Abstract
We have identified ScPex18p and ScPex21p, two novel S. cerevisiae peroxins required for protein targeting via the PTS2 branch of peroxisomal biogenesis. Targeting by this pathway is known to involve the interaction of oligopeptide PTS2 signals with Pex7p, the PTS2 receptor. Pex7p function is conserved between yeasts and humans, with defects in the human protein causing rhizomelic chondrodysplasia punctata (RCDP), a severe, lethal peroxisome biogenesis disorder characterized by aberrant targeting of several PTS2 peroxisomal proteins, but uncertainty remains about the subcellular localization of this receptor. Previously, we have reported that ScPex7p resides predominantly in the peroxisomal matrix, suggesting that it may function as a highly unusual intraorganellar import receptor, and the data presented in this paper identify Pex18p and Pex21p as key components in the targeting of Pex7p to peroxisomes. They each interact specifically with Pex7p both in two-hybrid analyses and in vitro. In cells lacking both Pex18p and Pex21p, Pex7p remains cytosolic and PTS2 targeting is completely abolished. Pex18p and Pex21p are weakly homologous to each other and display partial functional redundancy, indicating that they constitute a two-member peroxin family specifically required for Pex7p and PTS2 targeting.
Keywords: microbodies, peroxisomes, biogenesis, protein import, Saccharomyces cerevisiae
One of the central tenets of peroxisomal biogenesis is that soluble peroxisomal proteins are posttranslationally imported from the cytosol into preexisting organelles (Lazarow and Fujiki, 1985). The identification, both in yeasts and mammals, of multiple distinct genetic complementation groups defective in peroxisomal biogenesis (pex 1 mutants) has intimated that this is a complex process involving numerous cellular proteins (reviewed in Subramani, 1997; Erdmann et al., 1997; Subramani, 1998). Careful analysis of these mutants over recent years has revealed that peroxisomal protein import follows a branched pathway (Purdue and Lazarow, 1994), with each branch representing an import receptor specific for one of several types of peroxisome targeting sequences (PTS), followed by a shared membrane translocation process. So far, two classes of PTS have been well characterized; the COOH-terminal PTS1, which is generally a tripeptide conforming to the consensus S/A/C-K/R/H-L (Gould et al., 1989; although other variants exist, such as the tetrapeptide −KANL PTS1 of human catalase; Purdue and Lazarow, 1996), and the NH2-terminal PTS2 oligopeptide found on several peroxisomal proteins including thiolase (Osumi et al., 1991; Swinkels et al., 1991; Erdmann, 1994; Glover et al., 1994). Analysis of pex mutants that map to the specific branches of peroxisomal biogenesis, and thus fail to package only a subset of peroxisomal proteins, has proven particularly valuable, leading to the identification and characterization, in both yeasts and humans, of Pex5p (Van Der Leij et al., 1993; Dodt et al., 1995; Szilard et al., 1995; Terlecky et al., 1995; van der Klei et al., 1995) and Pex7p (Marzioch et al., 1994; Zhang and Lazarow, 1994; Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997; Elgersma et al., 1998), the import receptors for PTS1 and PTS2 type signals, respectively.
Pex7p, which has now been cloned from three yeasts and also from humans and mice, is of significant interest for two main reasons. First, human PEX7 has been recently shown to be the defective gene in rhizomelic chondrodysplasia punctata (RCDP), classifying this severe and relatively common disorder of peroxisomal biogenesis as a disease of protein targeting, and emphasizing the critical role of PTS2 targeting in peroxisomal biogenesis (Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997). Second, Pex7p is an unusual receptor in that it is soluble; this has led to considerable debate and interest regarding its subcellular distribution and function. Previously, we have reported that, when expressed in S. cerevisiae at a level equivalent to wild-type PEX7, a COOH-terminally tagged version of ScPex7p is functional (in that it rescues growth of a Δpex7 strain on oleic acid) and localized principally, if not exclusively, within the peroxisomal matrix (Zhang and Lazarow, 1994). This surprising finding suggests that Pex7p functions as a highly unusual, intraorganellar import receptor. Pex7p import into peroxisomes is saturable; overexpressed tagged Pex7p accumulates in the cytosol as well as peroxisomes. Other researchers have reported that Pex7p is both peroxisomal and cytosolic (Marzioch et al., 1994; Elgersma et al., 1998), and this has led to an alternative hypothesis for Pex7p function, which suggests that this is a mobile receptor that cycles between cytosol (where it can collect PTS2 cargo) and the peroxisomal surface, or matrix, where cargo is released, and Pex7p returns to the cytosol to begin another round of import.
