Summary
Fanconi anemia (FA) is a developmental and cancer-predisposition syndrome caused by mutations in genes controlling crosslink repair. Several FA proteins form a ubiquitin ligase that controls monoubiquitination of the FANCD2 protein in an ATR-dependent manner. Here we describe a new FA protein FANCI, identified as a ATM/ATR kinase substrate required for resistance to mitomycin C. FANCI shares sequence similarity with FANCD2, likely evolving from a common ancestral gene. The FANCI protein associates with FANCD2 and together, as the FANCI-FANCD2 (ID) complex, load onto chromatin in response to DNA damage. Like FANCD2, FANCI is monoubiquitinated and unexpectedly, ubiquitination of each protein is important for the maintenance of ubiquitin on the other, indicating the existence of a dual ubiquitin locking mechanism required for the ability of the ID complex to function. Mutation in FANCI is responsible for loss of a functional FA pathway in a patient with Fanconi anemia complementation group I.
Keywords: Fanconi anemia, FANCI, FANCD2, ubiquitination, phosphorylation, DNA repair, Mitomycin C
Introduction
The ability to sense and respond to DNA damage and DNA replication stress is critical for cellular and organismal survival. A failure to properly respond to genotoxic stress can lead to both developmental difficulties and tumorigenesis. Cells have evolved a complex signal transduction pathway that senses genotoxic stress and responds by activating specific types of repair, arresting the cell cycle and altering transcription. At the core of this signal transduction pathway are the ATM and ATR kinases (Bakkenist and Kastan, 2004; Bartek et al., 2004; Zhou and Elledge, 2000). These kinases phosphorylate over 20 known proteins in response to damage including the Chk1 and Chk2 kinases. While early theories on these pathways considered their major role to be controlling cell cycle transitions, it is now clear that they play critical roles in regulating essential functions in both DNA replication and DNA repair.
One pathway regulated by ATM/ATR is the Fanconi anemia (FA) crosslink repair pathway (Gurtan and D'Andrea, 2006). Patients with FA display multi-organ defects and most develop bone marrow failure in childhood (Butturini et al., 1994; Fanconi, 1967; Schmid and Fanconi, 1978). FA patients have a high incidence of hematological and nonhematological malignancies and their cells are hypersensitive to DNA interstrand crosslinking agents such as mitomycin C (MMC) (Alter et al., 2003). FA falls into 13 complementation groups and 12 FA genes have been cloned (Gurtan and D'Andrea, 2006; Reid et al., 2007; Taniguchi and D'Andrea, 2006; Xia et al., 2007; Xia et al., 2006). Eight of these proteins (all but D2, D1, J, and N) are subunits of a FA core complex, a nuclear E3 ubiquitin ligase (Machida et al., 2006; Meetei et al., 2004). A key substrate of this ligase is FANCD2, which is monoubiquitinated on lysine 561 (Garcia-Higuera et al., 2001). It has been hypothesized that there is another critical substrate for the ligase in addition to FANCD2 because fusion of ubiquitin to the chicken FANCD2 protein mutant for the lysine acceptor allows complementation of chicken FANCD2 mutants but not FA mutants defective for the ligase activity (Matsushita et al., 2005).
FANCD2 ubiquitination is critical for MMC-resistance and is required for the FANCD2 protein to form damage-induced foci on chromatin (Garcia-Higuera et al., 2001). How the FA pathway controls inter-strand crosslink repair is not clear but an important finding was that the FANCD1 gene is BRCA2, which has a known role in regulation of Rad51 loading and homologous recombination (Howlett et al., 2002).
Of all of the FA complementation groups, only FA-I remains uncharacterized at the molecular level (Levitus et al., 2004). FA-I mutant cells dot not ubiquitinate FANCD2, precluding its localization to repair foci. Like FA-D2 cells, FA-I cell lines have normal FA E3 ligase complex formation (Levitus et al., 2004).
ATR appears to directly regulate the FA pathway. ATR is required for monoubiquitination of FANCD2 (Andreassen et al., 2004) and phosphorylates FANCD2 on several sites required for FANCD2 function (Ho et al., 2006; Taniguchi et al., 2002). In this study we provide new data supporting a role for DNA damage signaling in the FA pathway. Through a proteomic screen for substrates for the ATM and ATR kinases (Matsuoka et al., submitted) combined with a DNA damage sensitivity screen, we identified the FANCI gene. FANCI is paralogous with FANCD2, is also monoubiquitinated on a lysine critical for its function and may be the second critical FA ligase substrate. FANCI binds FANCD2 to form the ID complex that loads onto chromatin in response to DNA damage.
Results
KIAA1794/FANCI is a phosphoprotein
KIAA1794/FANCI was identified as a protein whose phosphorylation was induced upon IR treatment (Matsuoka et al., submitted). In that study, SILAC (reviewed in (Mann, 2006)) and peptide immunoprecipitation (Rush et al., 2005) using phosphospecific antibodies followed by mass spectrometry before and after DNA damage was used to identify those proteins that were inducibly phosphorylated on SQ or TQ motifs. Three phosphorylation sites were detected in a human KIAA1794 protein: S730, T952, S1121, and two other sites in the mouse protein S555, T558. We renamed the KIAA1794 protein as FANCI, since, as shown below, the locus encoding this protein is mutated in an individual with Fanconi anemia complementation group I. Immunoblotting of FANCI after IR with a phospho-SQ antibody confirmed its inducible phosphorylation (Figure 1A), thus placing it in the ATM/ATR pathway.
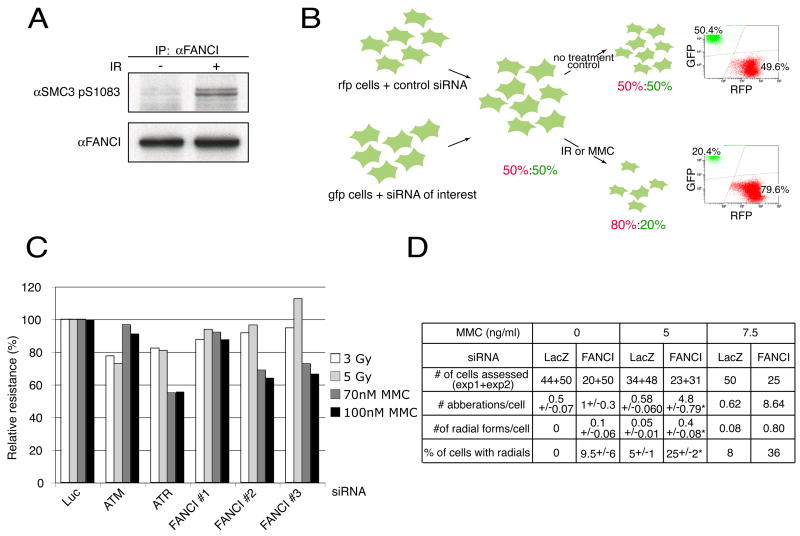
Figure 1. Identification of the KIAA1794/FANCI protein.
A. Western analysis with an antibody raised against a phosphorylated form of SMC3 (SMC3 pS1083) on immunoprecipitates performed with FANCI antibody (BL999) from 293T extracts before and after DNA damage.
