Abstract
We have investigated the intracellular roles of an Xklp2-related kinesin motor, KRP180, in positioning spindle poles during early sea urchin embryonic cell division using quantitative, real-time analysis. Immunolocalization reveals that KRP180 concentrates on microtubules in the central spindle, but is absent from centrosomes. Microinjection of inhibitory antibodies and dominant negative constructs suggest that KRP180 is not required for the initial separation of spindle poles, but instead functions to transiently position spindle poles specifically during prometaphase.
Keywords: Xklp2, kinesin, sea urchin, mitosis, γ-tubulin
Introduction
During mitosis, the mitotic spindle uses microtubules (MTs) and motors to segregate sister chromatids and to establish the position of the cleavage plane. These events depend upon the accurate positioning of spindle poles during spindle assembly, maintenance, and elongation, and current work is aimed at determining the precise functions of mitotic motors in spindle pole positioning (Hoyt and Geiser 1996; Sharp et al. 2000).
We have used cleavage stage echinoderm embryos to characterize the functions of mitotic motors, including motors that are involved in spindle pole positioning. The living echinoderm embryo is a classic developmental system for studies of mitosis and cell division as it offers excellent cytology and is amenable to micromanipulation (Wright et al. 1993; Morris and Scholey 1997; Scholey 1998). Members of two families of MT-based motors, the kinesins and dyneins, are known to function in positioning both chromosomes and spindle poles during mitosis in a variety of systems (Goldstein 1993; Vaisberg et al. 1993; Holzbaur and Vallee 1994; Vernos and Karsenti 1995; Echeverri et al. 1996; Karsenti et al. 1996; Starr et al. 1998; Sharp et al. 2000). In sea urchin embryos, experiments including microinjection of pan-kinesin antibodies revealed critical mitotic functions for kinesin-related motors in general (Wright et al. 1993). Furthermore, the microinjection of class-specific antibodies revealed that two kinesin-related protein (KRP) motors, KRP110 and KRP170, are required for spindle assembly and function, presumably by acting to position spindle poles (Wright et al. 1993; Chui, K.K., G.C. Rogers, A.M. Kashina, K.P. Wedaman, D.J. Sharp, D.T. Nguyen, and J.M. Scholey, manuscript submitted for publication). Finally, a calmodulin-binding COOH-terminal kinesin, Kinesin-C, is present in the early embryo where it is also suspected to participate in spindle pole positioning, but, so far, its function has not been experimentally tested (Rogers et al. 1999).
The functional inhibition of a number of different mitotic motors that are believed to position spindle poles in several systems results in a strikingly similar terminal phenotype: monoastral spindle formation, characterized by duplicated, but closely spaced, spindle poles surrounded by a MT aster attached distally to chromosomes (Vernos et al. 1995; Kashina et al. 1997; Molina et al. 1997; Sharp et al. 1999). Monoastral spindles can arise by one of two pathways: (a) a failure of duplicated spindle poles to separate during spindle assembly, or (b) a plateward collapse of separated poles due to a failure in maintaining spindle shape. Such a phenotype is observed by perturbing the function of the Xenopus laevis kinesin Xklp2 in vitro (Boleti et al. 1996). Xklp2 is a plus end–directed kinesin that localizes to centrosomes throughout the cell cycle, being localized to MT minus ends through its interaction with an accessory protein, TPX2 (targeting protein for Xklp2; Wittmann et al. 1998). Functional inhibition of Xklp2 before or after spindle assembly in Xenopus egg extracts results in the formation of monoastral spindles (Boleti et al. 1996), leading to the proposal that Xklp2, tethered near the centrosome, moves toward the plus ends of MTs attached to the opposite centrosome, thereby driving the separation of spindle poles during spindle assembly and maintenance.
In this report, we have identified and characterized a homologue of Xklp2, called KRP180, from sea urchin early embryos. Using quantitative, real-time imaging of embryos microinjected with inhibitors of KRP180, we show that this motor plays a critical role in positioning spindle poles during mitosis, specifically at prometaphase. Immunolocalization of KRP180 and sea urchin γ-tubulin reveals that this motor is not a component of the centrosome but instead concentrates in the region of the central spindle where it may position spindle poles by generating forces on microtubules that are attached to the poles. This study is the first functional analysis of the mitotic function of a member of the Xklp2 subfamily in a living system.
Materials and Methods
Materials
Strongylocentrotus purpuratus (collected from tidepools on the northern California coast) and Lytechinus pictus (Marinus, Inc.) sea urchins were used in this study and maintained as outlined by Morris et al. (1997). General reagents were from Sigma-Aldrich unless otherwise stated.
Cloning Sea Urchin KRP180 and γ-Tubulin Isoforms
A PCR screen of a S. purpuratus unfertilized egg λ ZAP library (Stratagene) with hyperconserved kinesin motor domain sequences yielded a partial cDNA clone encoding the KRP180 motor domain. This clone was used for additional library and PCR screens (Rogers et al. 1999) that identified a total of five different KRP180 cDNAs that collectively span the entire 4,392-bp ORF.
A partial cDNA encoding sea urchin γ-tubulin was identified in a nested PCR screen of the egg cDNA library using four degenerate primers to hyperconserved sequences found in higher eukaryotic γ-tubulins (Rogers 2000). This original clone (c9), encoding SpGamma1, was then used in a library screen and two additional γ-tubulin cDNAs were identified [c1 (1.38 kb) and c3 (0.94 kb)] whose nucleotide sequence were identical to each other but different from c9. These two new clones encode a different γ-tubulin isoform (SpGamma2); c1 encodes the entire SpGamma2 ORF. Sequence analysis and structural predictions were performed using the GCG sequence analysis software (Devereux et al. 1984) unless otherwise stated.
Recombinant Proteins and Antibody Production
Two expression plasmids for a GST fusion with the KRP180 NH2-terminal stalk region (named GST-Coil 1) and COOH terminus (named GST-Coil 2) were made by directionally subcloning KRP180 nucleotides 1,191–1,965 and 3,495–4,389 into pGEX-GST (Amersham Pharmacia Biotech). E.coli–expressed GST-Coil 2 was purified under nondenaturing conditions on glutathione-agarose (Amersham Pharmacia Biotech) and then injected into four female BALB/c mice. After an adequate immune response was reached, polyclonal ascites formation was induced by injection of T-180 sarcoma cells (ATCC) intraperitoneally. Four different batches of ascites fluid from the four mice are identified as polyclonal antibodies 180.1, 180.2, 180.3, and 180.4.
A full-length SpGamma2 cDNA was PCR amplified, subcloned directionally into a pGEX-GST expression plasmid, and expressed/purified under nondenaturing conditions as described for GST-Coil 2. Anti-human γ-tubulin peptide antibody (catalog no. T3559; Sigma-Aldrich) was affinity-purified against recombinant GST-SpGamma2 protein.
Polyclonal antibodies from the anti-KRP180 (180.1-180.4) and T3559 sera/crude IgG were affinity-purified against Affigel (Bio-Rad Laboratories) columns precoupled to their respective antigens (GST-Coil 2 and GST-SpGamma2 protein). All affinity-purified antibodies were acid eluted, neutralized in Tris buffer, dialyzed into either microinjection buffer (MIB: 150 mM K-aspartate and 10 mM K-phosphate, pH7.2) or TBS, and concentrated.
Hydrodynamic Studies of KRP180
Native KRP180 was partially purified from S. purpuratus egg cytosolic extract by a cycle of binding to, and subsequent low salt/ATP elution from, endogenous taxol-stabilized MTs (see Chui, K.K., G.C. Rogers, A.M. Kashina, K.P. Wedaman, D.J. Sharp, D.T. Nguyen, and J.M. Scholey, manuscript submitted for publication). To determine the Stokes radius and sedimentation coefficient of KRP180, MT elution fractions were evenly divided and placed over a gel filtration column or on linear 5–20% sucrose density gradients and all subsequent fractions screened for KRP180 by Western blot. The native molecular mass of KRP180 was calculated by the method of Siegel and Monty 1966 with the partial specific volume adjusted to 0.716 (Zamyatnin 1972).
