Abstract
Mammalian cell nucleoli disassemble at the onset of M-phase and reassemble during telophase. Recent studies showed that partially processed preribosomal RNA (pre-rRNA) is preserved in association with processing components in the perichromosomal regions (PRs) and in particles called nucleolus-derived foci (NDF) during mitosis. Here, the dynamics of nucleolar reassembly were examined for the first time in living cells expressing fusions of the processing-related proteins fibrillarin, nucleolin, or B23 with green fluorescent protein (GFP). During telophase the NDF disappeared with a concomitant appearance of material in the reforming nuclei. Prenucleolar bodies (PNBs) appeared in nuclei in early telophase and gradually disappeared as nucleoli formed, strongly suggesting the transfer of PNB components to newly forming nucleoli. Fluorescence recovery after photobleaching (FRAP) showed that fibrillarin-GFP reassociates with the NDF and PNBs at rapid and similar rates. The reentry of processing complexes into telophase nuclei is suggested by the presence of pre-rRNA sequences in PNBs. Entry of specific proteins into the nucleolus approximately correlated with the timing of processing events. The mitotically preserved processing complexes may be essential for regulating the distribution of components to reassembling daughter cell nucleoli.
Keywords: mitosis, nucleolus, prenucleolar body, pre-rRNA processing, rDNA transcription
Introduction
Of the numerous membraneless subcompartments of the interphase nucleus (Lamond and Earnshaw 1998; Cook 1999; Matera 1999), the nucleolus is the most prominent. The organization of the nucleolus around clusters of tandemly repeated genes for preribosomal RNA (Shaw and Jordan 1995) facilitates its ability to perform its major function, assembly of ribosomal subunits (Pederson 1998; Olson et al. 2000). The nucleolus is further organized by the process of transcription; i.e., the systems for folding, processing, modification and packaging of the pre-rRNA associate with the transcripts and become integral parts of the nucleolar structure (Scheer and Hock 1999; Olson et al. 2000). During mitosis this highly organized structure must disassemble, the subcomponents must be distributed to the daughter cells and then reassembled at the end of mitosis. A key event in this process is the repression of RNA pol I–driven pre-rRNA synthesis in early prophase (Prescott, and Bender 1962; Weisenberger and Scheer 1995) and its subsequent reactivation in the later stages of mitosis (Scheer et al. 1993). This cycle of transcriptional repression and reactivation is regulated at the level of initiation; i.e., the RNA pol I transcription factor SL1 and the upstream binding factor (UBF) are inactivated by phosphorylation (Heix et al. 1998; Kuhn et al. 1998; Klein and Grummt 1999). Both dephosphorylation of SL1 and UBF and rephosphorylation of UBF at a different site facilitate the reactivation of transcription (Voit et al. 1999). However, the regulation of the mitotic behavior of the other parts of the ribosome assembly machinery is less well understood.
The various classes of nucleolar components distribute to several different parts of the cell during mitosis. The transcriptional apparatus remains assembled during mitosis, with RNA pol I and its associated transcription factors located in the nucleolar organizer regions (NORs) of chromosomes (Weisenberger and Scheer 1995; Jordan et al. 1996; Roussel et al. 1996). In contrast, the pre-rRNA–processing components are found in at least two different sites. The first is the perichromosomal region (PR) where these components surround all chromosomes beginning in prometaphase and remain until early telophase (Hernandez-Verdun and Gautier 1994; Dundr et al. 1997). The second location is in numerous, relatively large cytoplasmic spherical particles termed nucleolus-derived foci (NDF), which first appear in anaphase, diminish in number during telophase and eventually disappear in G1 phase (Dundr et al. 1996, Dundr et al. 1997; Dundr and Olson 1998). The NDF do not contain components of the RNA pol I transcription machinery, confirming the spatial separation of pre-rRNA transcription machinery and pre-rRNA–processing components during mitosis. However, the NDF contain partially processed pre-rRNA transcripts lacking the 5′ETS leader sequence and having reduced levels of the 3′ETS segment; i.e., a mixture of 45S and 46S pre-rRNAs. The latter RNAs are associated with components implicated in pre-rRNA processing including fibrillarin, nucleolin, protein B23, hPop1 and U3 and U8 snoRNAs. The fact that these long pre-rRNAs are preserved in these complexes indicates that pre-rRNA processing as well as transcription is suppressed during mitosis. Their preservation throughout mitosis raises the intriguing possibility that they somehow participate in the rebuilding of the postmitotic nucleolus, possibly by maintaining the organization and structure of the processing machinery in the absence of a nucleolar structure.
The current view of nucleolar reformation at the end of mitosis is that the process begins in late anaphase or early telophase when RNA pol I transcription is reinitiated (Scheer et al. 1993; Fomproix et al. 1998). At the same time specific nucleolar components present in the perichromosomal regions are released from decondensing chromosomes and begin to associate with prenucleolar bodies (PNBs) in newly forming daughter nuclei. The PNBs appear to be similar to the NDF in composition in that they contain a number of nucleolar proteins and snoRNAs implicated in pre-rRNA processing (Azum-Gélade et al. 1994; Jiménez-García et al. 1994; Beven et al. 1996). Of special importance is the fact that PNBs do not contain any transcriptional components and transcription has never been shown to occur within them (Bell et al. 1992; Gébrane-Younès et al. 1997). The PNBs are believed to migrate towards the nucleolar organizer regions (NORs) where they fuse and become the dense fibrillar components of the newly built nucleolus. This fusion seems to be dependent on RNA pol I transcription, with the new transcripts capturing material from the PNBs (Scheer et al. 1993).
Although the past several years have brought progress in our understanding of nucleologenesis, the in vivo details of the process and the role of the partially processed pre-rRNA are far from clear. Therefore, we initiated studies to examine the pathway by which components of the NDF and PNBs are incorporated into nucleoli in living cells and to determine the locations of partially processed pre-rRNA during postmitotic assembly of nucleoli. We used two approaches for this. First, the behavior of nucleolar proteins was examined in vivo as cells advanced through telophase. To this end we visualized the movements of nucleolar proteins expressed as chimeras with green fluorescent protein (GFP) by time-lapse fluorescence microscopy in cells progressing through the later parts of mitosis. Second, cells in various stages of telophase were probed for pre-rRNA sequences using fluorescence in situ hybridization (FISH) and simultaneous localization of nucleolar proteins by immunofluorescence microscopy. We found that the NDF disappear during telophase and their dissociated components appear to enter nuclei. At the same time certain pre-rRNA sequences are found in telophase nuclei, where they are associated initially with decondensing chromosomes and later with PNBs. The PNBs are major sources of processing-related components for assembly of nucleoli in the late stages of mitosis. These studies describe for the first time the mitotic reassembly of nucleoli in living cells. Our results provide direct support for the concept that postmitotic nucleoli are partly constructed from assembled components derived from the maternal cell.
Materials and Methods
Engineering of the GFP Fusion Proteins
A full-length 963-bp clone of the human fibrillarin cDNA (Aris and Blobel 1991) was PCR-amplified with AmpliTaq DNA polymerase (Perkin Elmer) without a translation termination codon (TGA) using the following oligonucleotides: sense, 5′-GCCATGAAGCCAGGATTCAGTCCC-3′ and antisense, 5′-GTTCTTCACCTTGGGGGGTGGCC-3′. The amplified PCR product was directly subcloned into the pCR2.1 vector (Invitrogen) and then inserted as an EcoRI-EcoRI fragment into the pEGFP-N3 vector (CLONTECH Laboratories, Inc.). The fusion protein contained GFP at the COOH terminus of human fibrillarin. The full-length 876-bp sequence of rat protein B23 (Chang et al. 1988) was PCR amplified with AmpliTaq DNA polymerase (Perkin Elmer) without a translation termination codon (TGA) using the following oligonucleotides: sense, 5′-ATGGAAGATTCGATGGACATG-3′ and antisense, 5′-AAGAGACTTCCTCCACTGCCAG-3′. The amplified PCR product was directly subcloned into pCR2.1 vector (Invitrogen) and then inserted as an EcoRI-EcoRI fragment into pEGFP-N3 vector (CLONTECH Laboratories, Inc.). GFP was fused in frame to the COOH terminus of protein B23. Human nucleolin fused in frame to the COOH terminus of GFP and inserted into pAlter MAXAD vector was a generous gift from Dr. Dmitry Goldgaber (SUNY, Stony Brook, NY).
Cell Culture and Transfection
Monkey CMT3 cells (Gerard and Gluzman 1985) were grown on 18 × 18- or 22 × 22-mm poly-l-lysine–coated glass coverslips in DME (GIBCO BRL) supplemented with 10% FCS (GIBCO BRL), 1% glutamine, and penicillin and streptomycin at 37°C in 5% CO2 atmosphere. For the in situ hybridization studies the cells were synchronized at the G1/S transition by a double-thymidine block with 2.5 mM thymidine (Bootsma et al. 1964). The cells were released and allowed to proceed to mitosis (∼8 h). For visualization of GFP-protein chimeras the cells were plated some 12–24 h before transfection; at 75% confluency they were transfected using Fugene 6 (Roche Diagnostics) and 1 μg of plasmid DNA according to the manufacturer's instructions.