B. Schematic of the multicolor competition assay (MCA). See text for details. In this example, the knockdown of a protein of interest caused the gfp cells to become DNA damage sensitive without influencing their proliferative capacity in the absence of damage. The relative resistance to damage of the si-treated cells is 40% of the non-si treated cells.
C. MCA analysis in U2OS cells treated with siRNAs against ATM and ATR and three different siRNAs against FANCI.
D. Cytogenetic abnormalities in IMR90 cells transfected with siRNA against KIAA1794 or LacZ control and treated with 0, 5, or 7.5 ng MMC per ml. Asterisk indicates a statistically significant difference in means as calculated by the t-test. Experiment with 7.5 ng MMC per ml was performed once.
Multicolor Competition Assay (MCA)
To efficiently study DNA damage sensitivity of cells with a variety of genetic perturbations, we developed a simple competition assay that is both quantitative and fast (Figure 1B). Two populations of U20S (osteosarcoma) cells differing only in their color were created by expression of red (RFP) or green (GFP) fluorescence protein. siRNA depletion of the protein of interest was carried out in the green cells while the red cells were transfected with control siRNA. Equal numbers of green and red cells were mixed, left untreated or treated with gamma-irradiation or mitomycin C (MMC). After 7 days, cells were harvested and ratio of red to green cells determined using flow cytometry. The green to red ratio in untreated cells acted as a control for the relative cell growth. The assay was validated using siRNAs targeting ATM (IR-sensitivity) and ATR (MMC- and IR-sensitivity) (Figure 1C and Supplementary Figure 1).
MCA was applied to study a subset of ATM and ATR substrates (Matsuoka et al., submitted). Cells treated with a combination of three siRNAs against one of the tested proteins, FANCI (KIAA1794 a.k.a. FLJ10719), demonstrated 60% survival after 70 nM MMC treatment and 91 of survival after 3 Gy IR treatment relative to control siRNA transfected cells (data not shown). To exclude off target effects, three siRNAs were tested independently. Two of three siRNAs reproduced the phenotype of MMC-sensitivity with only a slight effect on the IR sensitivity (Figure 1C). This decreased survival is due to a DNA repair defect as metaphase spreads of primary fibroblasts transfected with FANCI siRNA and treated with MMC revealed frequent cytogenetic abnormalities including chromatid and chromosome breaks as well as radial forms (Figure 1D), hallmarks of Fanconi anemia.
FANCI displays homology to FANCD2
BLAST analysis with FANCI revealed high conservation among eukaryotes from human to Dictyostelium but not yeasts and limited conservation to a predicted partial S.purpuratus sequence similar to FANCD2 (Figure 2A). The homology region extended over 151 amino acids with 19% identity, 45% similarity. The coding region of FANCI was amplified from a human lymphocyte cDNA library (Elledge et al., 1991) and recovered an open reading frame of 3984 nucleotides, coding for a 1328 AA protein of a calculated molecular weight 150 kDa. This cDNA corresponds to a putative splice variant isoform 3 of the KIAA1794 (Q9NVI1) locus on chromosome 15q25-q26.
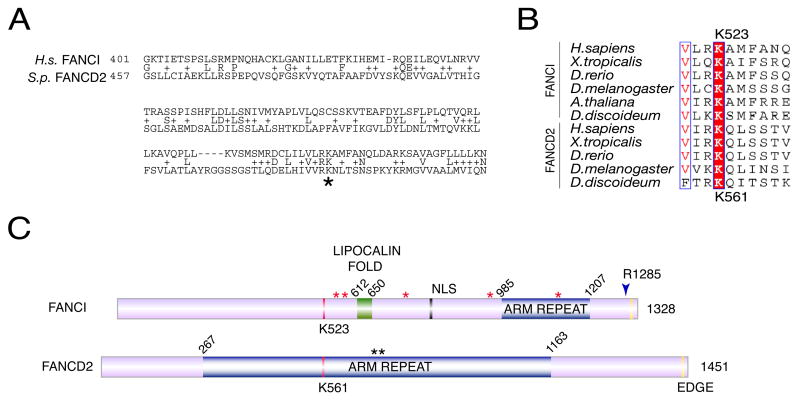
Figure 2. Identification of evolutionarily conserved regions of KIAA1794 /FANCI.
A. A BLAST alignment identifying human KIAA1794 conservation with a portion of the Strongylocentrotus purpuratus (S.p.) ortholog of FANCD2. A star indicates the lysine corresponding to K561 in FANCD2
B. Alignment of FANCI and FANCD2 identifies a conserved lysine K523.
C. Schematic of FANCI and FANCD2. Highlighted are two regions predicted by the SCOP database (Murzin et al., 1995) as ARM repeats which represent alpha-alpha superhelix folds (aa 985–1207 in FANCI and aa 267–1163 in FANCD2) and a lipocalin fold (aa 612–650), which is predicted to bind hydrophobic ligands in its interior. Also shown is putative bipartite NLS (aa 779–795) identified in FANCI. Red stars indicate phosphorylation sites identified in human or mouse proteins (Matsuoka et al., submitted). Black stars indicate the ATR sites in FAND2. The EDGE sequence is also conserved between the proteins. An arrowhead indicates the disease-causing mutation in a cell line of Fanconi anemia complementation group I (see Figure 6).
Alignment of FANCI and FANCD2 revealed a modest 13% identity and 20% similarity across the entire protein (Figure 2B, Supplementary Figure 3). Comparable levels of similarity were found between the FANCD2 and FANCI paralogs in other species including A. thaliana, and D. melanogaster. The most striking conservation between FANCI and FANCD2 across the species surrounded the site that has been previously shown to be monoubiquitinated in FANCD2 and to be essential for the functionality of the FA pathway, K523 in FANCI and K561 in FANCD2 (Figure 2B and 2C) (Garcia-Higuera et al., 2001).
FANCI participates in cell cycle checkpoints and DNA repair pathways
As the ATM/ATR pathways control multiple cellular responses, we asked if FANCI participates in cell cycle control, DNA synthesis control, or homologous recombination following DNA damage. siRNA against FANCI abrogated the G2/M checkpoint in U2OS cells (Figure 3A) and also had a small but reproducible effect in the intra-S phase checkpoint (Figure 3B). Interestingly, in unirradiated cells FANCI depletion caused an increased basal level of damage as judged by γ-H2AX (Figure 3C) indicative of a role in maintenance of genomic stability.
