Abstract
The spindle checkpoint ensures proper chromosome segregation by delaying anaphase until all chromosomes are correctly attached to the mitotic spindle. We investigated the role of the fission yeast bub1 gene in spindle checkpoint function and in unperturbed mitoses. We find that bub1 + is essential for the fission yeast spindle checkpoint response to spindle damage and to defects in centromere function. Activation of the checkpoint results in the recruitment of Bub1 to centromeres and a delay in the completion of mitosis. We show that Bub1 also has a crucial role in normal, unperturbed mitoses. Loss of bub1 function causes chromosomes to lag on the anaphase spindle and an increased frequency of chromosome loss. Such genomic instability is even more dramatic in Δbub1 diploids, leading to massive chromosome missegregation events and loss of the diploid state, demonstrating that bub1 + function is essential to maintain correct ploidy through mitosis. As in larger eukaryotes, Bub1 is recruited to kinetochores during the early stages of mitosis. However, unlike its vertebrate counterpart, a pool of Bub1 remains centromere-associated at metaphase and even until telophase. We discuss the possibility of a role for the Bub1 kinase after the metaphase–anaphase transition.
Keywords: Bub1, centromere, mitosis, Schizosaccharomyces pombe, spindle checkpoint
Accurate chromosome segregation through mitosis requires the proper attachment of kinetochores to the spindle before anaphase begins. Kinetochore capture by microtubules is an event which is primarily governed by chance and is therefore prone to error (for review see Nicklas, 1997). The spindle checkpoint ensures high fidelity chromosome segregation by delaying anaphase until all chromosomes are correctly attached to the spindle (for review see Elledge, 1996; Rudner and Murray, 1996; Wells, 1996; Allshire, 1997; Hardwick, 1998).
Genetic analyses in budding yeast have identified some of the genes involved in the spindle checkpoint pathway. Mutations in the three MAD (mitotic arrest-deficient) genes, MAD1-3, and the three BUB (budding uninhibited by benzimidazole) genes, BUB1-3 result in hypersensitivity to spindle poisons as mutant cells are unable to arrest mitotic progression in response to spindle damage (Hoyt et al., 1991; Li and Murray, 1991). Among these checkpoint components are protein kinases and phosphoproteins suggesting that checkpoint function requires a phosphorylation-based transduction pathway. Bub1 is a protein kinase that can bind and phosphorylate Bub3 (Roberts et al., 1994). Mad1 can bind Mad2 (Chen, R.-H., and K.G. Hardwick, unpublished observations) and is hyperphosphorylated upon checkpoint activation (Hardwick and Murray, 1995). The Mps1 kinase is required for the spindle checkpoint and appears to directly phosphorylate Mad1 (Hardwick et al., 1996; Weiss and Winey, 1996). A combination of genetics and biochemical analyses placed BUB1, BUB3, and MPS1 upstream of MAD1 and MAD2 whereas BUB2 and MAD3 would act downstream of these genes (Hardwick and Murray, 1995; Hardwick et al., 1996). A dominant allele of BUB1 has recently been identified which activates the spindle checkpoint when overexpressed. Analysis of its overexpression in mad, bub, and mps1 mutants suggests that the Bub1 and Mps1 kinases act together at an early step in the checkpoint pathway, upstream of all the other Bub and Mad proteins (Farr and Hoyt, 1998).
In addition to gross alterations in spindle structure, the budding yeast spindle checkpoint can also respond to low doses of microtubule depolymerizing drugs, to defects induced by mutations in centromere-binding proteins or in centromeric DNA, and to aberrantly segregating centromeres (for review see Rudner and Murray, 1996). All of these defects could interfere with kinetochore–microtubule attachment and a sensing system at kinetochores would allow the checkpoint to monitor the whole process of spindle assembly. Consistent with this idea, a phosphoepitope recognized by the monoclonal antibody 3F3 is only present at unattached kinetochores in mammalian somatic cell lines (Gorbsky and Ricketts, 1993). Furthermore, the vertebrate homologues of Mad2, Bub1, Mad3 and Bub3 all localize to unattached kinetochores (Chen et al., 1996; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998). It is not clear precisely how the spindle checkpoint monitors chromosome attachment. Checkpoints components could sense free microtubule binding sites at kinetochores or the tension exerted within the kinetochore by spindle poleward forces or a combination of both.
The most likely target of the spindle checkpoint is the anaphase-promoting complex (APC)1 or cyclosome. The APC acts as a ubiquitin-protein ligase to specifically mark proteins consequently targeted for destruction by the proteasome (for review see King et al., 1996). Activation of the APC triggers anaphase by degrading inhibitors of sister chromatid separation like budding yeast Pds1 and fission yeast Cut2 (Cohen-Fix et al., 1996; Funabiki et al., 1996) and allows cells to exit mitosis by degrading mitotic cyclins and regulators of spindle structure such as Ase1 (Juang et al., 1997). Recently, it was shown that Mad2 interacts directly with Slp1/Cdc20 (Hwang et al., 1998; Kim et al., 1998), which is thought to be a substrate-specific activator of APC-dependent proteolysis (He et al., 1997; Li et al., 1997; Visintin et al., 1997). It has been proposed that when the spindle checkpoint is activated Mad2, most likely in a complex with Mad1 and Mad3 (Hwang et al., 1998), represses the activity of Cdc20 and thus inhibits the metaphase–anaphase transition. The molecular mechanism of this repression is not understood.
The centromere/kinetochore complex is a key element in the process of mitosis. As well as the regulatory function of monitoring chromosome attachment to the spindle, the kinetochore carries out a number of mechanical functions. These include sister chromatid cohesion, attachment of sister chromatids to the spindle microtubules and the subsequent chromosome movements of metaphase and anaphase. In most eukaryotes, centromeric DNA consists of long arrays of repetitive DNA packaged into transcriptionally silent, recombination cold, late-replicating, heterochromatin. The exact function of heterochromatin is unknown but kinetochores in higher eukaryotes generally form within centromeric heterochromatin (Karpen and Allshire, 1997). In Schizosaccharomyces pombe three regions of the genome are known to assemble transcriptionally silent, heterochromatin-like structures. These are the centromeres, the telomeres, and the silent mating-type loci ( Allshire et al., 1994; Nimmo et al., 1994; Thon et al., 1994 ). Mutations in clr4, rik1, and swi6 genes were shown to affect repression at the silent mating-type loci, at centromeres, and to a limited extent at telomeres (Lorentz et al., 1992; Ekwall and Ruusala, 1994; Allshire et al., 1995). The swi6 + gene encodes a structural component of heterochromatin which localizes to centromeres, telomeres, and the mating-type region and functional Rik1 and Clr4 are required for this localization (Ekwall et al., 1995, 1996). Although Swi6 is present at telomeres it does not appear to play a crucial role in telomere function since mutations in the swi6 gene have little effect on telomere silencing and do not affect telomere length nor telomere clustering (Allshire et al., 1995; Ekwall et al., 1996). In addition, deletion of the swi6 gene induces an elevated loss rate of both linear and circular minichromosomes, suggesting that Swi6 acts primarily at the centromere. Indeed, the lack of a functional swi6 + gene causes centromeres to lag on anaphase spindles (Ekwall et al., 1995). From these observations, it has been proposed that cells devoid of a functional swi6 gene assemble a compromised centromere (Ekwall et al., 1996).
In order to uncover new centromere proteins, we looked for loci that genetically interact with swi6 through a synthetic lethal screen. One of the mutants identified was found to bear a mutation in a gene homologous to budding yeast BUB1. We show that S. pombe bub1 + is required to detect alterations in spindle structure or defects in kinetochore function. Activation of the checkpoint results in the recruitment of Bub1 to centromeres and the introduction of a delay in the completion of mitosis. Through genetic and cytological observations we show that the loss of the bub1 gene causes defects in chromosome segregation, providing clear evidence that Bub1 function is also required in a normal, unperturbed mitosis. Immunofluorescence analyses indicate that Bub1 is systematically recruited to centromeres during the prophase and prometaphase stages of normal mitosis. In metaphase, a fraction of the Bub1 pool is released from centromeres but surprisingly, some remains centromere associated until cytokinesis raising the possibility of a role for Bub1 at kinetochores past the metaphase–anaphase transition.