Immunofluorescence
Coverslips with attached cells were washed in PBS and fixed with 3% paraformaldehyde in PBS for 20 min at room temperature, rinsed in PBS, and subsequently permeabilized with 0.2% Triton X-100 in PBS for 5 min on ice, and then were washed extensively with 1% BSA in PBS. The cells were incubated with the primary antibody diluted in PBS for 1 h, washed in PBS, and incubated with appropriate secondary antibodies conjugated with either fluorescein or Texas red (Amersham Pharmacia Biotech) or TRITC (Sigma-Aldrich) for 50 min. The cells were washed extensively with PBS, briefly in H2O and ethanol, air dried, and mounted on the slides with Mowiol (Calbiochem-Novabiochem) containing 1 mg/ml p-phenylenediamine. Fibrillarin was detected with human autoimmune serum S4 (Deitz) at dilution 1:150 (kindly provided by Dr. R.L. Ochs, Precision Therapeutics, Pittsburgh, PA). Nucleolin was visualized with mouse mAb 7G2 at dilution 1:100 (a generous gift from Dr. S. Pinol-Roma, Mount Sinai School of Medicine, New York, NY). Protein B23 was detected using anti-B23 monoclonal antibody (mAb) at dilution 1:20 (kindly provided by Dr. P.K. Chan, Baylor College of Medicine, Houston, TX).
Analysis of GFP Fusions and Endogenous Proteins by Immunoblotting
The cells were grown to 50–75% confluency in 60-mm culture dishes and transfected with 2 μg of plasmid DNA using Fugene 6 (Roche Diagnostics) for 24 h. The cells were harvested with a cell scraper, collected in ice-cold PBS, and pelleted for 5 min at 4°C and 2,500 rpm in a Savant table top centrifuge. The cell pellets were resuspended in SDS-PAGE sample buffer (60 mM Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 14.4 mM β-mercaptoethanol, and 0.01% bromophenol blue), boiled for 4 min, resolved on 13.5% SDS–polyacrylamide gels, and transferred to nitrocellulose membranes (Costar) using the Polyblot™ transfer system (American Bionetics). The membranes were washed with buffer A (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 0.1% Triton X-100), blocked for 90 min with 5% nonfat dry milk powder in buffer A. The blots were incubated for 3 h with primary antibodies (human autoimmune serum S4 [Dietz] against fibrillarin at dilution 1:5,000, monoclonal antibody against protein B23 at dilution 1:2,000 and rabbit polyclonal antibody against the NH2-terminal end of nucleolin at dilution 1:5,000), diluted in buffer A containing 5% dry milk, washed with buffer A, and incubated for 1 h with secondary antibodies conjugated to alkaline phosphatase (Bio-Rad Laboratories) diluted in buffer A with 5% dry milk. After extensive washing in buffer A, the blots were washed in H2O and 1 M Tris-HCl, pH 8.6, and incubated in developing solution (0.3 mg/ml nitroblue tetrazolium and 50 μg/ml indoxyl phosphate in dimethylformamide) in 1 M Tris-HCl, pH 8.6, at 37°C. After color development, the blots were fixed with 10% acetic acid.
Time-Lapse Fluorescence Microscopy
Approximately 16–24 h after transfection the cells were trypsinized and transferred to Nalgene Lab Tek II chambers in DME with 25 mM Hepes without phenol red and imaged at 37°C on a Leica TCS-SP inverted confocal microscope with a 100×/1.4 NA planapochromat oil objective using immersion oil with refractive index and viscosity optimized for 37°C. Excitation was at 488 nm, detection between 500 and 575 nm. Optical scans were collected every 18 s. The focus, contrast, and brightness settings were constant during the course of image acquisition. The images were arranged sequentially in a movie sequence on NIH Image. Figures were assembled into composite images with Metamorph and Adobe Photoshop and printed on a Kodak 8670 PS Thermal printer.
Fluorescence Recovery after Photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were performed on a Leica TCS-SP inverted confocal microscope with 100×/1.4 NA planapochromat oil objective using immersion oil with refractive index and viscosity optimized for 37°C. Excitation was at 488 nm, detection between 500 and 575 nm. In transiently transfected CMT3 cells a spot of ∼1 μm in diameter was bleached for 0.5 s using the 488-nm laser line at 100% laser power. Cells were imaged at 2-s intervals. For quantification, fluorescence intensities of the entire nucleus and in regions of interest in the bleached area and outside of the nucleus were measured using Leica TCS-SP software. All intensities were background subtracted. To normalize for loss of fluorescence signal due to the bleach pulse and potential artifactual bleaching of GFP during the imaging scans, recovery intensities were normalized to the total fluorescence signal at each time point. Cells exhibiting a loss of signal of more than 10% during the imaging phase were discarded for analysis. The relative fluorescence intensity was calculated as described by Phair and Misteli 2000.
Hybridization Probes
The hybridization probes used in this study were intensively characterized in Dundr and Olson 1998. In brief, The 5′ETS core region of human pre-rRNA, which corresponded to a SacI-KpnI fragment of human pre-rRNA (nt +934/+1,444) in pBluescript SK(−) was linearized with XhoI and the antisense hybridization probe (+1,270/+1,444) was produced by in vitro transcription with biotin-16-UTP using T7 RNA polymerase. The linearized pTRI RNA 18S antisense template, which contain an 80-bp insert of the human 18S rRNA gene (nt +4,271/+4,349) in pTRIPLEscript vector, was obtained from Ambion. The hybridization antisense riboprobe (residues −43 to +5 relative to transcription start) was produced by in vitro transcription with biotin-16-UTP using T7 RNA polymerase. The linearized pTRI RNA 28S template containing a 115-bp cDNA fragment of the human 28S rRNA gene (+12,345/+12,458) inserted into the KpnI-XbaI sites of pTRIPLEscript vector was obtained from Ambion. The hybridization antisense probe (nt +37 relative to transcription start) was produced by in vitro transcription with biotin-16-UTP using T7 RNA polymerase.
In Situ Hybridization
Cells grown on poly-l-lysine–coated glass coverslips were washed with PBS and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After rinsing in PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min on ice, and then washed with PBS and finally with 2× SSC. The hybridization mixture was prepared as described by Jiménez-García et al. 1994. In brief, 100 ng of probe and 20 μg of yeast tRNA was dried under vacuum. 10 μl of deionized formamide was added, and the mixture was denatured for 10 min at 70°C. The probe was immediately chilled on ice and the hybridization mixture was made to a final concentration of 2× SSC, 1%BSA, and 10% dextran sulfate. A hybridization mixture (20 μl) was placed onto each coverslip and allowed to incubate in a chamber moistened with 2× SSC/50% formamide for 16–18 h at 42°C. The coverslips were rinsed with 2× SSC/50% formamide at 37°C, 2× SSC and 1xSSC at room temperature for 30 min each. The cells were incubated with avidin-DCS-conjugated with Texas red (Vector Laboratories; 2 μg/μl) in 4× SSC/0.25% BSA for 60–75 min, and then rinsed in 4× SSC, 4× SSC/0.1% Triton X-100, 4× SSC, and PBS. Coverslips were mounted in Mowiol (Calbiochem-Novabiochem) containing 1 mg/ml p-phenylenediamine. When in situ hybridization was followed by immunofluorescence, after rinsing the cells in PBS, the coverslips were incubated with anti-B23 mAb for 50 min at room temperature. The coverslips were then rinsed in PBS and incubated with sheep anti–mouse fluorescein-labeled secondary antibody (Amersham Pharmacia Biotech) for 45 min. The cells were washed several times with PBS, briefly with ethanol, air dried, and mounted in Mowiol (Calbiochem-Novabiochem) containing p-phenylenediamine. Samples were observed on a Nikon Eclipse 800 microscope using a MicroMax Interline (5 MHz) cooled CCD camera.
Ultrastructural Immunocytochemistry
The CMT3 cells were synchronized as above and fixed with 8% paraformaldehyde in 200 mM Pipes (pH 7.0) containing 5 mM MgCl2 and then in 0.01% glutaraldehyde in the same buffer for 2 min. The cells were washed in PBS, collected by scraping and pelleted. The cell pellet was initially embedded in 5% gelatin in PBS, dehydrated in increasing concentrations of ethanol, and finally embedded in Lowicryl K4M. Polymerization was performed under long-wave-length UV light (2× 15 W, Ted Pella) for 4 d at −20°C and for 2 d at room temperature. Sections were cut at a nominal setting of 95 nm on a Reichert Ultracut S ultramicrotome. The sections were placed on carbon-Parlodoin-coated copper grids. The sections were incubated with rabbit polyclonal antibody against protein B23 for 4 h. After washing with PBS, the grids were incubated with goat anti–rabbit IgG conjugated to 10-nm gold particles. After washing with PBS and water, the grids were stained with 5% aqueous uranyl acetate. Samples were observed on a Zeiss EM 10 electron microscope.
Online Supplemental Material
The following supplemental videos are available at http://www.jcb.org/cgi/content/full/150/3/433/DC1.
Video 1.
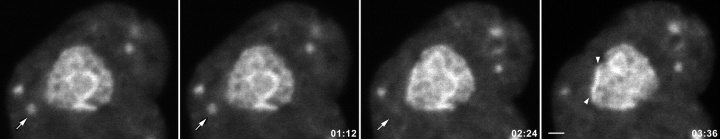
The dynamic behavior of NDF throughout the cytoplasm and the disappearance of NDF during telophase is visualized with protein B23-GFP. Movie frames were captured at 18-s intervals. See Fig. 2 for a static presentation at four selected time points.