Immunofluorescence Microscopy
Fixation and pre-extraction of sea urchin embryos were performed as previously described (Henson et al. 1995). In brief, dejellied eggs were washed in filtered natural sea water (FNSW) containing 10 mM PABA, fertilized, and monitored with a dissection microscope until first cleavage. Fertilization envelope stripped embryos were fixed with 100% methanol (−20°C), rehydrated stepwise into methanol solutions of TBS + 0.05% Tween (TBST), and blocked with 5% normal goat serum (NGS).
Pre-extraction of mitotic embryos was performed using either 1% NP-40 or 1% Triton X-100 detergent for either 10 or 15 min. Affinity-purified anti-KRP180 antibody was used at 1 μg/ml in TBST + 5% NGS. Two different anti–α/β-tubulin antibodies were used: an anti-bovine brain tubulin (Gelfand lab) and an FITC-conjugated anti–α-tubulin monoclonal DM1a (Sigma-Aldrich). Double-labeling with the DM1a and anti-KRP180 antibodies was performed as described in Rogers 2000. Secondary antibodies were purchased from Jackson ImmunoResearch Lab. Inc. DAPI (1 μg/ml) was added to the embryos and washed extensively before mounting on poly-l-lysine–coated glass coverslips in a 90% glycerol solution containing 20 mg/ml phenylenediamine in PBS. Indirect immunofluorescent micrographs were obtained as either single optical sections or projections using a Leica TCS SP confocal with each image resulting from 16–32 averaged scans and analyzed using Adobe Photoshop v5.0.
Antibody and Recombinant Protein Microinjection
Control mouse IgG or the four pooled anti-KRP180 polyclonal antibodies (180.1–180.4), were dialyzed into MIB and concentrated to 12.4 mg/ml (final intracellular [Ab] at 0.62 mg/ml). Glutathione–Sepharose-purified recombinant GST, GST-Coil 1, and GST-Coil 2 protein was purified in MIB buffer, concentrated, and microinjected at a range of concentrations (final intracellular [protein] from 0.96–9.1 μM). We observed an identical consistent phenotype after introducing GST-Coil 2 protein into early embryos regardless of the protein concentration used, suggesting that the lowest concentration of 0.96 μM was sufficient to inhibit KRP180 function. Embryo microinjections were performed by a procedure modified from Kiehart 1982 and Wright et al. 1993. Lateral injection chambers (Kiehart 1982) allowed high-resolution observation of injections and development by restraining the embryos during cleavage until the ciliated blastula stage. Fertilized eggs were injected with 5% cell volume between 20 min after fertilization until cytokinesis of first cleavage. Embryos were observed by differential interference contrast microscopy on an inverted microscope (model IM-35; Carl Zeiss, Inc.) using a Plan 40× objective and imaged as described (Wright et al. 1993; Morris and Scholey 1997). An Argus 10 Image Processor (Hamamatsu Photonics) was used for contrast enhancement.
Quantitation of spindle pole positioning and morphology was performed by capturing video printouts of injected embryos, using a Sony UP-880 video graphic printer, every 0.5–5 min at the time of nuclear envelope breakdown (NEBD) or after injection if a spindle was already present. Video prints were digitized using a flatbed scanner and processed using Adobe Photoshop v5.0 and NIH Image v1.62. Spindle pole-to-pole distances were measured as the distance between the center-points of each spindle pole. Changes in these movements over time were plotted using Cricket Graph v1.3.2.
Results
Molecular Analysis of KRP180, a Member of the Xklp2 Kinesin Subfamily
We initiated a PCR screen of an unfertilized sea urchin egg cDNA library that was designed to amplify motor domain-specific cDNAs encoding KRPs potentially involved in positioning centrosomes during sea urchin cleavage (Rogers et al. 1999). One of the KRPs obtained in this screen was cloned and sequenced using the strategy outlined in the Materials and Methods. We have named this motor KRP180 based on its apparent molecular mass (180 kD) as measured by Western blot of the native protein from sea urchin eggs.
The deduced primary sequence of KRP180 predicts a polypeptide 1,463 amino acids in length and a molecular mass of 166,590 Daltons (Fig. 1 A), with an NH2-terminal motor domain (amino acid residues 1–347) linked to an extensive segment of predicted α-helical coiled-coil, 106 nm long (residues 362-1463; Fig. 1 C). The presence of two predicted PEST sequences (at residues 393–426 and 709–742 with PEST scores +13.22 and 7.56, respectively) suggests that this protein is a target for proteolysis and rapid turnover (Rogers et al. 1986). In addition, two p34cdc2 kinase phosphorylation sites [(T/S)PX(K/R); Nigg 1993; Fig. 1 A], and a predicted tyrosine kinase phosphorylation site (residues 1,135–1141) were found in the stalk region.
Figure 1.
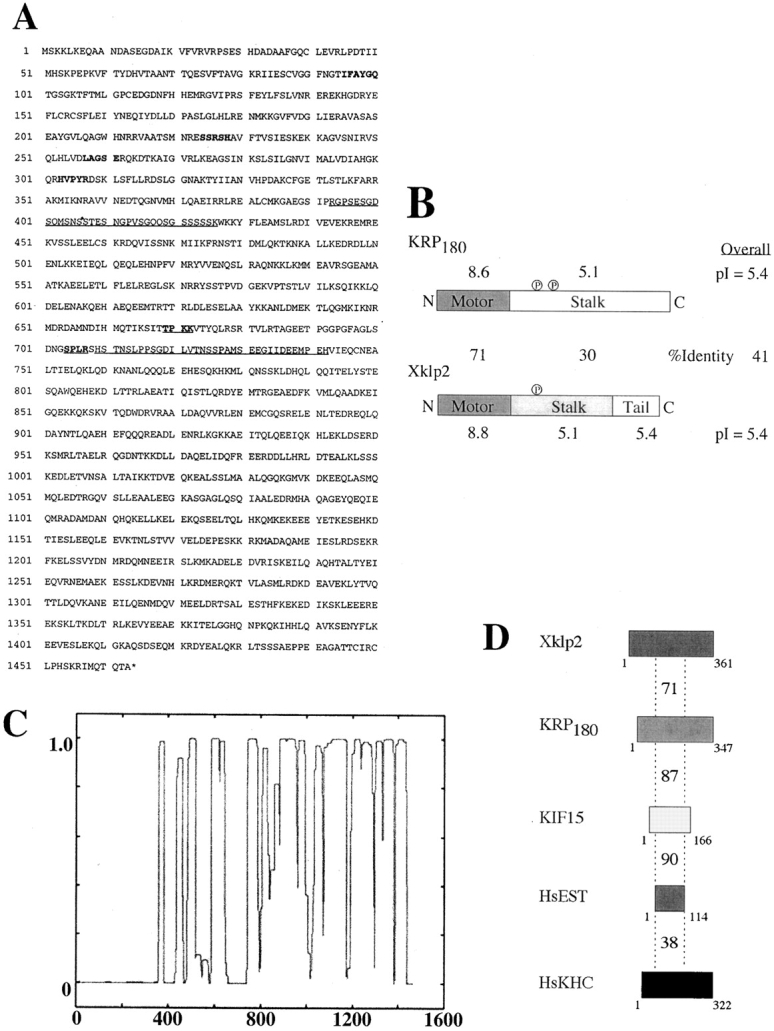
Molecular analysis of sea urchin KRP180. (A) The deduced amino acid sequence of KRP180 encodes an NH2-terminal motor polypeptide 1,463 amino acid residues long. Amino acids in bold correspond to the hyperconserved motor sequences found in kinesin motor domains. PEST sequences are shown as underscored amino acids. Underscored and bold amino acids correspond to consensus p34cdc2 kinase phosphorylation sites. The nucleotide sequence of KRP180 is available from GenBank under accession number AF28433. (B) Aligned linear maps of the motor polypeptides KRP180 and Xklp2 demonstrate the conservation of sequence identity and isoelectric point values in their domain organization. Consensus p34cdc2 kinase phosphorylation sites are shown as a circled letter P. Sequence identity scores are shown as percentages between the linear maps, isoelectric point values are shown above and below the linear maps, and overall scores are shown on the right. (C) KRP180 forms an extensive region of α-helical coiled-coil as determined in a Lupus plot using the Coils program (Lupus et al. 1991). (D) Linear maps of the full-length and partial motor domains of the Xklp2 members found in Xenopus (Xklp2), sea urchin (KRP180), mouse (KIF15), and human (HsEST), as well as the motor domain of human conventional kinesin heavy chain (HsKHC) demonstrate a region in the catalytic core of the motors, equal to the size of the smallest of the motors shown (HsEST), share identity to the Xklp2 motor domain. Identity scores compared with Xklp2 are shown above each motor domain.