Materials and Methods
Strains, Media, Transformation and Genetic Techniques
All the strains used in this study are listed in Table I. Media were essentially as described by Moreno et al. (1991) and Allshire et al. (1994). Synthetic minimal medium is PMG. PMGthi is PMG 20 μM thiamine. When appropriate, media were supplemented with 0.1 g/liter of leucine (L), uracil (U), adenine (A), and histidine (H). When required, Phloxin B was added to the media (2.5 mg/l) to stain colonies containing dead cells. YES refers to yeast extract medium supplemented with LUAH. YES 1/10A is supplemented with a limiting amount of adenine (0.01 g/liter) to allow the development of the red and pink colors of ade6-210 and ade6-216 colonies. Transformation and genetic techniques were as described (Moreno et al., 1991). Minichromosome loss rates were measured as described (Allshire et al., 1995). Briefly, cells from white colonies formed on a YES 1/10A plate were harvested, diluted in water, plated on YES 1/10A, and then incubated 4 d at 32°C. The rate of minichromosome loss per division is the number of colonies with a red sector covering at least half of the colony divided by the total number of white colonies plus half-sectored colonies.
Table I.
S. pombe Strains Used in This Study
| Strains | Genotype | Origin | ||
|---|---|---|---|---|
| 14 | h + leu1-32 ura4DS/E ade6-704 his1–102 CM3112 [sup3–5] | R. Allshire, Edinburgh, Scotland, UK | ||
| 57 | h − ars1-pREP41swi6-ars1 Δswi6::his1 + leu1-32 ura4DS/E ade6–210 his1–102 | This study | ||
| 77 | h − leu1–32 ura4DS/E ade6–210 his1–102 | R. Allshire | ||
| 139 | h − bub1–118 ars1-pREP41swi6-ars1 Δswi6::his1 + leu1–32 ura4DS/E ade6–210 his1–102 | This study | ||
| 277 | h + bub1–118 leu1–32 ura4DS/E ade6–210 his1–102 | This study | ||
| 379 | h − nda3KM311 leu1–32 ura4DS/E ade6–210 his1–102 | R. Allshire | ||
| 393 | h − Δbub1::ura4 + leu1–32 ura4DS/E ade6–216 his1–102 | This study | ||
| 399 | h − Δbub1::ura4 + ars1-pREP41swi6-ars1 Δswi6::his1 + leu1–32 ura4DS/E ade6–210 his1–102 | This study | ||
| 401 | h − nda3KM311 Δbub1::ura4 + leu1–32 ura4DS/E ade6–216 his1–102 | This study | ||
| 411 | h − bub1–6HA-ura4 + leu1–32 ura4DS/E ade6–210 his1–102 | This study | ||
| 415 | h − nda3KM311 bub1–6HA-ura4 + leu1–32 ura4DS/E ade6–210 his1–102 | This study | ||
| 423 | h − Δswi6::his1 + leu1–32 ura4DS/E ade6–210 his1–102 | R. Allshire | ||
| 428 | h − Δbub1::ura4 + leu1–32 ura4DS/E ade6–704 his1–102 CM3112 [sup3–5] | This study | ||
| 429 | h − Δswi6::his1 + bub1–6HA-ura4 + leu1–32 ura4DS/E ade6–210 his1–102 | This study | ||
| 499 | h − leu1–32 ura4DS/E ade6–210 his1–102 Ch16 [ade6–216 Δbub1::ura4 + ] | This study | ||
| 502 | h + Δbub1::LEU2 leu1–32 ura4DS/E ade6–210 his1–102 Ch16 [ade6–216 Δbub1::ura4 + ] | This study | ||
| 576 | ade6–210/ade6–216 diploid | This study | ||
| 578 | Δbub1::LEU2/Δbub1::ura4 + ade6–210/ade6–216 diploid | This study |
DNA Manipulations and Plasmid Constructions
DNA manipulations were performed according to standard procedures. Sequencing was done by Genome express S.A. (Grenoble, France) on double-stranded templates using an Applied Biosystems 373XL sequencer (Foster City, CA). The fission yeast B1 genomic library made in pUR19 (S. pombe ura4 + gene as selection marker) was provided by A.M. Carr (Medical Research Council Cell Mutation Unit, Brighton, UK).
pREP41swi6 Construction.
A SmaI-NsiI 2.35-kbp fragment from pAL2 (Lorentz et al., 1994) bearing the swi6 + gene was inserted between the HindII and PstI sites of pBluescript SK. The entire swi6 + open reading frame (ORF), including 5′ and 3′ untranslated regions, was cut out as a 1.3-kbp SspI-BamHI fragment and cloned between the NdeI filled-in/ BamHI sites of pREP41 (Basi et al., 1993) to yield pREP41swi6.
Construction of pbub1-6HA.
A 540-bp fragment was generated by PCR using oligonucleotides bub1-3′ in (5′-GGAATTCGCGGCCGCAAATTTTTCTTTTTTCGATGC-3′) and bub1-Xba out (5′-ACTTCTTCGTCTAGAAACCG-3′). This fragment contains a XbaI site at the 5′ end, the last 178 codons of bub1+ without the stop codon and a NotI site and a EcoRI site at the 3′ end. A 220-bp fragment was amplified using primers bub1-3′ out (5′-GGAATTCTAATTTTTTGCCTAGAGG-3′) and bub1-3′ out (5′-GCATGCATATTGCAGGTTTG-3′). This fragment contains an EcoRI site at the 5′ end, a STOP codon followed by 200 bp of bub1+ 3′ untranslated regions, and a NsiI site at the 3′ end. The two PCR products were cut with EcoRI, ligated together, and then cut with XbaI and NsiI and cloned between the XbaI and PstI sites of pUC19ura4 (S. pombe ura4 + cloned into the SphI site of pUC19). Finally, six copies of the hemagglutinin (HA) sequence were inserted into the NotI site to give pbub1-6HA.
pΔbub1::ura4+ and pΔbub1::LEU2 Construction.
The 3787-bp BamHI-SphI fragment containing the bub1 + gene was cloned into pBluescript SK to give pSKbub1+. The S. pombe ura4 + gene cloned into the HindIII site of pBluescript SK was cut out with SalI and SpeI and cloned into XhoI-SpeI–digested pSKbub1+ to give pΔbub1::ura4+. Similarly, the Saccharomyces cerevisiae LEU2 gene cloned into the HindIII site of pBluescript SK was cut out with SalI and SpeI and cloned into XhoI-SpeI–digested pSKbub1+ to give pΔbub1::LEU2.
Strain Constructions
Strain 57.
pREP41swi6 was linearized with MluI (within ars1 sequences in pREP41) and transformed into strain 423. Single copy integrants at the ars1 locus were screened by Southern blotting.
Δbub1::ura4 + strains were obtained by transformation with the 3.2-kb BamHI-SphI fragment from pΔbub1::ura4+. Δbub1::LEU2 strains were made by transformation with the BamHI-SphI fragment from pΔbub1:: LEU2. All strains were checked by Southern blotting.
Strain 411.
pbub1-6HA was cut with the single ClaI site lying within bub1 + sequences and transformed into strain 77. Southern blotting was used to identify a strain with the expected structure.
Isolation of the bub1-118 Mutant
Strain 57 was mutagenized with ethylmethane sulfonate as described (Moreno et al., 1991). Mutagenized cells were plated onto PMGLUA plates and incubated at 32°C until colony formation. Colonies were then replica-plated onto PMGLUAthi phloxinB and PMGLUA phloxinB and incubated at 32°C for 2–3 d. Colonies that stained dark red on the thiamine-containing plates were selected. Mutant strains were backcrossed at least three times. From ∼120,000 colonies screened, seven mutants were isolated. All seven mutants were recessive and defined five genes named ssl1 to ssl5 (swi6 synthetic lethal). The ssl5 gene was renamed bub1. It is represented by a single allele, ssl5-118 which was renamed bub1-118.
Cloning of bub1+
bub1-118 was found to be tightly linked to ade6 on chromosome III. To isolate the bub1 + gene, strain 139 was transformed with the genomic DNA library, selecting for adenine prototrophy. Ade+ transformants were screened for ability to grow onto thiamine containing medium. Among the complementing plasmids, p115bub1+ had the smallest genomic insert and was used for subcloning.