Figure 2.
Behavior of NDF during telophase in living cells. The CMT3 cells were transiently transfected with the protein B23-GFP vector. After 24 h, cells in telophase were selected for observation by time-lapse confocal fluorescence microscopy. The images were collected every 18 s over a 90-min period. See also video 1 available at http://www.jcb.org/cgi/content/full/150/3/433/DC1. Protein B23-GFP was present in several NDF in the cytoplasm and distributed throughout the nucleoplasm and in newly forming nucleoli. The disappearance of the NDF in the proximity of the nuclear envelope (arrows) was accompanied by a concomitant increase in the fluorescence of the adjacent nuclear interior (arrowheads). Bar, 2 μm.
Video 2.
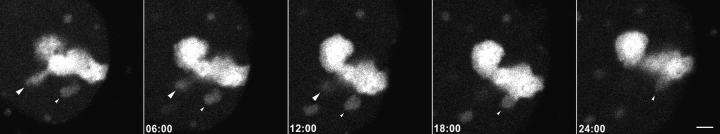
Dynamics of nucleolar reassembly in telophase were analyzed by the visualization of fibrillarin-GFP. See Fig. 3 for a static presentation at 20 selected time points.
Figure 3.
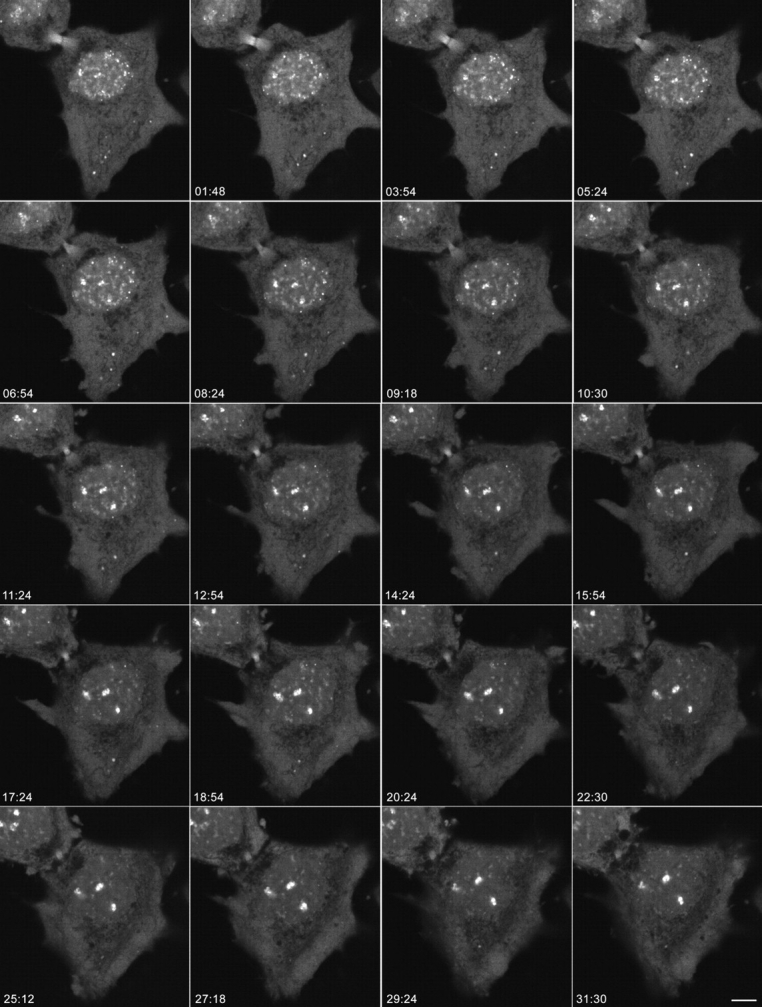
Dynamics of nucleolar reassembly in telophase. CMT3 cells in telophase, which were transiently expressing the fibrillarin-GFP protein, were subjected to time-lapse confocal fluorescence microscopy. The images were collected every 18 s over a 60-min period. See also video 2 available at http://www.jcb.org/cgi/content/full/150/3/433/DC1. The series of frames shows the progression of the cell from early to late telophase with transfer of material from PNBs to the newly forming nucleoli (the three brightest spots in the nuclei). Note the gradual disappearance of the PNBs in the nucleoplasm and the NDF in the cytoplasm with a concomitant increase in the fluorescent signal in the nucleoli. Bar, 10 μm.
Video 3.
Higher magnification of dynamics of nucleolar reassembly in the nucleoplasm of a telophase cell shown in Fig. 3. See Fig. 4 for a static presentation of 16 selected time points of the enlarged nucleoplasm and 20 selected time points of enlarged single nucleolus.
Figure 4.
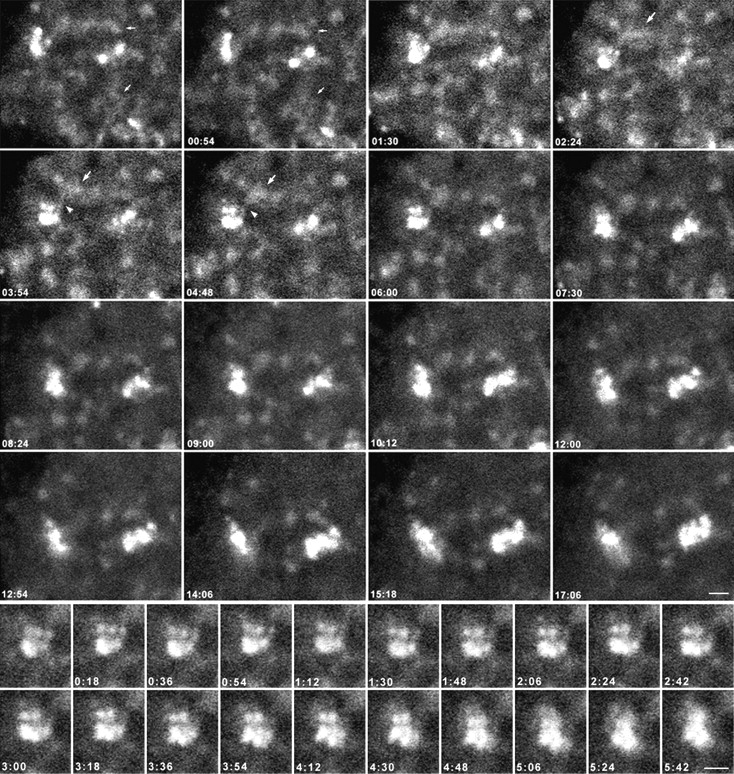
Higher magnification views of the nucleoplasm of the fibrillarin-GFP–expressing telophase cell in Fig. 3. The upper panels show that the PNBs are distributed throughout the nucleoplasm in a network-like organization during the early time points (small arrows). For the upper panels, see also video 3 available at http://www.jcb.org/cgi/content/full/150/3/433/DC1. As the PNBs (large arrows) move close to growing nucleoli, material appears to transfer between PNBs and nucleoli through streams of small particles (arrowhead). In the later time points the PNBs become distinct and are diminished in size and number. The lower panels show enlargements of images containing a single assembling nucleolus at early time points in telophase. Areas from the upper left-hand corners of the frames in the upper panels were enlarged to show a single nucleolus as it is formed. Each of the three distinct spots above the major spot resembles a dense fibrillar component (DFC) surrounding a fibrillar center (FC); i.e., a relatively bright ring of signal around a tiny open area. These units grow in size and brightness over the time of observation and eventually fuse with the main body of the nucleolus (see frame 5:42). Bars, 2 μm.
Video 4.
Behavior of nucleolin-GFP during telophase. The incorporation of the material from two PNBs into nucleoli is shown. See Fig. 5 for a static presentation at five selected time points.
Figure 5.
Behavior of nucleolin-GFP during telophase. CMT3 cells in telophase, which were transiently expressing the nucleolin-GFP fusion protein were subjected to time-lapse confocal fluorescence microscopy. The images were collected every 18 s over a 30-min period. See also video 4 available at http://www.jcb.org/cgi/content/full/150/3/433/DC1. In the time-lapse sequence, material from two PNBs (arrowheads) is incorporated into nucleoli. In frame 0 a stream of small particles appears between the PNB and the assembling nucleolus (large arrowhead). By 18 min the PNB as well as the connecting stream have disappeared. The second PNB (small arrowhead) approaches the nucleolus in the 6 min frame, forming a connection at 12 min and nearly disappears by 24 min. Bar, 2 μm.
Results
Subcellular Localization of Nucleolar GFP Chimeras
The reassembly of the nucleolus during mitosis was visualized by transiently expressing nucleolar proteins fused with GFP in CMT3 cells. To confirm that the subcellular locations of the transiently expressed GFP-proteins were similar to those of the endogenous proteins, transfected cells were subjected to immunolabeling with specific antibodies raised against the endogenous protein. The cells were then examined by fluorescence microscopy to determine the degree of colocalization of the endogenous and GFP fusion proteins. The levels of expression and the integrity of the GFP chimeras were determined by immunoblotting of cell extracts.
