Abstract
Goldfish reproduction is coordinated by pheromones that are released by ovulating females and detected by males. Two highly potent pheromones, a dihydroxyprogesterone and a prostaglandin, previously have been identified, and their effects on goldfish behavior have been studied in depth. We have cloned goldfish olfactory epithelium cDNAs belonging to two multigene G-protein coupled receptor families as a step toward elucidating the molecular basis of pheromone recognition. One gene family (GFA) consists of homologs of putative odorant receptors (≈320 residues) found in the olfactory epithelium of other fish and mammals. The other family (GFB) consists of homologs of putative pheromone receptors found in the vomeronasal organ (VNO) of mammals and also in the nose of pufferfish. GFB receptors (≈840 residues) are akin to the V2R family of VNO receptors, which possess a large extracellular N-terminal domain and are homologs of calcium-sensing and metabotropic glutamate receptors. In situ hybridization showed that the two families of goldfish receptors are differentially expressed in the olfactory epithelium. GFB mRNA is abundant in rather compact cells whose nuclei are near the apical surface. In contrast, GFA mRNA is found in elongated cells whose nuclei are positioned deeper in the epithelium. Our findings support the hypothesis that the separate olfactory organ and VNO of terrestrial vertebrates arose in evolution by the segregation of distinct classes of neurons that were differentially positioned in the olfactory epithelium of a precursor aquatic vertebrate.
A vast number of odorants can be discriminated by the olfactory system of vertebrates (1–3). Electrophysiological studies have revealed that olfaction is mediated by the interplay of neural signals arising from many different kinds of sensory neurons (4). The cloning of three large multigene families of putative odorant and pheromone receptors has revolutionized the field (5–9). What is the mechanism of this remarkable combinatorial recognition process? Goldfish are an attractive model system for the study of this intriguing problem for several reasons. First, the reproductive behavior of goldfish is coordinated by several pheromones of known structure. Preovulatory females release 17α,20β-dihydroxy-4-pregnen-3-one and related hormones a day before ovulation (10–11). The detection of these steroids by the olfactory system of males leads to the production of sperm and seminal fluid. Shortly after ovulation, females release prostaglandin F2α and derivatives, which immediately trigger courtship behavior by males (12). Second, these pheromones are effective at very low concentration. The threshold is in the picomolar to nanomolar range, compared with micromolar to millimolar for most olfactants. Third, olfaction in fish may be simpler than in mammals, given the 10-fold smaller size of their receptor repertoire (13).
Aquatic vertebrates have a single kind of olfactory epithelium, whereas terrestrial vertebrates possess a vomeronasal organ (VNO) in addition to a main olfactory epithelium (MOE) (14, 15). The VNO of rodents and other land animals plays a key role in detecting pheromones that govern mating behavior (16). However, the VNO does not have a monopoly on the sensing of pheromones nor does the MOE on all other odorants. Pigs, for example, detect the potent pheromone androstenone by using their MOE (17). Conversely, the VNO in garter snakes plays a role in feeding as well as courtship behavior (18). Understanding how olfactory recognition tasks in terrestrial vertebrates are partitioned between the VNO and MOE may be deepened by knowing how they are accomplished in the single olfactory apparatus of fish. It is also noteworthy that the olfactory epithelium of fish contains both ciliated and microvillar cells, whereas the MOE is predominantly ciliated and the VNO microvillar (15, 19). Delineating the development and function of ciliated and microvillar sensory neurons in fish is likely to shed light on how the MOE and VNO came into being in terrestrial vertebrates.
Electrophysiological and behavioral studies of goldfish olfaction are well advanced. In contrast, little is known about goldfish olfaction at the molecular level. We report here the cloning of two multigene families of G-protein-coupled receptors from goldfish olfactory epithelium as a step toward identifying pheromone receptors and understanding how their activation leads to specific recognition. One family (termed GFA) encodes homologs of putative odorant receptors that are present in the MOE of mammals, whereas the other (termed GFB) encodes homologs of putative pheromone receptors of the V2R class present in the VNO of mammals. Significantly, these two families are expressed in neurons whose nuclei are positioned at different vertical levels of the goldfish olfactory epithelium.