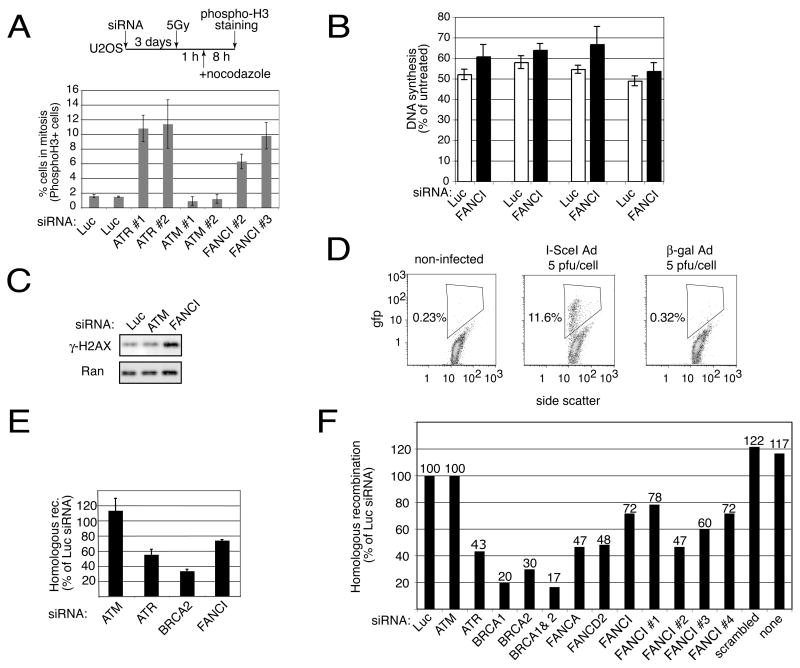
Figure 3. Checkpoint and repair defects in cells with reduced levels of FANCI.
A. Cells depleted for FANCI have checkpoint defects. U2OS cells were treated as shown in the schematic. Two separate fields of cells were examined. The mean and standard deviation from two fields are shown. Average of 1000 cells per siRNA were scored.
B. Effects of FANCI depletion on radio-resistant DNA synthesis. U2OS cells transfected with the indicated combination of three different siRNAs were irradiated with 5Gy or 10Gy of γ-IR depending on an experiment, allowed to recover for 30 minutes and assayed in triplicate for DNA synthesis. The means and standard deviations of four separate experiments are shown. For comparison, IR treatment of the ATM siRNA-transfected cells causes DNA synthesis to be 70–80% of the level found in the untreated cells.
C. Reduction of FANCI causes spontaneous DNA damage. U2OS cells transfected with the indicated combinations of three different siRNAs were collected three days later and the level of γ-H2AX was assayed without inflicting any exogenous damage. Western analysis with Ran antibody acted as a loading control.
D. Flow cytometric analysis of DR U2OS cells uninfected or infected with the AdNgus24i adenovirus carrying I-SceI (I-SceI-Ad) or AdCA36 carrying β-galactosidase (β-gal-Ad). Infections were carried out at an M.O.I. of 5 and analysis for gfp positive cells was performed at 36 hours after infection.
E&F. FANCI is required for homologous recombination. E. DR U2OS cells were transfected with the indicated combination of three different siRNAs and three days later were infected with 10 pfu/cell of adenovirus carrying I-SceI. Flow cytometric analysis of gfp positive cells was carried out 36 hours after infection. Mean and standard deviation of 8 experiments (ATM), 7 experiments (ATR), 4 experiments (Brca2) and 3 experiments (FANCI) are shown. F. DR U2OS cells were transfected with the indicated individual siRNAs, infected with 5 pfu/cell of adenovirus carrying I-SceI (AdNgus24i) and analyzed 24 hours later.
The FA pathway has been previously implicated in homologous recombination (HR) (Nakanishi et al., 2005; Niedzwiedz et al., 2004; Yamamoto et al., 2005) and therefore we examined FANCI for a potential role in HR repair. DR-U2OS cells used in this assay (Xia et al., 2006) have an integrated HR reporter. Induction of a double-strand break, resulted in a robust repair as indicated by the appearance of 12% GFP positive cells (Figure 3D). All four siRNAs to FANCI reduced recombination from 78% to 47 % of controls, similar to siRNAs to ATR, FANCA and FANCD2 (Nakanishi et al., 2005) but less than siRNAs to BRCA1 and BRCA2 which are thought to be more directly involved in the recombination process (Figure 3E and 3F). These results indicate that FANCI is an important component of the HR repair pathway.
FANCI localizes to damage-induced foci in multiple cell types
To assess FANCI localization, immunofluorescence experiments were performed on transformed (U2OS, HeLa, and 293T) and primary (BJ) cell lines. Analysis using two antibodies BL999 and BL1000 revealed foci in a subset of untreated cells and in nearly all cells after DNA damage. In some experiments, a nuclear rim staining was also detected. These FANCI foci corresponded to damage-induced foci as they colocalize with FANCD2 staining (Figure 4A) (Garcia-Higuera et al., 2001). Confirmation of the antibody specificity was achieved using transfected Myc-FANCI and anti-Myc antibodies (Supplementary Figure 4A). siRNA treated cells showed decreased damage-induced foci staining with BL999 and BL1000 antibodies after Triton pre-extraction (data not shown).
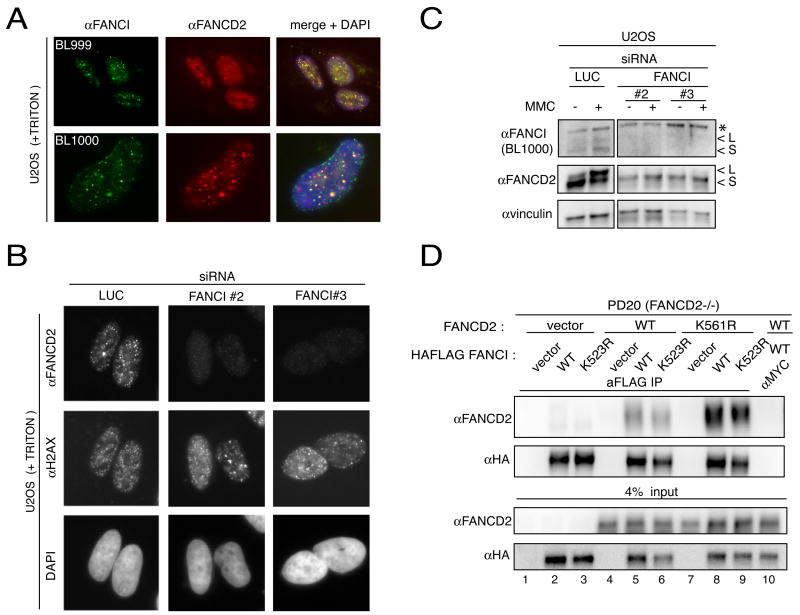
Figure 4. FANCI localizes and interacts with FANCD2.
A. Localization of the endogenous FANCI using BL999 and BL1000 antibodies. U2OS cells treated with 1 μM mitomycin C for 24 hours were triton-extracted before co-staining with anti-FANCI (BL999 or BL1000) and anti-FANCD2 antibodies.
B. Localization of FANCD2 in cells transfected with individual siRNAs against FANCI. U2OS cells were transfected with the indicated individual siRNAs against FANCI and treated with 1 μM mitomycin C. Twenty-four hours later, following triton extraction, the cells were co-stained with an antibody against FANCD2 and H2AX.
C. Western analysis of FANCD2 in U2OS cells transfected with individual siRNAs against FANCI. L is the long (monoubiquitinated) and S is the short form of the proteins. Asterisk indicates cross-reacting band.
D. Interaction of FANCD2 and FANCI. Total protein (0.5 mg) from PD20 fibroblasts expressing indicated constructs was immunoprecipitated with FLAG or control Myc antibodies under non-damaged conditions. The immunoprecipitates were analyzed by western blotting with a rabbit anti-FANCD2 or mouse anti-HA antibody.