Elutriation and H1 Kinase Assays
Cultures were grown overnight in YES to 5 × 106 cells/ml and then loaded onto a JE-5.0 Beckman elutriation rotor (Palo Alto, CA). Small G2 cells were then shifted to 18°C and time points taken at 1-h intervals. Cells were fixed with 70% ethanol, before washing with PEM (100 mM Pipes, pH 6.9, 1 mM EGTA, 1 mM MgSO4) and staining with 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor. Cell samples for kinase assays were taken and frozen in liquid nitrogen. Extracts were made by bead-beating in lysis buffer containing 50 mM Hepes, pH 7.6, 80 mM Na-β-glycerophosphate, 50 mM KCl, 15 mM MgCl2, 20 mM EGTA, 2 mM Na vanadate, 1 mM DTT, 0.5% Triton, 1 mM PMSF, and 1 μg/ml of leupeptin, pepstatin, and chymostatin. Extracts were then spun in a microfuge at 6,000 rpm for 5 min to remove beads and insoluble material, before protein concentrations were measured (Bio-Rad assay; Hercules, CA) and equalized. 1 μl of extract was added to a 10 μl kinase assay containing 80 mM Na-β-glycerophosphate, pH 7.4, 15 mM MgCl2, 20 mM EGTA, 2 μg histone H1 (Boehringer Mannheim, Mannheim, Germany), 1 μCi of γ-32P-ATP, 200 μM ATP, and 1 mM DTT. Kinase assays were incubated for 15 min at 30°C, stopped by adding an equal volume of 2× SDS sample buffer, and then separated by 15% SDS PAGE. Gels were stained, fixed, and then dried and the relative levels of H1 kinase activity determined by autoradiography.
Cytological Techniques
Immunostaining was as described previously (Ekwall et al., 1996) with some modifications. Briefly, cells were fixed in 1.2 M sorbitol, 1.7% paraformaldehyde for 40 min. Incubation with anti-HA antibodies and secondary antibodies were performed for 3–16 h in PEMBAL in which BSA was replaced with 5% (wt/vol) nonfat dry milk. Monoclonal anti-HA 16B12 (Berkeley Antibody, Berkeley, CA) was used at a 1:400 dilution, affinity-purified rabbit anti-HA Y-11 (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:50, and anti–α-tubulin monoclonal antibody TAT1 (Woods et al., 1989) at 1:20. Fluorescence in situ hybridization (FISH) was as described (Ekwall et al., 1996) using the centromere probe pRS140 (Takahashi et al., 1992). Fluorescence microscopy was done using a Leica DMRXA microscope (St. Gallen, Switzerland). Images were captured with a Princeton charge-coupled device camera (Princeton, NJ) and processed with the Metamorph software. Figures were mounted using Adobe Photoshop 4.0 (Adobe Systems, San Jose, CA). In the montages from Fig. 4, different exposure times were used for the Bub1 signal, depending on the brightness of the subject. In particular, the exposure time or signal intensity were reduced for prophase and prometaphase cells to minimize halo formation around intense Bub1 signals. Length measurements were done using the Metamorph software (Universal Imaging, West Chester, PA).
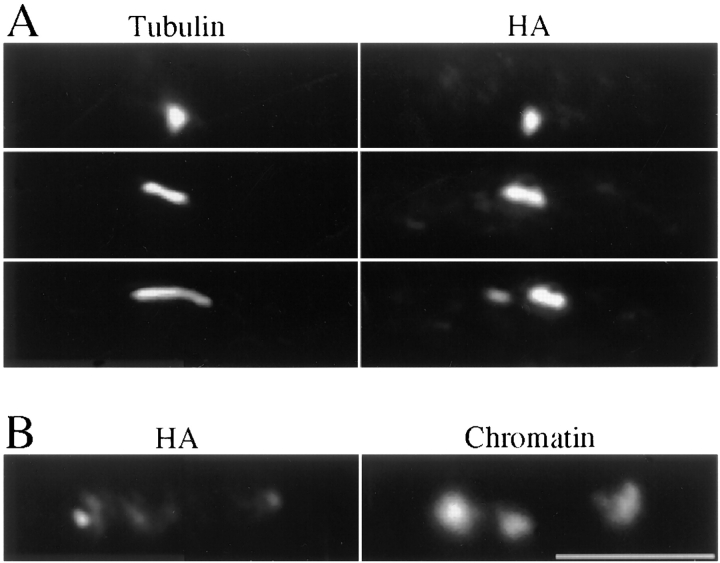
Figure 4.
Cellular localization of Bub1 through the mitotic cell cycle. Cells from strain 411 (bub1-HA) were grown to early log phase in YES medium at 26°C, fixed, and then processed for immunofluorescence microscopy. (A) Montage of cells stained with rabbit polyclonal anti-HA (rHA in red), anti-tubulin (green), and DAPI (blue). For each cell, the corresponding panels are placed below each other. From left to right, an example is shown of cells at various stages of the cell cycle, from interphase (a) through prophase (b), prometaphase (c and d), metaphase (e), anaphase A (f), anaphase B (g), and cytokinesis (h).Yellow is produced upon colocalization of red (Bub1) and green (tubulin) signals. (B) Bub1 colocalizes with centromeres throughout mitosis. Montage of bub1-HA cells stained with mHA (red) and the digoxigenin-labeled centromere probe (green). Blue, DAPI staining. Left to right: interphase (a), prophase (b–d), prometaphase (e), metaphase (f), anaphase A (g), anaphase B (h and i), and cytokinesis (j). Bottom, merged image where yellow is produced upon colocalization of red (Bub1) and green (centromeres) signals. Bars, 5 μm.
Results
Isolation of the bub1+ Gene
In order to identify genes involved in mitotic centromere function, we performed a screen for mutants inviable in the absence of the centromere protein Swi6. Seven mutants were isolated defining five complementation groups. The bub1-118 mutation was found to be tightly linked to ade6 on chromosome III and this linkage was exploited to clone the bub1 + gene (Materials and Methods). Bub1 + resides within a 3,769-bp BamHI-SphI fragment which was fully sequenced (GenBank/EMBL/DDBJ accession number AF064796). A start codon was found at position 433, followed by an uninterrupted ORF encoding a predicted 1,044-amino acid protein with a calculated molecular mass of 113,363 D and a pI of 6.22. Database searches using the BLAST program (Altschul et al., 1990) revealed that the predicted peptide was similar to the budding yeast spindle checkpoint serine/threonine protein kinase encoded by BUB1 (Roberts et al., 1994). A significant match was also found with Bub1 homologues from Drosophila (P = 6.5 × 10−30), human (P = 3.1 × 10−29), and mouse (P = 2.5 × 10−28). Fig. 1 A shows the alignment of fission yeast Bub1 with S. cerevisiae and mouse proteins. The best conserved region resides in the COOH terminus kinase domain. In particular, lysine 762 which was shown to be essential for kinase activity and Bub1 function in budding yeast is conserved in all three proteins (Roberts et al., 1994). In addition, there is a well-conserved region towards the NH2 terminus of Bub1 which is conserved in all three Bub1 proteins, and in the human Bub1-related protein and budding yeast Mad3 (Fig. 1 B). In fact it was this region of homology which led one of us (K.G. Hardwick) to first identify the fission yeast bub1 + gene in BLAST searches of sequence databases. The molecular function of this domain remains unclear: it is close to, but nevertheless distinct from the recently defined Bub3 interaction domain (Taylor et al., 1998) (Hardwick, K.G., unpublished observations on Mad3).
Figure 1.
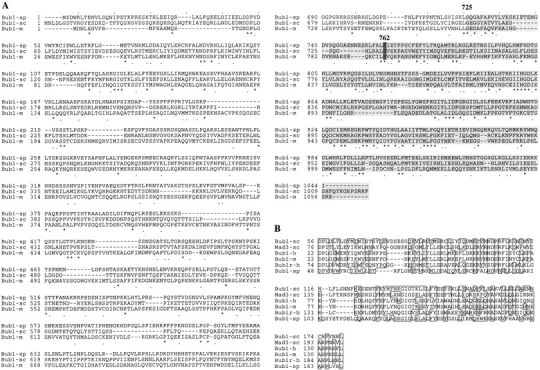
Sequence comparison of Bub1 homologues. (A) Alignment of Bub1 protein sequences from S. pombe (sp), S. cerevisiae (sc), and mouse (m). Asterisks, identical residues; dots, similar residues; shaded area, the extent of the kinase domain. (B) Sequence alignment of the NH2-terminal domains of the Bub1 and Mad3 homologues from S. cerevisiae (sc), human (h), mouse (m), and S. pombe (sp). Boxes, identical or similar residues.