KRP180 displays a strikingly high degree of homology (71% identity) with the motor of the NH2-terminal plus end–directed Xenopus laevis kinesin, Xklp2, which decreases to 30% outside their motor domains. KRP180 and Xklp2 are predicted to form coiled-coil rods that extend throughout the length of their non-motor domains, and they both lack a predicted tail domain, much like class II myosins (Warrick and Spudich 1987). Both motors share similar p34cdc2 kinase consensus phosphorylation sites within their stalk domains that exactly align (Fig. 1 B), suggesting a conserved method of regulation. Finally, we note that Xklp2 homologues have been found in mouse and human (Nakagawa et al. 1997) suggesting that KRP180 is a member of a phylogenetically diverse subfamily of kinesins, the Xklp2 subfamily (Fig. 1 D).
Native KRP180 from Sea Urchin Eggs Is a Homodimer
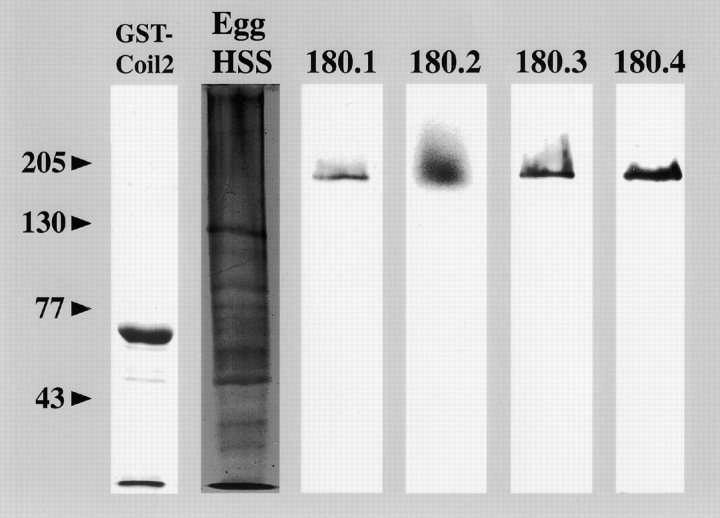
To determine the hydrodynamic properties and subunit composition of native KRP180, we raised four mouse polyclonal antibodies against a region of the motor's COOH terminus fused to GST (Fig. 2, lane GST-Coil 2). All four affinity-purified antibodies (180.1–180.4) specifically recognized a 180-kD polypeptide in unfertilized sea urchin egg extract (Fig. 2). Native KRP180 was partially purified from egg extract by a single cycle of AMP-PNP–induced binding to endogenous MTs and subsequent release with ATP in either a high or low salt buffer. This eluate was further fractionated by gel filtration and sucrose density gradient centrifugation to measure the hydrodynamic properties of KRP180 (Table ). KRP180 behaves as a homogeneous protein throughout the biochemical fractionation steps and has a Stokes radius (RS) of 10.07 nm and a sedimentation coefficient of 8.33 S. (Although, in high salt KRP180 displays a sedimentation coefficient of 5.8 S, suggesting that it exists in a folded conformation in low salt that then assumes an elongated conformation in high salt.) Based on these hydrodynamic properties, native KRP180 is calculated to have a molecular mass of 334 kD and an axial ratio of 20–40, indicative of a long rod-shaped molecule. Since the deduced mol wt of the cDNA-encoded polypeptide is 167 kD, we predict the native KRP180 quaternary structure to be a homodimer lacking any accessory proteins.
Figure 2.
Anti-KRP180 antibodies specifically recognize this motor in sea urchin egg extracts. A recombinant GST-KRP180 COOH-terminal coiled-coil fusion protein was purified on a glutathione–Sepharose column (lane GST-Coil 2) and used to raise mouse polyclonal antisera. The fusion protein runs at 60 kD. Affinity-purified anti-KRP180 antibodies from four different mice all specifically recognize a polypeptide ∼180 kD in mass (lanes 180.1, 180.2, 180.3, and 180.4) found in sea urchin high speed supernatant (HSS) unfertilized egg extract (lane Egg HSS; Coomassie-stained SDS-7.5% PAGE).
Table 1.
KRP180 Hydrodynamic Properties
| Property | Low salt | High salt |
|---|---|---|
| Stokes radius (Rs) | 10.07 nm | – |
| S value | 8.33 S | 5.8 S |
| Native mol wt | 334 kD | |
| Axial ratio | 20–40 | |
| Rod length | 106 nm | |
| Estimated mol wt from DNA | 167 kD | |
| Predicted structure | 2 × 167 kD = 334 kD homodimer |
KRP180 Is Concentrated on Microtubules in the Central Region of the Spindle
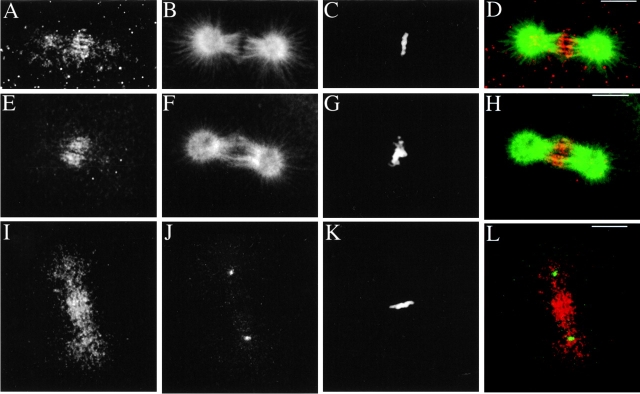
To determine the intracellular localization of KRP180, we immunolocalized this motor in first cleavage stage S. purpuratus whole embryos. During interphase, KRP180 localized along astral MT arrays (data not shown) and also displayed a random punctate staining in the cytoplasm during all stages of the cell cycle (Fig. 3A, Fig. E, Fig. I, Fig. M, and Fig. Q). During prophase, KRP180 decorated spindle poles in a punctate staining pattern that persisted throughout the completion of mitosis, as well as along MTs that extended around the nuclear envelope (Fig. 3, A–D). In prometaphase, KRP180 was concentrated in a filamentous pattern along spindle MTs that invaded the nuclear region (Fig. 3, E–H). KRP180 also localized to spindle MTs in metaphase embryos where it appeared asymmetrically distributed, concentrating on the distal ends of half spindle MTs, often as a filamentous pattern crossing the metaphase plate between the chromosomes (Fig. 3, I–L). During anaphase/telophase, KRP180 was enriched in the spindle midzone/midbody along MTs bridging the half spindles in this region (Fig. 3, M–T). All anti-KRP180 antibodies consistently gave identical staining patterns that were abolished by co-incubation with the antigenic GST-Coil 2 protein (data not shown).
Figure 3.
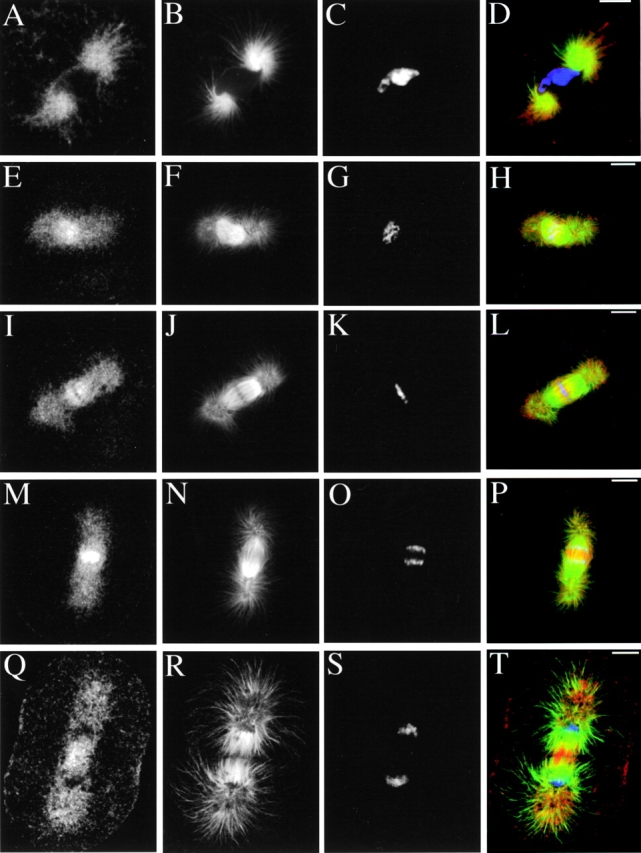
Immunolocalization of KRP180 in one-cell cleavage stage sea urchin (S. purpuratus) embryos. Whole embryos were methanol-fixed and stained with anti-KRP180 (A, E, I, M, and Q) and an FITC-conjugated anti–α-tubulin antibody (B, F, J, N, and R). Anti-KRP180 was recognized with Cy5-conjugated secondary antibody and DNA was visualized with (1 μg/ml) DAPI (C, G, K, O, and S). Merged images are shown with KRP180 in red, tubulin in green, and DNA in blue (D, H, L, P, and T). Prophase (A–D), prometaphase (E–H), metaphase (I–L), anaphase (M–P), and telophase/cytokinesis (Q–T). Bar, 10 μm.