FANCI and FANCD2 form a complex required for FANCD2 localization to damage-induced foci
Depletion of FANCI in U2OS using three separate siRNAs resulted in diminished ubiquitination of FANCD2 upon damage (Figure 4C) and the loss of this modification corresponded to a prominent reduction in FANCD2 signal at damage-induced foci as well as appearance of cells with no visible FANCD2 foci (Figure 4B). Moreover, the steady state level of FANCD2 was decreased upon depletion of FANCI (Figure 4C). There was also a reciprocal relationship between FANCD2 and FANCI since the knockdown of FANCD2 also led to decreased foci formation of FANCI (Supplementary Figure 4B, top panel). Loss of FANCD2 upon depletion of FANCI might be expected if the two proteins are found in a complex. Immunoprecipitation of HA-FLAG-tagged FANCI expressed in 293T cells with antibodies against either HA or FLAG, but not MYC, resulted in co-immunoprecipitation of endogenous FANCD2 (Supplementary Figure 4C). The interaction was independent of DNA damage and was robust with 15–20% of total FANCD2 immunoprecipitated. Immunoprecipitation of endogenous FANCI was also able to co-immunoprecipitate FANCD2 (Supplementary Figure 4D) and immunoprecipitation with FANCD2 antibodies recovered FANCI (Supplementary Figure 4E). To test if monoubiquitination of FANCD2 was required for this interaction, PD20 cells complemented with WT or the K561R mutant of FANCD2 that cannot be monoubiquitinated (Garcia-Higuera et al., 2001) were used in immunoprecipitation experiments. Immunoprecipitation of HA-FLAG-tagged FANCI expressed in these cells recovered both WT and the K561R mutant FANCD2 (Figure 4D, lanes 8 and 9) suggesting that ubiquitination of FANCD2 is not required for the interaction with FANCI.
FANCI becomes ubiquitinated after damage and during an unperturbed S phase
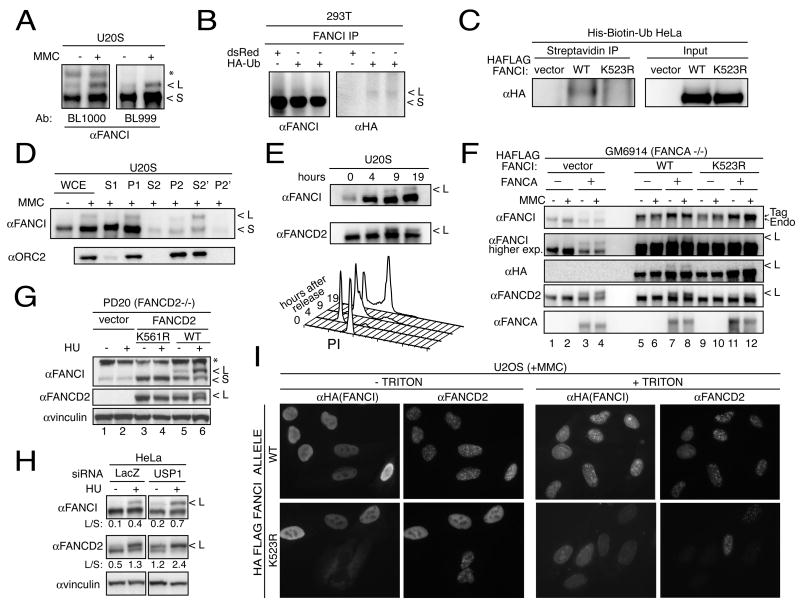
Presence of the conserved lysine in FANCI at a position corresponding to the FANCD2 ubiquitination site raised the possibility that FANCI is also ubiquitinated. Indeed, a slower migrating band was present on the western blots performed with two non-overlapping anti-peptide antibodies in U2OS cells (Figure 5A) as well as in other cell lines including primary BJ fibroblasts (Figure 5 and data not shown). The slower migrating band (long form, L), although present in the untreated cells, increased after DNA damage inflicted by MMC (Figure 5A) or HU (Figure 5G and H). The molecular weight difference between the long form and the short form (S) is consistent with monoubiquitination. To test this, FANCI was immunoprecipitated from 293T cells expressing HA-tagged ubiquitin and immunoblotted with HA-antibodies (Figure 5B). A band of appropriate size was identified corresponding to the long form of FANCI only in cells transfected with HA-tagged ubiquitin but not in control cells. To exclude the possibility that the monoubiquitinated protein seen in Figure 5B is a FANCI-associated protein, pulldowns from HeLa extracts expressing His-biotin-ubiquitin were performed with Streptavidin under fully denaturing conditions (Tagwerker et al., 2006). WT but not K523R FANCI mutant (see below) precipitated under these conditions (Figure 5C). Therefore, both, FANCI and FANCD2 are monoubiquitinated in vivo.
Figure 5. FANCI ubiquitination and its dependence on Fanconi anemia pathway.
A. Western blot analysis of FANCI in U20S cells. U2OS cells were treated with 1 μM MMC and 24 hour later cells were lysed directly in 2x Laemmlie buffer. Long (L) and short (S) forms of FANCI are shown. The asterisk indicates a cross-reacting band.
B. In vivo ubiquitination of FANCI. Whole cell extracts of 293T cells transiently transfected with HA-tagged ubiquitin or control plasmid carrying dsRed marker were immunoprecipitated using antibodies raised against FANCI and analyzed by western blot with a FANCI antibody (left) and antibody recognizing the HA tag (right).
C. In vivo ubiquitination of FANCI. HeLa cells expressing ubiquitin tagged with His and a biotynylation signal were treated with 2 mM HU for 16 hours, lysed in 8M urea and precipitated using Streptavidin beads under denaturing conditions.
D. Chromatin fractionation of FANCI in U2OS cells. Cells were treated with 1 μM MMC and 24 hours later cells were collected and processed into cellular fractions. Whole cell extract (WCE), cytoplasmic proteins (S1), intact nuclei (P1), soluble nuclear proteins (S2), chromatin-enriched pellet (P2), soluble and insoluble fractions after micrococcal nuclease treatment (S2’ and P2’) are indicated. Orc2 antibody was used to follow the chromatin fraction.
E. Cell cycle analysis of FANCI ubiquitination. After release from nocodazole, cells were collected at indicated times for the western analysis (top panel) and for cell cycle analysis using flow cytometry (lower panel).
F. Analysis of ubiquitination in GM6914 (FA-A) fibroblasts. Cells expressing vector or WT FANCA were stably transduced with empty vector, or HA-tagged WT FANCI. Twenty-four hours after 1 μM MMC treatment cells were collected and western blotting was performed with the indicated antibodies.
G. Analysis of ubiquitination in PD20 (FA-D2) fibroblasts. Cells expressing vector, K561R mutant or WT FANCD2, were treated with 2mM HU and collected 15 hours later. Western blotting was performed with the indicated antibodies including FANCD2 antibody to confirm absence (lane 1 and 2) or presence (lanes 3, 4, 5, and 6) of FANCD2 protein. The asterisk indicates a cross-reacting band.