Fission Yeast Bub1 Is an Essential Component of the Spindle Checkpoint
A null allele was made by replacing the entire bub1 + ORF either with the ura4 + or LEU2 genes. The resulting haploid strains are viable showing that bub1 + is not essential for cell viability. In budding yeast, Δbub1 strains are viable but hypersensitive to microtubule-depolymerizing agents such a nocodazole or benomyl owing to the loss of a functional spindle checkpoint (Hoyt et al., 1991; Roberts et al., 1994). As shown in Fig. 2 A, the bub1-118 and Δbub1 strains at 32°C are unable to form colonies on medium containing 7.5 μg/ml benomyl whereas the wild-type control strain still does. A Δswi6 strain was also included in the experiment. Cells devoid of Swi6 are known to be hypersensitive to benomyl presumably as a result of an altered interaction of weakened kinetochores with spindle microtubules (Ekwall et al., 1996). Interestingly, the Δbub1 strain showed the same pattern of benomyl sensitivity as the Δswi6 strain (Fig. 2 A). Sensitivity to a spindle poison can arise either from a structural defect in the spindle/ kinetochore or from the loss of the spindle checkpoint. The two classes of defects should be distinguishable since the former are checkpoint proficient and should be able to halt cell cycle progression in response to spindle damage whereas the latter should not. To investigate whether Δbub1 and Δswi6 have a functional mitotic checkpoint we asked whether mutant strains can arrest their cell cycle in the absence of a spindle. To prevent spindle formation, we used nda3KM311, a cold-sensitive mutation in the β-tubulin gene. When shifted to the restrictive temperature, nda3KM311 cells accumulate with condensed chromosomes, single spindle pole body, a high level of histone H1 kinase activity, and unseparated sister chromatids (Hiraoka et al., 1984; Kanbe et al., 1990; Moreno et al., 1989; Funabiki et al., 1993). We synchronized cultures of the different nda3 mutant strains by elutriation, and Fig. 2 B shows that when shifted to the cold, Δswi6 nda3KM311 cells arrest with high levels of H1 kinase activity whereas the kinase activity drops rapidly in Δbub1 nda3KM311 cells. The drop in kinase activity is accompanied by a wave of septation (Fig. 2 C), confirming that the Δbub1 nda3 KM311 cells are progressing through the cell cycle. The effects of such cell cycle progression in the absence of a spindle are readily apparent: several of the checkpoint-defective cells display the classic cut (cell untimely torn) phenotype because they fail to segregate their DNA to the two daughter cells prior to septation (Fig. 2 D). In stark contrast to this the Δswi6 nda3KM311 cells do not septate and remain arrested with hypercondensed chromosomes (data not shown). An elutriated nda3KM311 control culture behaved like the Δswi6 nda3KM311 strain (data not shown).
Figure 2.

bub1 + is required for spindle checkpoint function. (A) bub1 mutants are hypersensitive to benomyl. Strains were grown in YES medium to 0.5–2 × 107 cells/ml. About 200 cells were spotted onto YES plates containing the indicated concentrations of the anti-microtubule drug benomyl and incubated at 32°C for 3 d. (B–D) bub1 mutants are unable to maintain a mitotic arrest. Small G2 cells of nda3KM311 Δswi6 and nda3KM311 Δbub1 were shifted to 18°C, which disrupts spindle microtubules due to the nda3 defect, and samples were taken every hour. (B) Unlike the Δswi6 cells which arrest with high levels of histone H1 kinase activity (left), the Δbub1 cells fail to maintain such an arrest and the kinase activity drops after 3 h (right). DAPI and calcofluor staining shows that this is followed by a wave of septation (C) from 4–6 h as the Δbub1 cells (solid squares) progress through the cell cycle, and ultimately leads to a cut phenotype (D) as septation occurs without chromosome segregation. The Δswi6 cells maintain their arrest with hypercondensed chromosomes (data not shown) and do not septate (C, open circles). (E) Δbub1 cells are unable to prevent sister chromatid separation in the absence of a spindle. Cells from strain 379 (nda3KM311) and 401 (nda3KM311 Δbub1) were grown in YES medium at 32°C to 3 × 106 cells/ml, shifted to 18°C for 8 h, and then fixed and processed for FISH using a probe detecting all three centromeres (cenFISH). Nuclear DNA was stained with DAPI. Bar, 5 μm.
From this experiment we concluded that Δbub1 cells are deficient for the spindle checkpoint whereas the checkpoint is functional in Δswi6 cells. To further investigate the consequences of the loss of a functional bub1 + gene, we asked whether sister chromatids separate when Δbub1 nda3KM311 cells are shifted to the cold. Cells from bub1 + nda3KM311 and Δbub1 nda3KM311 strains were grown to early log phase at 32°C and then transfered to 18°C. Aliquots of the cultures where fixed at 1, 5, and 15 h and then processed for FISH using a probe detecting all three centromeres (cenFISH). Fig. 2 E shows an nda3KM311 cell displaying three separate cenFISH signals. Those cells represented only a fraction of cells with condensed chromatin (Table II) because the chromosomes have to diffuse away from each other to show separate cenFISH signals. No more than three spots could be detected in bub1 + nda3 KM311 cells even 15 h after the shift to 18°C, showing that sister chromatid cohesion is maintained in those cells. By contrast, cells with more than three spots were readily detected in Δbub1 nda3KM311 (Fig. 2 E, right). They first appeared 1 h after the temperature shift and rose throughout the course of the experiment (Table II). In cells with more than three spots the DNA always appeared decondensed, consistent with an exit from mitosis. It should be mentioned that the proportion of cells with separated sisters is likely to be underestimated since, as mentioned above, individual chromosomes or chromatids must diffuse away from each other to produce separate cenFISH signals. Although not quantitative, this experiment clearly demonstrates that sister chromatid separation occurs in Δbub1 cells in the absence of a spindle.
Table II.
Loss of Sister Chromatid Cohesion in nda3-KM311–arrested Δbub1 Cells
| Strain | Background | Time at 18°C | Number of cells examined | Number of cells with 1, 2, 3, and >3 cenFISH signals* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | >3 | |||||||||||
| h | ||||||||||||||
| 1 | 418 | 399 | 16 | 3 | 0 | |||||||||
| 379 | nda3KM311 | 5 | 421 | 273 | 115 | 33 | 0 | |||||||
| 15 | 422 | 179 | 179 | 64 | 0 | |||||||||
| 1 | 431 | 397 | 30 | 3 | 1 | |||||||||
| 401 | nda3KM311 Δbub1 | 5 | 428 | 300 | 71 | 42 | 15 | |||||||
| 15 | 408 | 241 | 96 | 46 | 25 | |||||||||
One cenFISH signal was defined as a well-individualized spot of fluorescence; doublets were scored as a single signal.
The above experiments show that Bub1 is an essential component of the fission yeast spindle checkpoint. In the absence of a mitotic spindle, cells with a nonfunctional bub1 gene fail to maintain sister chromatid cohesion and high levels of H1 kinase activity, and as a result they exit mitosis with unsegregated chromosomes.
bub1+ Function Is Required for High Fidelity Segregation of Chromosomes in Normal Mitosis
As shown above, Bub1 function is essential to prevent chromosome segregation in the absence of a spindle. Next, we asked whether Bub1 is also required for normal mitosis. This was assayed by measuring the fidelity of chromosome segregation. The rate of chromosome loss was estimated by scoring the mitotic loss rate of the 530-kb linear minichromosome Ch16 (Matsumoto et al., 1990) using the half-sectoring assay method (Allshire et al., 1995). In wild-type cells, Ch16 loss rate is less than 0.1% of cell divisions (Allshire et al., 1995). As a functional bub1 + gene was present on Ch16, we constructed Ch16Δbub1, a Ch16 derivative in which the bub1 + gene was deleted. Chromosome loss assays demonstrated that Ch16Δbub1 is as stable as the original Ch16 in an otherwise wild-type background (Table III). By contrast, the minichromosome was lost in 3.5% of cell divisions in Δbub1 cells, showing that the loss of bub1 + function causes a 70-fold increase in Ch16 loss over wild-type.
