Abstract
A multisubunit protein complex, termed cohesin, plays an essential role in sister chromatid cohesion in yeast and in Xenopus laevis cell-free extracts. We report here that two distinct cohesin complexes exist in Xenopus egg extracts. A 14S complex (x-cohesinSA1) contains XSMC1, XSMC3, XRAD21, and a newly identified subunit, XSA1. In a second 12.5S complex (x-cohesinSA2), XSMC1, XSMC3, and XRAD21 associate with a different subunit, XSA2. Both XSA1 and XSA2 belong to the SA family of mammalian proteins and exhibit similarity to Scc3p, a recently identified component of yeast cohesin. In Xenopus egg extracts, x-cohesinSA1 is predominant, whereas x-cohesinSA2 constitutes only a very minor population. Human cells have a similar pair of cohesin complexes, but the SA2-type is the dominant form in somatic tissue culture cells. Immunolocalization experiments suggest that chromatin association of cohesinSA1 and cohesinSA2 may be differentially regulated. Dissociation of x-cohesinSA1 from chromatin correlates with phosphorylation of XSA1 in the cell-free extracts. Purified cdc2-cyclin B can phosphorylate XSA1 in vitro and reduce the ability of x-cohesinSA1 to bind to DNA or chromatin. These results shed light on the mechanism by which sister chromatid cohesion is partially dissolved in early mitosis, far before the onset of anaphase, in vertebrate cells.
Keywords: Xenopus egg extract, sister chromatid cohesion, SMC proteins, chromosome structure, mitosis
Introduction
Sister chromatid cohesion is established along the entire length of the chromosome at the time of DNA replication and persists throughout the G2 phase (for reviews see Miyazaki and Orr-Weaver 1994; Biggins and Murray 1999; Nasmyth 1999). In most organisms, each chromatid undergoes dramatic structural changes, known as condensation, at the onset of mitosis (for reviews see Koshland and Strunnikov 1996; Hirano 2000). The condensation process is likely to accompany a partial release of cohesion, producing two recognizable chromatids within a metaphase chromosome. Nevertheless, the linkage between the sister chromatids is maintained to ensure proper alignment of the chromosomes on the metaphase plate. The final release of cohesion takes place at the onset of anaphase, leading to complete separation of the sister chromatids. Malfunction in any one of these processes would result in chromosome missegregation and aneuploidy, potentially contributing to birth defects or tumor development (for reviews see Hassold and Jacobs 1984; Lengauer et al. 1998).
Recent studies show that a multisubunit complex, termed cohesin, is likely to be a central player in sister chromatid cohesion. In Saccharomyces cerevisiae, Smc1p, Smc3p, and Scc1p (also known as Mcd1p) were identified in genetic screens for mutants that displayed premature separation of sister chromatids in mitosis (Michaelis et al. 1997). Scc1p/Mcd1p was also characterized as a gene product that interacts genetically and physically with Smc1p (Guacci et al. 1997). Smc1p and Smc3p belong to a large family of chromosomal ATPases (the structural maintenance of chromosomes [SMC] family), members of which are involved in many aspects of higher order chromosome architecture and dynamics (for reviews see Hirano 1999; Strunnikov and Jessberger 1999). In Xenopus laevis, a systematic search for SMC proteins led to the identification of a 14S protein complex in which XSMC1, XSMC3, and XRAD21, the homologues of Smc1p, Smc3p, and Scc1p/Mcd1p (Rad21 in Schizosaccharomyces pombe; Birkenbihl and Subramani 1995), respectively, associate with two unidentified subunits, p155 and p95. Immunodepletion of Xenopus cohesin from interphase egg extracts results in the assembly of chromosomes with unpaired sister chromatids in subsequent mitosis (Losada et al. 1998). More recently, a biochemical study in S. cerevisiae has further confirmed that Smc1p, Smc3p, and Scc1p/Mcd1p are present in a yeast cohesin complex, along with a new subunit called Scc3p (Toth et al. 1999).
In S. cerevisiae, the cohesin complex binds to chromatin in late G1, establishes sister chromatid cohesion in the S phase, and remains bound to chromatin by metaphase (Michaelis et al. 1997; Uhlmann and Nasmyth 1998). At the metaphase–anaphase transition, the Scc1p/Mcd1p subunit is proteolytically cleaved in an Esp1p/separin-dependent manner, which in turn triggers dissociation of cohesin from chromatin and promotes sister chromatid separation (Uhlmann et al. 1999). A similar, if not identical, mechanism may also operate in S. pombe (Yanagida 2000). However, in vertebrate cells, the timing of the dissociation of cohesin from chromatin is drastically different from that of yeast cohesin. In Xenopus, cohesin binds to chromatin during interphase, but most of it (∼95%) dissociates from chromatin upon entry into mitosis, when cdc2-cyclin B gets activated (Losada et al. 1998). Immunofluorescent staining of human and mouse cells also shows that the bulk of SMC1 and SMC3 relocates from chromatin to cytoplasm in early mitosis (Schmiesing et al. 1998; Darwiche et al. 1999; Stursberg et al. 1999).
To explain these results, two models have been proposed for the role of vertebrate cohesins in sister chromatid cohesion (Losada et al. 1998). In the first model, the cohesin complex participates in holding the sister chromatids together from the S phase to G2, but it does not play a major role in cohesion from prophase to metaphase. A different protein component, such as MEI-S332 protein in Drosophila melanogaster (LeBlanc et al. 1999), could function as a mitosis-specific chromatid glue. In the second model, the cohesin complex is released from chromatin sequentially at two different stages of mitosis in vertebrate cells. A partial release of cohesin, and thereby a partial dissolution of cohesion, occurs in early mitosis, concomitant with condensin-mediated chromatid compaction. A small amount of cohesin that remains bound to intersister regions of the condensed chromatids may keep functioning as a glue in metaphase. The recent discovery of a pair of separin and securin molecules from Xenopus and humans (Zou et al. 1999) implies that a separin-dependent cleavage of cohesin subunits (or other cohesion molecules) could trigger anaphase sister chromatid separation in vertebrates, as has been demonstrated in S. cerevisiae (Uhlmann et al. 1999). Even less is known about how dissociation of cohesins from chromatin in early mitosis might be regulated in vertebrate cells. Release at this stage could also be promoted by a mechanism involving protein cleavage (Orr-Weaver 1999). Alternatively, it could be regulated by a totally different postranslational modification of cohesin subunits.
To address these unresolved issues, we have first identified p155, a previously uncharacterized subunit of the 14S cohesin complex, as the Xenopus orthologue of mammalian stromal antigen (SA) 1 (Carramolino et al. 1997), and named it XSA1. We have also found that a minor population of XSMC1, XSMC3, and XRAD21 associates with XSA2, a different member of the SA protein family, forming a novel cohesin complex of ∼12.5S. Both XSA1 and XSA2 share similarity to yeast Scc3p (Toth et al. 1999). The SA1-type (cohesinSA1) and SA2-type (cohesinSA2) complexes are also present in human cells. Unlike in Xenopus egg extracts, however, cohesinSA2 is more abundant than cohesinSA1 in somatic tissue culture cells. Immunolocalization experiments have shown that cohesinSA1 and cohesinSA2 are differentially regulated at the level of nuclear transport in Xenopus cell-free extracts. Finally, we provide evidence that phosphorylation of the SA subunits could play a key role in dissociation of cohesins from chromatin in early mitosis.
Materials and Methods
Cloning of the 155-kD Subunit of the Xenopus Cohesin Complex
Xenopus cohesin complexes were immunoprecipitated with anti-XSMC3 antibody from a Xenopus egg high speed supernatant, separated by 7.5% SDS-PAGE, and stained with Coomassie blue G. A gel fragment containing p155 was excised and digested in situ with lysylendopeptidase. The resulting peptides were fractionated by reverse phase chromatography and sequenced by Edman degradation as described previously (Bell et al. 1993). The following sequences were obtained: X(F)PR(N)D(P)QAE (E)ALA; (M)YSDAFLNTS(Q/Y)(L)K; XLDSLLK. A database search revealed that highly homologous sequences are present in a human protein called SA1 of unknown function (Carramolino et al. 1997). Taking advantage of this information, a human cDNA fragment was amplified by PCR using a λgt10 library as a template. Oligonucleotides used for the PCR were as follows: hSA1-6, 5′-GTGGAATTCTTTGGCTGTTTGCCAGCAGTG-3′ (EcoRI tag sequence is underlined); and hSA1-8, 5′-GCAGGATCCACCTTGCTCTTGAACAAGTTC-3′ (BamHI tag sequence is underlined). The two primers amplified a 520-bp fragment containing a part of human SA1 cDNA (corresponding to amino acids 773–939), which was used as a hybridization probe to screen a Xenopus oocyte cDNA library (Stratagene). After two rounds of screening, 15 independent clones were obtained that could be classified into two groups. The major group (11/15 clones) encoded a protein most homologous to human and mouse SA1 (designated XSA1 for Xenopus SA1), whereas the minor group (4/15) encoded a protein most homologous to human and mouse SA2 (designated XSA2). A full-length sequence for XSA1 was assembled from four overlapping clones. For XSA2, a full-length sequence was determined after amplifying the 5′ end sequence by nested PCR. The peptide sequences derived from microsequencing are all found in XSA1, but not in XSA2. Therefore, we conclude that the p155 cohesin subunit is identical to XSA1.