H. Ubiquitination of FANCD2 and FANCI in HeLa cells transfected with siRNA against USP1 and LacZ control, treated with 2 mM HU and collected 15 hours later. L/S indicates the ratio of the monoubiquitinated to non-ubiquitinated FANCI or FANCD2.
I. Localization of FANCI and FANCD2 in WT and K523R FANCI-expressing U20S cells. Cells stably transduced with the HA-tagged WT or K523R mutant allele of FANCI were treated with 1 μM MMC and processed 24 hours later for immunofluorescence. Note that cells not expressing K523R in the lower panels (K523R - triton) are included as controls for FANCD2 staining. Two FANCD2 positive cells in the lower right panel (+triton) are presumed not to have K523R FANCI expression although that cannot be tested directly since triton removes nucleoplasmic FANCI. Similar results were observed in U2OS cells expressing the K523R mutant treated with HU.
Chromatin fractionation experiments revealed that the ubiquitinated form of FANCI, like FANCD2 (Montes de Oca et al., 2005), is enriched in chromatin (Figure 5D). To ask whether FANCI is modified during the cell cycle, U2OS cells were synchronized and released from a mitotic block. Cells in mitosis and G1 phase of cell cycle lacked ubiquitinated FANCI or FANCD2 (Figure 5E). By 9 hours after release, when most cells were in early S phase, FANCI appeared ubiquitinated. Since the experiment was done in the absence of exogenous damage, we conclude that endogenous FANCI is modified in an unperturbed S phase.
Ubiquitination of FANCI is FANCA and FANCD2 dependent
To search for the E3 ubiquitin ligase for FANCI, we examined FANCI modification in FANCA mutants defective for the core E3 ligase complex. FA-A cells GM6914 cells lacking FANCA showed no ubiquitination of the endogenous or HA-tagged FANCI (Figure 5F, lanes 1, 2, 5, and 6) but ubiquitination was restored after complementation with WT FANCA (Figure 5F, lanes 3, 4, 7, 8).
FANCD2 and FANCI show reciprocal ubiquitination dependencies. PD20 fibroblasts, which lack FANCD2 (Jakobs et al., 1996), when transfected with the ubiquitination-defective FANCD2 K561R mutant also fail to ubiquitinate FANCI (Figure 5G, lanes 3 and 4). The same cells complemented with WT FANCD2 restore FANCI modification (Figure 5G, lanes 5 and 6). PD20 cells expressing WT or K561R FANCD2 also showed increased levels of FANCI (Figure 5G) consistent with the notion that the non-ubiquitinated forms of the protein bind constitutively in a heterodimeric (or multimeric) Fanconi anemia ID complex.
USP1 is the deubiquitinating enzyme for FANCD2 (Nijman et al., 2005). To test whether USP1 also can affect FANCI monoubiquitination, HeLa cells were transfected with siRNA against USP1. Reduction of USP1 increased the L to S ratio for both FANCI and FANCD2 under basal conditions and after HU treatment. (Figure 5H).
Lysine 523 of FANCI is critical for its ubiquitination
To determine whether the conserved lysine 523 of FANCI was required for ubiquitination, a WT or K523R mutant HA-tagged FANCI were stably expressed in GM6914 (Figure 5F) and in 293T cells (Supplementary Figure 5A). Only in cells that expressed the WT FANCI but not the K523R mutant was the L form detected with the HA antibody (Figure 5F, lanes 7, 8, 11, 12 and Supplementary Figure 5A). Interestingly, cells overexpressing the FANCI K523R mutant, but not WT, showed diminished monoubiquitination of FANCD2 (Figure 5F, lanes 11, 12 and Supplementary Figure 5A) suggesting that the mutant FANCI displays a dominant negative activity.
Consistent with the role of ubiquitination of FANCD2, the FANCI K523R mutant failed to form DNA damage foci (Figure 5I +TRITON) despite its overproduction (data not shown). Cells expressing K523R FANCI allele showed pan-nucleoplasmic FANCD2 staining and greatly diminished localization to DNA damage-induced foci best visualized after triton pre-extraction (Figure 5I). These data show that the K523R mutant has a dominant negative effect on FANCD2 foci formation.
FANCI is mutated in cells from the Fanconi anemia complementation group I
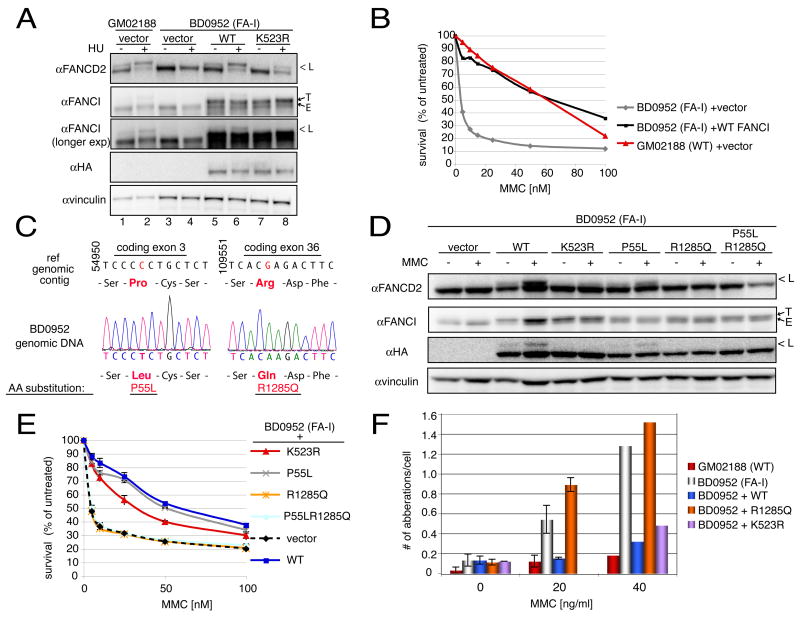
Phenotypic similarities of cells with reduced levels of FANCI to cells from Fanconi anemia patients, including marked MMC but only mild IR sensitivity (Figure 1B) alerted us to the possibility that mutations in FANCI might be responsible for human disease. Published reports included only one remaining complementation group for which the responsible gene was unknown, Fanconi anemia complementation group I (Levitus et al., 2004). We worked with one of the cell lines from this group, BD0952, EBV-transformed peripheral lymphocytes from a patient with a classic presentation of Fanconi anemia. BD0952 express a full-length FANCI protein at normal levels relative to control cells (GM03288) (Figure 6A). However, this protein is not ubiquitinated in BD0952 cells, even after Mitomycin C treatment (Figure 6A and data not shown).
Figure 6. Complementation of BD0952 (FA-I) cells with the KIAA1794/FANCI gene.
A. Complementation of FANCD2 ubiquitination defects in FA-I cells by expression of WT FANCI. Cells stably transduced with empty vector, HA-tagged WT or K523R FANCI, were untreated or treated with 100 nM MMC and collected 24 hours later by lysis in Laemmli buffer. Western analysis with FANCD2, FANCI, and HA antibodies was performed. GM02188 (WT control) cells acted as a control for the presence of long (L, ubiquitinated) forms of FANCD2 and FANCI, which are absent in the uncomplemented BD0952 cells. The transduced form of the protein is identified as T (tagged) since it runs slightly slower than the endogenous (E) form. Also see Supplementary Figure 5B. B. Complementation of MMC sensitivity of BD0952 cells by expression of WT FANCI but not empty vector. Logarithmically growing cells of indicated genotypes were treated in triplicate with different levels of MMC ranging from 0 to 100 nM. They were allowed to grow for 6 days at which time they were harvested and total cell number was counted using a coulter counter. Total cell numbers at each dose were divided by the number of cells in the untreated sample to arrive at percent survival.