The Xenopus Xklp2 motor is unique in its localization in that it concentrates on centrosomes throughout the cell cycle (Boleti et al. 1996); more specifically to MT minus ends in a variety of in vitro structures with or without centrosomes (Wittmann et al. 1998). Although KRP180 staining was enriched in the central spindle, a weaker punctate staining pattern on spindle poles was observed. To more clearly determine whether KRP180 localized to centrosomes, we obtained an antibody against γ-tubulin, a well characterized component of the centrosome.
A PCR screen identified two different isoforms of sea urchin γ-tubulin: a partial cDNA encoding SpGamma1 and a full-length cDNA encoding SpGamma2 (Fig. 4 A). Both γ-tubulin isoforms share significant overall identity (72–89%) with Drosophila melanogaster, Xenopus, and human γ-tubulins (Fig. 4A and Fig. B). We obtained a commercial anti–γ-tubulin antibody, T3559, because its peptide antigen (residues 38–53 of human γ-tubulin) differed from a sequence found in SpGamma2 by only two conservative substitutions (both Glu to Asp; Fig. 4 A). After affinity-purification against recombinant GST-SpGamma2 (Fig. 4 C, lane 1), the T3559 antibody specifically recognized a single polypeptide in egg high speed supernatant (HSS) that we assumed to be either one or more maternally loaded γ-tubulins (Fig. 4 C, lane 2), as well as recombinant GST-SpGamma2 (Fig. 4 C, lane 3). In addition, T3559 antibody stained centrosomes in pre-extracted mitotic embryos (Fig. 4 D). Pre-extraction of one-cell stage metaphase embryos did not affect the bright filamentous staining of KRP180 that colocalized with MTs in the central region of the spindle between the aligned chromosomes (Fig. 5, A–D). However, pre-extraction significantly diminished the punctate staining of KRP180 on the spindle poles; longer periods of pre-extraction eliminated nearly all pole staining (Fig. 5, E–H). In embryos that were pre-extracted for an amount of time where the motor was still associated with the poles, KRP180 did not colocalize with γ-tubulin at the centrosome (Fig. 5, I–L). The localization of KRP180 to the central region of the spindle where MTs are predicted to overlap into antiparallel arrangements suggests this motor could crossbridge antiparallel MTs to promote MT-sliding.
Figure 4.
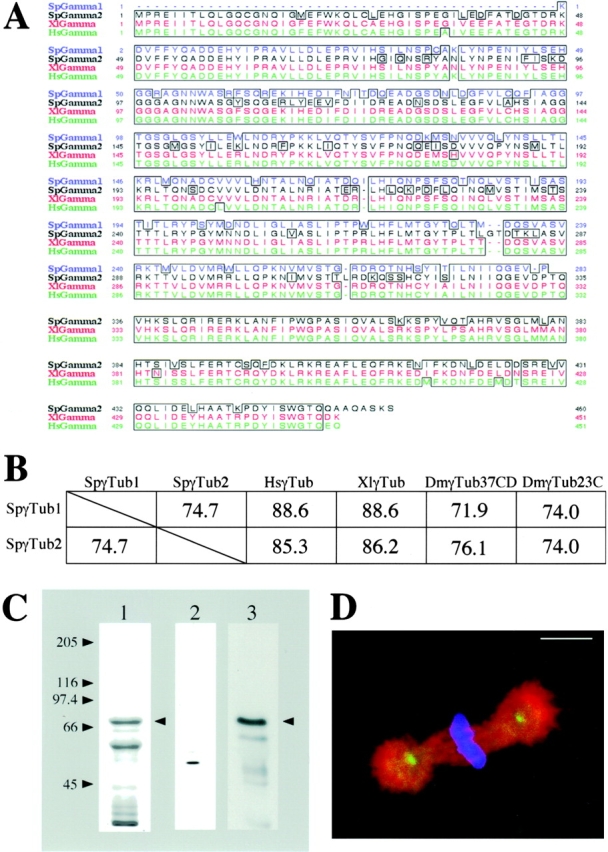
Two different γ-tubulin isoforms (SpGamma1 and SpGamma2) are found in sea urchin (S. purpuratus) eggs and early embryos. (A) A lineup of the deduced amino acid sequences of four different γ-tubulins found in sea urchin (SpGamma1 and SpGamma2), Xenopus (XlGamma), and human (HsGamma). Identical amino acids between the four γ-tubulins are surrounded with black boxes. The nucleotide sequences of SpGamma1 and 2 are available from GenBank under accession numbers AF284334 and AF284335. (B) Both sea urchin γ-tubulin isoforms (SpγTub1 and SpγTub2) are related to the γ-tubulins found in human (HsγTub), Xenopus (XlγTub), and the two isoforms in Drosophila (DmγTub37CD and DmγTub23C). The overall percentage of identity that these tubulins share are compared. (C) Purified, recombinant full-length GST-SpGamma2 fusion protein runs at 70 kD upon Coomassie-stained SDS-7.5% PAGE (lane 1). Affinity-purified anti–γ-tubulin antibody specifically recognized γ-tubulin in sea urchin (HSS) egg extract (lane 2) and the SpGamma2 fusion protein (lane 3) by Western blot. The GST-SpGamma2 fusion protein is indicated by the arrowheads and the molecular mass markers correspond to all three lanes. (D) A triple-labeled merged confocal micrograph shows a pre-extracted metaphase sea urchin embryo fixed and stained with anti–γ-tubulin (green), anti–α-tubulin (red) antibodies, and DAPI (blue). Bar, 10 mm.
Figure 5.
KRP180 does not coimmunolocalize to the centrosome with γ-tubulin in one-cell cleavage stage sea urchin (S. purpuratus) embryos. Pre-extracted metaphase embryos were fixed and stained with anti-KRP180 (A, E, and I) and either an FITC-conjugated anti–α-tubulin antibody (B and F) or an anti–γ-tubulin peptide antibody (J). DNA was visualized with (1 μg/ml) DAPI (C, G, and K). Merged images are shown with KRP180 in red and tubulin in green. Bar, 10 μm.