MATERIALS AND METHODS
Cloning of Homologs of Mammalian MOE Receptors.
Molecular cloning experiments were carried out by using standard procedures (20, 21). Two pairs of degenerate primers based on sequences in transmembrane segments 3 and 7 of putative rat odorant receptors (13) were used in PCR to isolate partial-length homologs from goldfish genomic DNA. One microgram of genomic DNA isolated from goldfish liver served as the template for each reaction. The first cycle of PCR was carried out at 94°C for 4 min, 45°C for 2 min, and 72°C for 3 min, and the next 35 cycles at 94°C for 1 min, 45°C for 2 min, and 72°C for 3 min. One-fifth of the first PCR served as the template for the second PCR using the same primers and conditions. The predicted-sized PCR products (≈520 nt) were obtained and subcloned for sequencing. Seven distinct partial-length goldfish receptor (GFA) genes belonging to three divergent subfamilies were isolated. A goldfish olfactory epithelium cDNA library was constructed in λZIP (GIBCO). Five hundred thousand plaques were screened at high stringency with a pool of three partial GFAs representing different subfamilies. Both strands of each candidate gene were sequenced by an Applied Biosystems automated sequencer. Four full-length cDNAs were obtained.
Cloning of Homologs of Mammalian VNO Receptors.
A pair of degenerate primers was designed based on sequences of putative rat and mouse pheromone receptors belonging to the V2R class (7–9). The 5′ primer was: 5′-ACNCCNAT(T/C/A)GTNAA(A/G)GCNAA(T/C)AA-3′, corresponding to the amino acid sequence TPIVKANN in the first intracellular loop of the receptor. The 3′ primer was: 5′-(T/C)TTNGC(T/C)TC(A/G)TT(A/G)AANG(C/T)(A/G)TC-3′, corresponding to D(A/T)FNEAK in the third intracellular loop. Eight distinct PCR fragments were identified by sequencing. Screening of the goldfish olfactory epithelum cDNA with a mixture of these PCR fragments yielded two full-length cDNA clones and seven partial-length cDNA clones.
Genomic Southern Blots.
Ten micrograms of genomic DNA isolated from goldfish liver was digested with either EcoRI, HindIII, or HaeIII, and electrophoresed on 0.8% agarose gels. The digested DNA samples then were transferred to nylon membranes for hybridization. 32P-labeled probes were synthesized by random priming of each cloned gene. About 5 × 107 cpm of each probe in 5 ml was used for hybridization at high stringency.
Northern Blots.
RNA was extracted and purified from goldfish olfactory epithelium, brain, heart, liver, intestine, and eggs. One microgram of each poly(A)+ RNA was applied to a blot. Hybridization at high stringency was carried out by using 32P-labeled DNA probes made from a pool of GFA cDNAs (GFA2, GFA25, and GFA28) or of GFB cDNAs (GFB1 and GFB8). The blot was stripped and then probed with goldfish β-actin DNA as a quantitation standard.
In Situ Hybridization.
Goldfish were anesthetized with 0.5% 3-aminobenzoic acid ethyl ester (MS222, Sigma) and then sacrificed. The olfactory epithelium was dissected and fixed with 4% fresh paraformaldehyde/PBS overnight. Olfactory epithelium then was transferred into 30% sucrose/PBS for 3 hr, embedded in Tissue-Tek OCT compound, and sectioned at 14 μm on a Cryostat. In situ hybridization was carried out essentially as described (22). Digoxigenin-labeled antisense and sense RNA probes were generated from full-length GFA and GFB cDNAs and from partial-length GFB cDNAs. The probes were fragmented to 100- to 200-bp pieces by alkaline hydrolysis before hybridization.