Preparation of Antibodies
Rabbit polyclonal antisera were raised against synthetic peptides corresponding to the COOH-terminal amino acid sequences of XSA1 (EDDSGFGMPMF) and XSA2 (DPASIMDESVLGVSMF). Immunization and affinity purification of antibodies were performed as described previously (Hirano et al. 1997). We also raised antisera against a recombinant XSA1 protein and obtained results similar to those with the peptide antibodies.
Extract Preparation
Low speed supernatants (LSSs) of Xenopus eggs were prepared in XBE2 buffer (10 mM potassium-Hepes, pH 7.7, 0.1 M KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA) as described previously (Losada et al. 1998). LSSs were further fractionated by centrifugation at 50,000 rpm at 4°C for 2 h in a Beckman TLS55 rotor to yield high speed supernatants (HSSs). HSSs do not contain membrane fractions and, thereby, cannot initiate DNA replication in vitro (Sheehan et al. 1988). HeLa cell nuclear extracts were prepared in buffer B (20 mM potassium-Hepes, pH 8.0, 0.1 M KCl, 2 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 1 mM β-mercaptoethanol, 20% glycerol), as described by Mayeda and Krainer 1999 with minor modifications: nuclear pellets were resuspended in 0.3 M KCl instead of 0.2 M KCl; and a sonication step was included after homogenization of nuclei.
Immunoprecipitation, Immunodepletion, and Immunoaffinity Purification
Immunoprecipitations were performed as described previously (Hirano et al. 1997). For immunodepletions, 25 μl of protein A agarose beads (Life Technologies Inc.) or Affi-Prep protein A support (Bio-Rad Laboratories) were coated with 5 μg each of anti-XSMC1 and anti-XSMC3, 10 μg of anti-XSA1, 10 μg of anti-XSA2, or 10 μg of control rabbit IgG. The antibody-coupled beads were incubated with 50 μl of extract (HSS or LSS) at 4°C for 1 h. The extent of each depletion was estimated by quantitative immunoblotting using internal standards. For affinity purification, interphase HSS was first fractionated with 35% ammonium sulfate. Most of the 9S cohesin complex remains soluble, whereas the cohesin holocomplexes precipitate after incubation on ice for 30 min. The pellet was resuspended in XBE2-gly (to half the initial volume of extract), and mixed in batches with 1/10 vol of protein A agarose beads to which anti-XSMC3 antibodies had been covalently coupled (0.5–1 mg antibody/ml beads; Hirano et al. 1997). After incubation at 4°C for 1 h, the mixture was poured into a column and washed consecutively with 80 vol of buffer XBE2 containing 10% glycerol (XBE2-gly), 10 vol of XBE2-gly containing 0.4 M KCl, and 10 vol of XBE2-gly supplemented with 0.5% Triton X-100. Cohesins were eluted from the column with XBE2-gly containing 0.5 mg/ml XSMC3-tail peptide. Peak fractions were pooled, supplemented with 0.1 mg/ml casein and 1 mM DTT, and concentrated fourfold in Microcon-30 tubes (Millipore Corp.). For the filter-binding assay, the salt concentration of the protein solution was reduced to 50 mM KCl during the concentration step. Immunoaffinity purification of h-cohesinSA1 and h-cohesinSA2 from HeLa nuclear extracts was carried out in the same way, using anti-XSA1 and anti-XSA2, respectively, and buffer B containing 10% glycerol instead of XBE2-gly. Ammonium sulfate fractionation was omitted in this case.
Immunofluorescent Staining
Demembranated sperm nuclei (1,000 nuclei/μl) were incubated with cell cycle–specific Xenopus egg HSS to assemble interphase chromatin and mitotic chromosomes in vitro. 10-μl samples were fixed with 100 μl of 2% paraformaldehyde in XBE2 for 10 min and centrifuged onto coverslips through a 5-ml cushion of 30% glycerol in XBE2. For nuclei or chromosomes assembled in LSS, 0.25% Triton X-100 was included in the fixation step. HeLa cells were grown on poly-l-lysine–coated coverslips, fixed with 2% paraformaldehyde, and permeabilized with 0.2% Triton X-100. Fixed samples were processed for immunofluorescence as described previously (Losada et al. 1998) and analyzed using a Zeiss Axiophot microscope equipped with a cooled charge-coupled device camera (Photometrics Ltd.). Grayscale images were pseudocolored and merged using Adobe Photoshop 5.5 software. Similar results were obtained when HeLa cells were fixed in methanol at −20°C for 5 min.
Solid-phase Chromatin Assembly
Solid-phase DNA templates were prepared according to Sandaltzopoulos and Becker 1999. A 10.5-kb plasmid DNA (pWH-5) was digested with SmaI and HindIII, and the HindIII site was labeled with Klenow enzyme in the presence of biotin-16-dUTP. Unincorporated biotinylated nucleotides and the excised SmaI-HindIII fragment were removed by gel filtration (Chromaspin 1000; CLONTECH Laboratories, Inc.). The labeled DNA was coupled to streptavidin-coated paramagnetic beads (model M-280; Dynal Inc.) following the manufacturer's instructions. To assemble solid-phase chromatin, 100 μl of twofold-diluted interphase HSS was supplemented with an ATP regeneration system (1 mM MgATP, 10 mM creatine phosphate, and 50 μg/ml creatine kinase) and 0.002% NP-40, and incubated with 1 μg of DNA (coupled to 50 μg of beads) at room temperature for 1–2 h on a rotating wheel. To analyze chromatin components, the beads were separated from the extract, washed with XBE2 containing 0.2% NP-40, and incubated at 37°C for 10 min in SDS buffer (62.5 mM Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 0.002% bromophenol blue, 10 mM DTT). The eluted fraction was analyzed by immunoblotting. For the cohesin dissociation assay (Fig. 7 B), solid-phase chromatin was assembled on the DNA-coupled magnetic beads in interphase HSS, washed with XBE2-0.05% NP-40, and resuspended in XBE2-gly containing 0.1 mg/ml ovalbumin and 1 mM MgATP. 1/20 vol of a cdc2-cyclin B fraction (see below) or of control buffer was added, and the mixture was incubated at room temperature for 1 h. Released fractions were saved, and unreleased fractions were recovered from the beads after washing them with XBE2-0.2% NP-40. The two fractions were analyzed by immunoblotting.
Figure 7.
Phosphorylation of x-cohesin by cdc2-cyclin B and its effect on DNA binding. (A) Xenopus cohesins were immunoprecipitated from an interphase HSS with anti-XSMC3, and incubated with purified cdc2-cyclin B (lanes 1 and 3) or no kinase (lanes 2 and 4) in the presence of γ-[32P]ATP. After washing, the immunoprecipitates were separated by SDS-PAGE and analyzed by autoradiography (lanes 1 and 2) or by immunoblotting (lanes 3 and 4). (B) Solid-phase chromatin was assembled on DNA-coupled magnetic beads in an interphase HSS, washed, and treated with cdc2-cyclin B (lanes 1 and 2) or no kinase (lanes 3 and 4) in the presence of γ-[32P]ATP. After incubation at 22°C for 1 h, proteins remaining bound to the chromatin beads (lanes 1 and 3; unreleased [U]) and those dissociated from the beads (lanes 2 and 4; released [R]) were analyzed by immunoblotting with antibodies against cohesin subunits (top) or by autoradiography (bottom). (C) Filter binding assay. Purified cohesins were first treated with no kinase (lane 2), cdc2-cyclin B (lane 3), or cdc2-cyclin B in the presence of the CDK inhibitor roscovitine (lane 4), and then incubated with plasmid DNA in a low salt buffer. Buffer alone (lane 1) or cdc2-cyclin B alone (lane 5) were also mixed with plasmid DNA. The binding reactions were passed through glass fiber filters, and washed sequentially with low salt buffer, high salt buffer, and buffer containing SDS. The ratio of bound DNA to total DNA was calculated as described previously (Kimura et al. 1999). The average of values from three independent experiments was plotted with error bars. (D) Bead-binding assay. Unphosphorylated (lanes 1, 3, and 5) or phosphorylated (lanes 2, 4, and 6) cohesins were mixed with no-DNA beads (lanes 1 and 2; control beads), DNA-coupled beads (lanes 3 and 4; DNA beads), or solid-phase chromatin beads assembled in a cohesin-depleted interphase HSS (lanes 5 and 6; chromatin beads). After incubation for 1 h, the beads were washed and proteins bound to the beads were analyzed by immunoblotting.