C. Sequence analysis of the FANCI genomic locus in BD0952 (FA-I) cells. Sequence of the genomic contig (ref|NT_010274.16|Hs15_10431:4714523–4889523 Homo sapiens chromosome 15 genomic contig, reference assembly), and sequence and sequence traces of genomic DNA from BD0952 cells, shown together with the resulting amino acid sequence deduced from the DNA sequence data.
D. Complementation of FANCD2 ubiquitination by expression of WT FANCI or P55L FANCI, but not R1285Q or P55L, R1285Q FANCI mutants. Cells stably transduced with the indicated alleles of FANCI were left untreated or were treated with 100 nM of MMC and processed 24 hours later as indicated in panel A.
E. Complementation of MMC sensitivity of BD0952 cells by expression of WT FANCI or P55L FANCI, but not R1285Q or P55L, R1285Q FANCI mutants. Experiments were done as indicated in Figure legend 6B.
F. Cytogenetic abnormalities in BD0952 cells cells expressing WT, K523R or R1285Q FANCI alleles. Indicated cells were treated with 0, 20 or 40 ng MMC per ml of media and analyzed for presence of chromosomal aberrations 48 hours later. K523R mutant was not assessed at 20 ng of MMC per ml. Analysis was done only once at 40ng of MMC per ml. 30–50 metaphases were evaluated for each cell line.
FANCD2 is not ubiquitinated in FA-I cells (Figure 6A and (Levitus et al., 2004)), thus restoration of FANCD2 ubiquitination acts as a surrogate marker for the functional complementation of the Fanconi anemia pathway. Expression of the FANCI cDNA in BD0952 cells restored FANCD2 ubiquitination (Figure 6A). This exogenous FANCI was also monoubiquitinated (Figure 6A and Supplementary Figure 5B). Appearance of the monoubiquitination was not due to changes in the cell cycle of the cells expressing FANCI (Supplementary Fig. 5C). Also, the levels of expression of the exogenous proteins were comparable to the endogenous protein (compare T [tagged] vs. E [endogenous], Figure 6A and Supplementary Figure 5B). Expression of WT FANCI in BD0952 also complemented their MMC resistance to WT levels (Figure 6B).
To look for FANCI mutations in BD0952 cells, we amplified and sequenced the cDNA from BD0952 mRNA and found two base substitutions as candidates for the Fanconi anemia-causing mutation in BD0952 cells. They included a C to T transition which resulted in Pro to Leu change at amino acid 55, and a G to A transversion which resulted in Arg to Glu substitution in an absolutely conserved Arg1285 at the C-terminus of the protein. We confirmed these mutations by amplifying exon 3 and exon 36 from genomic DNA. Sequencing confirmed presence of both mutations in genomic DNA in homozygous form (Figure 6C). Homozygosity was expected at the disease locus since the patient comes from a consanguineous family with both parents expected to contribute the disease allele of FANCI to their child.
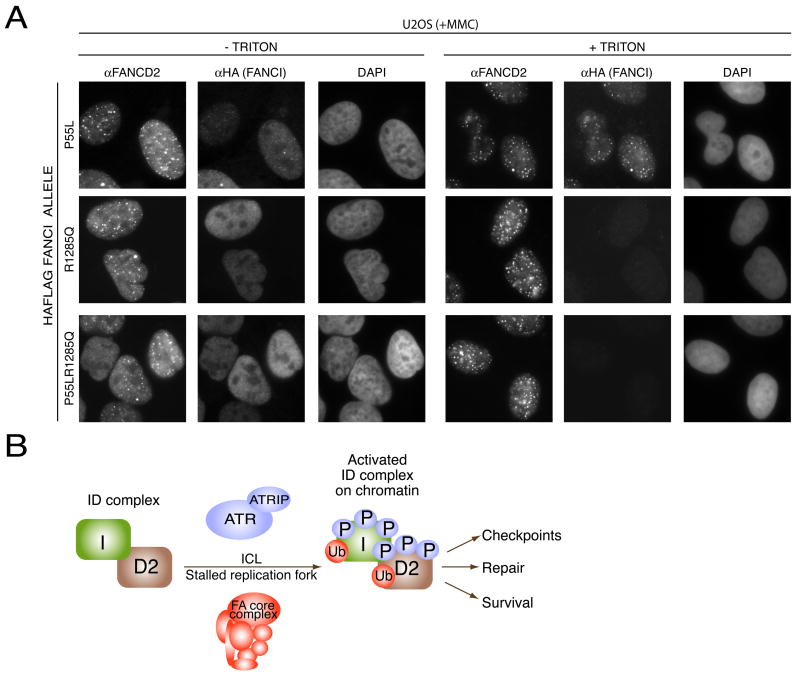
To test which mutation causes the disease, we made expression constructs containing P55L, R1285Q, and P55L&R1285Q substitutions. Only the WT and the P55L allele were able to complement the FANCD2 monoubiquitination defect (Figure 6D) and MMC sensitivity (Figure 6E) of BD0952 cells. These two proteins were also themselves monoubiquitinated in BD0952 cells. When introduced into U2OS cells or into BD0952 cells, the P55L allele was found in foci together with FANCD2 (Figure 7A and Supplementary Figure 6). Cells expressing R1285Q or P55L R1285Q alleles showed no monoubiquitination of FANCD2 (Figure 6D) and failed to restore MMC-resistance (Figure 6E). Unlike WT FANCI allele, which could complement breakage phenotype seen in BD0952 cells, the R1285Q allele expressing cells showed high number of aberrations after treatment with MMC (Figure 6F). When introduced into U2OS or BD0952 cells, the R1285Q or P55L R1285Q alleles failed to localize to damage-induced foci (Figure 7A, +TRITON panel, Supplementary Figure 6) despite robust expression levels of the mutant proteins as judged by the immunofluorescence staining in the absence of triton extraction (Figure 7A –TRITON panel). Together, these studies indicate that the R1285Q change is the disease causing mutation in BD0952 cells.
Figure 7. Localization of mutant FANCI alleles.
A. Localization of WT, P55L, R1285Q, and P55L, R1285Q mutant proteins in U2OS cells. U20S cells transduced with the indicated alleles of FANCI were treated with 100 nM MMC and 24 hours later were processed for immunofluorescence.
B. Model of Fanconi anemia ID complex regulation and function. The phosphorylation-ubiquitination cascade culminates in chromatin loading of the Fanconi anemia ID complex, which directs downstream repair events.