Sequence Analyses.
DNA and protein sequences were analyzed by using GCG programs (Wisconsin Package Version 9.1, GCG), blastp (Version 2.0.4, National Center for Biotechnology Information) (23), Kyte-Doolittle hydrophobicity analysis in dnasis version 2.0, and clustalw multiple sequence alignment (24).
RESULTS
Cloning of a Multigene Family of MOE Receptor Homologs.
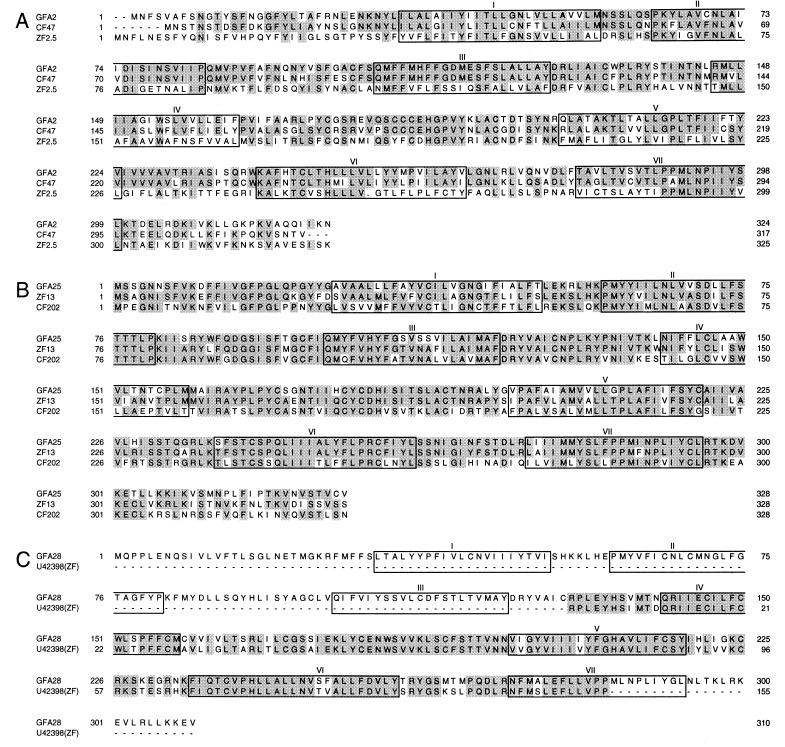
Degenerate PCR performed with two pairs of primers derived from transmembrane segments 3 and 7 of rat putative odorant receptors led to the isolation of seven different clones from goldfish genomic DNA. Sequence and Southern blot analyses showed that these clones belong to three subfamilies. A goldfish olfactory epithelium cDNA library then was screened at high stringency with probes representing these three subfamilies. Four full-length clones (GFA2, GFA12, GFA25, and GFA28) were obtained. GFA12 and GFA25 have the same coding sequence but distinct 5′ and 3′ untranslated sequences. GFA2, GFA25, and GFA28 contain coding sequences 324, 328, and 310 residues long, respectively (Fig. 1). The amino acid sequences of GFA2 and GFA25 are 29% identical and quite different from that of GFA28 (<20% identity). These goldfish receptors are clearly homologs of olfactory receptors expressed in the olfactory epithelia of other fish and mammals. They contain seven transmembrane segments flanked by short N-terminal and C-terminal regions, lack a discernible N-terminal signal sequence, and display significant sequence similarity to known odorant receptors.
Figure 1.
Comparison of the deduced amino acid sequences of three putative goldfish olfactory receptors with the sequences of their closest homologs (CF and ZF denote catfish and zebrafish, respectively). In this and subsequent comparisons, identical residues are shaded. The predicted positions of the seven transmembrane domains are boxed. Dots indicate gaps that were inserted for optimal alignment. (A) GFA2. (B) GFA25. (C) GFA28.