KRP180 Is Required To Maintain Spindle Shape At Prometaphase, but Not for the Initial Separation of Spindle Poles
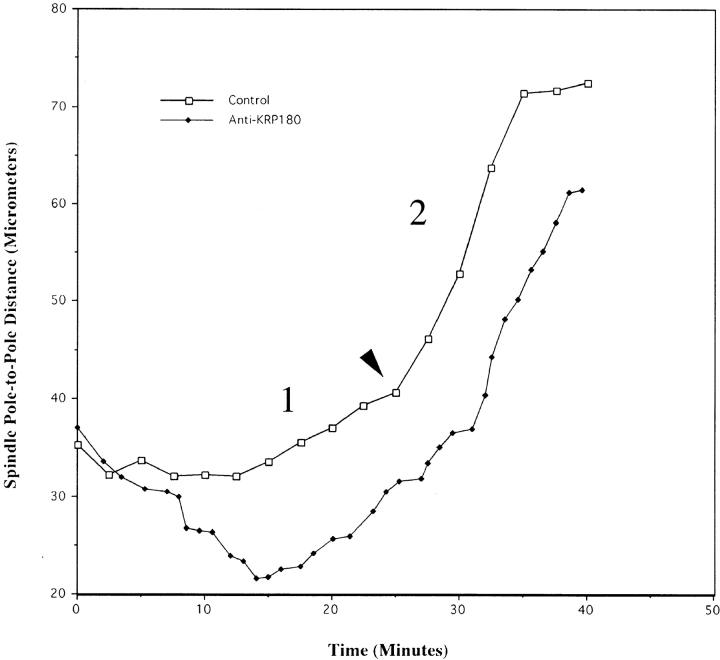
To probe KRP180's function during embryonic cell division, we microinjected affinity-purified anti-KRP180 antibodies or control mouse IgG into fertilized sea urchin eggs and observed their effects on spindle pole positioning using quantitative, real-time analysis. The relative movement of spindle poles in control embryos was analyzed by determining the distance between the center points of opposite spindle poles as a function of time (see Fig. 7). One-cell stage embryos microinjected with control nonspecific mouse IgG progressed normally through mitosis and cytokinesis (Fig. 6 A), as well as through subsequent cleavages until ciliogenesis at the blastula stage of development, when observation was terminated (n = 19). Control injected embryos completed mitosis in 40 min on average, from NEBD (time 0) to the completion of cytokinesis (time, 40 min). As shown in Fig. 7 (open squares), pole separation does not occur at a linear rate, but rather proceeds in a complex fashion. After NEBD, spindle poles maintained a metaphase spacing of ∼33 μm for an average of 14.6 ± 4.3 min (n = 6) before initiating anaphase. During anaphase B, these embryos displayed two distinct phases of spindle pole separation: an initial slow phase lasting an average of 12.2 ± 1.9 min (n = 6) and moving at an average rate of 1.08 ± 0.45 μm/min (n = 5) and a fast second phase (8.8 ± 1.7 min [n = 6], 3.55 ± 0.38 μm/min [n = 5]) (Fig. 7; Table ).
Figure 7.
KRP180 is required to maintain spindle shape in prometaphase sea urchin embryos. Spindle pole-to-pole distances were measured over time in control and anti-KRP180 antibody injected embryos. Time 0 in each graph represents NEBD and the final time point in each graph indicates the completion of cytokinesis. Bold type numbers 1 and 2 indicate two distinct phases of anaphase B spindle elongation. The arrowhead indicates the transition from the first slow phase of spindle pole elongation to the second fast phase.
Figure 6.
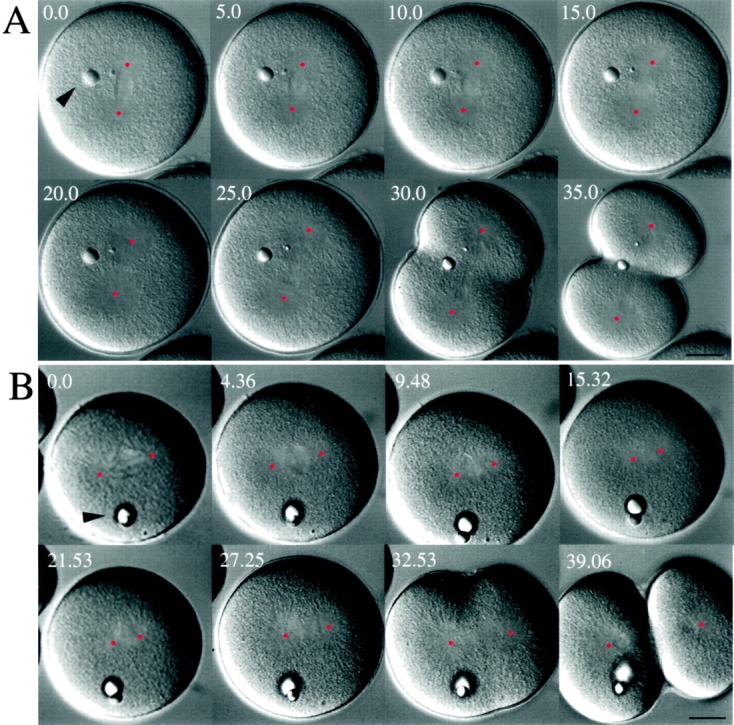
Effects of anti-KRP180 and control antibody microinjected into mitotic sea urchin (L. pictus) embryos. Video frames of representative control IgG (A) and anti-KRP180 (B) injected embryos are shown. The video frames (with time shown as minutes and seconds) show the effects of microinjection on first cleavage in both embryos during NEBD (time 0.0 min) until the completion of cytokinesis (time 35.0 min for control and 39.06 for experimental embryo). Both embryos were microinjected in interphase, shortly after fertilization. The center of each pole is marked with a red dot to assist in measuring spindle pole-to-pole distances. The intracellular oil droplets (arrowheads) confirm that both embryos were microinjected successfully. Bar, 20 μm.
Table 2.
Spindle Dynamics in Antibody and Recombinant Protein-injected Embryos
| Reagent | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse IgG | n | Anti-KRP180 | n | GST | n | GST-Coil 1 | n | GST-Coil 2 | n | |
| Average velocity | μm/min | |||||||||
| Rate of slow phase | 1.08 ± 0.45 | 5 | 1.06 ± 0.06 | 3 | 1.41 ± 0.32 | 4 | 1.30 ± 0.1 | 5 | 1.04 ± 0.38 | 9 |
| Rate of fast phase | 3.55 ± 0.38 | 5 | 3.22 ± 0.60 | 7 | 3.80 ± 0.75 | 4 | 3.90 ± 0.97 | 5 | 3.37 ± 0.53 | 10 |
| Rate of spindle collapse | NA | 2.00 ± 0.41 | 4 | NA | NA | 0.87 ± 0.2 | 7 | |||
| Average time | min | |||||||||
| Initiation of anaphase | 14.6 ± 4.3 | 6 | 16.0 ± 3.2 | 6 | 12.8 ± 4.3 | 4 | 13.0 ± 3.5 | 5 | 15.2 ± 3.5 | 8 |
| Duration of slow phase | 12.2 ± 1.9 | 6 | 10.5 ± 1.3 | 3 | 11.4 ± 2.9 | 4 | 13.3 ± 3.3 | 5 | 12.5 ± 1.9 | 9 |
| Duration of fast phase | 8.8 ± 1.7 | 6 | 11.3 ± 4.8 | 7 | 7.3 ± 2.5 | 3 | 8.3 ± 2.5 | 4 | 7.8 ± 1.8 | 9 |
NA, Not applicable because this event did not occur.
We observed an appearance of mitotic defects in anti-KRP180 injected embryos that was dependent upon the timing of the injection. Antibody introduced into the embryo during mitosis (i.e., immediately after NEBD) had no effect on the first mitotic division; quantitation of spindle pole separation appeared identical to control embryos (n = 6; data not shown). However, embryos that were injected during interphase displayed one of three distinct phenotypes. (a) Embryos that were delayed in initiating NEBD (50 min on average) and displayed a prophase morphology with separated spindle poles on opposite sides of the nucleus (8 of 18 embryos). (b) Embryos that arrested in the cell cycle in a prophase morphology (6 of 18 embryos). These embryos had arrested for 37 min on average with an intact nucleus and then died. Embryonic cell death is hallmarked by a dynamic ruffling of the cell cortex as the cytoplasm turns granular followed by cell lysis; such behavior is characteristic of apoptosis as proposed by Hinchcliffe et al. 1998. (c) Embryos that displayed no defects or delays before NEBD (4 of 18 embryos).
Of the anti-KRP180 antibody containing embryos that were injected in interphase and entered mitosis, all clearly displayed two separated spindle poles positioned on opposite sides of the nucleus before NEBD, suggesting that KRP180 is not required for the initial separation of spindle poles at prophase (12 of 12 embryos; categories a and c above). However, all of these embryos underwent a dramatic spindle collapse immediately upon NEBD where separated poles moved plateward at an average rate of 2.0 ± 0.41 μm/min (n = 4; Fig. 6 B; Table ). Measurements of spindle pole separation as a function of time of a representative embryo is shown in Fig. 7. Although the spindle had collapsed, poles were clearly visible in a side-by-side position (Fig. 6 B; 15.32 min) with an average pole-to-pole distance of 20.4 μm ± 0.25 (n = 6). Most collapsed spindles continued into anaphase displaying both slow and fast phases of anaphase B at rates and durations observed in controls (8 of 12 embryos; Fig. 7; Table ). However, 4 of 7 embryos did display subtle defects in the rates and length of time spent in the first slow phase of anaphase B, and slight defects in cytokinesis were observed as well (3 of 8).