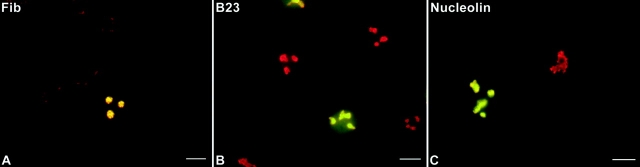
Fibrillarin-GFP displayed an immunofluorescence pattern nearly identical to that of endogenous fibrillarin as detected by the human autoimmune serum S4 (Deitz). Both signals colocalized in compact central intranucleolar regions, which were sometimes connected (Fig. 1 A, yellow); these corresponded to the nucleolar dense fibrillar components (DFCs) as described by Ochs et al. 1985. The signal was also seen in coiled bodies (not shown). Protein B23-GFP and the endogenous protein also colocalized in similar patterns as visualized by a mouse monoclonal antibody against protein B23 as shown by Spector et al. 1984. The predominant signal was in the peripheral parts of the nucleoli, against a background of general nucleolar and diffuse nucleoplasmic staining (Fig. 1 B, yellow). Endogenous nucleolin detected by a mouse monoclonal anti-nucleolin antibody colocalized with nucleolin-GFP in the nucleolus with predominant staining of peripheral parts of nucleoli and diffuse staining of the nucleoplasm (Fig. 1 C, yellow), similar to the pattern seen by Spector et al. 1984. Colocalization of GFP-fusion proteins and endogenous proteins was also seen in mitotic cells with all three of the expressed chimeric proteins (not shown). These initial studies suggested that the presence of the GFP moiety did not significantly affect the subcellular location of the fusion proteins.
Figure 1.
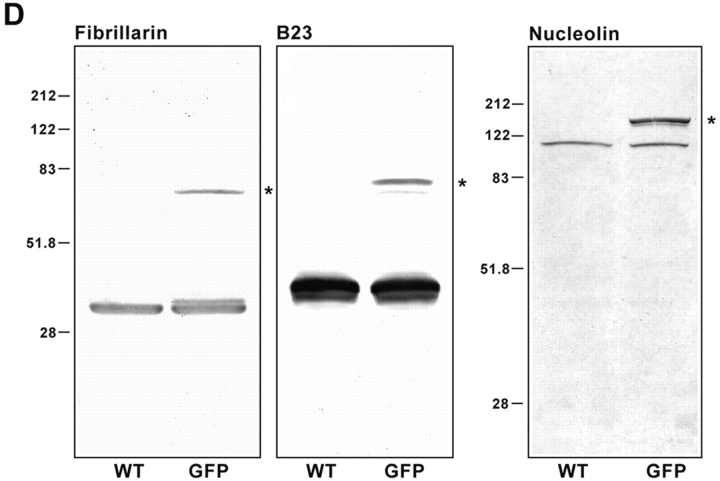
Fibrillarin-GFP, protein B23-GFP, and nucleolin-GFP chimeras colocalize with endogenous proteins in nuclear and nucleolar regions of CMT3 cells. The cells were transiently transfected with pEGFP-fibrillarin (A), pEGFP-protein B23 (B), or pEGFP-nucleolin (C) and fixed 24 h after transfection. The endogenous protein was detected by staining with the corresponding antibody followed by treatment with a specific secondary antibody conjugated with Texas red. The merged images show the colocalization of fibrillarin-GFP with endogenous fibrillarin in the dense fibrillar components (A, yellow). Protein B23-GFP (B, yellow) and nucleolin-GFP (C, yellow) were distributed throughout the nucleolus with higher accumulation in the nucleolar periphery and traces of signal in the nucleoplasm. Bars, 10 μm. (D) SDS-PAGE analysis of whole cell lysates from CMT3 cells transiently transfected with pEGFP-fibrillarin, pEGFP-protein B23 or pEGFP-nucleolin. Electrophoresis and immunoblotting with the corresponding antibody was done 40 h after transfection. The control lanes (WT) contain lysates of cells exposed to the FuGene 6 reagent without DNA. Each of the WT lanes shows only one band corresponding to the endogenous protein. In the samples from the transfected cells (GFP) both the wild-type protein and the GFP fusion protein (star), migrating at a slower rate, were detected. The molecular masses (in kD) of marker proteins are indicated on the left-hand sides of the blots.
To verify that full-length GFP-fusion proteins were expressed, CMT3 cells were transiently transfected with the expression plasmids and whole cell extracts were prepared from transfected and control cells and equal aliquots were subjected to SDS-PAGE and immunoblotting using specific antibodies. With autoimmune serum S4, fibrillarin was seen in extracts from transfected and untransfected cells as a 34-kD band, whereas the fibrillarin-GFP fusion protein was seen as a band of 62 kD apparent molecular in transfected cells (Fig. 1 D, star). Similarly, protein B23 was detected by a mouse monoclonal antibody, which showed the endogenous protein migrating as a band at 38 kD in transfected and untransfected cells. As expected, B23-GFP was detected as a band of 66 kD apparent molecular mass (Fig. 1 D, star). When nucleolin was detected by a rabbit polyclonal antibody against its NH2-terminal end, a band of ∼110 kD was observed in transfected cells and untransfected cells, corresponding to endogenous nucleolin; the nucleolin-GFP fusion protein (star) as a band of 138 kD apparent molecular weight was seen only in transfected cells (Fig. 1).
NDF during Telophase
To investigate the dynamics of NDF in living cells during the later stages of mitosis, CMT3 cells were transiently transfected with plasmids expressing fibrillarin-GFP, protein B23-GFP or nucleolin-GFP and observed by time-lapse fluorescence microscopy. The monkey kidney CMT3 cells were used because of their large nucleoli and the presence of numerous, very large NDF during mitosis (Dundr et al. 1997). Cells that significantly overexpressed GFP-fusion protein were not selected for microscopic observation. To examine the possible transfer of material from the NDF to nuclei, telophase cells containing prominent NDF were observed by fluorescence microscopy. Images were collected from a single focal plane every 18 s for periods of up to two hours without significant fading of the signal. In cells transfected with the B23-GFP vector, some of the NDF, especially those close to the nuclear envelope disappeared over a period of 3–4 min (Fig. 2, arrow). Others remained stable and showed no major change in intensity during that period of time. Observed cells were viable and underwent normal cytokinesis, indicating that no significant photodamage had occurred (see Fig. 3). NDF often exhibited rapid random movements and some of them appeared to fuse. In early telophase the number of NDF observed in a single optical section was between less than a dozen and more than 50, but by the end of telophase they were reduced to a few or none (not shown). As the signal from the NDF decreased there was a concomitant increase in fluorescence in a region of the nucleus close to the nuclear envelope (Fig. 2, arrowheads), suggesting that fluorescent material was transferred to the nucleus. Other NDF seemed to remain relatively stable throughout the period of observation.
In cells transfected with the fibrillarin-GFP construct, the NDF appeared as bright, compact and highly mobile structures throughout a weakly fluorescing cytoplasm (Fig. 3). As with the NDF identified by B23-GFP those seen with fibrillarin-GFP exhibited a variety of movements; some moved apparently randomly, others moved unidirectionally and still others in circles. The velocities of the NDF movements ranged from 1.8 to 15 μm/min. Individual NDF frequently moved towards one another, seemingly associating with each other, but eventually separating again. Fusion of NDF was seen only rarely. Some NDF seemed to disintegrate and disappear over a period of time. These live cell fluorescence studies illustrate the dynamic nature of the NDF and suggest that dissociated components from them enter telophase nuclei.
PNBs and Nucleolar Reassembly during Telophase
The formation of nucleoli in live telophase cells was readily observable by time-lapse fluorescence microscopy using cells transfected with the fibrillarin-GFP construct. In the nuclei of early telophase cells two classes of fluorescent spots of approximately the same size were recognizable against a generally fluorescent background (Fig. 3). PNBs were seen as spots of lower intensity in larger numbers near the nuclear envelope than in the center of the nucleus, especially in the early time points. The less abundant, brighter spots were nucleoli in their very early stages of assembly (Fomproix et al. 1998). As the cells proceeded through telophase the signal from the PNBs decreased with a concomitant increase in the intensity of the nucleolar fluorescence. Within 30 min, the overall signal from the nucleoplasm was nearly equivalent in intensity to that of the cytoplasm, with the nucleoli standing out as the predominant fluorescent structures in the nuclei (compare the first with the last frames of the video sequence). The decrease in the signal from the PNBs was unlikely to be due to photobleaching since there was an increase in the nucleolar signal during the same time period. At the end of the sequence only a few PNBs persisted. Interestingly, the PNBs that were present in the later time points of the time-lapse sequence exhibited very little movement in the nucleoplasm during the time of observation. These data strongly support the idea that the reassembling nucleoli are at least partly constructed from material derived from the PNBs.
At higher magnification, the behavior of PNBs could be analyzed in detail (Fig. 4). In early stages of telophase the shapes of the PNBs were highly irregular, and many of them were linked (large panels, small arrows). At very early time points, when newly formed nucleoli and the PNBs were essentially the same size, some of the PNBs came very close to each other (large arrow) and eventually fused into larger PNBs; a thin line of fluorescence often connected these with nucleoli (Fig. 4, large panels, arrowheads). Distinct substructures could be seen in some of the forming nucleoli (see the three small bright spots adjacent to the intense spot in the first six frames of the small panels). These substructures occasionally appeared as a ring of fluorescence; this suggests that these spots are newly forming DFCs, since fibrillarin preferentially localizes to the DFC. The nucleolar substructures gradually increased in intensity and then fused with the main nucleolar body (Fig. 4, small panels, frame 5:42). The connections between the PNBs and the growing nucleolar bodies remained visible during the later time points, but they decreased in fluorescence intensity as the PNBs disappeared and the nucleoli enlarged (Fig. 4, large panels, frame 17:06). At the same time, the remaining PNBs stood out as more discrete structures. In contrast to their rapid rates of movement in early telophase (up to 2.4 μm/min), the PNBs in late telophase became essentially immobile.