Table III.
Effect of bub1 Loss of Function on the Fidelity of Minichromosome Transmission through Mitosis
| Strain | Background | Chromosome loss per division | Fold increase in loss rate | |||||
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| 530-kb linear | 499 | Wild-type | <0.05 (0 out of 2,187) | 1 | ||||
| minichromosome | 502 | Δbub1 | 3.5 (81 out of 2,302) | 70 | ||||
| Ch16Δbub1 | ||||||||
| 30-kb circular | 14 | Wild-type | 1.6 (46 out of 2,875) | 1 | ||||
| minichromosome | 428 | Δbub1 | 20.4 (470 out of 2,302) | 12.75 | ||||
| CM3112 |
Similarly, loss of bub1 + function induced an elevated loss rate of CM3112, a circular minichromosome bearing a functional centromere derived from chromosome III (Matsumoto et al., 1990). The loss rate was up to 20% (Table III), a value similar to the loss rate of acentric or ars plasmids in fission yeast (Heyer et al., 1986; Matsumoto et al., 1990). Therefore, the mitotic segregation of CM3112 is almost completely disrupted in cells lacking bub1 + function.
To investigate whether the segregation of regular chromosomes was also affected, we looked at the rate of breakdown of a diploid homozygous for the bub1 deletion. Chromosome missegregation events in a diploid generate aneuploid cells. As fission yeast cells are highly sensitive to aneuploidy (Niwa and Yanagida, 1985), aneuploids are unstable and return rapidly to the haploid state. Thus the rate of diploid breakdown can be used to estimate the rate of whole chromosome loss (Allshire et al., 1995). Diploids were forced by intragenic complementation between the ade6-210 (red) and ade6-216 (pink) alleles. Ade+ cells were harvested from a plate lacking adenine and plated onto YES medium containing a limiting amount of adenine to allow the development of colored (haploid) colonies. As shown in Fig. 3 A, cells from the bub1 + /bub1 + strain formed pure white (diploid) colonies and very few colored (haploid) colonies (the rate of breakdown is <10−3/cell division, Allshire et al., 1995). By contrast, cells from the Δbub1/Δbub1 strain never formed white colonies but exclusively pink (ade6-216), red (ade6-210), or mixed pink/red haploid colonies. The extreme instability of the diploid precluded any precise measurement of the rate of breakdown. However, since white colonies never formed in the absence of selection for Ade+ cells, one can conclude that bub1 + function is essential for chromosome stability in diploid cells.
Figure 3.

bub1 + function is essential for high fidelity chromosome transmission through mitosis. (A) The diploid state can not be maintained in the absence of a functional bub1 + gene. Diploids were forced by intragenic complementation between the red (ade6-210) and pink (ade6-216) alleles of the ade6 locus. Ade+ cells were harvested from medium lacking adenine and plated onto YES 1/10 ade medium to allow the development of red and pink colors. The bub1 +/bub1 + (wild-type) diploid was stable, producing pure white colonies whereas the Δbub1/Δbub1 strain readily broke down to the haploid state, producing red, pink, and sectored colonies. (B) Chromosomes lag on the anaphase spindle in Δbub1 cells. Exponential growing cells in YES medium at 26°C were fixed and processed for tubulin (green) and DAPI (red, pseudocolor) staining. Right: In wild-type haploid and diploid anaphases, the chromatin masses are at the spindle poles. Left: Δbub1 haploid and diploid cells with chromosomes remaining in the midzone of the anaphase spindle. Bar, 10 μm.
We next investigated whether defects in chromosome segregation could be seen by cytological observation of mitotic cells. Strains 77 (bub1 +) and 393 (Δbub1) were grown to early log phase at 26°C, fixed, and then processed for immunofluorescence microscopy. DAPI and anti-tubulin antibody staining visualized nuclear chromatin and microtubules, respectively. The most striking observation in bub1 mutants was the presence of unsegregated chromosomes at anaphase (Fig. 3 B, top left). In ∼10% of late anaphase cells (Table IV), a chromatin mass remained near the spindle midzone whereas the bulk of chromatin had already reached the spindle poles. Lagging chromosomes were never observed in wild-type anaphase B cells.
Table IV.
Frequency of Abnormal Anaphases in bub1 Mutants
| Strain | Background | Number of late-anaphase cells analyzed* | Late-anaphase cells with lagging chromosomes‡ | |||
|---|---|---|---|---|---|---|
| 77 | Wild-type | 300 | 0 | |||
| 393 | Δbub1::ura4 + | 406 | 52 (12.8%) | |||
| 277 | bub1-118 | 404 | 38 (9.4%) |
Late-anaphase cells were defined as cells with a spindle length greater than 5 μm.
Cells were classified as having lagging chromosomes when DAPI-stained material was observed on the spindle, at least 1.5 μm away from one spindle pole. Parentheses, fraction of anaphase cells with lagging chromosomes.
A similar experiment was performed with diploid strains. Cells from bub1 + /bub1 + and Δbub1/Δbub1 Ade+ colonies were inoculated into complete medium, grown to early log phase and processed for immunofluorescence. As Δbub1/ Δbub1 diploids were extremely unstable, the cell population was composed of a majority of haploid cells and very few diploid cells. Nevertheless, the latter could be identified unambiguously by their larger size. Among late-anaphase Δbub1/Δbub1 cells, 45% (52 out of 116) showed lagging chromosomes (Fig. 3 B, bottom left) whereas cells with lagging chromosomes were rarely (5 out of 600) observed in bub1 + /bub1 + late-anaphase cells.
The above results clearly demonstrate that bub1 + function is required for high fidelity chromosome segregation during normal mitosis. It is unclear why diploid cells are so sensitive to the loss of bub1 + function. One possibility is that diploid mitosis relies more heavily on the spindle checkpoint than haploid mitosis simply because in a diploid there are more chromosomes and hence more kinetochores to be captured by the mitotic apparatus.
Cellular Localization of Bub1
Strain 411 was constructed in which the bub1 + gene is appended with six copies of the HA epitope. The strain displayed the same benomyl sensitivity as a bub1 + strain, showing that the tagged protein was functional (data not shown). To localize the protein in the cell, two anti-HA antibodies were used: a mouse monoclonal antibody (mHA) and an affinity-purified rabbit polyclonal serum (rHA). The monoclonal always gave the best staining and allowed the finest observations whereas rHA was used for double staining with the anti-tubulin antibody to identify the cell cycle stage. Whatever the anti-HA antibody used, no staining was seen on untagged fixed cells (data not shown).
Fig. 4 A shows cells stained with rHA and tubulin. A series of frames are shown which recapitulate the stages of the cell cycle from interphase through mitosis. In interphase cells (panel a), rHA staining was diffused through the nucleus with some faint spots around the nuclear periphery. When cells enter mitosis (panel b), rHA systematically produced a single bright spot of fluorescence colocalizing with the prophase spindle (spindle length <1 μm). In prometaphase cells (spindle length <2 μm), two bright Bub1 spots were seen, always positioned along the spindle axis (panels c and d).
In most cases when cells approached metaphase (spindle length >2 μm and unsegregated DNA) there was an abrupt reduction in the Bub1 staining intensity (Fig. 4 A, panel e), although intense signals were sometimes observed. Among 150 metaphase cells examined, five retained an intense staining pattern (one, two, or three bright spots) but the vast majority (145) showed weakly stained foci (Fig. 4 A, panel e) and a diffuse labelling of the chromatin. Therefore, in a normal mitosis the intense staining pattern of Bub1 is restricted to prophase and prometaphase and is lost when cells reach metaphase. In anaphase cells (Fig. 4 A, f and g), some of the signal appeared to trail behind the separating chromatin masses but discrete spots could also be seen leading the separating chromatin masses, suggesting that a fraction of the Bub1 pool is at the kinetochores in anaphase. At telophase (Fig. 4 A, panel h) the interphase pattern reappeared.