A blastp search of GenBank showed that GFA2 is 71% identical in amino acid sequence to CF47, a full-length catfish gene (accession no. F45774, ref. 13), and 38% identical to ZF2.5, a full-length zebrafish gene (accession no. AF012759, ref. 25) (Fig. 1A). Likewise, GFA25 has odorant receptor counterparts in zebrafish and catfish (Fig. 1B). GFA25 is 78% identical to ZF13 (accession no. AF012746, ref. 25) and 59% identical to CF202 (accession no. G45774, ref. 13). GFA28 has no full-length homolog in the database but is 85% identical to a zebrafish sequence (accession no. U43298, ref. 26) in a 155-residue overlap region (Fig. 1C).
Cloning of a Multigene Family of VNO Receptor Homologs.
The cloning of two families of putative pheromone receptors, termed V1R (also called VNR or Gi2α-VN; ref. 6) and V2R (also called VR or Go-VN; refs. 7–9) from the VNO of mice and rats stimulated us to search for their homologs in goldfish. We looked in the olfactory epithelium because goldfish, like other aquatic vertebrates, lack a distinct VNO. PCR of goldfish genomic DNA using several pairs of degenerate primers based on conserved V1R sequences did not yield any products. In contrast, PCR using primers based on V2R sequences gave fragments showing significant sequence similarity to calcium-sensing receptors, metabotropic glutamate receptors, and mammalian V2Rs. A goldfish olfactory epithelium cDNA library then was screened with a mixture of these PCR fragments. Two full-length genes (GFB1 and GFB8, and seven partial-length genes (GFB2, 5, 7, 9, 10, 12, and 14) were isolated. GFB2 has the same coding sequence as GFB14 but has a 430-nt longer 3′ untranslated region. The deduced amino acid sequences of the two full-length genes and six partial-length genes are compared in Fig. 2. GFB1 and GFB8 encode 844- and 848-residue proteins, respectively, which are 53% identical. They have a very long N-terminal sequence (≈570 residues) followed by a seven-transmembrane motif (≈270 residues) and a very short C-terminal region (≈20 residues). A canonical N-terminal signal sequence is present in both. The sequences of GFB1 and GFB8 are very similar in the transmembrane region (77% identity) and considerably less so in the long N-terminal region (42% identity). The high degree of sequence conservation of the transmembrane region is also evident in the partial-length sequences.
Figure 2.
Comparison of the deduced amino acid sequences of two full-length (GFB1 and GFB8) and six partial-length (GFB5, 7, 9, 10, 12, and 14) goldfish homologs of mammalian V2R vomeronasal receptors.
A blastp search of GenBank showed that GFB1 and GFB8 are most similar to pufferfish homologs of rodent V2R receptors (e.g., accession nos. AB008858 and AB008860, ref. 28), mammalian extracellular calcium-sensing receptors (e.g., accession no. A56715, ref. 29), rodent V2R receptors (accession no. AF053986, ref. 9; accession no. AF011413, ref. 7; accession no. AF016182, ref. 8), and mammalian metabotropic glutamate receptors (e.g., accession no. P23385, ref. 27). The goldfish and pufferfish receptors exhibit a high degree of sequence identity, 50–54% for the entire sequence, 40–47% for the N-terminal domain, and 70–76% for the transmembrane domain. The degree of sequence identity of the mouse V2R2 receptor to the fish homologs is lower, 35–36% for the entire sequence, 31–33% for the N-terminal domain, and 41–44% for the transmembrane domain. The conservation of cysteine residues is striking. Of the 29 cysteines common to GFB1 and GFB8, 23 are also present in V2R2 and the two pufferfish homologs.
Southern Blots.