A similar pattern of behavior was found for nucleolin-GFP. In the time-lapse sequence shown in Fig. 5, material from two PNBs (arrows) is incorporated into nucleoli. In frame 0 a line of fluorescent material appears between the PNB and the assembling nucleolus (arrowhead). By 18 min the PNB as well as the connecting material have disappeared. The second PNB approaches the nucleolus at 6 min, forming a connection at 12 min and nearly disappears by 24 min. These data further support the idea that as PNBs dissociate into their subcomponents, they become major suppliers of material for rebuilding nucleoli at the end of mitosis.
FRAP Analyses of NDF and PNBs
An unanswered question is whether the NDF and PNBs are stable structures that tightly retain their components until their time of release or if there is rapid turnover of these components within the structures. This question was approached by subjecting cells expressing fibrillarin-GFP to fluorescence recovery after photobleaching (FRAP). NDF and PNBs as well as selected areas of newly reassembled nucleoli were photobleached for 0.5 s using the 488-nm laser line of a confocal microscope at 100% laser power. Cells were then imaged at two second intervals. The fluorescence in all three structures in telophase cells recovered to 85–90% of its prebleach intensity within 20–30 s (Fig. 6). The recovery curves appeared to be biphasic with an initial rapid phase occurring in one or two seconds followed by a slower phase for the next 18–20 s. The overall half-time of recovery was less than one second. The recovery curves of the NDF or the PNBs were nearly identical with a mobile fraction of about 90% for either structure. This indicates that 90% of fibrillarin-GFP in the NDF and PNBs is rapidly dissociating from and reassociating with them, suggesting that both structures are in continuous flux. Interestingly, the mobile fraction of fibrillarin in the newly forming nucleolus during telophase is slightly smaller, although the recovery profile was similar to that of the NDF or PNBs. This suggests that there is a slightly greater retention of fibrillarin in telophase nucleoli compared with the NDF and PNBs. In the interphase nucleolus fibrillarin has a much smaller mobile fraction and a slower rate of recovery than that of the telophase nucleolus (Phair and Misteli 2000).
Figure 6.
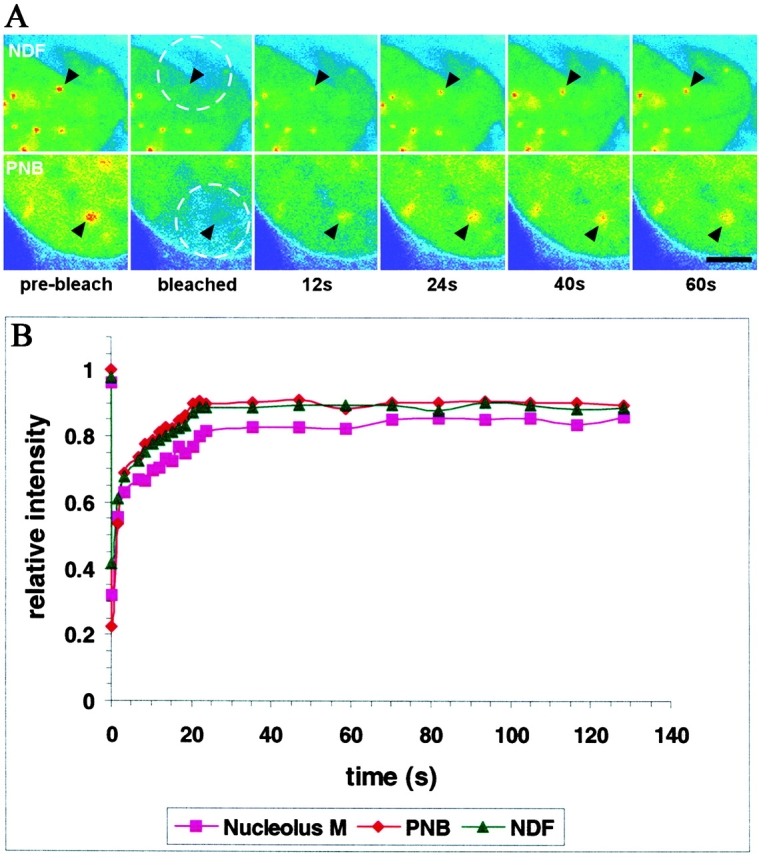
FRAP on fibrillarin-GFP in NDF, PNB and nucleoli during telophase. An area in a telophase cell including either an NDF or a PNB was bleached for 0.5 s using the 488-nm laser line of a confocal microscope at high power. The cells were then observed under normal fluorescence microscopy conditions, with images collected at 2-s intervals and the kinetics of recovery measured. (A) Images before and immediately after the bleach pulse and during recovery were taken at the indicated times. The bleached area is indicated by a dashed circle and the positions of the bleached NDF or PNB are indicated by arrowheads. Bar, 4 μm. (B) Quantitative measurements of fluorescence recovery. Values represent the relative recovery relative to total cellular fluorescence. The values were averages of determinations from six different cells for each subcellular compartment.
The Ultrastructures of NDF and PNBs Are Similar
The similar recovery kinetics of NDF and PNBs in the FRAP experiments suggests that the are structurally similar. To test this possibility more directly the ultrastructural features of the two particles were compared. The CMT3 cells were synchronized, mitotic cells were collected after mechanical shake-off and embedded in Lowicryl. After sectioning and immunogold labeling using an anti-B23 polyclonal antibody the sections were observed by electron microscopy. The NDF in anaphase cells identified by the immunogold labeling showed the same general fibrogranular structure as the PNBs detected by the same antibody in the nuclei of telophase cells (Fig. 7, arrows). This supports the idea that the two classes of particles have similar structures and that they are built from the same nucleolar material.
Figure 7.
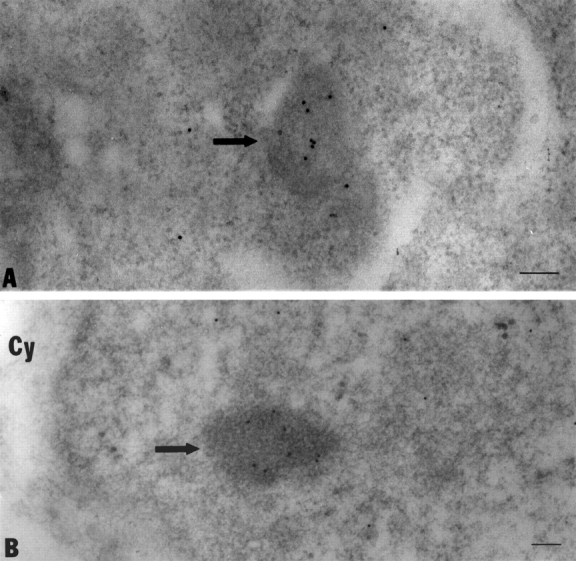
The ultrastructures of the NDF and PNBs have similar features. The CMT3 cells were synchronized and mitotic cells were harvested and embedded in Lowicryl. After sectioning and immunogold labeling with an anti-B23 polyclonal antibody the sections were viewed under the electron microscope. The NDF (A) in the cell plasms of anaphase cells identified by the 10-nm immunogold particle labeling (arrow) showed the same general fibrogranular structure as seen in the PNBs (B) in the nuclei of telophase cells (arrow). Bars, 100 nm.
Timing of Entry of Nucleolar Components into Nuclei and Nucleoli during Telophase
An important issue is whether the separate components of NDF and PNBs are released simultaneously as nucleoli are reassembled or whether they are delivered at different times as they are needed for the assembly process. Initial attempts were made to answer these questions in live cells by colocalizing B23-blue fluorescent protein (BFP) and fibrillarin-GFP; however, technical problems prevented simultaneous detection of the two fusion proteins. Therefore, this problem was approached by double immunofluorescence using antibodies to fibrillarin, protein B23 and nucleolin. In early anaphase protein B23 and fibrillarin had nearly identical patterns of localization (Fig. 8, A–C). Both proteins were localized in perichromosomal regions (PRs) and NDF as well as being distributed generally in the cell plasm. However, in late anaphase cells, the relative distributions of fibrillarin and B23 began to diverge (Fig. 8, D–F). The two proteins colocalized in the PRs and in the NDF, but only fibrillarin was found in tiny dots in the interiors of the regions surrounded by the decondensing chromosomes (Fig. 8, arrowheads in D and F). These dots appear to be nucleoli in their earliest stages of assembly, which is consistent with the findings of Fomproix et al. 1998 that pre-rRNA transcription begins in late anaphase.
Figure 8.