In addition to antibody microinjection, we studied this motor's mitotic function by introducing recombinant truncated KRP180 proteins into one-cell stage embryos. Such constructs might act as dominant negative proteins by competing with the endogenous motor complex for binding to cargo or transiently-associated accessory polypeptides. To this end, we designed NH2- and COOH-terminal GST-tagged coiled-coil encoding constructs designated GST-Coil 1 and GST–Coil 2 (Fig. 8 A and 2). The sequence for the GST-Coil 1 and Coil 2 domains was based on the work of Boleti et al. 1996 who found that GST constructs of the exactly homologous Xklp2 domains functioned as a negative control and as a dominant negative, respectively, in in vitro spindle assembly assays.
Figure 8.
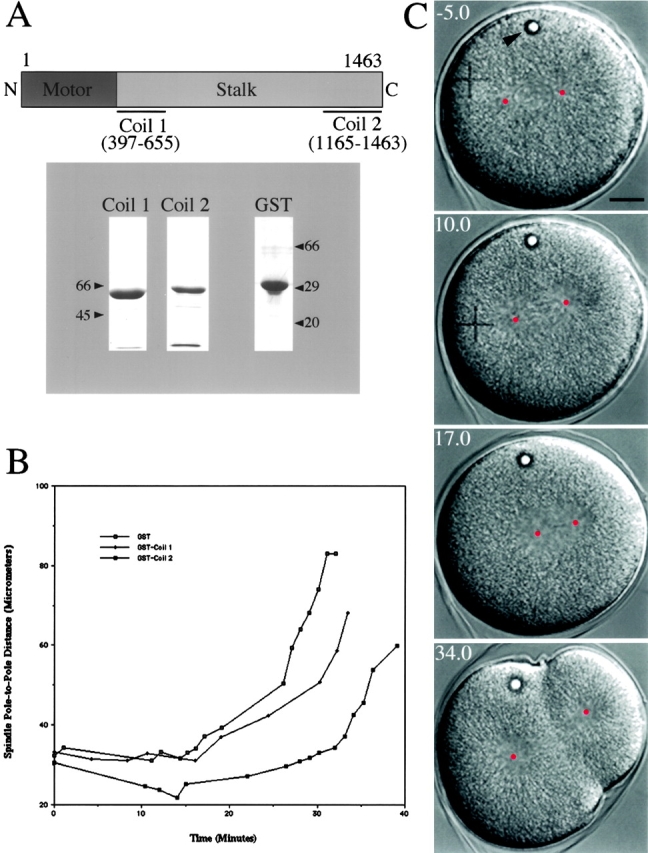
Effects of GST-KRP180 dominant/negative protein microinjected into mitotic sea urchin (L. pictus) embryos. (A) Purified GST-KRP180 stalk fusion proteins (Coil 1 and Coil 2) were used as dominant/negative proteins. The numbers below the linear map of the KRP180 polypeptide correspond to the amino acids that comprise the fusion proteins. Molecular mass markers indicate the size of the glutathione–Sepharose-purified fusion proteins (lane Coil 1 and 2; Coomassie-stained 7.5% and lane GST; 12% SDS-PAGE). (B) Microinjection of GST-Coil 2 into mitotic embryos perturbs the maintenance of prometaphase spindle shape, whereas microinjection of GST-Coil 1 and control GST does not. Spindle pole-to-pole distances were measured over time for individual spindles in GST-, Coil 1–, and Coil 2–injected embryos. Time 0 represents NEBD and the final time point indicates the completion of cytokinesis. Anaphase B initiates ∼15 min post-NEBD in the three spindles shown. (C) Video frames of a representative GST-Coil 2 injected embryo are shown. The video frames (with time in minutes) show the effects of microinjection on first cleavage five minutes before NEBD (time −5.0) until near the completion of cytokinesis (time 34.0). This embryo was microinjected in interphase, shortly after fertilization. The center of each spindle pole is marked with a red dot to assist in spindle length measurements while the intracellular oil droplet (arrowhead) indicates a successful microinjection. This embryo divides asymmetrically due to mispositioning of the spindle after displaying a prometaphase spindle collapse.
We have found that microinjection of GST-Coil 2 into one-cell stage embryos completely phenocopied the results obtained with the antibody injection experiments. As in anti-KRP180–injected embryos, GST-Coil 2 injected embryos displayed mitotic defects dependent upon the timing of the injection. Recombinant protein injected during mitosis (i.e., after NEBD) had no effect on the first embryonic cleavage as determined by quantitation of spindle pole separation over time compared with controls (n = 13; data not shown). However, every GST-Coil 2 containing embryo that was injected either during interphase (n = 9) or immediately at NEBD (n = 2), displayed spindle poles on opposite sides of the nucleus that moved plateward after completion of NEBD: a phenotype strikingly similar to anti-KRP180 injected embryos. We observed that GST-Coil 2 was a less potent inhibitor of KRP180 function as compared with the anti-KRP180 antibody because the GST-Coil 2–induced spindle collapse occurred at a rate less than half that observed in anti-KRP180 injected embryos (Table ). In addition, the extent of spindle collapse (depicted by the length of the collapsed metaphase spindles) was less in embryos injected with GST-Coil 2 than anti-KRP180 (25.1 ± 3.0 μm [n = 10] as opposed to 20.4 μm). Measurements of spindle pole separation as a function of time and video frames of a representative GST-Coil 2 injected embryo are shown in Fig. 8B and Fig. C. Mitotic arrest was not observed in embryos whose spindles had collapsed (n = 11), and anaphase B was initiated at rates and durations similar to those observed in control injected embryos (Table ). As was observed with the anti-KRP180 injections, a small percentage of GST-Coil 2 injected embryos displayed defects in the duration and rate of the initial slow phase of anaphase B (18%) or divided asymmetrically (9%; Fig. 8 C). Finally, as observed in antibody injected embryos, injection of GST-Coil 2 before NEBD resulted in one of three phenotypes: a prophase delay for an average of 40.5 min (2 of 11 embryos), a prophase arrest leading to cell death (2 of 11 embryos), or no defects or delays before NEBD (7 of 11 embryos).
As was found for control IgG injected embryos, microinjection of either GST-Coil 1 (n = 9) or control GST protein (n = 11) during interphase or mitosis had no deleterious effect on first cleavage embryos. Kinetic analysis of pole-to-pole positioning revealed that GST-Coil 1– and GST-injected embryos maintained average metaphase spindle lengths of 31.8 μm and 30.9 μm, respectively, before initiating anaphase (Fig. 8 B; Table ). Likewise, during anaphase B, these embryos displayed a characteristic biphasic separation of spindle poles: an initial slow phase followed by a fast second phase (Fig. 8 B; Table ).
Thus, the injection of KRP180 inhibitors, either antibody or a dominant negative construct, results in the collapse of bipolar spindles. Taken together, these data suggest that KRP180 functions to position spindle poles and thus to transiently maintain prometaphase spindle shape in early sea urchin embryos.
Discussion
KRP180 and the Xklp2 Subfamily of KRPs
The molecular data on KRP180 from sea urchin embryos suggests that this motor protein belongs to a phylogenetically diverse subfamily of KRPs, whose founding member is the centrosome-associated motor, Xklp2 (Boleti et al. 1996), which also has homologues in humans, mice, Arabidopsis thailiana and Oryza sativa (Nakagawa et al. 1997; Lee, Y.-R., and B. Liu, personal communication). Hydrodynamic data suggest that KRP180, like Xklp2, is a homodimer of two motor subunits (Wittmann et al. 1998). Functional data suggest that sea urchin KRP180 serves to position spindle poles during prometaphase in early embryos. Similarly, Xklp2 serves to separate spindle poles in spindle assembly assays using Xenopus egg extracts (Boleti et al. 1996). Thus, members of the Xklp2 subfamily of KRPs may have important mitotic roles involving spindle pole positioning in a wide range of systems.