To better understand the PTS2 branch of peroxisomal biogenesis, and in doing so help unravel the mysteries of Pex7p localization and function, we have initiated a search for other proteins involved in this pathway. In this paper, we report the identification and initial characterization of Pex18p and Pex21p, a structurally and functionally related pair of peroxins that interact with Pex7p, and are specifically required for both peroxisomal biogenesis of Pex7p and PTS2 targeting.
Materials and Methods
Yeast Strains, Media, and Growth Conditions
Yeast strains are shown in Table I. For transformation, genomic DNA extraction, and preparation of yeast proteins for affinity chromatography (see below), yeast were grown in synthetic complete (SCD) medium (0.67% yeast nitrogen base, 2% dextrose supplemented with amino acids, uracil and adenine) to a density of 2 × 107 cells/ml. For two-hybrid library screening, bait plasmid was introduced first, and these cells were grown in SCD medium lacking tryptophan (to maintain the plasmid) to a density of 2 × 107 cells/ml, then diluted fourfold into YPD (1% yeast extract, 2% peptone, 2% dextrose) and regrown to 2 × 107 cells/ml before transformation with library DNA. For induction of peroxisomes, cells were grown as previously described (Purdue and Lazarow, 1996), except that preculturing was routinely performed in SCD lacking appropriate amino acids and/ or uracil, to maintain plasmids introduced into the yeast cells, before growth in YPGO medium (1% yeast extract, 2% peptone, 3% glycerol, 0.1% oleic acid, 0.25% Tween 40) or SCEO (0.67% yeast nitrogen base, 2% ethanol, 0.1% oleic acid, 0.25% Tween 40 supplemented with amino acids, uracil and adenine) for 18 h. Growth with oleic acid as sole carbon source was in SCO (0.67% yeast nitrogen base, 0.1% oleic acid, 0.25% Tween 40 supplemented with amino acids, uracil and adenine) liquid and solid media.
Table I.
Yeast Strains Used in This Study
| W303 | MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112 | |
| W303Δpex18 | MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112, pex18::TRP1 | |
| W303Δpex21 | MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112, pex21::URA3 | |
| W303Δpex18Δpex21 * | MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112, pex18::TRP1, pex21::URA3 | |
| W303Δpot1 | MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112, pot1::HIS3 | |
| PJ69-4A | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacz | |
| PJ69-4AΔpex7 | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacz, pex7::HIS3 | |
| PJ69-4AΔpot1 | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacz, | |
| pot1::HIS3 | ||
| PJ69-4AΔpex18Δpex21 | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, | |
| met2::GAL7-lacz, pex18::TRP1, pex21::URA3 | ||
| PJ69-4AΔpex13 | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, | |
| met2::GAL7-lacz, pex13::URA3 | ||
| PJ69-4AΔpex14 | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, GAL2-ADE2, LYS2::GAL1-HIS3, | |
| met2::GAL7-lacz, pex14::URA3 |
For expression of URA3 based plasmids in W303Δpex18Δpex21, the URA3 gene integrated at the PEX21 locus was first disrupted with HIS3, using the marker-swap plasmid pUH4 (Cross, 1997).