In Vitro Phosphorylation of Cohesins
An active cdc2-cyclin B fraction was purified from Xenopus interphase HSS as described previously (Solomon et al. 1992). The final protein solution contained 600 nM cdc2-cyclin B in EB buffer (80 mM β-glycerophosphate, pH 7.3, 15 mM MgCl2, 20 mM EGTA, 0.1 mg/ml ovalbumin, 0.05% NP-40, 8% glycerol, 0.05 mM ATP, 5 mM glutathione, 10 mM DTT, 0.5 mM PMSF). Phosphorylation of immunoprecipitated cohesins was performed as follows (see Fig. 7 A). Xenopus cohesins were recovered on protein A agarose beads from interphase HSS using anti-XSMC3 antibodies, and the beads were resuspended in XBE2-gly containing 0.1 mg/ml ovalbumin and 1 mM MgATP. 1/20 vol of cdc2-cyclin B solution or EB buffer alone was added, and the mixture was incubated at 22°C for 1 h. In some reactions, γ-[32P]ATP (6,000 Ci/mmol) was included at a final concentration of 0.5 μCi/μl. After washing the beads with XBE2, protein samples were separated by SDS-PAGE and analyzed by immunoblotting or autoradiography. For phosphorylation of cohesins in solution (see Fig. 7C and Fig. D), purified cohesins (50–100 nM) were supplemented with 0.1 mg/ml ovalbumin and 1 mM MgATP, and incubated with 1/10 vol of cdc2-cyclin B or EB buffer alone at 22°C for 1 h. When necessary, roscovitine (Calbiochem) was added to the phosphorylation reactions at a final concentration of 0.3 mM to block the cyclin-dependent kinase (CDK) activity (Rudolph et al. 1996).
DNA- or Chromatin-binding Assays for Purified Cohesins
The filter-binding assay (see Fig. 7 C) was performed as described previously (Kimura et al. 1999). The binding reaction mixture (5 μl) contained 10 ng of supercoiled plasmid DNA (JHX-1, 3.2 kb) and ∼90 nM affinity-purified cohesins (phosphorylated or unphosphorylated). For the bead-binding assay (see Fig. 7 D), cohesins were mixed with three different kinds of beads: (1) beads with no coupled DNA; (2) beads with coupled DNA; and (3) beads on which interphase chromatin had been assembled in a cohesin-depleted HSS. The binding mixtures (10 μl) contained 50 μg of magnetic beads (with or without 1 μg of DNA), 60 nM cohesins (phosphorylated or unphosphorylated), 1 mg/ml casein, 1 mM MgATP, and 50 mM β-glycerophosphate. 0.3 mM roscovitine was also added into the binding reactions to stop further phosphorylation by cdc2-cyclin B, which had been carried over from the previous treatment. After incubation at room temperature for 1 h, the beads were washed with XBE2-0.2% NP-40, and bound proteins were analyzed by immunoblotting.
Results
The p155 Subunit of Xenopus Cohesin Is the Orthologue of Mammalian SA1
We cloned a cDNA encoding the 155-kD subunit of the Xenopus 14S cohesin complex on the basis of amino acid sequence obtained by microsequencing (see Materials and Methods). The cDNA predicted a 1,265–amino acid residue polypeptide with a calculated molecular mass of 145.2 kD. A database search revealed that it was highly homologous to a group of mammalian proteins, called SAs, of unknown functions. Three members of the SA family have been found in mouse and humans (Carramolino et al. 1997). The Xenopus p155 sequence is most homologous to the SA1-type proteins (89% identical; Fig. 1), suggesting that the 155-kD cohesin subunit is the Xenopus orthologue of SA1 (XSA1). During the cloning procedure, we isolated another homologous cDNA that encodes the Xenopus orthologue of mammalian SA2 (termed XSA2). XSA2 is 1,194 amino acids long and has a calculated molecular mass of 137.7 kD. XSA1 has an NH2-terminal extension of 78 residues that is not found in XSA2 (Fig. 1). Although the central core regions of the XSA1 and XSA2 sequences are ∼75% identical to each other, the COOH-terminal domain (∼200 amino acids long) is considerably different between them. Consistent with our finding, Toth et al. 1999 have recently reported that Scc3p, a subunit of the Saccharomyces cerevisiae cohesin complex, shows similarity to members of the mammalian SA family.
Figure 1.

Sequence alignment of Xenopus and human SA1 and SA2. Identical amino acid residues are shaded. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers AF255017 and AF255018.
XSA1 and XSA2 Are Present in Different Cohesin Complexes in Xenopus Egg Extracts
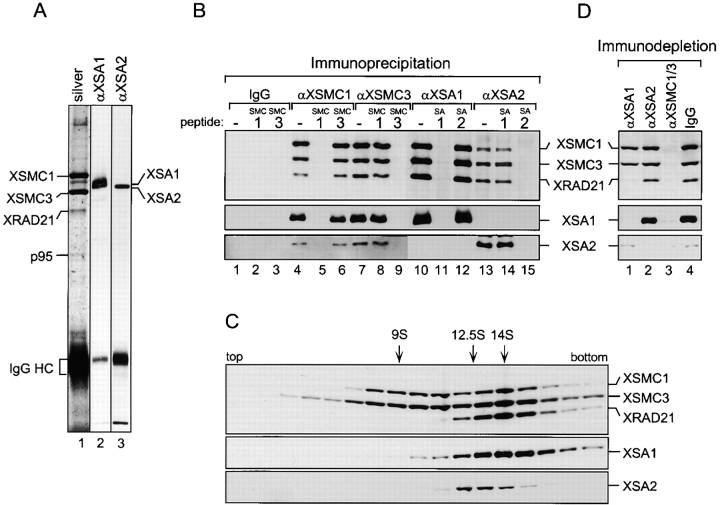
To confirm that XSA1 is indeed a cohesin subunit and to determine the relationship between XSA1 and XSA2, rabbit antibodies were prepared against synthetic peptides corresponding to their COOH-terminal sequences. By immunoblotting against a Xenopus egg extract, anti-XSA1 recognized a polypeptide of 155 kD, whereas anti-XSA2 reacted with a different polypeptide of 150 kD (data not shown). Each antibody was specific for each polypeptide and no cross-reaction was observed. We found that XSA1 and XSA2 were present in the immunoprecipitates obtained with anti-XSMC1 or anti-XSMC3 (Fig. 2A and Fig. B, lanes 4 and 7). However, anti-XSA1 coimmunoprecipitated XSMC1, XSMC3, and XRAD21, but not XSA2 (Fig. 2 B, lane 10). Conversely, anti-XSA2 did not coimmunoprecipitate XSA1 (Fig. 2 B, lane 13). The specificity of all the immunoprecipitation reactions was demonstrated further by the addition of competing peptides (Fig. 2 B, lanes 5, 6, 8, 9, 11, 12, 14, and 15). These results indicate that XSA1 and XSA2 associate with XSMC1, XSMC3, and XRAD21 in two different complexes. Consistently, when a total egg extract was fractionated by sucrose gradient centrifugation, XSA1 and XSA2 sedimented in different positions (Fig. 2 C). The XSA1-containing complex had a sedimentation coefficient of ∼14S, whereas the XSA2-containing complex migrated to a position of ∼12.5S. The egg extract also contained a 9S heterodimer of XSMC1 and XSMC3 as reported previously (Losada et al. 1998). To assess the relative amounts of the three different forms, immunodepletion experiments were performed using antibodies against the different cohesin subunits. Anti-XSA1 depleted ∼90% of XRAD21 and ∼40–50% of both XSMC1 and XSMC3 from the extracts (Fig. 2 D, lane 1), whereas anti-XSA2 depleted only ∼10% of XRAD21 and ∼5% of XSMC1 and XSMC3 (Fig. 2 D, lane 2). A combination of anti-XSMC1 and anti-XSMC3 depleted all the subunits, including the 9S population of XSMC1-XSMC3 (Fig. 2 D, lane 3). These results suggest that, besides the SMC heterodimer, Xenopus egg extracts contain two different cohesin holocomplexes. The 14S complex consists of XSMC1, XSMC3, XRAD21, and XSA1, whereas the 12.5S complex contains XSMC1, XSMC3, XRAD21, and XSA2. In this manuscript, we refer to the XSA1- and XSA2-containing complexes as x-cohesinSA1 and x-cohesinSA2, respectively. x-cohesinSA1 is ten times as abundant as x-cohesinSA2 in the egg extracts. The exact stoichiometry of the subunits in each complex remains to be determined.