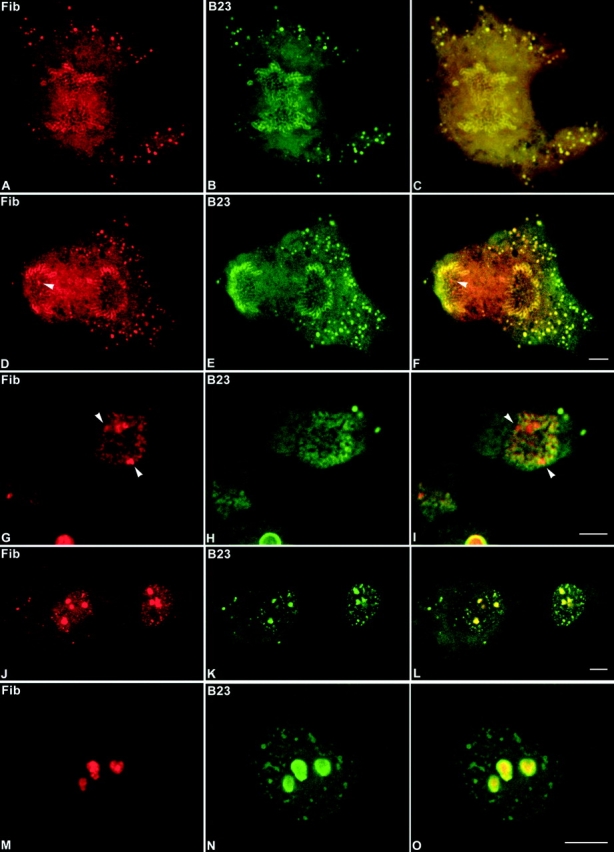
Fibrillarin and protein B23 enter reassembling nucleoli at different times during mitosis. The CMT3 cells were double-labeled with anti-fibrillarin autoimmune serum (red) and mouse monoclonal antibody against protein B23 (green). Fibrillarin and protein B23 colocalized in the perichromosomal region and in numerous NDF (arrowheads) during early (A–C) and late (D–F) anaphase. From late anaphase (D) to early telophase (G) fibrillarin is detectable in tiny newly formed nucleoli (arrowheads), which are negative for protein B23 (E and H). In late telophase fibrillarin (J) and protein B23 (K) colocalized in PNBs and postmitotic nucleoli (L). In early G1 phase fibrillarin (M) is exclusively localized in postmitotic nucleoli in contrast to protein B23 (N), which is still detectable in persisting PNBs (O). Bars, 10 μm.
The differences in the locations of the two proteins became more apparent in early telophase cells. The developing nucleoli became clearly visible (Fig. 8, arrowheads in G and I) among the decondensing chromosomes. As in the anaphase cells fibrillarin, but not protein B23 was present in the reforming nucleoli. However, the two proteins colocalized in the NDF and in the region of the decondensing chromosomes (Fig. 8 I). By late telophase when the nucleoli appeared as prominent structures, protein B23 and fibrillarin were both present in nucleoli (Fig. 8, J–L) and colocalized in the NDF and in the PNBs distributed throughout the nucleoplasm. The PNBs at this stage occurred more frequently near the nuclear envelope, where the nucleolar components were apparently being released from decondensing chromosomes. When the cells progressed to early G1 phase fibrillarin became exclusively localized in nucleoli (Fig. 8, M–O). In contrast, protein B23 localized not only in nucleoli but it also was present in persisting PNBs scattered throughout the nucleoplasm. When cells were examined in a similar series of studies using an antibody to nucleolin, the results were virtually the same as those obtained with fibrillarin (not shown). Thus, both fibrillarin and nucleolin are released from the PNBs and appear in nucleoli earlier than protein B23. These results are summarized in Table . The data support the idea that individual nucleolar components are disengaged from the PNBs at different times as they are needed for constructing nucleoli.
Table 1.
Presence of Absence of Nucleolar Proteins and Preribosomal RNAs in Cellular Compartments at the End of Mitosis
| Nucleolar component | Early anaphase | Late anaphase | Early telophase | Late telophase | Early G1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PR | NDF | PR | NDF | No | NDF | PNB | No | NDF | PNB | No | PNB | No | |
| Fibrillarin | + | + | + | + | + | + | + | + | + | + | + | − | + |
| Nucleolin | + | + | + | + | + | + | + | + | + | − | + | − | + |
| Protein B23 | + | + | + | + | − | + | + | − | + | + | + | + | + |
| 5′ETS leader | − | − | − | − | + | − | − | + | − | − | + | − | + |
| 5′ETS core | + | + | + | + | + | + | + | + | + | − | + | − | + |
| 18S | + | + | + | + | + | + | + | + | + | − | + | − | + |
| 28S | + | + | + | + | − | + | + | * | + | + | + | + | + |
Data indicate the presence (+) or absence (−) of proteins or pre-rRNA segments in cellular substructures determined by immunofluorescence microscopy and fluorescence in situ hybridization (FISH), respectively. The asterisk indicates presence not determined. PR, perichromosomal region; NDF, nucleolus-derived foci; No, nucleoli; PNB, prenucleolar bodies. The probes for pre-rRNA segments (5′ETS leader, 5′ETS core, 18S and 28S) examined by FISH are described in Materials and Methods.
Preribosomal RNA Sequences Are Present in Telophase Nuclei
Previous work from this laboratory showed the presence of partially processed pre-rRNA transcripts in the NDF (Dundr and Olson 1998). Because NDF and PNBs have very similar characteristics and contain many of the same components, it was important to determine whether PNBs also contained the partially processed pre-rRNA. To answer this question, we employed fluorescence in situ hybridization (FISH) to probe for various segments of pre-rRNA in the PR and PNBs. These included probes to sequences in the 5′ETS leader and core regions and to segments of 18S and 28S rRNAs. As expected from previous studies (Dundr and Olson 1998) the 5′ETS leader sequence was only detected in the newly forming nucleoli and not in the NDF or PNBs in all of the mitotic cells examined (not shown). In contrast, during anaphase the 5′ETS core sequence was detected in PRs and in numerous NDF where it colocalized with protein B23 (Fig. 9, A–C). The distribution of this sequence in early telophase (Fig. 9, D–F) was similar to that seen in anaphase, with the signal present in NDF and in newly forming PNBs in the vicinity of the PRs. In addition, the 5′ETS core sequence was seen in newly forming nucleoli (Fig. 9D and Fig. F, arrowheads). The protein B23 signal did not colocalize with the 5′ETS core sequence in nucleoli in cells at this stage of telophase (Fig. 9E and Fig. F). Except for a high cytoplasmic background, the 18S sequence exhibited behavior almost identical to that of the 5′ETS core sequence (not shown).
Figure 9.
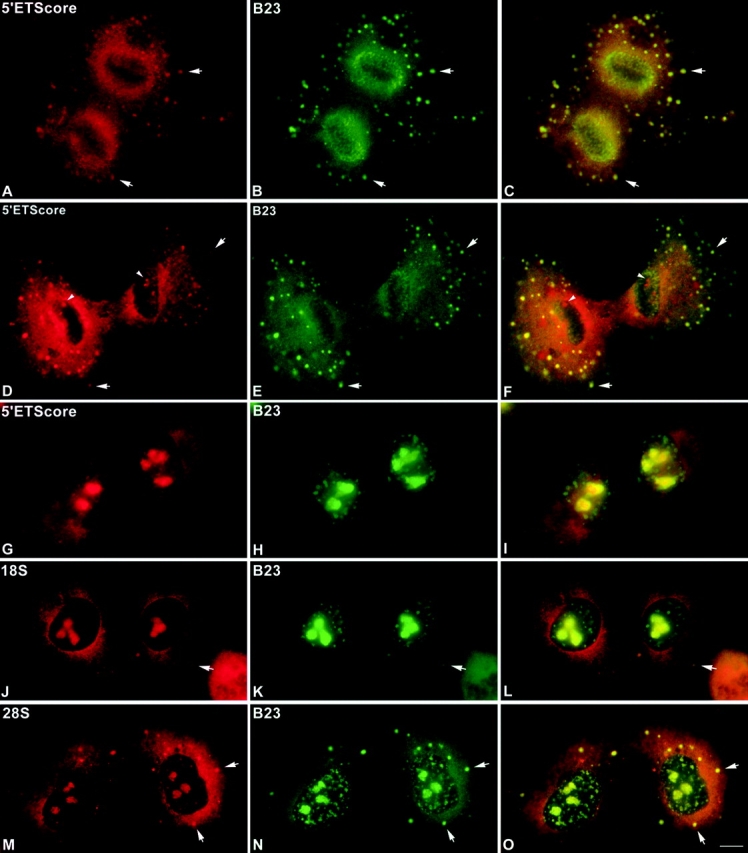
Preribosomal RNA sequences are present in telophase nuclei. CMT3 cells were subjected to fluorescence in situ hybridization (FISH) to probe for various segments of pre-rRNA including sequences in the 5′ETS core region and in 28S rRNA. During anaphase the 5′ETS core sequence was detected in the perichromosomal regions (PRs) and in numerous NDF (arrows) where it colocalized with protein B23 (A–C). The distribution of this sequence in early telophase (D–F) was similar to that seen in anaphase, with the signal present in NDF (arrows) and in the vicinity of the PRs (arrowheads). The 5′ETS core sequence was also seen in newly forming nucleoli (D and F, arrowheads). Protein B23 did not colocalize with the 5′ETS core sequence in nucleoli in cells at this stage of telophase (E and F). In late telophase/early G1 cells the 5′ETS core sequence was present primarily in newly forming nucleoli, with no detectable signal for this sequence in the PNBs labeled by anti-B23 antibody (G–I). The signal for the 18S rRNA sequence was seen in newly forming nucleoli of late telophase/early G1 cells, but not in the PNBs labeled by anti-B23 antibody (J–L). In late telophase cells the FISH signal for a 28S rRNA sequence was clearly visible in PNBs (M), where it colocalized with protein B23 (N and O). Bar, 10 μm.