Two-Hybrid Library Screening
Cells were grown as described above. To maximize transformation efficiencies, cells pretransformed with bait were processed using a carefully optimized transformation procedure (Gietz and Schiestl, 1995). Transformation efficiency was assessed by plating small aliquots onto SCD plates lacking tryptophan and leucine (to select for bait and library plasmids, respectively), and the remainder was screened on SCD plates lacking tryptophan, leucine, and adenine. Growth on these plates indicated the presence of bait and library plasmids and production of a functional Gal4p hybrid, since the ADE2 gene in PJ69-4A (which lacks Gal4p) is under the control of the Gal4p dependent GAL2 promoter (see Table I; James et al., 1996). Positives were rescreened for activation of expression of HIS3 (by plating on media lacking tryptophan, leucine and histidine) and β-galactosidase (by enzymatic assays [Ausubel et al., 1987] on glass-bead homogenates), which are under the regulation of the alternative Gal4p-dependent promoters GAL1 and GAL7 in PJ69-4A, and therefore serve as additional markers of interaction (James et al., 1996). To isolate library plasmids from positive clones, cells were grown in SCD liquid media lacking leucine (to induce loss of bait, but not library plasmid), and plasmid DNA was prepared and transformed at low dilution into competent E. coli XL1-Blue cells for amplification and purification. Dideoxy sequencing was performed using oligonucleotides GAD424A (5′-TCGATGATGAAGATACCCC-3′) and GAD424B (5′-GCACGATGCACAGTTGAAGTG-3′), which flank the cloning site of the library vectors. Clone identification was performed by searching the entire yeast genome database with these sequences using BLAST.
Plasmids, Cloning, and Gene Disruption
After determination that their gene products were peroxins, yeast open reading frames YHR160C and YGR239C were assigned the names ScPEX18 and ScPEX21, respectively, and their gene products are referred to as ScPex18p and ScPex21p.
PEX18 was PCR amplified from W303 (wild-type) genomic DNA with primers that map to the extreme 5′ end of the PEX18 coding region (5′-GGATCCAAACAATGAATAGTAAC-3′; initiation ATG is underlined) and 49-28 nucleotides 3′ to the termination codon (5′-GTTATATATCTGAAATTCATGG-3′). The PCR product was cloned, sequenced, and subcloned under control of the POX1 promoter of the acyl-CoA oxidase gene into plasmids pRS315 (Sikorski and Hieter, 1989) and Yep351 (Hill et al., 1986) to produce pYcp-PEX18 and pYep-PEX18, respectively. PEX21 was amplified in a similar way, using PCR primers mapping to the extreme 5′ end of the PEX21 coding region (5′-TAGAATACCATGCCCAGTGTCT-3′; initiation ATG is underlined) and 397-378 nucleotides 3′ to the stop codon (5′-AGCAAGCAACACCAAAACCA-3′) and cloned to produce pYcp-PEX21 and pYep-PEX21. Epitope tagging of PEX18 and PEX21 was achieved by replacing the downstream PCR primers with primers designed to extend the reading frames of PEX18 (5′-TGGGACGTCGTATGGGTAAGCAATTCTGTCTTCAACATC-3′) and PEX21 (5′-TGGGACGTCGTATGGGTAATCAAGTATGTCTTTGTGAAT-3′) with a partial copy of the sequence encoding the hemagglutinin nonapeptide epitope (YPYDVPDYA), incorporating an AatII restriction site positioned to allow subsequent in-frame cloning of an AatII fragment encoding the remaining portion of the triple HA epitope tag. Cloning into yeast expression vectors was as for the untagged genes. His6-tagged PEX18 and PEX21 bacterial overexpression constructs were generated by subcloning the two-hybrid library clone inserts encoding the COOH-terminal 176 residues of Pex18p (as a BamHI-BglII fragment) into BamHI-digested pPET15b (to create pPET15b-PEX18), and the COOH-terminal 96 residues of Pex21p (as a SmaI-SmaI fragment) into pPET15b digested with XhoI and filled in with Klenow DNA Polymerase and deoxynucleotides (to create pPET15b-PEX21).