Figure 2.
Biochemical characterization of Xenopus cohesin complexes containing XSA1 and XSA2. (A) Cohesins were immunoprecipitated from a Xenopus egg HSS using anti-XSMC3 antibodies and analyzed by silver staining (lane 1) and immunoblotting with anti-XSA1 (lane 2) or anti-XSA2 (lane 3). (B) Immunoprecipitations were performed using antibodies against XSMC1 (lanes 4–6), XSMC3 (lanes 7–9), XSA1 (lanes 10–12), XSA2 (lanes 13–15), or control rabbit IgG (lanes 1–3). The antigen peptides for XSMC1 (lanes 2, 5, and 8), XSMC3 (lanes 3, 6, and 9), XSA1 (lanes 11 and 14), or XSA2 (lanes 12 and 15) were added at 0.4 mg/ml to demonstrate the specificity of these reactions. The immunoprecipitates that were recovered on protein A agarose beads were analyzed by immunoblotting using the antibodies indicated. (C) An interphase HSS was fractionated in a 5–20% sucrose gradient centrifuged at 36,000 rpm for 15 h in an SW50.1 rotor (Beckman), and fractions were analyzed by immunoblotting. The peaks corresponding to x-cohesinSA1 (14S), x-cohesinSA2 (12.5S), and the XSMC1-XSMC3 heterodimer (9S) are indicated. (D) Aliquots of an HSS were immunodepleted using anti-XSA1 (lane 1), anti-XSA2 (lane 2), and a mixture of anti-XSMC1 and anti-XSMC3 (lane 3) or control rabbit IgG (lane 4), and analyzed by immunoblotting.
The SA2-type Cohesin Complex Is Predominant in Human Somatic Tissue Culture Cells
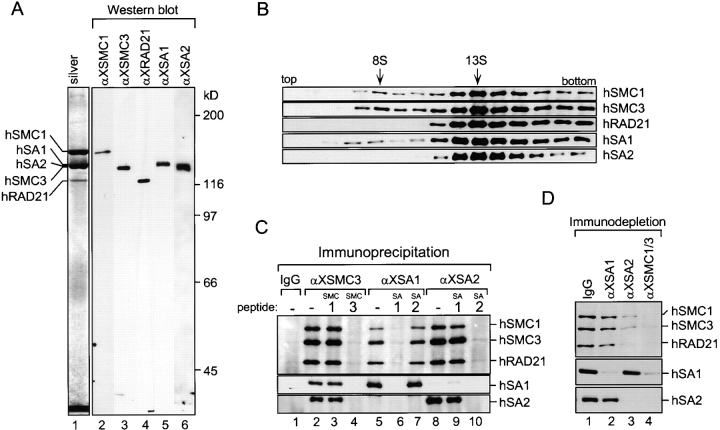
The antibodies raised against the Xenopus cohesin subunits recognize their human counterparts, allowing us to characterize the human cohesin complexes from HeLa cell extracts (Fig. 3 A). A human cohesin fraction affinity-purified using anti-XSMC3 contained hSMC1 (160 kD), hSMC3 (140 kD), and hRAD21 (125 kD). hSA1 and hSA2, the human homologues of XSA1 and XSA2, respectively, were also detected in this fraction by immunoblotting. Sucrose gradient centrifugation of a HeLa cell extract showed that most of hSMC1, hSMC3, hRAD21, hSA1, and hSA2 cosedimented in a major peak of ∼13S (Fig. 3 B). As judged by immunoprecipitation, however, hSA1 and hSA2 are present in different complexes that share hSMC1, hSMC3, and hRAD21 as common subunits (Fig. 3 C). Immunodepletion experiments showed that anti-XSA1 and anti-XSA2 removed ∼25 and 75%, respectively, of hSMC1, hSMC3, and hRAD21 from an extract (Fig. 3 D). Thus, in HeLa cell extracts, the ratio between the hSA1-containing complex (h-cohesinSA1) and the hSA2-containing complex (h-cohesinSA2) is about 1:3. This is in striking contrast to Xenopus egg extracts, in which the ratio between x-cohesinSA1 and x-cohesinSA2 is 10:1.
Figure 3.
Biochemical characterization of the human cohesin complexes. (A) Human cohesins were affinity-purified from HeLa cell nuclear extracts using anti-XSMC3 antibodies and analyzed by silver staining (lane 1) and by immunoblotting with anti-XSMC1 (lane 2), anti-XSMC3 (lane 3), anti-XRAD21 (lane 4), anti-XSA1 (lane 5), and anti-XSA2 (lane 6). (B) A HeLa cell nuclear extract was fractionated in a sucrose gradient as described in Fig. 2 C. The two peaks containing SMC subunits are indicated. (C) Immunoprecipitations from HeLa cell nuclear extracts were performed using control rabbit IgG (lane 1) or antibodies against XSMC3 (lanes 2–4), XSA1 (lanes 5–7) and XSA2 (lanes 8–10). The antigen peptides for XSMC1 (lanes 3), XSMC3 (lanes 4), XSA1 (lanes 6 and 9), or XSA2 (lanes 7 and 10) were added at 0.4 mg/ml to demonstrate the specificity of these reactions. The immunoprecipitates that were recovered on protein A agarose beads were analyzed by immunoblotting. (D) Aliquots of a HeLa cell nuclear extract were immunodepleted using control rabbit IgG (lane 1), anti-XSA1 (lane 2), anti-XSA2 (lane 3), or a mixture of anti-XSMC1 and anti-XSMC3 (lane 4), and analyzed by immunoblotting.
Localization of CohesinSA1 and CohesinSA2 in Xenopus Egg Extracts
Previous experiments using Xenopus cell-free extracts have shown that XSMC1, XSMC3, and XRAD21 bind to chromatin during interphase, and that most of them (∼95%) dissociate from chromatin at the onset of mitosis (Losada et al. 1998). The identification of XSA1 and XSA2 as subunits of two different cohesin complexes prompted us to test whether they have different functional characters in terms of chromatin binding. First, HSSs were used to assemble interphase chromatin and single chromatid mitotic chromosomes from sperm nuclei, and they were analyzed by immunofluorescence. We found that both XSA1 and XSA2 were bound to the interphase chromatin in an indistinguishable manner, and neither of them was detectable in the condensed chromosomes (Fig. 4 A).
Figure 4.

Localization of cohesin complexes in nuclei and chromosomes assembled in Xenopus egg extracts. (A) Xenopus sperm chromatin was incubated in interphase or mitotic HSS. The assembled interphase chromatin (I) and mitotic chromosomes (M) were fixed, isolated through a glycerol cushion, and immunostained with anti-XSA1 (e and f, green) or anti-XSA2 (g and h, green). The DNA was counterstained with DAPI (a–d, red). Merged images are also shown (i–l). (B) Xenopus sperm chromatin was incubated at 22°C for 90 min in interphase LSS, where it was replicated. Half of this interphase mixture (I) was fixed and processed for immunostaining with anti-XSA1 (a, f, and k) or anti-XSA2 (d, i, and n). The other half was driven into mitosis (M) by the addition of a mitotic LSS, incubated for another 2 h, fixed, and immunostained. A mass of entangled chromosomes (b, g, l, e, j, and o) and an individual chromosome (c, h, and m) are shown. (a–e, red) DNA; (f–h, green) anti-XSA1; (i and j, green) anti-XSA2; (k–o) merged images. Stronger staining with anti-XSA1 was often observed on a bent region of the chromosome, most likely corresponding to the pericentromeric region (arrowhead). The images of antibody staining were captured using different exposure times to visualize weak signals on the mitotic chromosomes and, therefore, the label intensities cannot be compared quantitatively. (C) Xenopus sperm nuclei were incubated in an interphase LSS depleted of x-cohesins and supplemented with h-cohesinSA1 and h-cohesinSA2. After incubation at 22°C for 90 min, the nuclei were fixed, and hSA1 and hSA2 were detected with anti-XSA1 and anti-XSA2, respectively. (a and b, red) DNA; (c and d, green) antibody staining; (e and f) merged images. Bars, 5 μm.