In late telophase/early G1 phase cells the 5′ETS core sequence was present primarily in newly forming nucleoli, but very little or no signal for this sequence was detected in the PNBs (Fig. 9, G–I). Although there was a general background signal for the 5′ETS core in the nucleoplasm, there were no distinct spots that colocalized with the protein B23 signal in the PNBs (Fig. 9I). Similarly, the 18S rRNA signal was not present in the PNBs but it was clearly present in the nucleoli of cells at this stage (Fig. 9, J–L). In contrast, in late telophase cells the FISH signal for a 28S rRNA sequence was clearly visible in PNBs, where it colocalized with protein B23 (Fig. 9, M–O). Thus, there is a marked difference between the behaviors of the 5′ half of the pre-rRNA transcript (including the 5′ETS core and 18S rRNA sequences) and the 3′ half, which includes the 28S rRNA sequence (Table ). On the one hand, the 5′ETS core and 18S rRNA sequences are enriched in the material associated with decondensing chromosomes in early telophase, but they are undetectable in the PNBs in later telophase cells. On the other hand, the 28S rRNA sequence is found in PNBs throughout telophase and into early G1 phase.
Discussion
Over the past several years a few key observations have advanced our understanding of the process of nucleolar reassembly at the end of mitosis (Dundr et al. 1997; Dundr and Olson 1998; Fomproix and Hernandez-Verdun 1999; Savino et al. 1999; Sirri et al. 1999, Sirri et al. 2000; Verheggen et al. 1998). First, material derived from maternal cell nucleoli is maintained in an assembled state. Second, partially processed pre-rRNA is preserved during mitosis. Finally, some of this material is used for rebuilding the daughter cell nucleoli. This work provides further insights into nucleologenesis by examining the dynamic behavior of complexes containing pre-rRNA processing components associated with partially processed pre-rRNA in living cells and by characterizing the timing of events related to nucleolar assembly during telophase.
The reassembly of nucleoli has been described as a process involving the fusion of PNBs with the nucleolar organizer regions and developing nucleoli after RNA pol I transcription is reactivated (Benavente, et al. 1987; Scheer et al. 1993). The studies reported here demonstrate that this is not a simple fusion process whereby whole PNBs are engulfed into the telophase nucleoli. On the contrary, the time-lapse studies show that it is material dissociated from the PNBs that enters the growing nucleoli and actual fusion events are very rare. When the PNBs are in close proximity to the nucleoli, narrow connections can be seen between the PNBs and nucleoli. However, the proximity of the two structures is probably not essential for transfer of material, as illustrated by the fact that PNBs relatively distant from the nucleoli also disappear during the process of nucleolar formation (see Fig. 3 and Fig. 4).
The mechanism of transfer of material from the PNBs to nucleoli remains unclear. Because the PNBs undergo rapid dissociation and reassociation as indicated by the FRAP studies it cannot be ruled out that the material is transferred to nucleoli by diffusion and mass action, rather than by an active process. Previous studies showed that RNA pol I transcription is essential for nucleolar formation (Benavente et al. 1987) and it has been suggested that the material in the PNBs is captured by the nascent pre-rRNA transcripts. Recent work in this laboratory confirmed that blockage of RNA pol I transcription by a low dose of actinomycin D prevents nucleolar formation in telophase (Dundr, M., and M.O.J. Olson, unpublished observations). This treatment does not inhibit the formation of PNBs, but it prevents the transfer of material from them to the nucleoli. It has been shown that the reactivation of transcription at the end of mitosis is accomplished by dephosphorylation of cdc2/cyclin B type sites in transcription factors (Heix et al. 1998; Klein and Grummt 1999; Voit et al. 1999). Since similar phosphorylation sites are also present in several of the nucleolar proteins present in the PNBs; e.g., B23 and nucleolin, it is conceivable that their release could be facilitated by the same mechanism. In other words, dephosphorylation events could shift the equilibrium toward dissociation of the components from the PNBs.
Although the current studies provide insights into how the PNBs disappear from nuclei, the mechanism of their formation is less clear. PNBs have been shown to originate in the perichromosomal regions (Fomproix et al. 1998). However, material may also be transferred from the NDF into telophase nuclei to supply components to PNBs. The latter possibility is supported by the fact that the NDF and PNBs have several components in common; e.g., fibrillarin, nucleolin, protein B23 and U3 and U8 snoRNAs (Dundr et al. 1997; Dundr and Olson 1998). The studies reported here indicate that both types of particles also contain sequences from pre-rRNA. Except for location and size, could the NDF be essentially the same as the PNBs? Electron microscopic studies indicate that the two types of particles have virtually identical fibrogranular structures. In addition, the fibrillarin-GFP FRAP curves for the NDF and PNBs are very similar.
In spite of their structural similarities, there are important differences between the NDF and the PNBs. First, the NDF are much more mobile than the PNBs; i.e., the NDF seem to have freedom of movement over much larger distances than the PNBs. Second, the PNBs are much smaller and nearly uniform in size, whereas the NDF vary in diameter from 0.1 to 3 μm. Finally, and most importantly, the PNBs seem to differ from the NDF in pre-rRNA and protein content. Except for the 5′ETS leader sequence, the NDF contain the full-length pre-rRNA transcript (Dundr and Olson 1998). In the current study the 28S-region sequence was also clearly present in the PNBs of cells from telophase to early G1 phase. This is consistent with the findings of Medina et al. 1995 showing the presence of preribosomal RNA sequences in the PNBs of onion cell telophase nuclei. However, we never detected the 5′ETS core or 18S sequences in PNBs that were located by the anti-B23 antibody in the numerous telophase cells examined. In contrast, FISH analysis of early telophase cells showed that the nucleolar material associated with the chromosome periphery contained sequences from the 5′ETS core and the 18S and 28S pre-rRNA segments. Thus, it seems likely that as the PNBs form, the 5′ETS core and 18S pre-rRNA sequences are removed, but the 28S rRNA segment is preserved and transferred to the PNBs. This suggests that some processing steps in the pre-rRNA maturation pathway have begun when the PNBs begin to form; i.e., the 5′ region of the pre-rRNA transcript, including the 18S sequence, seems to have split from the 3′ segment.
This and previous work also revealed differences in the protein content between PNBs and NDF. For example, fibrillarin is always seen in the NDF and is present in telophase PNBs, but it disappears from them in early G1 cells. At the same time protein B23 is found in the PNBs that persist into G1 phase. Nucleolin exhibits a pattern of behavior very similar to that of fibrillarin. This makes the PNBs appear to be heterogeneous; however, this heterogeneity may be related to the differences in RNA content. It seems likely that the early release of fibrillarin from PNBs reflects its association with segments of pre-rRNA, which are also released early (possibly regions of the 5′ half of the transcript). By the same token, the late release of protein B23 from PNBs may be related to its possible interaction with segments in the 3′ half of pre-rRNA.
The above observations of the presence of fibrillarin in the telophase PNBs and the absence of 5′ETS core and 18S sequences in them appears to be inconsistent with the idea that fibrillarin is released from the PNBs simultaneously with the pre-rRNA segments. However, it is possible that fibrillarin is not only associated with the 5′ETS core and 18S segments, but also with sequences further downstream in the transcript. Indeed, U3 and U8 snoRNAs associated with fibrillarin are also involved in the cleavage activities in the center region of the transcript (Maxwell and Fournier 1995; Sollner-Webb et al. 1996). It is also possible that much of the fibrillarin in PNBs is not directly associated with pre-rRNA. The early release of nucleolin from the PNBs (Table ) correlates with its involvement in the early stages of pre-rRNA processing (Ginisty et al. 1998). Recent work by Savino et al. 1999 also showed that fibrillarin and nucleolin were released from the PNBs much earlier than Nop52 and B23 during telophase. This agrees with our findings that protein B23 persists in PNBs much longer than fibrillarin and nucleolin. Savino et al. 1999 also suggest that the timing of release of PNB components approximately follows the order of processing. Although our data generally support that view, the absence of the 5′ half of the transcript in the PNBs makes it unlikely that the fibrillarin and nucleolin present in PNBs are associated with that portion of pre-rRNA. It is certainly possible that the pre-rRNA segments are released from the PNBs in the same order as in the processing pathway (Sollner-Webb et al. 1996) and that the proteins associated with those segments follow the same order. However, the deficiency in sequences from the 5′ half of the pre-rRNA transcript in PNBs suggests that this RNA processing or degradation actually begins before the PNBs are formed.