For gene disruption, pPEX18::TRP1 was generated by replacement of an EcoRI-EcoRI fragment of PEX18 (coding nucleotides 119–797) with an EcoRI-EcoRI fragment carrying the TRP1 gene. pPEX21::URA3 was prepared by replacement of an EcoRI-NsiI fragment (coding nucleotides 363-end) of PEX21 with an EcoRI-PstI fragment carrying the URA3 gene. A BamHI-SalI fragment of pPEX18::TRP1 and a HindIII-EcoRI (partial) fragment of pPEX21::URA3 were purified after digestion, and used for transformation, individually or consecutively of W303 and PJ69-4A. pPEX7::HIS3, for the disruption of PEX7 from PJ69-4A, was made by replacing a BglII-BglII portion of PEX7, which includes the entire open reading frame, with a BamHI-BamHI fragment containing HIS3. pPOT1:: HIS3, for the disruption of thiolase from PJ69-4A and W303, was made by replacing an SphI-StuI portion of POT1, which includes the promoter and first 776 coding nucleotides, with an SphI-SmaI fragment containing HIS3. pPEX13::URA3 and pPEX14::URA3, for the disruption of PEX13 and PEX14 from PJ69-4A, were made by replacement of a KpnI-SalI fragment of PEX13 (encoding residues 69–145) and a BclI-XhoI fragment of PEX14 (encoding residues 140-201) with URA3-containing KpnI-SalI and BamHI-SalI fragments, respectively. All disruptions were confirmed by PCR on genomic DNA.
For construction of the two-hybrid bait plasmid pGBT9-PEX7, an NcoI site was introduced around the PEX7 initiation codon by PCR, and the entire PEX7 coding region was then purified as an NcoI(filled in)- BamHI(partial) digestion product, which was then cloned into the SmaI-BamHI sites of pGBT9 to produce an in-frame fusion with the Gal4p DNA-binding domain. pGBT9-thio (complete thiolase coding sequence) and pGBT9-Δthio (thiolase coding sequence lacking amino acids 1–16, which constitute the PTS2) were generated by subcloning the appropriate regions of thiolase into pGBT9.
Plasmid pChAT-KANL, encoding a version of bacterial chloramphenicol acetyltransferase appended at the COOH terminus with −KANL, which is targeted to yeast peroxisomes in a Pex5p-dependent fashion, has been described previously (Purdue and Lazarow, 1996), as have the plasmids encoding epitope-tagged Pex7p at high (pBXNPEB1-HA3 in Zhang and Lazarow [1994], now referred to as pBXNPEX7-HA3) and low (YipPEB1-HA3 in Zhang and Lazarow [1994], now referred to as YipPEX7-HA3) expression levels, and the plasmid encoding a fusion between the NH2-terminal 16 amino acid PTS2 of S. cerevisiae thiolase and GFP (PTS2-GFP; Huang and Lazarow, 1996). The plasmid encoding acyl-CoA oxidase fused to GFP was constructed by cloning the POX1 gene (complete with its promoter) into the episomal plasmid Yep351 (Hill et al., 1986), followed by replacement of the termination codon with an AatII restriction site by site-directed mutagenesis, digestion with AatII, blunt-ending, and in-frame cloning of a blunt-ended XhoI-SacI fragment containing GFP.