To estimate the size of the gene families, blots of genomic DNA digested with EcoRI, HindIII, or HaeIII were hybridized at high stringency with individual GFA and GFB cDNAs. The hybridization patterns of the three GFA cDNAs are different (Fig. 3A), indicating that they represent three divergent subfamilies. Also, the nine GFB cDNAs gave seven different hybridization patterns (Fig. 3B), indicating that they represent seven divergent subfamilies. The number of bands seen in the Southern blots of the GFBs is significantly greater than that of the GFAs under conditions of equal stringency. The GFB family contains at least 30 members.
Figure 3.
Genomic Southern blot analysis of goldfish homologs of mammalian odorant receptor and vomeronasal receptor genes. Goldfish genomic DNA digested with EcoRI (lanes 1), HindIII (lanes 2), or HaeIII (lanes 3) was electrophoresed, blotted, and hybridized with 32P-labeled probes prepared from GFA and GFB cDNAs. The positions (kb) of HindIII-digested λDNA markers are shown on the left.
Tissue Specificity of Expression.
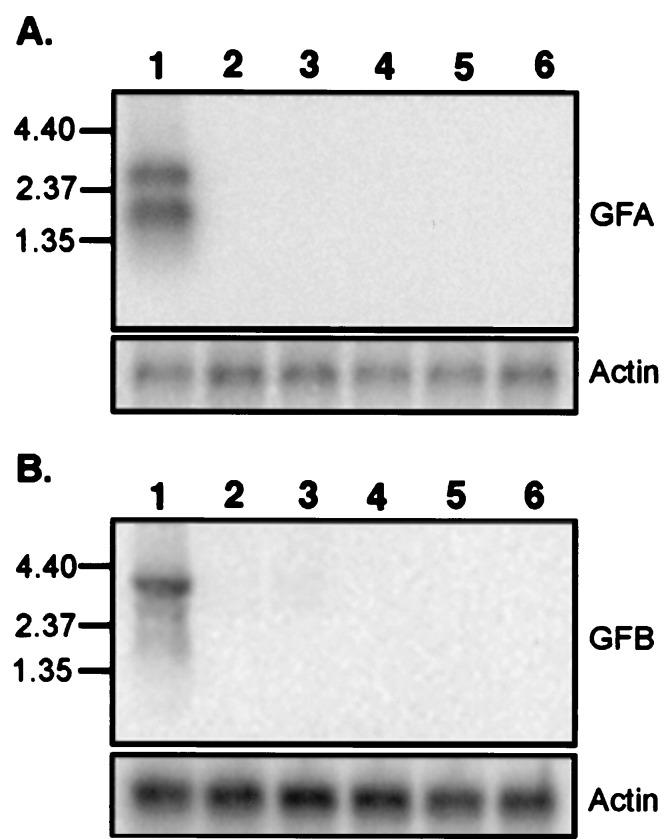
Northern blots were performed to determine the tissue distribution of the two gene families. A pool of GFA probes and a pool of GFB probes were used for the two sets of Northern blots. Hybridization at high stringency showed that both receptor families are expressed in olfactory epithelium, but not in brain, heart, liver, intestine, and eggs (Fig. 4).
Figure 4.
Northern blot analysis of the expression of the two families of goldfish receptors. Poly(A)+ RNAs isolated from goldfish (lanes 1) olfactory epithelium, (lanes 2) brain, (lanes 3) heart, (lanes 4) liver, (lanes 5) intestine, and (lanes 6) eggs were fractionated on a 1% gel, blotted, and hybridized to a mixture of radiolabeled probes prepared from (A) GFA2, GFA25, and GFA28 and (B) GFB1 and GFB8. β-Actin served as a control. The positions (in kb) of markers are shown on the left.
Differential Expression of the GFA and GFB Gene Families.