An interphase LSS was used to assemble interphase nuclei with a nuclear envelope, in which a complete round of DNA replication took place. The staining of interphase nuclei with anti-XSA1 overlapped completely with the DNA staining (Fig. 4 B, a, f, and k). In contrast, anti-XSA2 primarily labeled the nuclear envelope, and there was a little sign of colocalization to chromatin inside the nucleus (Fig. 4 B, d, i, and n). We converted the reaction mixture into a mitotic state, producing mitotic chromosomes consisting of two paired sister chromatids. Weak labeling on the chromosomes was detected with anti-XSA1 (Fig. 4 B, b, g, and l), but not with anti-XSA2 (Fig. 4 B, e, j, and o). The immunofluorescent signals of anti-XSA1 were located between the sister chromatids (Fig. 4 B, c, h, and m) and often enriched on subchromosomal regions with a high tendency to bend, possibly corresponding to centromeric regions (Fig. 4 B, h and m, arrow). Although similar staining was sometimes observed with anti-XSMC3, anti-XSA1 gave us more consistent results. Immunoblotting experiments show that ∼95% of XSA1 is released from chromatin upon mitotic activation (data not shown), as reported previously for XSMC1, XSMC3, and XRAD21 (Losada et al. 1998). These results suggest that a small population of x-cohesinSA1 (∼5%) remains on the chromatin during metaphase, and may contribute to holding the two sister chromatids together.
The preferential association of XSA2 with the nuclear envelope could indicate that x-cohesinSA1 and x-cohesinSA2 are differentially regulated at the level of nuclear transport in this cell-free extract. To test whether this property is also common to the human complexes, affinity-purified h-cohesinSA1 and h-cohesinSA2 were introduced into Xenopus interphase LSS that had been immunodepleted of the endogenous complexes. Consistent with the behavior of XSA1 and XSA2, hSA1 localized to interphase chromatin (Fig. 4 C, a, c and e), whereas hSA2 stained the nuclear surface (Fig. 4 C, b, d, and f).
Localization of CohesinSA1 and CohesinSA2 in Human Tissue Culture Cells
The localization of human cohesin complexes in HeLa cells was examined by immunofluorescence (Fig. 5). In interphase, both hSA1 and hSA2 localized to the nucleus. There was no sign of an enrichment of hSA2 on peripheral regions of the nucleus. Detergent extraction of the cells before fixation did not affect the staining of interphase cells (data not shown), suggesting that these proteins actually bind to chromatin. After nuclear envelope breakdown, the proteins were distributed throughout the whole cell, and they appeared to be excluded from the condensed chromosomes. We have been unable to detect hSA1, hSA2, or hSMC3 in mitotic chromosomes either between the sister chromatids or at centromeric regions. In telophase, hSA1 and hSA2 started to accumulate inside the newly formed daughter cell nuclei. Thus, h-cohesinSA1 and h-cohesinSA2 appear to behave very similarly throughout the cell cycle in HeLa cells: the two complexes associate with chromatin in telophase; and the bulk of them dissociate from chromatin at the onset of mitosis.
Figure 5.
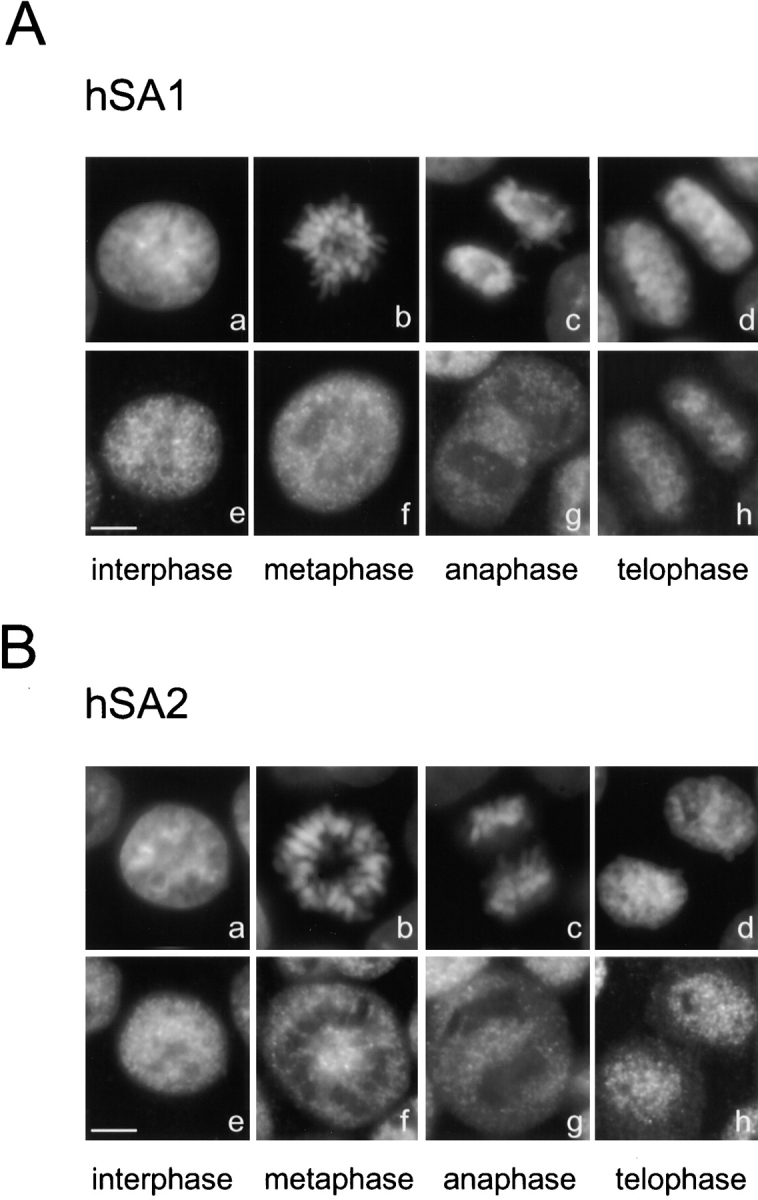
Localization of cohesin complexes in human tissue culture cells. HeLa cells were fixed with paraformaldehyde, and hSA1 (A) and hSA2 (B) were immunolocalized using anti-XSA1 and anti-XSA2, respectively. (a–d) DNA; (e–h) antibody staining. Bars, 5 μm.
A Cohesin Dissociation Assay Using a Solid-phase Chromatin Assembly System
To examine how dissociation of cohesins from chromatin might be regulated in early mitosis, an in vitro assay was developed using DNA-coupled paramagnetic beads (Sandaltzopoulos and Becker 1999). Both interphase and mitotic extracts support efficient assembly of nucleosomes around the immobilized DNA, as judged by micrococcal nuclease digestion (data not shown). The advantage of this solid-phase chromatin over sperm chromatin is that it can be easily separated from soluble components in a magnetic field. We found that cohesins, but not condensins, were recovered on chromatin assembled in an interphase extract (Fig. 6 A, lane 4). Conversely, mitotic chromatin contained condensins but not cohesins (Fig. 6 A, lane 3). Neither cohesins nor condensins were detected when uncoupled control beads were used (Fig. 6, lane 1 and 2). When immobilized chromatin containing cohesins was exposed to a mitotic extract, most cohesins dissociated and were replaced with condensins (Fig. 6 A, lane 5). These results show that this solid-phase chromatin assembly system is able to faithfully reproduce the cell cycle–specific targeting of cohesins and condensins previously described using sperm-derived chromatin (Hirano et al. 1997; Losada et al. 1998).
Figure 6.