Bacterial Overexpression and Affinity Chromatography
E. coli BL21(DE3) cells were transformed with plasmids pPET15b-PEX18 and pPET15b-PEX21, grown to an OD600 of 0.8, and then grown for an additional three hours in the presence of 1 mM IPTG. Cells were then harvested, resuspended in Binding Buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.9), sonicated, and centrifuged at 14,000 g for 10 min at 4°C. The resultant supernatant was loaded onto His•Bind Resin columns (Novagen Inc., Madison, WI), which were then washed extensively before recovery of bound proteins in elution buffer (1 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.9), in accordance with the manufacturer's recommendations. To test the binding of Pex7p to Pex18p/Pex21p, yeast cells expressing epitope tagged Pex7p were homogenized (Thieringer et al., 1991) in Binding Buffer (containing protease inhibitors) and loaded at a concentration of 5 mg protein/100 μl column bed volume onto His•Bind Resin columns that had been preloaded with either Pex18p, Pex21p, or no bait, and extensively washed. The unbound yeast proteins were recovered, and the columns were then rewashed and eluted as usual. Equivalent proportions of the unbound and eluted protein fractions were then analyzed by SDS–polyacrylamide gel electrophoresis followed by either Coomassie blue staining or immunoblotting.
Miscellaneous
Standard techniques of yeast genetics (Sherman et al., 1986) and molecular biology (Ausubel et al., 1987) were used throughout. Cellular fractionation, density gradient centrifugation, preparation of glass-bead homogenates, ECL immunoblotting, immunoprecipitation, and immunofluorescence were as described previously (Thieringer et al., 1991; Zhang and Lazarow, 1996; Purdue and Lazarow, 1996). Protease protection assays using Proteinase K-agarose beads were as described (Zhang and Lazarow, 1996), except that incubations were at 4°C and were performed on purified peroxisomes as well as crude organellar pellets.
Results
Pex18p and Pex21p, Novel Peroxins Capable of Interacting with ScPex7p: Two-Hybrid Analysis
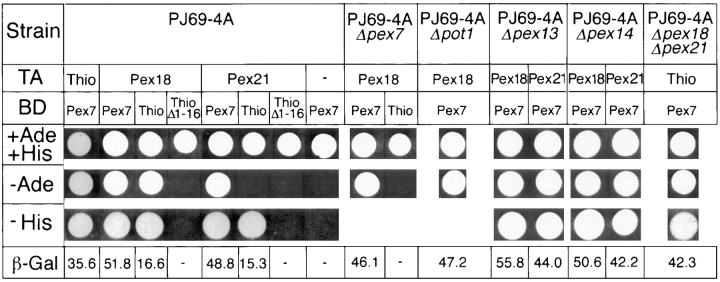
The two-hybrid host strain PJ69-4A and yeast genomic libraries representing all three reading frames, cloned in derivatives of the two-hybrid Gal4p transactivation domain plasmid pGAD424, were generously supplied by Dr. Philip James (University of Wisconsin; James et al., 1996). PJ69-4A represents an improved strain for two-hybrid screens, in that it contains three distinct inducible markers, each regulated by a different Gal4p-responsive promoter (see Materials and Methods). We and others have found the GAL2-ADE2 marker to be the most stringent, giving very few (if any) false positives, whereas the GAL1-HIS3 and GAL7-lacz markers are more sensitive (see Fig. 1) but also give many more false positives. Therefore, it was determined that library screening should be carried out on adenine omission plates, with positives subsequently being screened for HIS3 and lacz expression. To screen for Pex7p interacting proteins, the full-length ScPEX7 coding sequence (Zhang and Lazarow, 1994) was cloned as an in-frame fusion with the Gal4p DNA-binding domain in plasmid pGBT9 (Clontech Laboratories, Palo Alto, CA) to create the “bait” plasmid pGBT9-PEX7. To validate the usefulness of this bait in PJ69-4A, it was initially tested for interaction with thiolase, which has previously been shown to interact with Pex7p in two-hybrid analyses (Rehling et al., 1996; Zhang and Lazarow, 1996). As expected, interaction of bait with full-length thiolase resulted in an Ade+, His+, β-gal+ phenotype (Fig. 1). Deletion of the thiolase PTS2 (amino acids 1–16) abrogated activation of all three of these markers. For screening, PJ69-4A was transformed first with pGBT9-PEX7, and then with the transactivation domain libraries. 