We next looked at the localization of Bub-HA relative to centromeres (Fig. 4 B). Cells were stained with mHA and centromeres were detected by cenFISH. In interphase fission yeast cells, all three centromeres are clustered close to the spindle pole body at the nuclear periphery (Funabiki et al., 1993). mHA in those cells (Fig. 4 B, panel a) produced multiple faint spots at the nuclear periphery. Most cells examined (41 out of 49) did not show any colocalization of Bub1 with the cenFISH signal. In prophase cells, three major patterns of cenFISH staining were observed (Fig. 4 B, panels b–d). The most common pattern (39 out of 60) was a single cenFISH signal colocalizing with the HA spot (Fig. 4 B, panel b). Less frequently (19 out of 60) two FISH signals were seen with the HA spot localized in between the two FISH signals (Fig. 4 B, panel c). Rarely (2 out of 60), the three centromeres could be detected separately and in those cases they were located around the HA spot (Fig. 4 B, panel d). In cells with two intense HA signals (Fig. 4 B, panel e), the distance between the two spots was always short (mean value 0.47 ± 0.06 μm). For these cells, the major pattern (37 out of 47) was a single cenFISH signal between the two HA spots. In some cells (10 out of 47), two cenFISH signals were observed, overlapping with the HA signals (data not shown). In metaphase cells (Fig. 4 B, panel f), the intense pattern of staining was lost but some punctate staining was still present and colocalized with the centromeres. In cells having completed anaphase (Fig. 4 B, panels g–i), some HA staining colocalized with the centromeres at the spindle poles but the bulk of the signal was not centromere-associated and formed a string-like structure between the separating chromatin masses (Fig. 4 B, panel h) which eventually disappeared in late anaphase B cells (Fig. 4 B, panel i). In septated cells (Fig. 4 B, panel j) the interphase pattern reappeared.
To summarize, Bub1 in interphase is distributed in several foci at the nuclear periphery which are distinct from centromeres. When cells enter mitosis Bub1 is recruited to the centromeres, which are still clustered close to the SPBs at this early stage, and the staining becomes much more intense. In prometaphase, Bub1 usually forms two masses aligned along the spindle with the centromeres located in between. In metaphase, Bub1 appeared more diffuse but a fraction of Bub1 is still associated with the centromeres. In anaphase A and B the bulk of Bub1 is distinct from centromeres and follows the movement of the chromatin masses but a fraction of Bub1 remains centromere-associated until telophase.
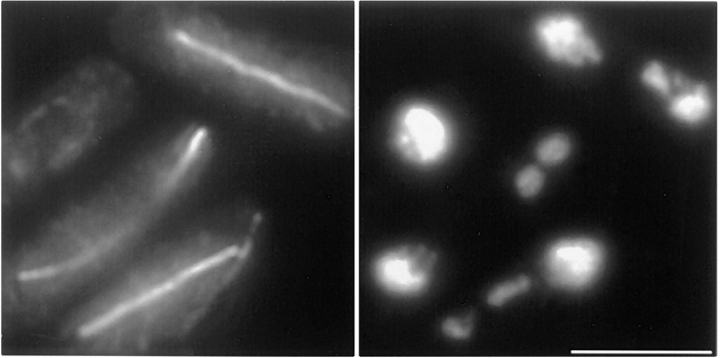
Next we looked at the localization of Bub1 in cells arrested in prometaphase in response to the absence of a spindle. Cells from strain 415 (nda3KM311 bub1-6HA) were grown to early log phase at 32°C, shifted to 18°C for 8 h, and then fixed and processed for anti-HA immunofluorescence and cenFISH. After 8 h at restrictive temperature, cells were arrested with condensed chromosomes. In some instances, the chromosomes had diffused away from each other, allowing individual chromosomes to be seen by DAPI staining (Fig. 5). The cell at the top of the image shows three intense Bub1-HA signals colocalizing with the centromeres. Strikingly, some cells displayed paired foci of Bub1-HA staining rather than well-defined spots. In the bottom of the two cells shown in Fig. 5, all three spots were actually doublets. This pattern of staining was routinely seen in nda3-arrested cells but most often, only one or two chromosomes displayed paired Bub1 foci. Cells with three single spots represented ∼63%, cells with one doublet and two spots 24%, two doublets one spot 25%, and three doublets 2%. The appearance of a doublet was not due to sister chromatid separation since the centromeric signal remained single, located in between the paired Bub1 foci. Such a staining pattern is consistent with Bub1 being associated with sister kinetochores. The mean distance between the two signals in a doublet was 0.42 ± 0.04 μm, which is strikingly similar to the distance separating the two Bub1 foci during the prometaphase stage of cycling cells. This suggests that the paired Bub1 structures observed in cycling cells are also formed by the close proximity of the sister kinetochores from all three chromosomes which are aligned on the spindle axis.
Figure 5.
Bub1 localizes at kinetochores in nda3KM311-arrested cells. Cells from strain 415 (nda3KM311 bub1-HA) were grown to early log phase at 32°C and shifted to 18°C for 8 h. Fixed cells were stained with mHA (red) and centromeric DNA was detected by FISH (green). Blue, DAPI staining. Bottom right, merged image. Two cells are shown. In some instances, Bub1 staining produces three doublets (the cell at the bottom). The merged image reveals that centromeric DNA is located precisely in between the paired dots, consistent with Bub1 being located on each sister kinetochore. Bar, 10 μm.
Bub1 Is Required to Detect and Correct Defects in Centromere Function
The bub1-118 mutant was identified in a screen for colethality with Δswi6. The Swi6 protein is a structural component of fission yeast heterochromatin. As pointed out in the introduction, the Swi6 protein has only a minor role if any at telomeres but appears crucial for normal centromere function since the lack of a functional swi6 + gene is correlated with a defective movement of centromeres at anaphase and an elevated rate of chromosome loss (Allshire et al., 1995; Ekwall et al., 1996). Therefore, the lethality of the Δswi6 Δbub1 double mutant suggests that the Bub1 checkpoint pathway is required to monitor defects in centromere function. If this is indeed the case, the lethality of the double mutant is expected to arise from a complete failure to segregate chromosomes. To test this prediction, we performed a cytological analysis of Δbub1 cells in which the synthetic lethality was induced by depletion of Swi6.
Strain 57 carries the swi6 + ORF under the control of the nmt promoter (nmtswi6) which is repressible by addition of thiamine to the culture medium (Maundrell, 1990). In the absence of thiamine, Swi6 is expressed and the strain behaves as a swi6 + strain. Addition of thiamine into the medium represses nmt and after 8–10 generations, Swi6 is depleted and defective anaphases appear (Table V) with a frequency similar to that observed in a Δswi6 strain grown at 32°C (data not shown). Strain 399 carries the Δbub1 allele in a nmtswi6 background. When Swi6 is produced, the frequency of defective anaphases was ∼7.4% (Table V), close to the value obtained for the bub1-deleted strain. 20 h after the addition of thiamine, when Swi6 was depleted, the fraction of abnormal anaphases represented up to 50% of late anaphase cells. The image shown in Fig. 6 shows a field of cells in which all anaphase cells display lagging chromosomes and/or obvious unequal segregation of the chromatin masses. Therefore, the lethality of the Δswi6 Δbub1 double mutant is likely to be a consequence of massive chromosome missegregation events. As a functional bub1 + gene is required for the viability of a Δswi6 cell, this suggests that Bub1 is somehow able to compensate for the centromere defect induced by the lack of Swi6. How can this be achieved? To address this question, we looked at Bub1 localization in a Δswi6 background. Cells from strain 429 (Δswi6 bub1-6HA) were grown to early log phase at 26°C and processed for anti-tubulin and anti-HA immunofluorescence. The Bub1 staining patterns in prophase and prometaphase were similar to that observed in a wild-type background (Fig. 7 A, top and middle) showing that the lack of Swi6 does not grossly affect the localization of Bub1. However, a large fraction of metaphase cells displayed an intense punctate staining (Fig. 7 A, bottom). Examination of 103 metaphase Δswi6 cells (spindle length greater than 2 μm and unsegregated DNA) revealed that 49 (47.5%) were devoid of intense Bub1-HA staining but 54 (52.5%) displayed one, two, or three intense Bub1-HA signals. This is in sharp contrast with what was observed in a swi6 + background where only five metaphase cells from 150 examined (3.4%) retained an intense Bub1-staining pattern.
Table V.