KRP180, a Mitotic Motor Required to Maintain Prometaphase Spindle Shape
Our immunolocalization data suggest that KRP180 is localized to interzonal MTs where it could serve to generate forces that position the attached spindle poles during prometaphase. In contrast, Xklp2 concentrates on centrosomes during all cell cycle stages (Boleti et al. 1996). Thus, although both KRP180 and Xklp2 are found to position spindle poles, the mechanisms by which they are proposed to do so differ.
Based on its localization to a region of the spindle where MTs overlap in an anti-parallel fashion, it is tempting to hypothesize that KRP180 functions to crossbridge anti-parallel MTs and drive MT-sliding. Assuming that KRP180 is a plus end–directed motor as was found for Xklp2 (Boleti et al. 1996), it could drive the separation of spindle poles by exerting force on antiparallel spindle MTs to which the poles are attached, as has been suggested with members of the bimC/bipolar kinesin subfamily (Sharp et al. 2000).
Microinjection of sea urchin embryos with KRP180-specific antibodies and recombinant dominant/negative constructs demonstrates that KRP180 is required to maintain the separation of spindle poles during first cleavage. Immediately after NEBD, every anti-KRP180 and GST-Coil 2 injected embryo we observed underwent a dramatic spindle collapse: poles moved plateward at a linear rate and assumed a juxtaposed position, giving rise to monoastral spindles. Such monoastral spindles are a hallmark of cells that fail to divide. For example, similar structures have been seen after the inhibition of either Xklp2 or Eg5 in Xenopus extracts suggesting that both these motors contribute to the formation of bipolar spindles (Sawin et al. 1992; Boleti et al. 1996). However, since the studies in Xenopus were performed using fixed images, it was unclear exactly when these motors exert their effects. In contrast, the live cell imaging described here strongly suggests that KRP180 serves to generate outward forces on spindle poles specifically after the initial separation of the spindle poles during prometaphase.
Quantitation of spindle pole movements in both anti-KRP180 and dominant negative protein injected embryos revealed that most collapsed spindles were able to initiate anaphase and complete mitosis. Occasionally we observed slight defects in spindle elongation and cytokinesis. We attribute these observations to structural defects in the organization of the spindle midzone/midbody on which these events depend, and that perhaps these structures had not assembled properly due to the close proximity of the poles after the spindle collapse. However, due to its midzone/midbody localization, we cannot rule out the possibility that KRP180 might function to assemble or promote MT-sliding during late mitosis in a semi-redundant fashion with other mitotic motors as well (see below).
Proposed Model of Mitotic Motor Functions
How is the mitotic function of KRP180 integrated into the mitotic function of other motors that operate in the early sea urchin embryo? Assuming that the functional inhibition of KRP180 by specific antibody and dominant/negative protein microinjection reveals the full spectrum of this motor's mitotic function, then these results suggest a specific role for KRP180 in maintaining prometaphase spindle shape. In addition to KRP180, we have also characterized new sea urchin motors that play important roles in positioning spindle poles during embryonic cleavage. These include the spindle-associated motors, KRP110 and KRP170 (members of the plus end–directed MKLP1 and bipolar kinesin subfamilies, respectively), a cortically localized cytoplasmic dynein, and a COOH-terminal kinesin, Kinesin-C, that is related to a unique class of Ca2+/calmodulin–regulated motors and whose function is unknown (Chui, K.K., G.C. Rogers, A.M. Kashina, K.P. Wedaman, D.J. Sharp, D.T. Nguyen, and J.M. Scholey, manuscript submitted for publication; Rogers et al. 1999; Rogers 2000). Functional analysis of KRP110, KRP170, and cortical dynein, by microinjection of antibodies and dominant/negative inhibitors, revealed that perturbation of any one of these three motors results in a prophase cell cycle arrest, implying a role for these motors in the initial assembly of the spindle. Similar studies also demonstrated a metaphase or anaphase cell cycle arrest when KRP110 and KRP170 function was perturbed, respectively. In addition, both KRP170 and cortical dynein are required for specific phases of spindle elongation at anaphase.
We propose that the predominant force driving spindle pole positioning is through the MT-sliding activity of these mitotic motors. Native KRP110 and KRP170 are homotetramers with a predicted bipolar ultrastructure (i.e., two motor domains on opposite ends of a coiled-coil rod), whereas native KRP180 is homodimeric with an extensive 106-nm rod. Interestingly, KRP180 has an axial ratio of 20 like KRP110, whereas KRP170 has a longer axial ratio of 80. Given the differences in axial ratios, these motors could crossbridge adjacent MTs with varying space between them. For example, adjacent MTs with a long spacing would be cross-linked by KRP170 that would then be linked into tight MT bundles by the action of KRP110 and KRP180. Such an activity of anti-parallel MT capture and bundling by MT cross-linking motors, combined with their subsequent force generating properties to promote MT sliding, would result not only in the ability to position spindle poles, but to establish the structural integrity of the spindle. Not surprisingly, functional inhibition of these motors results in cell cycle arrest phenotypes, possibly due to direct interference with spindle mechanics. We also propose that minus end–directed, cortical dynein and Kinesin-C function to position spindle poles using a MT-sliding mechanism. COOH-terminal kinesins function to counterbalance the pole separating forces of the bipolar kinesins, whereas dynein slides MTs relative to an actin-rich cortical network to which it localizes (Saunders and Hoyt 1992; Pidoux et al. 1996; O'Connell et al. 1993; Mountain et al. 1999; Sharp et al. 1999, Sharp et al. 2000; Rogers 2000).
These results suggest a plausible model for the pathway of spindle pole positioning during embryonic cell division through the coordinating functions of five different plus and minus end–directed MT-based motors (Fig. 9). Before NEBD, cortical dynein, KRP110, and KRP170 work together to coordinate the initial separation of spindle poles. Pole separation would result from the combined activities of cortical dynein pulling on astral MTs and the homotetrameric (and predicted bipolar) motors, KRP110 and KRP170, cross-linking and sliding anti-parallel MTs. KRP170, which has a greater axial ratio than KRP110, would cross-link adjacent anti-parallel spindle MTs with a long spacing that would then be linked into tighter MT bundles by the action of KRP110, serving to stabilize the connections between the poles as they are pushed/pulled around the nucleus.
Figure 9.
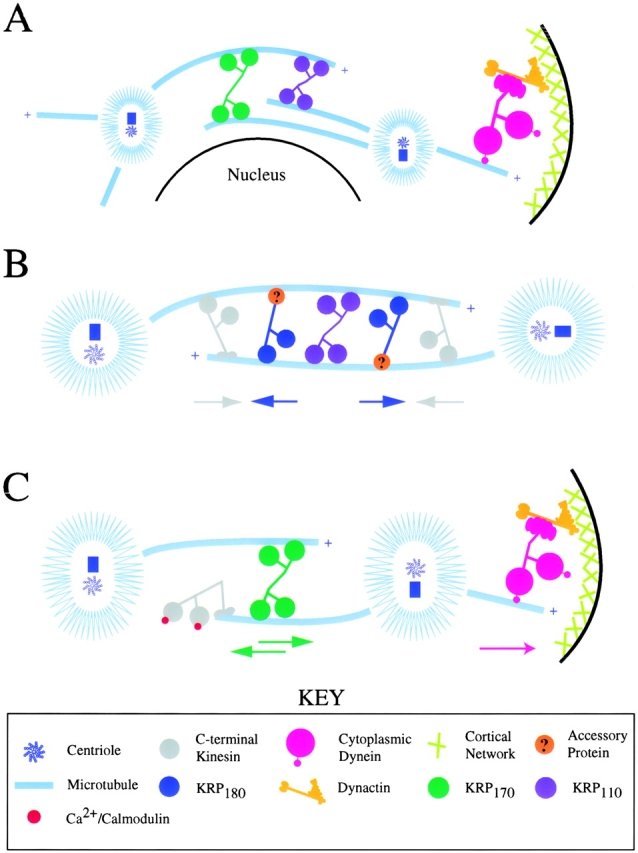
Proposed model describing the pathway of spindle pole positioning during embryonic cell division using the five MT-based motors: KRP180, the COOH-terminal kinesin (Kinesin-C), cytoplasmic dynein, KRP110, and KRP170. Diagrams are shown for the proposed timing and mechanisms of action during (A) prophase/spindle assembly, (B) (pro)metaphase/spindle maintenance, and (C) anaphase B/spindle elongation. Colored arrows indicate the direction of force exerted by the motor of the same color.