2 × 106 transformants were obtained for each library, and these were plated directly onto adenine omission plates. A total of 11 Ade+ colonies were identified, all of which were then shown also to display activation of the HIS3 and β-galactosidase genes. The library plasmids were rescued from these Ade+ clones, propagated in bacteria, purified and retransformed into PJ69-4A with and without bait, to confirm that interaction was dependent upon the simultaneous presence of both fusions. The library plasmids were sequenced, revealing that three different yeast genes, each encoding a protein of unknown function, were represented amongst the eleven positive clones. One of these genes, represented by a single positive, will be discussed in a later communication. 6 of the other 10 positive clones were for gene YHR160C, which encodes a protein of 283 amino acids, and included fusions between the Gal4p transactivation domain and the COOH-terminal 176 (5 clones) or 200 (1 clone) residues. The remaining four clones were for gene YGR239C, which encodes a protein of 288 amino acids, and included fusion between the Gal4p transactivation domain and the COOH-terminal 134 (3 clones) or 96 (1 clone) residues. Based on the data presented in this paper regarding the involvement of these genes in peroxisomal biogenesis, YHR160C and YGR239C will be referred to hereafter as PEX18 and PEX21, respectively. The interactions between Pex7p bait and one library clone for each of Pex18p and Pex21p are illustrated in Fig. 1.
Figure 1.
Pex18p and Pex21p interact with Pex7p in the yeast two-hybrid assay. Host strain PJ69-4A was transformed with two plasmids, one encoding the Gal4p transactivation domain (TA) alone (−) or fused to either residues 108–283 of Pex18p (Pex18), residues 155-288 of Pex21p (Pex21), or full-length thiolase (Thio), and the other encoding the DNA binding domain (BD) of Gal4p fused to full-length Pex7p (Pex7), full-length thiolase (Thio) or thiolase lacking the NH2-terminal 16 amino acids (ThioΔ1-16). Transformants were tested for growth on synthetic complete medium with (+Ade +His) or without adenine (−Ade) or histidine (−His). Total cell homogenates were also prepared, and assayed for β-galactosidase activity (β-Gal), which is expressed in nmol/min/mg protein. − = <0.5 units of activity. Some experiments were performed using PJ69-4A from which the PEX7, POT1, PEX13, PEX14, or PEX18/PEX21 genes had been deleted (PJ69-4AΔpex7, PJ69-4AΔpot1, PJ69-4AΔpex13, PJ69-4AΔpex14, and PJ69-4AΔpex18Δpex21, respectively). PJ69-4AΔpex7 and PJ69-4AΔpot1 transformants were not tested for activation of the GAL1-HIS3 marker, since these strains had been converted to His+ during gene disruption.
Since Pex7p has previously been shown to interact not only with the PTS2 of thiolase (Rehling et al., 1996; Zhang and Lazarow, 1996; Elgersma et al., 1998), but also with the peroxins Pex13p (Girzalsky et al., 1998) and Pex14p (Albertini et al., 1997; Brocard et al., 1997; Erdmann et al., 1997), we extended our two-hybrid analysis to ask whether these proteins may have an involvement in the observed interaction between Pex7p and Pex18p/Pex21p. To do this, the genes encoding thiolase, Pex13p, and Pex14p, were each deleted from the two-hybrid host PJ69-4A to create strains PJ69-4AΔpot1, PJ69-4AΔpex13 and PJ69-4AΔpex14, respectively, and the Pex7p-Pex18p/Pex21p interactions were reassessed in these disruption strains. As shown in Fig. 1, none of these deletions had any observable effect on the Pex7p-Pex18p/Pex21p interactions.
