figure 5. Model depicting two molecular contributions to release catalysis by RF1.
P-site bound tRNA carrying a tripeptide (colored circles) is protected in the absence of an A-site substrate by ribosomal RNA including the A and P loops (Schmeing et al., 2005b). RF1 (lavender) contributes to catalysis of peptide release by 1) “opening up” the ribosomal active site to allow solvent entry and 2) specific activation of a water molecule (red) controlled by the GGQ glutamine (Q).
Experimental Procedures
Purification of RF1 mutants and ribosomes
Site-directed mutagenesis of pET15b-RF1 vector was carried out using Quik Change XL (Stratagene) to create mutant plasmids. These plasmids encoding N-terminally His-tagged RF1 were transformed into BL21 DE3 pLysS cells, expression was induced and protein was purified on a 5 ml His-Trap FF FPLC column (GE Healthcare). Purified RF1 was dialyzed against storage buffer (30 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 5 mM β–mercaptoethanol, 50% glycerol) and stored at −20°C.
E. coli MRE600 (ATCC 29417) was grown in LB to an A600 of 0.6–0.8, pelleted and resuspended in buffer A (20 mM Tris-HCl pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA and 6 mM β-mercaptoethanol), and the cells were cracked using a French press. Clarified lysates were pelleted over sucrose cushions (1.1 M sucrose, 20 mM Tris-HCl [pH 7.5], 500 mM NH4Cl, 10 mM MgCl2 and 0.5 mM EDTA) in a Beckman Ti-45 rotor for 16h at 37000 rpm. and resuspended in buffer A. Samples were loaded onto a 10–40% sucrose gradient and separated by zonal centrifugation in a Ti-15 rotor for 19h at 28000 rpm. 70S ribosome peaks were pelleted and resuspended to 15 μM in buffer A and stored at −80°C. MS2-tagged ribosome mutants were expressed in DH10 cells as described previously (Youngman et al., 2004; Youngman and Green, 2005).
Formyl-Methionine Release Assay
To assay peptide release, complexes containing wild type ribosomes, f-[35S]-Met-tRNAfMet and mRNA with AUG and UAA codons in the P and A sites, respectively, were formed as described previously in a buffer containing 50 mM Hepes (pH 7.5) instead of Tris-HCl (Buffer B, 50 mM Hepes, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 1 mM DTT) (Youngman et al., 2004). For reactions with tagged mutant ribosomes, purified MRE600 30S subunits were added to a final concentration of 0.125 μM to compensate for loss of 30S subunits during purification. Complexes (0.2 μM) were reacted with a final concentration of 5 μM RF1 wild type or variant (25 μM in the case of N266A) at 37°C. Ribosome release complexes and release factor were reacted on a quench-flow apparatus (RQF-3 quench-flow, KinTek corporation), and the reactions were quenched in 25% formic acid. Slower reactions were carried out on the bench-top. Time points were spotted on TLC-cellulose plates and f-[35S]-Met was separated from f-[35S]-Met-tRNAfMet by electrophoresis at 1250 V for 30 min in pyridine acetate buffer (pH 2.8). The fraction of f-[35S]-Met released was quantitated by Phosphorimager. For each reaction the background fraction of f-[35S]-Met released in the absence of RF1 was measured at four time points and the average was subtracted from each time point in the RF1 reaction. Curves were fit to single exponential equations to obtain a rate constant for the reaction.
Release Specificity Assay
Release complexes for specificity experiments were formed as above with either UUU or UAA codons in the A site. Complexes were pelleted over the 1.1 M sucrose cushion described above for 2 h at 69000 rpm in a Beckman TLA-100.3 rotor and pellets were resuspended in buffer B. A 5 M hydroxylamine stock solution was adjusted to pH 7.5 with concentrated HCl. Hydroxylamine was added to a mix containing Buffer B and the A-site substrate (RF1 or deacylated tRNA) to yield a final reaction concentration of 1M hydroxylamine and either 20 μM tRNAPhe (Sigma), 5 μM RF1 or RF1 mutant, or buffer alone. The hydroxylamine containing A-site substrate mix was added to the release complex (0.25 μM) with an appropriate A-site codon at 25°C. Reactions with tRNA free in solution were initiated by addition of f-[35S]-Met-tRNAfMet to a concentration of 0.25 μM into a buffered 1 M hydroxylamine solution. Reactions were quenched as above and both the aminolysis and hydrolysis f-[35S]-Met products were separated from f-[35S]-Met-tRNAfMet and each other by electrophoresis on TLC-cellulose plates at 1250 V for 35 min. The background fraction of aminolysis and hydrolysis was determined for each reaction in the absence of hydroxylamine or A-site substrate at early time points and the average was subtracted from each time point in the experimental reaction. Curves obtained from either the aminolysis or hydrolysis reaction were fit to single exponential equations to give the rate constant kobs for f-[35S]-Met release which is the sum of kobs-HA + kobs-water. The contribution of the individual aminolysis and hydrolysis reactions to the release rate were calculated from the endpoints of each reaction; kobs-HA= (fMet-HA)/(fMet-HA + fMet-water) × kobs and kobs-water= (fMet-water)/(fMet-HA + fMet-water) × kobs.