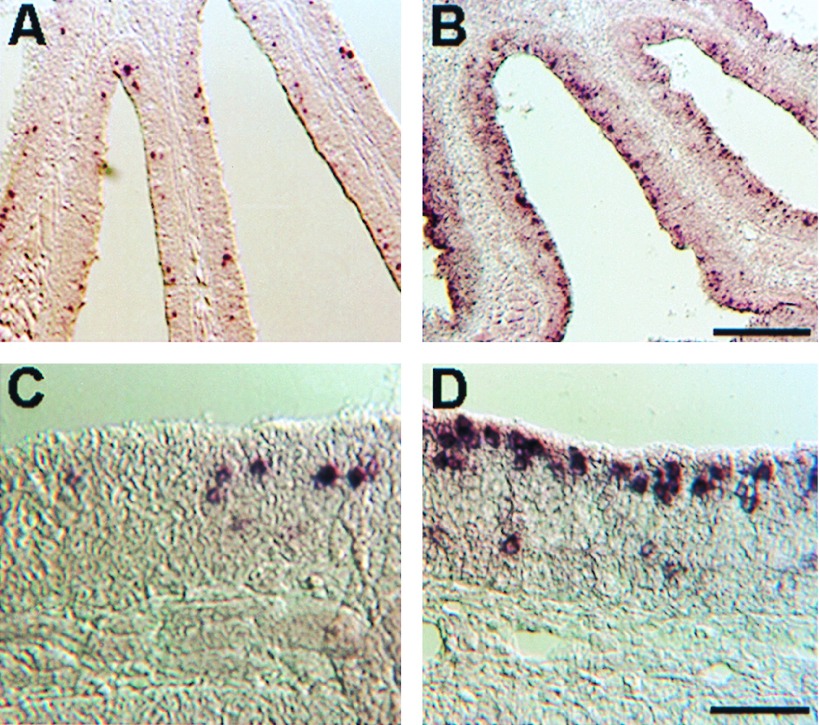
The expression patterns of these two gene families were visualized by in situ hybridization with fragmented digoxigenin-labeled RNA probes. The expression patterns of the three GFAs were essentially the same. As shown for GFA25 (Fig. 5 A and C), the positive neurons appeared to be randomly distributed in the lamellar plane. The cell bodies of GFA-positive cells were strongly labeled and positioned well below the apical surface. A strikingly different pattern was seen for the GFBs. As shown for GFB14 (Fig. 5 B and D), the nuclei of most GFB-positive cells were near the apical surface of the epithelium. The few GFB-positive cells located deeper in the epithelium may be newly formed cells en route to a more apical position. Moreover, the label in GFB-positive cells, in contrast with that in GFA-positive cells, was diffusely distributed around the cell body and sometimes extended to the apical surface. A high proportion of apical cells were labeled with GFB14, most likely because of the use of fragmented probe. The highly conserved transmembrane sequences of GFB14 probably cross-hybridized to RNA of cells expressing other members of the GFB family.
Figure 5.
In situ analysis of the expression of the two classes of receptors in the goldfish olfactory epithelium. Hybridization was carried out at high stringency on coronal sections of the epithelium by using fragmented antisense digoxigenin-labeled RNA probes. (A and C) Low-power and high-power views of GFA25. (B and D) Low-power and high-power views of GFB14. [Scale bar for A and B is shown in B (200 μm), and that for C and D is shown in D (50 μm).]
DISCUSSION
We have cloned goldfish olfactory epithelium cDNAs belonging to two multigene G-protein coupled receptor families. One gene family (GFA) encodes homologs of putative odorant receptors (≈320 residues) that are generally present in the olfactory epithelium of mammals, fish, and other vertebrates (5, 13, 30–32). Three of the four full-length members of the GFA family cloned by us (Fig. 1) are quite different from one another (<30% amino acid sequence identity). Their closest homologs are catfish and zebrafish olfactory receptors. It is evident that GFA2, GFA25, and GFA28 are members of divergent olfactory receptor subfamilies that recur in other fish. The other family (GFB) cloned from the goldfish olfactory epithelium (Fig. 2) consists of homologs of putative pheromone receptors that are present in the VNO but not in the olfactory epithelium of mammals. GFB receptors (≈840 residues) are akin to the V2R family of VNO receptors (7–9), which are homologs of calcium-sensing (29) and metabotropic glutamate receptors (27). They possess a large extracellular N-terminal domain in addition to a C-terminal seven-transmembrane-helix domain. This class of receptors also has recently been found in the olfactory epithelium of pufferfish (28).