Unexpectedly, we found the K523R FANCI allele was able to partially complement the FANCD2 monoubiquitination defect (Figure 6A, lanes 7 and 8) and MMC sensitivity defect in BD0952 cells (Figure 6F). This is in contrast to the findings that FANCI K523R mutant fails to be ubiquitinated or form damage-induced foci and that K523R allele when overexpressed acts as a dominant negative against FANCD2 ubiquitination and foci formation. These results suggest either that this allele is only partially defective or more likely that it is displaying interalleleic complementation with the FANCI R1285Q mutant present in BD0952 cells. Further studies will be needed to resolve this issue.
Discussion
The response to DNA damage and DNA replication stress is orchestrated by the ATM and ATR kinases that direct the phosphorylation of key proteins that carry out this stress response. Knowledge of their substrates is key to both identifying and elucidating DNA repair responses. In this study we examine one such recently identified substrate of these kinases (Matsuoka et al, submitted), the KIAA1794 protein, which we discovered to be encoded by the FANCI gene.
FANCI is a FANCD2 paralog required for crosslink repair
FANCI depletion caused a phenotype consistent with a role in DNA crosslink repair. FANCI is highly conserved from humans to Dictyostelium, but absent in budding and fission yeast. The interior of FANCI, like that of FANCD2 is composed of ARM repeats which fold into superhelical helices, suggesting that these proteins have an extended structure. A crucial finding was a short stretch of similarity to Sea Urchin FANCD2, a central component of the Fanconi anemia pathway. This conserved region contained a key lysine known to be monoubiquitinated in FANCD2 that is critical for FANCD2 function. FANCI contains a weak but significant similarity throughout the length of the human FANCD2 protein. As FANCI and FANCD2 are approximately the same size, show similarity along their length, and both have an extended region of super helical ARM repeats, it is likely that these proteins were derived from a common ancestral gene duplication event that subsequently diverged through evolution to produce two functionally distinct paralogs.
FANCI is mutated in FA-I cells
As KIAA1794/FANCI showed characteristics of a Fanconi anemia protein we anticipated that previously unassigned Fanconi patients might harbor mutations in our gene. We were able to obtain BD0952 cells from the FA-I complementation group. FA-I cells are MMC-sensitive and fail to monoubquitinate FANCD2 or form FANCD2 foci in response to DNA damage. Retroviral expressed FANCI complemented FA-I mutants for all three phenotypes. DNA sequence analysis revealed two amino acid changes in the FANCI protein. Complementation analysis reveals that the R1285Q mutation is responsible for the defect in BD0952 cells and fails to restore MMC-resistance and FANCD2 regulation. The R1285Q mutation is present in both copies of FANCI in these cells consistent with family consanguinity. The complementation, coupled with the identification of inactivating mutations in BD0952 cells, unambiguously identifies KIAA1794 as the FANCI gene. FANCI in the three other FA-I cell lines (Levitus et al., 2004) remains to be examined for the presence of mutations to confirm the cell-fusion data that placed them in the same complementation group as BD0952 cells.
FANCI is monoubiquitinated
Like FANCD2, FANCI has a slower migrating form that several lines of evidence suggest is due to monoubiquitination. First, the key lysine responsible for FANCD2 ubiquitination is conserved in FANCI. Secondly, FANCA mutant cells defective for the FA ubiquitin ligase complex fail to generate the slower migrating form. Third, mutation of this lysine, K523R, in FANCI prevents formation of the slower migrating form. Finally, FANCI antibodies specifically immunoprecipitate epitope tagged ubiquitin covalently attached to FANCI. Like FANCD2, the ubiquitinated form of FANCI is enriched on chromatin and the K523R FANCI mutant does not get loaded onto chromatin
The significance of FANCI monoubiquitination is illustrated by the fact that the K523R mutant of FANCI does not itself become monoubiquitinated and does not form damage foci. When introduced into U2OS cells, which express a WT FANCI allele, this mutant reduced FANCD2 monoubiquitination and foci formation. Furthermore, the K523R mutant fails to fully complement FANCD2 ubiquitination or MMC sensitivity of BD0952 cells indicating that ubiquitination is important for FANCI function. As this line is not a null mutant, it is possible that this intermediate FANCD2 monoubiquitination and MMC-resistance phenotype might represent interallelic complementation.
FANCI and D2 form an interdependent ID complex that is required for ubiquitination and chromatin association
We anticipated that FANCI would associate with FANCD2 because FANCI protein formed foci that colocalized with FANCD2, depletion of FANCI significantly reduced the monoubiquitination of FANCD2 preventing its inclusion into DNA damage induced foci, and FA-I cells have completely lost their ability to monoubiquinate FANCD2 despite fully formed FA complex (Levitus et al., 2004). Indeed, FANCI forms a complex with FANCD2, with approximately 20% of FANCD2 coimmunoprecipitating with tagged FANCI protein. The FANCI-FANCD2 complex, which we propose to call the Fanconi anemia ID complex, thus joins the Fanconi anemia core complex as a new downstream complex necessary for ICL repair and homologous recombination.
Just as the FANCI protein monoubiquitination is required for FANCD2 ubiquitination and foci formation, FANCD2 and FANCD2 ubiquitination is required for FANCI ubiquitination. Thus, these two paralogs are interdependent and employ a dual ubiquitination mechanism to affect downstream effector function. Furthermore, phosphorylation of FANCD2 is required for its own efficient ubiquitination and therefore for the efficient ubiquitination of FANCI. By extension, FANCI phosphorylation is also expected to be an essential event for the FA pathway following DNA damage and during normal S phase progression. This phosphorylation-ubiquitination cascade culminating in chromatin loading of the ID complex offers an exquisite control at sites of stalled forks. The ID complex, when correctly placed, can direct repair pathways to remove the crosslinks and repair the DNA so that replication can resume and cells can survive. Without this key event, cells are prone to genomic instability that can lead both to increased cell death, stem cell depletion, and tumorigenesis.
Among the key questions that remain to be answered is how the ID complex is recognized for ubiquitination. Is it directly ubiquitinated by the FANCL ligase complex or is there a more complex relationship among different ligases converging on this pathway. What is the function of ID ubiquitination and why must both components be monoubiquitinated? Is it merely required for chromatin loading or does it direct repair in the same way that PCNA ubiquitination directs repair? Does it represent a ubiquitin code that directs specific repair pathways? The identification of FANCI and the Fanconi anemia ID complex should now allow these issues to be approached with greater clarity.
Experimental Procedures
Cell lines
Complemented Cell lines PD20 and GM6914 were described previously (Taniguchi et al., 2002), DR-U2OS were provided by Maria Jasin (Xia et al., 2006). GM02188 was obtained from Coriell, BD0952 from European Collection of Cell Cultures (www.ecacc.org.uk), and U20S from American Cell Culture Collection (ATCC).
Antibodies
Antibodies were as follows: KIAA1794; BL999 and BL1000 (Bethyl), rabbit FANCD2 (Novus), mouse FANCD2 (Santa Cruz), FANCA (Rockland), ORC2 (BD Bioscience), Vinculin (Sigma), HA (Covance), MYC (Covance), SMC3pS1083 (Bethyl), PhosphoH3 (Upstate), Ran (BD Bioscience), γH2AX (Upstate). For the IPs, we used anti-HA affinity matrix (Roche), anti-FLAG M2 agarose (Sigma), c-MYC (Santa Cruz) and Protein A/G PLUS-Agarose (Santa Cruz). Secondary antibodies for IF were from Molecular Probes and Amersham and for western blots were from Jackson Laboratories.