At prometaphase, separated poles are held apart and spindle shape maintained by KRP180 through its putative ability to crossbridge antiparallel MTs of short spacing (via interaction with an unidentified MT-binding accessory protein) and to slide MTs (and poles) apart. KRP180's poleward force would be counterbalanced by a plateward force, attributed to Kinesin-C (Fig. 9 B). Collapse of the spindle in the absence of KRP180 function, although not always lethal, would decrease the fidelity of later mitotic events such as proper spindle elongation and cytokinesis, due to the inability of subsequent MT structures to form properly in the central spindle. In addition, KRP110 would also cross-link spindle MTs into tight bundles to enhance the integrity of the spindle.
Spindle elongation could be triggered by the inactivation of Kinesin-C. Plant homologues of Kinesin-C are inactivated when their motor domains bind Ca2+/calmodulin (Song et al. 1997). In sea urchin embryos, [Ca2+]i drops after NEBD and then rises globally at the metaphase-anaphase transition (Groigno and Whitaker 1998). In our model, Kinesin-C motor activity is turned on after NEBD when it would crossbridge MTs and maintain spindle shape by exerting a plateward force on the poles throughout (pro)metaphase. When [Ca2+]i increases at the beginning of anaphase, Kinesin-C is turned-off, releasing the plateward force on the spindle poles and allowing spindle elongation to occur in two extensive phases driven by the MT-MT sliding bipolar kinesin, KRP170, and cortical dynein pulling on astral MTs (Fig. 9 C). Additional studies performed in echinoderm embryos (Scholey 1998) combined with complementary studies performed in organisms amenable to genetic manipulation (Sharp et al. 2000) will be required to test this model and elucidate the precise roles that multiple mitotic motors play in spindle pole positioning.
In conclusion, the data described in this paper identify KRP180 as a member of the Xklp2 subfamily of mitotic motors and provide the first evidence of this motor's activity in vivo. A combination of function-blocking reagents was used to show that KRP180 serves to position spindle poles specifically during prometaphase. The notion that this motor functions during such a specific phase of mitosis underscores the subtle roles that some mitotic motors may perform in spindle function.
Acknowledgments
We would like to thank all of the members of the Scholey Lab and especially Drs. Daniel Buster, Stephen Rogers, Frank McNally, Lesilee Rose, and Bo Liu for helpful scientific discussions and critical reading of the manuscript. We would also like to thank Dr. Heiner Matthies for technical help with bacterial expression and discussions. Also, we thank the Gelfand lab for the anti-tubulin antibody.
This work was supported by a grant from the National Institutes of Health no. GM55507 to J.M. Scholey.
Footnotes
Abbreviations used in this paper: DAPI, 4′-6-diamidino-2-phenylindole; GST, glutathione-S-transferase; KRP, kinesin-related protein; MAP, microtubule-associated protein; MTs, microtubules; NEBD, nuclear envelope breakdown; ORF, open reading frame.
References
- Boleti H., Karsenti E., Vernos I. Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell. 1996;84:49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- Chui, K.K., G.C. Rogers, A.M. Kashina, K.P. Wedaman, D.J. Sharp, D.T. Nguyen, and J.M. Scholey. 2000. Roles of two kinesins in sea urchin embryonic cell division. Submitted. [DOI] [PubMed]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acid Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal B.M., Vaughan K.T., Vallee R.B. Molecular characterization of the 50-kd subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L.S.B. With apologies to Scheherazade - tails of 1001 kinesin motors. Annu. Rev. Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- Groigno L., Whitaker M. An anaphase calcium signal controls chromosome disjunction in early sea urchin embryos. Cell. 1998;92:193–204. doi: 10.1016/s0092-8674(00)80914-9. [DOI] [PubMed] [Google Scholar]
- Henson J.H., Cole D.G., Terasaki M., Rashid D., Scholey J.M. Immunolocalization of the heterotrimeric kinesin-related protein KRP((85/95)) in the mitotic apparatus of sea urchin embryos. Dev. Biol. 1995;171:182–194. doi: 10.1006/dbio.1995.1270. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E., Cassels G.O., Rieder C.L., Sluder G. The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J. Cell Biol. 1998;140:1417–1426. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur E.L.F., Vallee R.B. Dyneinsmolecular structure and cellular function. Annu. Rev. Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., Geiser J.R. Genetic analysis of the mitotic spindle. Annu. Rev. Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Boleti H., Vernos I. The role of microtubule dependent motors in centrosome movements and spindle pole organization during mitosis. Semin. Cell Dev. Biol. 1996;7:367–378. [Google Scholar]
- Kashina A.S., Rogers G.C., Scholey J.M. The bimC family of kinesinsessential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta. 1997;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Kiehart D.P. Microinjection of echinoderm eggsapparatus and procedures. Methods Cell Biol. 1982;25:13–31. doi: 10.1016/s0091-679x(08)61418-1. [DOI] [PubMed] [Google Scholar]
- Lupus A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Molina I., Baars S., Brill J.A., Hales K.G., Fuller M.T., Ripoll P. A chromatin-associated kinesin-related protein required for normal mitotic chromosome segregation in Drosophila . J. Cell Biol. 1997;139:1361–1371. doi: 10.1083/jcb.139.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R.L., Scholey J.M. Heterotrimeric kinesin-II is required for the assembly of motile 9+2 cilliary axonemes on sea urchin embryos. J. Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D.A. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Tanaka Y., Matsuoka E., Kondo S., Okada Y., Noda Y., Kanai Y., Hirokawa N. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc. Natl. Acad. Sci. USA. 1997;94:9654–9659. doi: 10.1073/pnas.94.18.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. Cellular substrates of p34cdc2 and its companion cyclin dependent kinases. Trends Cell Biol. 1993;3:296–300. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- O'Connell M.J., Meluh P.B., Rose M.D., Morris N.R. Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans . J Cell Biol. 1993;120:153–162. doi: 10.1083/jcb.120.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Ledizet M., Cande W.Z. Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol. Biol. Cell. 1996;7:1639–1655. doi: 10.1091/mbc.7.10.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C. The functional coordination of three different microtubule-based motors in positioning centrosomes during sea urchin embryogenesis. Ph.D. Thesis. University of California; Davis, CA: 2000. [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteinsthe PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Hart C.L., Wedaman K.P., Scholey J.M. Identification of Kinesin-C, a calmodulin-binding carboxy-terminal kinesin in animal (Strongylocentrotus purpuratus) cells. J. Mol. Biol. 1999;294:1–8. doi: 10.1006/jmbi.1999.3249. [DOI] [PubMed] [Google Scholar]
- Saunders W.S., Hoyt M.A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Sawin K.E., Leguellec K., Philippe M., Mitchison T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Scholey J.M. Functions of motor proteins in echinoderm embryosan argument in support of antibody inhibition experiments. Cell Motil. Cytoskel. 1998;39:257–260. doi: 10.1002/(SICI)1097-0169(1998)39:4<257::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Yu K.R., Sisson J.C., Sullivan W., Scholey J.M. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Brown H.M., Kwon M.K., Rogers G.C., Holland G., Scholey J.M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel L., Monty K. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Song H., Golovkin M., Reddy A.S.N., Endow S.A. In vitro motility of AtKCPB, a calmodulin-binding kinesin protein of Arabidopsis . Proc. Natl. Acad. Sci. USA. 1997;94:322–327. doi: 10.1073/pnas.94.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., Goldberg M.L. ZW10 helps to recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg E.A., Koonce M.P., McIntosh J.R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos I., Karsenti E. Chromosomes take the lead in spindle assembly. Trends Cell Biol. 1995;5:297–301. doi: 10.1016/s0962-8924(00)89045-5. [DOI] [PubMed] [Google Scholar]
- Vernos I., Raats J., Hirano T., Heasman J., Karsenti E., Wylie C. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Warrick H.M., Spudich J.A. Myosin structure and function in cell motility. Annu. Rev. Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Boleti H., Antony C., Karsenti E., Vernos I. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 1998;143:673–685. doi: 10.1083/jcb.143.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright B.D., Terasaki M., Scholey J.M. Roles of kinesin and kinesin-like proteins in sea urchin embryonic cell divisionevaluation using antibody microinjection. J. Cell Biol. 1993;123:681–689. doi: 10.1083/jcb.123.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyatnin A.A. Protein volume in solution. Prog. Biophys. Mol. Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]