The GFA and GFB families of receptors are expressed in the goldfish olfactory epithelium (Fig. 4), whereas their mammalian homologs are expressed separately in the MOE and VNO, respectively. Most intriguing, GFA and GFB are expressed in what appear to be different kinds of sensory neurons in the goldfish olfactory epithelium (Fig. 5). The nuclei of cells expressing GFA are further from the apical surface than are the nuclei of cells expressing GFB. The cells are also morphologically distinct but their identities are uncertain at the resolution of this study. The olfactory epithelia of fish contain both ciliated and microvillar sensory neurons, whereas the MOE of terrestrial vertebrates contains mostly ciliated cells, and the VNO contains mostly microvillar cells (14, 15). In the catfish olfactory epithelium, the nuclei of microvillar cells are nearer the apical surface than are the nuclei of ciliated cells (33). Hence, we surmise that GFA is expressed in ciliated cells and GFB in microvillar cells. Our finding that GFA and GFB are expressed in different kinds of cells supports the proposal that the MOE and VNO of terrestrial vertebrates arose in evolution by the segregation of distinct classes of sensory neurons that were differentially positioned in a precursor aquatic vertebrate (15, 33).
Retrograde fluorescent labeling studies have revealed that ciliated and microvillar cells in the catfish olfactory epithelium project to different parts of the olfactory bulb (33). Furthermore, calcium imaging studies have shown that different olfactants activate different regions of the olfactory bulb (34, 35). Thus, the ciliated and microvillar sensory neurons in the olfactory epithelia of fish recognize different kinds of olfactants. What might they be? After sectioning of the goldfish olfactory nerve, ciliated cells in the olfactory epithelium regenerate first, followed weeks later by microvillar cells (36). These regenerating goldfish regain the capacity to detect foods weeks before they can again sense pheromones, suggesting that ciliated sensory neurons recognize foods, whereas microvillar sensory neurons recognize pheromones. That study taken together with ours implies that GFA receptors sense foods and GFB receptors sense pheromones. It will be interesting to test this hypothesis by using recently devised strategies (37, 38) for determining the specificity of functionally expressed olfactory receptor genes.
Acknowledgments
We thank Dr. Thomas Finger, Dr. Ben Barres, and Dr. Denis Baylor for stimulating discussions and Dr. Peter Sorensen for providing goldfish tissues. Y.C. is the recipient of a National Research Service Award postdoctoral fellowship (1F32DC00318-01) from the National Institute on Deafness and Other Communication Disorders. This research was supported by Stanford University patent funds.