Although progress has been made in determining the composition of the PNBs (Bell et al. 1992; Bell and Scheer 1996; Jiménez-García et al. 1994; Scheer and Hock 1999; this work), very little is known about their structure or the organization of their components. Are they assembled around complexes of partially processed pre-rRNA and processing components or are they poorly organized aggregates of nucleolar proteins and RNAs? Supporting the former proposition are recent studies by Pinol-Roma 1999 indicating particles immunoprecipitated from mitotic cells by an antinucleolin antibody contain pre-rRNA sequences and a discrete set of proteins. These particles are also very similar in composition to the corresponding interphase particles. Thus, the NDF and PNBs could be higher order aggregates of processing complexes. These complexes could polymerize into larger structures with the aid of certain nucleolar proteins such as B23, which tends to form oligomers (Herrera and Olson 1996) and also interacts with multiple protein substrates (Szebeni and Olson 1999). In contrast, earlier studies using Xenopus extracts to assemble PNB-like bodies (Bell and Scheer 1996) suggest the PNBs are not well organized. Immunodepletion experiments showed that neither nucleolin, xNopp180, B23/NO38 nor fibrillarin are required for the self-assembly process in vitro. Xenopus extracts deprived of any of these proteins were capable of promoting formation of nuclear bodies, which, as judged from immunofluorescence microscopy analyses, lacked nucleolin, xNopp180, B23/NO38 or fibrillarin. Only in the case of fibrillarin there was a difference between the depleted and control extract; i.e., nuclei formed in fibrillarin-depleted extracts generally contained fewer nuclear bodies compared with controls. Thus, although nuclear bodies will form without fibrillarin, its presence seems to facilitate the assembly process. It is conceivable that in the absence of fibrillarin the concentration of the other components of the nuclear bodies must rise to a higher level in order to nucleate the self-assembly process.
Based on current and previous studies we propose the following model for nucleologenesis in mammalian cells. When transcription is shut down in early M-phase the initiated pre-rRNA transcripts are completed and released from the nucleolar dense fibrillar component in association with processing components. These complexes then associate with the peripheral regions of all chromosomes. In anaphase, some of these complexes remain with the chromosomes while others become packaged into large cytoplasmic particles called NDF. During telophase these processing complexes, or their subcomponents, enter nuclei by two pathways. The first is through their passive association with the perichromosomal regions, which automatically become part of the nucleus when the nuclear envelope is formed. Second, they may be transported into nuclei as small particles dissociated from the NDF. Once inside the nucleus, the components of these processing complexes are eventually incorporated into PNBs, which provide material for building postmitotic nucleoli. The incorporation of the processing components into newly forming nucleoli is dependent on the reactivated transcription in the nucleoli. The order in which this material is released from the PNBs and permitted to enter nucleoli seems to be dependent on the pathway of processing of the pre-rRNA molecules contained in the complexes. The transfer of these components between PNBs and nucleoli is probably by diffusion rather than by an active transport process. The availability of current technology should make it possible to confirm this model and to define the regulatory details of these processes in the near future.
Supplemental Material
Acknowledgments
The authors thank Drs. Pui K. Chan, Seraphin Pinol-Roma and Robert L. Ochs for providing antibodies; Dmitry Goldgaber for the nucleolin-GFP–expressing plasmid and John Aris and James Sylvester for specific DNA-containing plasmids.
This work was supported in part by grants to M.O.J. Olson from the National Institutes of Health and by the University of Mississippi Medical Guardian Society.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: DFC, dense fibrillar component; ETS, external transcribed spacer; FC, fibrillar center; NDF, nucleolus-derived foci; NOR, nucleolus organizer region; PNB, prenucleolar body; PR, perichromosomal region; RNA pol I, RNA polymerase I; UBF, upstream binding factor.
References
- Aris J.P., Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA. 1991;88:931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azum-Gélade M.C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gas N. Cell cycle redistribution of U3 snRNA and fibrillarin. Presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. J. Cell Sci. 1994;107:463–475. doi: 10.1242/jcs.107.2.463. [DOI] [PubMed] [Google Scholar]
- Bell P., Scheer U. Prenucleolar bodies contain coilin and are assembled in Xenopus egg extract depleted of specific nucleolar proteins and U3 RNA. J. Cell Sci. 1996;109:43–54. doi: 10.1242/jcs.110.1.43. [DOI] [PubMed] [Google Scholar]
- Bell P., Dabauvalle M.-C., Scheer U. In vitro assembly of prenucleolar bodies in Xenopus egg extract. J. Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven A.F., Lee R., Razaz M., Leader D.J., Brown J.W.S., Shaw P.J. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J. Cell Sci. 1996;109:1241–1251. doi: 10.1242/jcs.109.6.1241. [DOI] [PubMed] [Google Scholar]
- Bootsma D., Budke L., Vos O. Studies on synchronous division of tissue culture cells initiated by excess thymidine. Exp. Cell Res. 1964;33:301–309. doi: 10.1016/s0014-4827(64)81035-1. [DOI] [PubMed] [Google Scholar]
- Chang J.-H., Dumbar T.S., Olson M.O.J. cDNA and deduced primary structure of rat protein B23, a nucleolar protein containing highly conserved sequences. J. Biol. Chem. 1988;263:12824–12827. [PubMed] [Google Scholar]
- Cook P.R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Dundr M., Olson M.O.J. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol. Biol. Cell. 1998;9:2407–2422. doi: 10.1091/mbc.9.9.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Leno G.H., Lewis N., Rekosh D., Hammarskjöld M.-L., Olson M.O.J. Location of the HIV-1 Rev protein during mitosisinactivation of the nuclear export signal alters the pathway for postmitotic reentry into nucleoli. J. Cell Sci. 1996;109:2239–2251. doi: 10.1242/jcs.109.9.2239. [DOI] [PubMed] [Google Scholar]
- Dundr M., Meier U.T., Lewis N., Rekosh D., Hammarskjöld M.-L., Olson M.O.J. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–417. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- Fomproix N., Hernandez-Verdun D. Effects of anti-PM-Scl 100 (Rrp6p exonuclease) antibodies on prenucleolar body dynamics at the end of mitosis. Exp. Cell Res. 1999;251:452–464. doi: 10.1006/excr.1999.4578. [DOI] [PubMed] [Google Scholar]
- Fomproix N., Gébrane-Younès J., Hernandez-Verdun D. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J. Cell Sci. 1998;111:359–372. doi: 10.1242/jcs.111.3.359. [DOI] [PubMed] [Google Scholar]
- Gébrane-Younès J., Fomproix N., Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J. Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- Gerard R.D., Gluzman Y. New host cell system for regulated simian virus 40 DNA replication. Mol. Cell. Biol. 1985;5:3231–3240. doi: 10.1128/mcb.5.11.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Amalric F., Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heix J., Vente A., Voit R., Budde A., Michaelidis T.M., Grummt I. Mitotic silencing of human rRNA synthesisinactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D., Gautier T. The chromosome periphery during mitosis. Bioessays. 1994;16:179–185. doi: 10.1002/bies.950160308. [DOI] [PubMed] [Google Scholar]
- Herrera J., Olson M.O.J. Sedimentation analysis of the salt- and divalent metal ion-induced oligomerization of nucleolar protein B23. Biochemistry. 1996;35:2668–2673. doi: 10.1021/bi9523320. [DOI] [PubMed] [Google Scholar]
- Jiménez-García L.F., Segura-Valdez M.L., Ochs R.L., Rothblum L.I., Hannan R., Spector D.L. NucleologenesisU3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P., Mannervik M., Tora L., Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J., Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcriptionthe nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Vente A., Dorée M., Grummt I. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol. 1998;284:1–5. doi: 10.1006/jmbi.1998.2164. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Matera A.G. Nuclear bodies. Multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Maxwell E.S., Fournier M.J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Medina F.J., Cerdido A., Fernández-Gómez M.E. Components of the nucleolar processing complex (pre-rRNA, fibrillarin, and nucleolin) colocalize during mitosis and are incorporated to daughter cell nucleoli. Exp. Cell Res. 1995;221:111–125. doi: 10.1006/excr.1995.1358. [DOI] [PubMed] [Google Scholar]
- Ochs R., Lischwe M., Spohn W. Fibrillarina new protein of the nucleolus identified by autoimmune sera. Biol. Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Olson M., Dundr M., Szebeni A. The nucleolusan old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Pederson T. Survey and summary. The plurifunctional nucleolus. Nucleic Acids Res. 1998;17:1871–1876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott D.M., Bender M.A. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Roussel P., André C., Comai L., Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino T.M., Bastos R., Jansen E., Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Scheer U., Hock R. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Scheer U., Thiry M., Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Shaw P.J., Jordan E.G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Sirri V., Roussel P., Hernandez-Verdun D. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J. Cell Sci. 1999;112:3259–3268. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- Sirri V., Roussel P., Hernandez-Verdun D. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 2000;148:259–270. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tyc K., Steitz J.A. Ribosomal RNA processing in eukaryotes. In: Zimmerman R., Dahlberg A., editors. Ribosomal RNAStructure, Evolution, Processing and Function in Protein Synthesis. CRC Press; Boca Raton, FL: 1996. pp. 469–490. [Google Scholar]
- Spector D., Ochs R., Busch H. Silver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C23. Chromosoma. 1984;90:139–148. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- Szebeni A., Olson M.O.J. Nucleolar protein B23 has molecular chaperone activities. Prot. Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Le Panse S., Almouzni G., Hernandez-Verdun D. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis . J. Cell Biol. 1998;142:1167–1180. doi: 10.1083/jcb.142.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R., Hoffman M., Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D., Scheer U. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J. Cell Biol. 1995;129:561–575. doi: 10.1083/jcb.129.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.