Frequency of Abnormal Anaphases in Δbub1 Cells upon Depletion of Swi6
| Strain | Relevant background | Swi6* | Number of late-anaphase cells analyzed‡ | Late-anaphase cells with lagging chromosomes§ | ||||
|---|---|---|---|---|---|---|---|---|
| + | 203 | 1 (0.5%) | ||||||
| 57 | nmtswi6, bub1 + | |||||||
| − | 300 | 8 (2.7%) | ||||||
| + | 202 | 15 (7.4%) | ||||||
| 399 | nmtswi6, Δbub1 | |||||||
| − | 100 | 50 (50%) |
Cells were grown to early log phase at 32°C on minimal medium without thiamine (Swi6 +), then thiamine was added and cells grown for 8–10 generations (Swi6 −). Samples were fixed and processed for tubulin and chromatin staining.
Late-anaphase cells were defined as cells with a spindle length greater than 5 μm.
Cells were classified as having lagging chromosomes when DAPI-stained material was observed on the spindle, at least 1.5 μm away from one spindle pole. Parentheses, fraction of anaphase cells with lagging chromosomes.
Figure 6.
The synthetic lethality between Δbub1 and Δswi6 mutants is characterised by a high incidence of anaphases with lagging chromosomes. Strain 399 (nmtswi6 Δbub1) was grown to early log phase in minimal medium, then thiamine was added to repress transcription from the nmt promoter and cells were cultured for an additional 20 h to allow Swi6 to be depleted. Fixed cells were subjected to tubulin (left) and DAPI (right) staining. All the anaphase cells in the field showed chromosomes still remaining in the midzone of the spindle. Bar, 5 μm.
Figure 7.
Bub1 localization in Δswi6 cells. Cells from strain 429 (Δswi6 bub1-HA) were grown to early log phase in YES medium at 26°C, fixed, and then stained with rHA and tubulin. (A) Top, cell in prophase; middle, cell in prometaphase; bottom, cell in metaphase. Bub1 staining in prophase and prometaphase is as in a wild-type background but the staining remains intense and punctate in about half of Δswi6 metaphase cells. (B) Bub1 appears diffuse on lagging chromosomes. An anaphase B cell is shown with a lagging chromosome lying in between the two separated chromatin masses (right). The same cell stained with mHA (left) shows a faint punctate signal leading the main chromatin masses. The lagging chromosome is diffusely stained with mHA and does not show any punctate staining.
Examination of Bub1-stained postmetaphase cells did not reveal any significant difference between swi6 + and Δswi6 strains. Of interest was the Bub1 staining status of lagging chromosomes in Δswi6 anaphase cells. In all cells examined (100), lagging chromosomes did not show any intense Bub1 staining but a rather diffuse labelling of the lagging chromatin (Fig. 7 B). The weak, diffuse signal on the lagging chromosomes is not due to poor immunofluorescence since, like in wild-type anaphases, Bub1 can be seen leading the separated main chromatin masses (Fig. 7 B).
Taken together, these observations are consistent with the idea that, in early mitosis, Bub1 is recruited to centromeres not yet bound to microtubules but this ability is lost past the metaphase–anaphase transition since aberrantly segregating centromeres at anaphase do not recruit Bub1.
Next we asked whether the dramatic recruitment of Bub1 in Δswi6 metaphases could be correlated with the introduction of a cell cycle delay. As shown in Table VI, the fraction of metaphase cells is doubled in a Δswi6 background. This increase in metaphase cells is statistically highly significant (P < 0.01 by the Chi squared test). Therefore, cells harboring a compromised centromere spent twice the normal amount of time in metaphase. It should be noted that there is also an increase in the fraction of late mitotic cells in Δswi6 compared to wild type. Although these data are at the borderline of statistical significance (P < 0.05) they suggest that the duration of anaphase and/or telophase is also increased in Δswi6 cells.
Table VI.
Distribution of Mitotic Cells in Δswi6 and Δbub1 Strains
| Strain | Background | Number of cells analyzed | Cells in prophase and prometaphase* | Cells in metaphase‡ | Cells in anaphase and telophase§ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 77 | Wild-type | 3,583 | 69 (1.9%) | 62 (1.7%) | 312 (8.7%) | |||||
| 423 | Δswi6::his1 + | 3,090 | 42 (1.4%) | 115 (3.7%) | 322 (10.4%) | |||||
| 393 | Δbub1::ura4 + | 3,047 | 46 (1.5%) | 67 (2.2%) | 305 (10%) |
Prophase and prometaphase were defined as cells with a spindle length shorter than 2 μm.
Metaphase cells were defined as cells with unsegregated DNA and a spindle length greater than 2 μm and shorter than 5 μm.
Anaphase and telophase cells were defined as cells with segregated DNA and spindle length greater than 5 μm. Parentheses, fraction of each cell population.
From these observations, we conclude that cells in which centromeres are weakened by the absence of the Swi6 protein are delayed in metaphase and this delay correlates with the persistence of an intense punctate Bub1 staining. This suggests that misattached kinetochores activate the checkpoint through the recruitment of Bub1 and as a consequence anaphase is delayed.
The above observations suggest that Bub1 is able to slow cell cycle progression when it is recruited to centromeres. In wild-type cycling cells, intense Bub1 staining is observed in prophase and prometaphase. Thus, if Bub1 acts to control the timing of normal mitosis, one might expect Δbub1 cells to pass more quickly through mitosis. As shown in Table VI, there is a modest decrease in prophase and prometaphase cells in the Δbub1 strain as compared to wild type, but the difference is not statistically significant. Therefore, the loss of bub1 + function does not appear to grossly alter the timing of mitosis.
Discussion
We have shown that fission bub1 + encodes a mitotic centromere protein which is an essential component of the spindle checkpoint. Vertebrate homologues of several checkpoint proteins, namely Mad2, Mad3, Bub1, and Bub3 have all been shown to localize to unattached kinetochores leading to the idea that microtubule-free kinetochores attract checkpoint components which in turn inhibit or delay cell cycle progression. Our data fully support this idea. Immunofluorescence analysis reveals that Bub1 is massively recruited to centromeres during the early stages of mitosis when kinetochores are not yet captured by microtubules, and when microtubule polymerization is prevented by a mutation in β-tubulin. In cells lacking the centromere protein Swi6, kinetochores are thought to be less efficient in capturing microtubules, and we found that in those cells the intense Bub1 staining persists in metaphase. Conversely, in wild-type cells the Bub1 staining is diminished in metaphase when all kinetochores have been captured. Thus, intense Bub1 staining at centromeres correlates well with the presence of microtubule-free kinetochores. There is also a good correlation between the centromeric recruitment of Bub1 and the introduction of a cell cycle delay. The prometaphase arrest of nda3KM311 cells at 18°C is dependent upon a functional bub1 + gene and is characterized by a strong Bub1 staining at kinetochores. Similarly, cells devoid of a functional swi6 + gene show metaphases with intense Bub1 staining and spend twice the normal amount of time in metaphase. Therefore, our data suggest that microtubule-free kinetochores attract Bub1 which in turn delays the onset of anaphase.
Although the above is true when kinetochore or spindle function is compromised, the situation is less clear in an unperturbed mitosis since the lack of a functional bub1 + gene does not appear to shorten the early stages of mitosis. It is possible that kinetochore capture in mitosis is normally a very efficient process which is completed well before full activation of the APC. However, we found that in cells lacking bub1 + function ∼13% of anaphases displayed lagging chromosomes, suggesting that stable kinetochore capture had not been achieved in a significant fraction of metaphases. In addition, it has been reported that overexpression of a dominant-negative form of mouse Bub1 in HeLa cells shortens mitosis by 25 min (Taylor and McKeon, 1997). Live analysis of mitosis in individual fission yeast cells will be necessary to address this proposed mitotic timing function for Bub1 more carefully.
Overexpression of Bub1 in budding yeast results in a nuclear accumulation, but unfortunately the protein could not be detected in wild-type cells (Roberts et al., 1994). The mouse Bub1 protein is recruited to kinetochores in early mitosis but dissociates in metaphase and is thus absent from kinetochores later in mitosis (Taylor and McKeon, 1997). Similarly in S. pombe, kinetochores are brightly stained with Bub1 in prophase and prometaphase and the intense signal is then lost in metaphase. However, some staining remains centromere associated in metaphase and throughout the final stages of mitosis, raising the possibility that Bub1 might have a role at kinetochores in anaphase. Such staining is reminiscent of that recently reported for p55Cdc20, the vertebrate homologue of budding yeast Cdc20 and fission yeast Slp1, both of which have been proposed to be effectors of the spindle checkpoint (Hwang et al., 1998; Kallio et al., 1998; Kim et al., 1998). Cdc20/Slp1 are essential genes required for the proteolytic destruction of a number of proteins including regulators of sister-chromatid separation (Pds1/Cut2) and the mitotic cyclins. Their precise roles and interaction with the APC remain unclear, but it has been reported (Kallio et al., 1998) that p55Cdc20 can be immunoprecipitated with components of the APC (Cdc27) and the spindle checkpoint (Mad2). Further biochemical work is necessary to determine whether this is true in fission yeast and if so whether Bub1 is also present in such complexes.