ABBREVIATIONS
- MOE
main olfactory epithelium
- VNO
vomeronasal organ
Footnotes
References
- 1. Lancet D. Annu Rev Neurosci. 1986;9:329–355. doi: 10.1146/annurev.ne.09.030186.001553. [DOI] [PubMed] [Google Scholar]
- 2.Axel R. Sci Am. 1995;273:154–159. doi: 10.1038/scientificamerican1095-154. [DOI] [PubMed] [Google Scholar]
- 3.Buck L B. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- 4.Sicard G, Holley A. Brain Res. 1984;292:283–296. doi: 10.1016/0006-8993(84)90764-9. [DOI] [PubMed] [Google Scholar]
- 5.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 6.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 7.Herrada G, Dulac C. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- 8.Matsunami H, Buck L B. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- 9.Ryba N J, Tirindelli R. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- 10.Sorensen P W, Hara T J, Stacey N E. J Comp Physiol A. 1987;160:305–313. [Google Scholar]
- 11.Sorensen P W, Caprio J. In: The Physiology of Fishes. 2nd Ed. Evans D H, editor. Boca Raton, FL: CRC; 1998. pp. 375–405. [Google Scholar]
- 12.Sorensen P W, Hara T J, Stacey N E, Goetz F W. Biol Reprod. 1988;39:1039–1050. doi: 10.1095/biolreprod39.5.1039. [DOI] [PubMed] [Google Scholar]
- 13.Ngai J, Dowling M M, Buck L, Axel R, Chess A. Cell. 1993;72:657–666. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- 14.Eisthen H L. Microsc Res Tech. 1992;23:1–21. doi: 10.1002/jemt.1070230102. [DOI] [PubMed] [Google Scholar]
- 15.Dulka J G. Brain Behav Evol. 1993;42:265–280. doi: 10.1159/000114166. [DOI] [PubMed] [Google Scholar]
- 16.Halpern M. Annu Rev Neurosci. 1987;10:325–362. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- 17.Dorries K M, Adkins-Regan E, Halpern B P. Brain Behav Evol. 1997;49:53–62. doi: 10.1159/000112981. [DOI] [PubMed] [Google Scholar]
- 18.Kubie J L, Halpern M. J Comp Physiol Psychol. 1979;93:648–667. doi: 10.1037/h0077061. [DOI] [PubMed] [Google Scholar]
- 19.Bargmann C I. Cell. 1997;90:585–587. doi: 10.1016/s0092-8674(00)80518-8. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fristsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 21.Sambrook J, Fristsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab.Press; 1989. [Google Scholar]
- 22.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipmann D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barth A L, Dugas J C, Ngai J. Neuron. 1997;19:359–369. doi: 10.1016/s0896-6273(00)80945-9. [DOI] [PubMed] [Google Scholar]
- 26.Barth A L, Justice N J, Ngai J. Neuron. 1996;16:23–34. doi: 10.1016/s0896-6273(00)80020-3. [DOI] [PubMed] [Google Scholar]
- 27.Masu M, Tanabe Y, Tsuchida K, Shigemoto R, Nakanishi S. Nature (London) 1991;349:760–765. doi: 10.1038/349760a0. [DOI] [PubMed] [Google Scholar]
- 28.Naito T, Saito Y, Yamamoto J, Nozaki Y, Tomura K, Hazama M, Nakanishi S, Brenner S. Proc Natl Acad Sci USA. 1998;95:5178–5181. doi: 10.1073/pnas.95.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrett J E, Capuana I V, Hammerland L G, Hung B C, Brown E M, Hebert S C, Nemeth E F, Fuller F. J Biol Chem. 1995;270:12919–12925. doi: 10.1074/jbc.270.21.12919. [DOI] [PubMed] [Google Scholar]
- 30.Ressler K J, Sollian S L, Buck L. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 31.Issel-Tarver L, Rine J. Proc Natl Acad Sci USA. 1996;93:10897–10902. doi: 10.1073/pnas.93.20.10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weth F, Nadler W, Korsching S. Proc Natl Acad Sci USA. 1996;93:13321–13326. doi: 10.1073/pnas.93.23.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita Y, Finger T E. J Comp Neurol. 1998;398:539–550. doi: 10.1002/(sici)1096-9861(19980907)398:4<539::aid-cne6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 34.Friedrich R W, Korsching S I. Neuron. 1997;18:737–752. doi: 10.1016/s0896-6273(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 35.Bozza T C, Kauer J S. J Neurosci. 1998;18:4560–4569. doi: 10.1523/JNEUROSCI.18-12-04560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zippel H P, Sorensen P W, Hansen A. J Comp Physiol A. 1997;180:39–52. [Google Scholar]
- 37.Zhang Y, Chou J H, Bradley J, Bargmann C I, Zinn K. Proc Natl Acad Sci USA. 1997;94:12162–12167. doi: 10.1073/pnas.94.22.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Ivic L, Otaki J M, Hashimoto M, Mikoshiba K, Firestein S. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]