Regulation of cohesin by phosphorylation in the cell-free extracts. (A) DNA-coupled paramagnetic beads were incubated with mitotic (M) or interphase (I) HSS to assemble solid-phase chromatin (lanes 3 and 4). Uncoupled beads were treated in the same way as controls (lanes 1 and 2). After washing the beads, bound proteins were analyzed by immunoblotting with antibodies against cohesin (upper) and condensin subunits (lower). Alternatively, solid-phase chromatin was assembled in an interphase HSS, washed, and then placed in a mitotic HSS (lane 5; I > M) or a cohesin-depleted mitotic HSS (lane 6; I > ΔM). After incubation for 2 h, proteins bound to the beads were analyzed as above. The fraction released from the I > ΔM chromatin beads was immunoprecipitated with anti-XSMC3 and analyzed by immunoblotting (lane 7). (B) DNA-coupled beads were incubated with an interphase HSS (I) for 1 h (lane 1). Half of the reaction mixture was incubated for another 1 h after addition of okadaic acid to 1 μM (lane 2; OA). The DNA beads were also incubated in a mitotic HSS (M) without (lane 3) or with (lane 4) 5 mM 6-dimethylaminopurine (DMAP) for 2 h. The chromatin beads were separated from the extract and washed. Cohesin fractions, which were present in chromatin (top) or extracts (middle and bottom), were detected by immunoblotting with the indicated antibodies. (C) A Xenopus interphase LSS was immunodepleted of x-cohesins using anti-XSMC3. An aliquot of the depleted extract was saved to check the absence of x-cohesins (lane 1). The remainder was divided into two parts, and one of them was converted into a mitotic state by addition of cyclin BΔ90 (Glotzer et al. 1991). Both LSSs depleted of x-cohesins were supplemented with purified h-cohesinSA1 and h-cohesinSA2 (∼7.5 nM each). After incubation at 22°C for 90 min, aliquots were taken from the interphase (lane 2) and the mitotic (lane 3) extracts. The human cohesin complexes were recovered from the LSS by immunoprecipitation with anti-XSMC3, and treated with buffer alone (lane 4) or λ-phosphatase (lane 5) at 30°C for 1 h. The aliquots of the extracts and the immunoprecipitates were separated by SDS-PAGE, and hSA1 and hSA2 were detected by immunoblotting with anti-XSA1 (top) and anti-XSA2 (bottom), respectively.
To test whether the mitotic dissociation of cohesins from chromatin might be accompanied by proteolytic cleavage of the XRAD21 subunit, cohesin-bound interphase chromatin was incubated with a cohesin-depleted mitotic extract (Fig. 6 A, lane 6), and the released fraction was immunoprecipitated with anti-XSMC3. The four subunits of x-cohesinSA1, which were released from the chromatin, remained intact, and their relative abundance was indistinguishable from that of the chromatin-bound complex (Fig. 6 A, compare lanes 4 and 7). This result suggests that dissociation of cohesins from chromatin is likely to involve a postranscriptional modification other than proteolytic cleavage.
Phosphorylation of Cohesins in the Cell-free Extracts
As an initial attempt to test whether dissociation of cohesin from chromatin may be regulated by mitotic phosphorylation, we took a pharmacological approach. Interphase chromatin was assembled on DNA-coupled beads to allow cohesins to bind (Fig. 6 B, top, lane 1) and then okadaic acid (an inhibitor of the type 1 and type 2A phosphatases) was added into the assembly mixture. This treatment caused dissociation of cohesins from the solid-phase chromatin (Fig. 6 B, top, lane 2). Under this condition, the electrophoretic mobility of the XSA1 subunit in the extract was changed, whereas those of the other subunits were unaffected (Fig. 6 B, middle and bottom, compare lanes 1 and 2). A less prominent shift of XSA1 was also detectable in mitotic extracts (Fig. 6 B, bottom, lane 3). The shift was not observed when the kinase inhibitor 6-dimethylaminopurine was added into the extracts (Fig. 6 B, bottom, lane 4), and this treatment partially restored the ability of cohesins to bind to chromatin (Fig. 6 B, top, lanes 3 and 4). Identical binding properties were observed using demembranated sperm nuclei instead of the solid-phase DNA templates (data not shown). These results suggest that chromatin association/dissociation of cohesins is regulated by a kinase/phosphatase balance in the extracts, and that a major target of phosphorylation is likely to be the XSA1 subunit.
The low amount of XSA2 in the egg extracts precluded a similar analysis of its phosphorylation. However, when purified human cohesin complexes were added into Xenopus egg extracts that had been previously depleted of the endogenous cohesin complexes (Fig. 6 C, lane 1), both hSA1 and hSA2 exhibited an electrophoretic mobility shift in a mitosis-specific manner (Fig. 6 C, lanes 2 and 3). Treatment of the human complexes with λ-phosphatase after reisolation from the mitotic extracts eliminated the shift (Fig. 6 C, lanes 4 and 5). Taken together, these results suggest that the SA1 and SA2 subunits become phosphorylated during mitosis, concomitant with their release from chromatin.
Phosphorylation of Cohesins by Cdc2 In Vitro
It has been shown that cdc2-cyclin B can directly phosphorylate condensin subunits in vitro and activate the positive supercoiling activity of the complex (Kimura et al. 1998). Similarly, cdc2-cyclin B may also phosphorylate cohesin subunits and modulate cohesin functions in vitro. When immunoprecipitated cohesins were incubated with cdc2-cyclin B in the presence of γ-[32P]ATP, only XSA1 was heavily labeled (Fig. 7 A, lanes 1 and 2). Moreover, this subunit exhibited an electrophoretic mobility shift upon phosphorylation (Fig. 7 A, lanes 3 and 4). The radiolabeling and the mobility shift were both sensitive to roscovitine, a CDK inhibitor (data not shown). A weak phosphorylation of XSA2 by cdc2-cyclin B could be detected, but it was difficult to study this event further because of its low abundance in the egg extracts.
One attractive possibility is that cdc2 phosphorylation of XSA1 induces dissociation of the cohesin complex from chromatin in early mitosis. To test this model, interphase chromatin was assembled on DNA-coupled beads, purified in a magnetic field, and incubated with or without cdc2-cyclin B. We were unable to detect cdc2-dependent dissociation of cohesin from chromatin in this purified system. A small population of cohesin was released into the soluble fraction, but this occurred even in the absence of cdc2 (Fig. 7 B, upper). Interestingly, only the soluble XSA1 in the released fraction, but not chromatin-bound XSA1, was phosphorylated by cdc2 under those conditions (Fig. 7 B, lower). This observation led to another possibility that phosphorylation of XSA1 by cdc2 might prevent reassociation of cohesins once they are released from chromatin during early mitosis. To investigate this, a filter-binding assay was first performed using affinity-purified Xenopus cohesins and naked DNA as a substrate. We found that cohesins can bind directly to DNA in this assay (Fig. 7 C, lane 2). Pretreatment of the cohesin fraction with cdc2-cyclin B significantly reduced its DNA-binding activity (Fig. 7 C, lane 3), and this effect was reversed when roscovitine was added into the phosphorylation reaction (Fig. 7 C, lane 4). In a complementary assay, DNA-coupled magnetic beads were used as binding substrates. Purified cohesins were found to efficiently bind to the DNA-coupled beads (Fig. 7 D, lane 3) but not to uncoupled control beads (Fig. 7 D, lanes 1 and 2). The amount of cohesins bound to the DNA beads was drastically reduced when cohesins were prephosphorylated with cdc2-cyclin B (Fig. 7 D, lane 4). Finally, we tested the binding of cohesins to the solid-phase chromatin assembled on the DNA-coupled beads. Again, the unphosphorylated form of cohesins exhibited a higher affinity to chromatin compared with the phosphorylated form (Fig. 7 D, lanes 5 and 6). These results show that phosphorylation of the XSA1 subunit by cdc2 decreases binding of cohesins to DNA and chromatin in vitro.