FANCI Cloning
PCR was performed using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) on a human cDNA library (Elledge et al., 1991). The total RNA from BD0952 cells was isolated using Trizol (Invitrogen). The RNA was reverse transcribed with Superscript III (Invitrogen) and dT primers. The PCR step was performed using Platinum Pfx DNA Polymerase (Invitrogen). Genomic DNA was prepared using DNeasy Tissue kit (Qiagen). See Supplementary Experiemental Procedures for primer sequences.
Mutagenesis
The QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene) or QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene) was used to make mutation in FANCI. See supplementary experiemental procedures for primer sequences.
siRNAs
Stealth siRNAs (Invitrogen) were transfected using Oligofectamine (Invitrogen) at final concentration of 85 nM total siRNAs. Assays were done 48–72 hours after transfection. Unless indicated otherwise, combination of three siRNAs against the same gene were used. siRNAs used in experiments shown in Figure 1D, and 5I were purchased from Qiagen. In these experiments, cells were transfected at concentration of 20nM using Hyperfect according to manufacturer’s instructions. See supplementary experiemental procedures for siRNA sequences.
Immunofluorescence
Cells grown on autoclaved cover slips were processed as described in the supplementary experimental procedures. For the IF on lymphoblastic cell lines, coverslips were treated with sterile Poly-D-lysine hydrobromide, molecular weight >300,000 (Sigma) and the cells were allowed to attach for several hours.
Chromatin Fractionation and Immunoprecipitations
Chromatin fractionation was preformed as described (Mendez and Stillman, 2000; Zou et al., 2002). Benzonase (Novagen) was included in immunoprecipitation reactions. Details may be found in supplementary experimental procedures.
Multicolor Competition Assay
SiRNA transfections were performed as described above with gfp cells being transfected with a control siRNA (luciferase) and rfp cells with an siRNA of interest. On the third day after transfections, gfp and rfp cells were counted and mixed in 1to 1 ratio and were left untreated or were treated with IR or MMC. After 7 days of culture, all cells were collected and analyzed using Cytomix FC500 Analyzer (Beckman Coulter). Relative survival of Luc siRNA-treated cells after damage was set to 100%.
G2/M checkpoint assay
U2OS cells were transfected with individual siRNAs for three days in a 96 well format. Cells were irradiated with 5 Gy and allowed to recover for 1hr before the addition of 100 ng nocodazole per ml of media to trap cells that bypass the G2/M checkpoint. Cells were fixed and stained with an antibody against Phospho-H3 9 hrs after irradiation.
Radioresistant DNA synthesis assay
RDS assays to evaluate the intra-S phase checkpoint were done as described previously (Silverman et al., 2004). For details see supplementary experimental procedures.
Homologous recombination assay
HR assay was performed as described (Nakanishi et al., 2005; Xia et al., 2006), except instead of transfecting cells with an I-SceI expressing plasmid we used an adenovirus AdNGUS24i (kindly provided by Frank Graham, McMaster University) expressing the I-SceI enzyme. Control adenovirus AdCA36 (Addison et al., 1997) expressed β-galactosidase.
Cell cycle synchronization
U2OS cells were treated with 2.5 mM thymidine for 24 hours, washed three times and released into 100 ng nocodazole per ml of media, incubated for 12 hours and collected by mitotic shakeoff. Cells were washed three times, counted and plated for collection at different times.
Mitomycin C sensitivity assay
Logarithmically growing cells were counted and diluted to 2x105 cells per ml, plated in triplicate for each drug dose and treated with different concentrations of freshly made Mitomycin C. After 6 days in culture, cells were harvested and counted using a Z2 Coulter Counter (Beckman Coulter). Cell numbers in the samples treated with the drug were normalized to the cell numbers in the untreated sample.
Bioinformatics
BLAST was used for homology searches (http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al., 1997). The SCOP database can be found at http://scop.mrc-lmb.cam.ac.uk/scop/(Murzin et al., 1995). Alignments were performed in ClustalX and were rendered using ESPript 2.2 (http://espript.ibcp.fr) (Gouet et al., 1999). GenBank accession number for FANCI is EF469766
Supplementary Material
Acknowledgments
We are grateful to K. Nakanishi and M. Jasin for DR U2OS, J. Lin and J.W. Harper for expression constructs, H. Takai and T. de Lange for protocols, F. Graham for the AdNGUS24i and AdCA36 adenoviruses, J. Parvin for His-biotin-Hela. A. Gurtan for reagents, C. Mathew for helpful discussions, A. Brass, M. Schlabach, G. Hu, H.Yen, F. Stegmeier, T. Westbrook, D. Chou, N. Solimini for constructs and advice, J. Daly and S. Lazo-Kallanian for cell sorting, L. Moreau for the cytogenetic analysis, and A. Liang for technical assistance. AS is supported by T32CA09216 to the MGH Pathology Department at the Massachusetts General Hospital. PV is a Swiss National Science Foundation Fellow. KEH is a Leukemia and Lymphoma Society Special Fellow. This work was supported by grants from the NIH to S.J.E. and from NIAID 1U19A1067751 to A.D.D. and S.J.E. S.J.E. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addison CL, Hitt M, Kunsken D, Graham FL. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J Gen Virol. 1997;78(Pt 7):1653–1661. doi: 10.1099/0022-1317-78-7-1653. [DOI] [PubMed] [Google Scholar]
- Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101:2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Butturini A, Gale RP, Verlander PC, Adler-Brecher B, Gillio AP, Auerbach AD. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood. 1994;84:1650–1655. [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci U S A. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanconi G. Familial constitutional panmyelocytopathy, Fanconi's anemia (F.A.). I. Clinical aspects. Semin Hematol. 1967;4:233–240. [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Gurtan AM, D'Andrea AD. Dedicated to the core: understanding the Fanconi anemia complex. DNA Repair (Amst) 2006;5:1119–1125. doi: 10.1016/j.dnarep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Jakobs PM, Sahaayaruban P, Saito H, Reifsteck C, Olson S, Joenje H, Moses RE, Grompe M. Immortalization of four new Fanconi anemia fibroblast cell lines by an improved procedure. Somat Cell Mol Genet. 1996;22:151–157. doi: 10.1007/BF02369905. [DOI] [PubMed] [Google Scholar]
- Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, Hoatlin ME, Waisfisz Q, Arwert F, de Winter JP, Joenje H. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood. 2004;103:2498–2503. doi: 10.1182/blood-2003-08-2915. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Kitao H, Ishiai M, Nagashima N, Hirano S, Okawa K, Ohta T, Yu DS, McHugh PJ, Hickson ID, et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol Cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, Houghtaling S, Grompe M, D'Andrea AD. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Schmid W, Fanconi G. Fragility and spiralization anomalies of the chromosomes in three cases, including fraternal twins, with Fanconi's anemia, type Estren-Dameshek. Cytogenet Cell Genet. 1978;20:141–149. doi: 10.1159/000130845. [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, Kimura M, Matsushita N, Arakawa H, Buerstedde JM, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.