Supplementary Material
Acknowledgments
We thank J. Brunelle and E. Youngman for preparation of ribosome mutants; M. Kim, S. Djuranovic, M. Acker, E. Youngman and J. Lorsch for comments and suggestions on the manuscript; C. Merryman, E. Youngman, J. Lorsch and other lab members for discussions and technical advice. This work was supported by funding from the NIH and salary support to R.G. from HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beringer M, Rodnina MV. Importance of tRNA interactions with 23S rRNA for peptide bond formation on the ribosome: studies with substrate analogs. Biol Chem. 2007a;388:687–691. doi: 10.1515/BC.2007.077. [DOI] [PubMed] [Google Scholar]
- Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol Cell. 2007b;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Brunelle JL, Youngman EM, Sharma D, Green R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA (New York, NY . 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey CT, Beaudet AL, Scolnick EM, Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:3163–3167. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das GK, Bhattacharyya D, Burma DP. A possible mechanism of peptide bond formation on ribosome without mediation of peptidyl transferase. J Theor Biol. 1999;200:193–205. doi: 10.1006/jtbi.1999.0987. [DOI] [PubMed] [Google Scholar]
- Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. The EMBO journal. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S, Panuschka C, Schmid W, Barta A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic acids research. 2003;31:6536–6542. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Pearson R. The Factors Determining Nucleophilic Reactivities. J Am Chem Soc. 1962;84:16–24. [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Champ S, Mora L, Merkulova-Rainon T, Kisselev LL, Buckingham RH. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J Biol Chem. 2005;280:2439–2445. doi: 10.1074/jbc.M407252200. [DOI] [PubMed] [Google Scholar]
- Jencks W, Carriuolo J. Reactivity of Nucleophilic Reagents toward Esters. J Am Chem Soc. 1960;82:1778–1786. [Google Scholar]
- Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- Lieberman KR, Dahlberg AE. Ribosome-catalyzed peptide-bond formation. Progress in nucleic acid research and molecular biology. 1995;50:1–23. [PubMed] [Google Scholar]
- Ma B, Nussinov R. Release factors eRF1 and RF2: a universal mechanism controls the large conformational changes. J Biol Chem. 2004;279:53875–53885. doi: 10.1074/jbc.M407412200. [DOI] [PubMed] [Google Scholar]
- McCaughan KK, Poole ES, Pel HJ, Mansell JB, Mannering SA, Tate WP. Efficient in vitro translational termination in Escherichia coli is constrained by the orientations of the release factor, stop signal and peptidyl-tRNA within the termination complex. Biol Chem. 1998;379:857–866. doi: 10.1515/bchm.1998.379.7.857. [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K, Kubo N, Narita S, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Polacek N, Gomez MJ, Ito K, Xiong L, Nakamura Y, Mankin A. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. Journal of molecular biology. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- Robinson R, Bower V. The Ionization Constant of Hydroxylamine. J Phys Chem. 1961;65:1279–1280. [Google Scholar]
- Scarlett DJ, McCaughan KK, Wilson DN, Tate WP. Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. Implications for macromolecular mimicry and structural changes in RF2. J Biol Chem. 2003;278:15095–15104. doi: 10.1074/jbc.M211024200. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005a;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005b;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic acids research. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. Journal of molecular biology. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12395–12400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell. 2007 doi: 10.1016/j.molcel.2007.06.032. In Press. [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nature structural & molecular biology. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- Wilson KS, Ito K, Noller HF, Nakamura Y. Functional sites of interaction between release factor RF1 and the ribosome. Nature structural biology. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Green R. Methods. Vol. 36. San Diego; Calif: 2005. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis; pp. 305–312. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- Zoldak G, Redecke L, Svergun DI, Konarev PV, Voertler CS, Dobbek H, Sedlak E, Sprinzl M. Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic acids research. 2007;35:1343–1353. doi: 10.1093/nar/gkl696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.