Discussion
Two Different Cohesin Complexes in Vertebrate Cells
Cohesins are multisubunit protein complexes that play a major role in sister chromatid cohesion. In this paper, we report the identification and characterization of two new cohesin subunits from Xenopus egg and human cell extracts. Both of them belong to the mammalian SA protein family, whose cellular functions were previously unknown (Carramolino et al. 1997). In Xenopus egg extracts, XSA1 and XSA2 associate with XSMC1, XSMC3, and XRAD21 to form a 14S complex (x-cohesinSA1) and a 12.5S complex (x-cohesinSA2), respectively. The difference between the sedimentation coefficients of the two complexes could be explained if x-cohesinSA1 had an additional subunit. In fact, our previous study showed that a 95-kD polypeptide (p95), whose identity remains to be determined, cosediments with the 14S complex by sucrose gradient centrifugation (Losada et al. 1998). The low abundance of x-cohesinSA2 (∼1/10 of x-cohesinSA1) explains why this population has been overlooked before. HeLa cell extracts also contain the SA1- and SA2-type cohesin complexes (h-cohesinSA1 and h-cohesinSA2, respectively). The two complexes share human homologues of XSMC1, XSMC3 (Schmiesing et al. 1998), and XRAD21 (McKay et al. 1996) as common subunits, and have similar sedimentation coefficients of ∼13S. There is no evidence for the presence of p95 in h-cohesinSA1, which is consistent with its S value being smaller than that of x-cohesinSA1. The ratio between cohesinSA1 and cohesinSA2 is 10:1 in Xenopus egg extracts, and 1:3 in HeLa cell extracts. The dominance of the SA1-type complex in Xenopus eggs is likely to be characteristic of early embryonic systems, because the SA1:SA2 ratio in Xenopus somatic tissue culture cells XL177 is very similar to that observed in HeLa cells (data not shown).
Recently, Toth et al. 1999 reported that Scc3p, a gene product required for sister chromatid cohesion in S. cerevisiae, has similarity to mammalian SA proteins, and is a subunit of the yeast cohesin complex containing Smc1p, Smc3p, and Scc1p/Mcd1p/Rad21. Thus, the subunit composition of the cohesin complexes is highly conserved from yeast to humans. Since Scc3p is the only SA-like sequence present in the genome of S. cerevisiae, it is most likely that a single cohesin complex functions in the mitotic cycle in this organism.
Chromatin Association of CohesinSA1 and CohesinSA2
Are there any functional differences between the SA1-type and SA2-type complexes? In nuclei assembled in an LSS, which are surrounded by a nuclear membrane, x-cohesinSA1 and x-cohesinSA2 exhibit different localization. XSA1 localizes to interphase chromatin within the nuclei, whereas XSA2 is apparently present on the nuclear envelope. Interestingly, when purified human cohesins are introduced into a Xenopus LSS, hSA1 and hSA2 behave like XSA1 and XSA2, respectively. These observations suggest that cohesinSA1 and cohesinSA2 are differentially regulated at the level of nuclear transport in this cell-free system. The p95 subunit present in x-cohesinSA1 could play an active role in this process. When added into a Xenopus egg extract, only h-cohesinSA1, but not h-cohesinSA2, may recruit p95 and thereby form a transport-competent complex. The NH2-terminal tail sequence conserved among SA1 proteins could offer a binding site for p95. The rapid transport of the abundant SA1-type complex across the nuclear membrane may be a strategy unique to the early embryonic cell cycle. It is known that, during early development of Xenopus, a nuclear envelope assembles around each individual chromosome shortly after anaphase (forming the so-called karyomeres) to allow rapid initiation of DNA replication before full reconstitution of the nucleus (Lemaitre et al. 1998). Later in development, a slower reformation of the nuclear membrane may provide enough time for cohesins to directly bind to the chromatin at the end of mitosis. In fact, no significant differences were observed in the timing of association of SA1 and SA2 with chromatin in human or Xenopus somatic tissue culture cells, as judged by immunofluorescent staining (Fig. 5 and data not shown). Consistently, if nuclear envelope assembly is suppressed by using HSS, both XSA1 and XSA2 can associate with interphase chromatin with similar efficiency in the Xenopus cell-free system (Fig. 4 A).
It is formally possible that cohesinSA1 and cohesinSA2 have distinct properties in terms of chromatin binding. A recent study in S. cerevisiae has shown that a protein complex containing Scc2p and Scc4p is required to load cohesin onto chromatin (Ciosk et al. 2000). In vertebrate cells, specific loading factors could exist for the two different cohesin complexes and determine their specialized functions by recruiting them to different chromosomal loci.
Do Cohesins Dissociate from Chromatin in Two Steps in Vertebrate Cells?
In S. cerevisiae, dissociation of cohesin from chromatin in anaphase leads to a single-step separation of sister chromatids (Uhlmann et al. 1999). However, in vertebrate cells, dissolution of sister chromatid linkage may be regulated by a two-step mechanism (Hirano 2000). The first step initiates at the onset of mitosis when tight cohesion between interphase chromatids is partially released, allowing each chromatid to condense. The second step occurs at the metaphase-anaphase transition and results in complete separation of the sister chromatids. Our current results suggest that dissociation of cohesins from chromatin may accompany the two-step resolution of cohesion in vertebrate cells. When the nuclei that are assembled in Xenopus LSS are converted into chromosomes consisting of two paired sister chromatids, most XSA1 and XSA2 dissociate from the nuclear structures, a result consistent with previous analysis of the other cohesin subunits (Losada et al. 1998). A small population of XSA1, however, is detectable between the two sister chromatids, suggesting that a small amount of cohesins could, in fact, play a role in holding the sister chromatids in metaphase. The apparent enrichment of x-cohesinSA1 in centromeric regions is consistent with the recent identification of cohesin association sites in S. cerevisiae by chromatin immunoprecipitation experiments (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). However, it should be added that we have been unable to detect any of the cohesin subunits on metaphase chromosomes in tissue culture cells. The intersister signals may be very sensitive to staining conditions and difficult to detect on chromosomes assembled in the cell. Alternatively, a cohesion molecule other than cohesin could play a predominant role in holding sister chromatids together in somatic metaphase chromosomes.
Regulation of Cohesin by Mitotic Phosphorylation In Vitro
If cohesins are released from chromatin at two different stages of mitosis, how are they regulated differentially? A recent study shows that vertebrate cells have a pair of separin and securin molecules, which may play a role in anaphase sister chromatid separation (Zou et al. 1999). However, it remains to be determined, whether separin-mediated cleavage of the RAD21 subunit triggers the release of cohesin at the onset of anaphase in vertebrate cells, as has been shown in S. cerevisiae (Uhlmann et al. 1999).
The current study focuses on the mechanism that promotes dissociation of cohesin from chromatin in early mitosis, and provides evidence that it may involve protein phosphorylation. We have developed an in vitro assay using DNA-coupled paramagnetic beads that faithfully recapitulates this process. Binding of cohesins to the solid-phase chromatin is interphase-specific. Dissociation of cohesins from chromatin can be induced by adding a phosphatase inhibitor into the assembly mixture, or by exposing purified interphase chromatin to a mitotic extract. XRAD21 is not proteolytically cleaved under this condition. Purified cdc2-cyclin B phosphorylates cohesin in its soluble form, but not in its chromatin-bound form. We have also found that purified x-cohesin can directly bind to DNA and chromatin, and that phosphorylation by cdc2-cyclin B in vitro reduces the affinity of the complex for both substrates. On the basis of these results, two scenarios can be considered for the possible role of cdc2-mediated phosphorylation of cohesins. First, cdc2 could directly phosphorylate cohesin subunits, and thereby actively induce release of the complex from chromatin in early mitosis. Our inability to reconstitute this reaction in vitro using purified chromatin and cdc2-cyclin B could be explained if additional factors were required to make the chromatin-bound form of cohesin accessible to phosphorylation by cdc2. Second, a mitotically activated factor other than cdc2 could play a primary role in the dissociation of cohesins. Cdc2-dependent phosphorylation then could contribute to preventing the released cohesins from reassociating with chromatin.
XSA1 is the major phosphate acceptor both in the cell-free extracts and in the cdc2 phosphorylation reaction. The low abundance of x-cohesinSA2 has hindered similar studies with XSA2. Nevertheless, both human SA1 and SA2 are phosphorylated when they are introduced in Xenopus egg extracts, suggesting that cohesinSA1 and cohesinSA2 may be regulated by a similar phosphorylation process. It is currently unknown whether Scc3p is phosphorylated in S. cerevisiae. On the other hand, Rad21 is phosphorylated in a cell cycle–specific manner in S. pombe (Birkenbihl and Subramani 1995), but the functional impact of this modification is not clear. In contrast, there is no evidence for phosphorylation of the XRAD21 subunit in Xenopus cohesin. It is reasonable to speculate that different organisms use a different repertoire of regulation. For example, the activities of the Xenopus condensin complex are modulated by phosphorylation of its non-SMC subunits (Kimura et al. 1998), whereas the equivalent complex in S. pombe is regulated at the level of nuclear transport by phosphorylation of one of its SMC subunits (Sutani et al. 1999).