One explanation for the anaphase localization of Bub1 at kinetochores could be that it also has a nonessential, structural function within the kinetochore. For example, Bub1 might increase the kinetochore's affinity for microtubules thereby promoting kinetochore capture and strengthening the attachment of chromosomes to the spindle. Lagging chromosomes in cells lacking bub1 + could arise from the combined effects of the lack of a functional checkpoint and a weakened kinetochore. Alternatively, a late role for Bub1 could also be restricted to a checkpoint function. Intriguingly, cells deleted for swi6 + do show an increased proportion of anaphase cells, suggesting that they activate a checkpoint which operates past the metaphase–anaphase transition. For instance, lagging chromosomes in anaphase cells could activate a checkpoint to delay cytokinesis. A cytokinesis checkpoint has recently been reported in budding yeast (Muhua et al., 1998). However, the existence of such a checkpoint in fission yeast and any involvement of Bub1 remain speculative. In addition, lagging chromosomes in Δswi6 cells were not intensely Bub1 stained, suggesting that unattached kinetochores are unable to activate the spindle checkpoint past the metaphase–anaphase transition.
The existence of lagging chromosomes in Δswi6 cells raises the question of the effectiveness of the checkpoint in preventing missegregation events. The survival of Δswi6 cells is dependent upon a functional bub1 gene showing that the checkpoint is required to correct the defect induced by the lack of Swi6. However, the rescue is only partial since a fraction of anaphase cells show lagging chromosomes. Why do these missegregations events escape the checkpoint? Experiments in budding yeast have shown that mutations in the essential kinetochore component Ndc10 abrogated the spindle checkpoint (Tavormina and Burke, 1998), leading to the idea that a kinetochore has to be formed to recruit checkpoint components. It is possible that the lack of Swi6 produces a heterogenous population of centromeres with respect to kinetochore function. One class would have compromised centromeres with some residual kinetochore and checkpoint function and the other class would have disrupted kinetochores which, like ndc10 kinetochores, would remain undetected by the checkpoint and would generate lagging chromosomes at anaphase. This scenario could also explain why we did not observe any recruitment of Bub1 to the centromeres of lagging chromosomes. Further experiments are thus needed to investigate a possible role for Bub1 during the final stages of mitosis.
In light of recent results, it has been suggested that mutational inactivation of the spindle checkpoint might play an important role in the progression of human cancer. Some cancers are associated with a chromosomal instability phenotype (CIN) leading to aneuploidy. It has been proposed that checkpoint defects might lead to aneuploidy (Hartwell and Kastan, 1994) and that this would then be instrumental in tumorigenesis through the loss of tumor-suppressor genes (Orr-Weaver and Weinberg, 1998). Cahill and colleagues showed that CIN in colorectal cancers is correlated with the loss of the spindle checkpoint and, in a number of cases, is associated with a mutation in hBUB1, the human homologue of budding yeast BUB1 (Cahill et al., 1998). These results suggest that aneuploidy is due to the lack of a functional checkpoint. Δbub1 spores in budding yeast grow very slowly and often fail to form colonies, but when propagated this growth phenotype reverts possibly due to the accumulation of suppressor mutations (Roberts et al., 1994). To our knowledge a quantitative rate of chromosome loss has not been reported for such budding yeast strains. Overexpression of the dominant-negative mouse Bub1 construct may have affected mitotic timing, but it did not appear to have a significant effect on chromosome segregation (Taylor and McKeon, 1997). We have shown that haploid S. pombe Δbub1 strains lose a 530-kb linear minichromosome 70 times more frequently than wild-type strains, and that homozygous bub1Δ diploids are extremely genetically unstable. Thus, our fission yeast results strongly support the idea of a causal relationship between the lack of a functional bub1 + gene, the loss of the spindle checkpoint, and the appearance of chromosome instability and aneuploidy in human cancers.
Acknowledgments
We thank R. Allshire, A. Pidoux (Medical Research Council, Edinburgh, Scotland, UK), and J. Bégueret (Centre National de Recherche Scientifique, Bordeaux, France) for advice and helpful discussion, K. Gull (School of Biological Sciences, Manchester, UK) for the gift of TAT1 antibody, A. Carr (Medical Research Council, Brighton, UK) for providing the S. pombe genomic library, A. Murray (University of California, San Francisco, CA) in whose lab the cloning of bub1 was initiated, and S. MacNeill (Institute of Cell and Molecular Biology, University of Edinburgh, Edinburgh, Scotland, UK) for help with the elutriation.
This work was supported by the Centre National de la Recherche Scientifique and the Wellcome Trust. P. Bernard was supported by a fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur.
Abbreviations used in this paper
- APC
anaphase-promoting complex
- BUB
budding uninhibited by benzimidazole
- CIN
chromosomal instability phenotype
- DAPI
4′,6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- HA
hemagglutinin
- MAD
mitotic arrest-deficient
- mHA
anti-HA mouse monoclonal antibody
- ORF
open reading frame
- rHA
anti-HA rabbit polyclonal serum
- ssl
swi6 synthetic lethal
References
- Allshire RC. Centromeres, checkpoints and chromatid cohesion. Curr Opin Genet Dev. 1997;7:264–273. doi: 10.1016/s0959-437x(97)80137-2. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat J-P, Granston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Javerzat J-P, Redhead NJ, Granston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Altshul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JKV, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Chen R-H, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J-M, Kirshner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiaeis controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;1:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat J-P, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat J-P, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4 + and rik1 +disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiaespindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis is required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ, Ricketts WA. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. J Cell Biol. 1993;122:1311–1321. doi: 10.1083/jcb.122.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG. The spindle checkpoint. Trends Genet. 1998;14:1–4. doi: 10.1016/S0168-9525(97)01340-1. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombespindle checkpoint protein Mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Sipiczki M, Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986;6:80–89. doi: 10.1128/mcb.6.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: a cold sensitive nda3mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiaegenes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe T, Hiraoka Y, Tanaka K, Yanagida M. The transition of cells of the fission yeast β-tubulin mutant nda3-311as seen by freeze-substitution electron microscopy: requirement of functional tubulin for spindle pole body duplication. J Cell Sci. 1990;96:275–282. doi: 10.1242/jcs.96.2.275. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters J-M, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz A, Heim L, Schmidt H. The switching gene swi6 affects recombination and gene expression in the mating-type region of Schizosaccharomyces pombe. . Mol Gen Genet. 1992;233:436–442. doi: 10.1007/BF00265441. [DOI] [PubMed] [Google Scholar]
- Lorentz A, Ostermann K, Fleck O, Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophilaand mammals. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Murakami S, Niwa O, Yanagida M. Construction and characterization of centric and acentric linear minichromosomes in fission yeast. Curr Genet. 1990;18:323–330. [Google Scholar]
- Maundrell K. nmt1of fission yeast. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Muhua L, Adames NR, Murphy MD, Shields CR, Cooper JA. A cytokinesis checkpoint requiring the yeast homologue of an APC-binding protein. Nature. 1998;393:487–491. doi: 10.1038/31014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nimmo E, Cranston G, Allshire R. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO (Eur Mol Biol Organ) J. 1994;16:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Yanagida M. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr Genet. 1985;9:463–470. [Google Scholar]
- Orr-Weaver TL, Weinberg TA. A checkpoint on the road to cancer. Nature. 1998;392:223–224. doi: 10.1038/32520. [DOI] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PA, Burke DJ. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiaeis independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 1998;148:1701–1713. doi: 10.1093/genetics/148.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localisation of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a human Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Cohen A, Klar A. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. . Genetics. 1994;138:29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WAE. The spindle-assembly checkpoint: aiming for a perfect mitosis, every time. Trends Cell Biol. 1996;6:228–234. doi: 10.1016/0962-8924(96)10018-0. [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma bruceiby a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]