Dynamic Exchange of Non-SMC Subunits in Mitotic and Meiotic Cohesins
Emerging lines of evidence suggest that the SMC subunits can associate with a variety of non-SMC partners, thereby fulfilling unique functions at different developmental stages of an organism or acquiring new functions during evolution (Hirano 1999; Strunnikov and Jessberger 1999). The current finding of two different SA subunits in vertebrate cohesins offers additional support to this notion. Even more drastic examples of the exchange of non-SMC partners recently have been inferred from studies on meiotic chromosome dynamics. Rec8p is a meiosis-specific gene product homologous to Scc1p/Mcd1p/Rad21, and is required for meiotic sister chromatid cohesion in both S. pombe and S. cerevisiae (Molnar et al. 1995; Klein et al. 1999; Parisi et al. 1999; Watanabe and Nurse 1999). Rec8p colocalizes with Smc3p in meiotic cells (Klein et al. 1999), suggesting that Rec8p may replace Scc1p/Mcd1p/Rad21 and act as a meiosis-specific subunit of the cohesin complex. The genome of S. pombe has two SA/Scc3p-like sequences and one of them, Rec11, is involved in meiotic cohesion and recombination (Krawchuk et al. 1999). Likewise, the human and mouse genomes contain a third member of the SA family of proteins, SA3. A recent study shows that mammalian SA3 (also called STAG3) is involved in meiotic chromosome pairing (Pezzi et al. 2000), possibly along with SMC1 and SMC3 (Eijpe et al. 2000). Thus, Rec11 and SA3 could also function as components of the meiotic cohesin complex. The subunit composition of these hypothetical cohesin complexes is summarized in Fig. 8. Specialization of cohesins in higher eukaryotes probably reflects an increase of complexity in the regulation of cohesion functions. Further investigation will be required to fully understand how different signals act on different regulatory subunits of the cohesin complexes, and how this class of highly sophisticated protein machines control chromosome morphogenesis both in mitosis and meiosis.
Figure 8.
Mitotic and meiotic cohesins in different organisms. The subunit composition of S. pombe mitotic cohesin and the three meiotic cohesin complexes are hypothetical. The Scc1p/Rad21/Rec8-like subunits are shown by white rectangles, whereas the Scc3p/Rec11/SA-like subunits are indicated by black ovals. For simplicity, no distinction is made between SMC1 and SMC3, and the heterodimers are represented as symmetrical V-shaped structures.
Acknowledgments
We thank J. Méndez, J. Nakayama, and members of the Hirano lab for critically reading the manuscript, M. Murray and J. Méndez for instruction of HeLa cell extract preparation, and K. Kimura (all from Cold Spring Harbor Laboratory) for advice on the immobilized chromatin assembly and in vitro phosphorylation experiments.
This work was supported by grants from the Human Frontier Science Program and the Pew Scholars Program in the Biomedical Sciences (to T. Hirano) and the National Institutes of Health (to R. Kobayashi). A. Losada was the recipient of a fellowship from the Robertson Research Fund.
Footnotes
T. Yokochi's present address is Laboratory of Molecular Embryology, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892.
Abbreviations used in this paper: CDK, cyclin-dependent kinase; HSS, high speed supernatant; LSS, low speed supernatant; SA, stromal antigen; SMC, structural maintenance of chromosomes.
References
- Bell S.P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A. Sister chromatid cohesion in mitosis. Curr. Opin. Cell Biol. 1999;9:230–236. doi: 10.1016/s0959-437x(99)80034-3. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R.P., Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- Blat Y., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Carramolino L., Lee B.C., Zaballos A., Peled A., Barthelemy I., Shav-Tal Y., Prieto I., Carmi P., Gothelf Y., González de Buitrago G. SA-1, a nuclear protein encoded by one member of a novel gene familymolecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. doi: 10.1016/s0378-1119(97)00121-2. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K. Cohesins binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Darwiche N., Freeman L.A., Strunnikov A. Characterization of the components of the putative mammalian sister chromatid cohesion complex. Gene. 1999;233:39–47. doi: 10.1016/s0378-1119(99)00160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M., Heyting C., Gross B., Jessberger R. Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J. Cell Sci. 2000;113:673–682. doi: 10.1242/jcs.113.4.673. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae . Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T.J., Jacobs P.A. Trisomy in man. Annu. Rev. Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- Hirano T. SMC-mediated chromosome mechanicsa conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation and separation. Annu. Rev. Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi R., Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Kimura K., Hirano M., Kobayashi R., Hirano T. Phosphorylation and activation of 13S condensin by cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K., Rybenkov V.V., Crisona N.J., Hirano T., Cozzarelli N.R. 13S condensin actively reconfigures DNA by introducing global positive writheimplications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S.B.C., Michaelis C., Nairz K., Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Koshland D., Strunnikov A. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Krawchuk M.D., DeVeaux L.C., Wahls W.P. Meiotic chromosome dynamics dependent upon the rec8(+), rec10(+) and rec11(+) genes of the fission yeast Schizosaccharomyces pombe . Genetics. 1999;153:57–68. doi: 10.1093/genetics/153.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc H.N., Tang T.T.-L., Wu J.S., Orr-Weaver T.L. The mitotic centromeric protein MEI-S332 and its role in sister-chromatid cohesion. Chromosoma. 1999;108:401–411. doi: 10.1007/s004120050392. [DOI] [PubMed] [Google Scholar]
- Lemaitre J.M., Géraud G., Méchali M. Dynamics of the genome during early Xenopus leavis developmentkaryomeres as independent units of replication. J. Cell Biol. 1998;142:1159–1166. doi: 10.1083/jcb.142.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K.W., Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- McKay M.J., Troelstra C., van der Spek P., Kanaar R., Smit B., Hagemeijer A., Bootsma D., Hoeijmakers J.H.J. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Mistrot C., Guacci V., Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesinschromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miyazaki W.Y., Orr-Weaver T.L. Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Molnar M., Bahler J., Sipiczki M., Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Separating sister chromatids. Trends Biochem. Sci. 1999;24:98–104. doi: 10.1016/s0968-0004(99)01358-4. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. The difficulty in separating sisters. Science. 1999;285:344–345. doi: 10.1126/science.285.5426.344b. [DOI] [PubMed] [Google Scholar]
- Parisi S., McKay M.J., Molnar M., Thompson M.A., van der Spec P.J., van Drunen-Schoenmaker E., Kannar R., Lehmann E., Hoejimakers J.H.J., Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzi N., Prieto I., Kremer L., Pérez Jurado L.A., Valero C., del Mazo J., Martínez-A C., Barbero J.L. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB (Fed. Am. Soc. Exp. Biol.) J. 2000;14:581–592. doi: 10.1096/fasebj.14.3.581. [DOI] [PubMed] [Google Scholar]
- Rudolph B., Saffrich R., Zwicker J., Henglein B., Muller R., Ansorge W., Eilers M. Activation of cyclin-dependent kinases by Myc mediates induction of cyclin A, but not apoptosis. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:3065–3076. [PMC free article] [PubMed] [Google Scholar]
- Sandaltzopoulos R., Becker P.B. A solid-phase approach for the analysis of reconstituted chromatin. In: Becker P.B., editor. Chromatin Protocols. Humana Press; Totowa, NJ: 1999. pp. 195–206. [DOI] [PubMed] [Google Scholar]
- Schmiesing J.A., Ball A.R., Gregson H.C., Alderton J.M., Zhou S., Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc. Natl. Acad. Sci. USA. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan M.A., Mills A.D., Sleeman A.M., Laskey R.A., Blow J.J. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J. Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M.J., Lee T., Kirschner M.W. Role of phosphorylation in p34cdc2 activationidentification of an activating kinase. Mol. Biol. Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A.V., Jessberger R. Structural maintenance of chromosomes (SMC) proteinsconserved molecular properties for multiple biological functions. Eur. J. Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- Stursberg S., Riwar B., Jessberger R. Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complexes RC-1. Gene. 1999;228:1–12. doi: 10.1016/s0378-1119(99)00021-9. [DOI] [PubMed] [Google Scholar]
- Sutani T., Yuasa T., Tomonaga T., Dohmae N., Takio K., Yanagida M. Fission yeast condensin complexessential roles of non-SMC subunits for condensation and cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Cosma M.P., Wirth K., Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Toth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Cell cycle mechanisms of sister chromatid separation; roles of Cut1/separin and Cut2/securin. Genes Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- Zou H., McGarry T.J., Bernal T., Kirschner M.W. Identification of a vertebrate sister chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]