In Vitro Binding
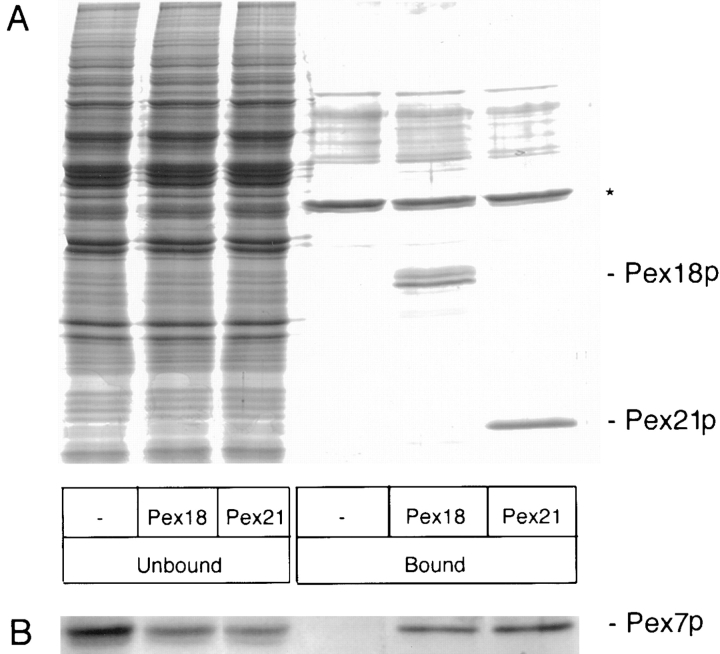
To complement the in vivo two-hybrid interaction analyses, we sought to investigate the ability of Pex7p to bind Pex18p and Pex21p in vitro. To achieve this, the COOH-terminal regions of Pex18p and Pex21p that interact with Pex7p in the two-hybrid system were overexpressed as polyhistidine (His6) tagged proteins in bacteria, and subsequently purified on polyhistidine affinity columns. The His6-tagged proteins were specifically retained on the columns, to which were then added protein extracts from yeast cells expressing hemagglutinin epitope-tagged Pex7p. Coomassie blue staining (Fig. 2 A) revealed that almost all of the yeast proteins failed to bind to the columns to any great extent, and none showed any discernible difference in binding to columns loaded with Pex18p, Pex21p, or neither. However, as shown in Fig. 2 B, immunoblot analysis revealed that the epitope-tagged Pex7p was specifically retained on the Pex18p and Pex21p columns, but not on columns lacking either of these peroxins. This supports the two-hybrid data indicating that Pex18p and Pex21p can specifically interact with Pex7p.
Figure 2.
Pex18p and Pex21p interact with Pex7p in vitro. Bacterial extracts from E. coli BL21 (DE3) cells overexpressing His6-tagged Pex18p (Pex18), Pex21p (Pex21), or neither (−) were first applied to His6-affinity columns, which were then loaded with glass bead homogenates of yeast expressing epitope tagged Pex7p (plasmid pBXNPEB1-HA3 in Zhang and Lazarow [1994], now referred to as pBXNPEX7-HA3). The unbound yeast proteins were recovered and, after extensive washing, the bound proteins were eluted together with the His6-tagged Pex18p/Pex21p. Aliquots of the unbound and bound fractions were resolved in duplicate by SDS PAGE, and subjected to (A) Coomassie blue staining, which detected the eluted Pex18p (which migrates as a dimer) and Pex21p, and (B) immunoblotting with monoclonal antibody 12CA5 to detect the epitope-tagged Pex7p. An abundant unidentified yeast protein that adheres to the columns even in the absence of His6-tagged bait is marked by an asterisk.
Pex18p and Pex21p Are Weakly Homologous to Each Other
Sequence comparison of Pex18p and Pex21p revealed a weak but significant level of homology (Fig. 3), raising the possibility that the ability of both these proteins to interact with Pex7p may result from their sharing a common structural domain. Whereas the observed homology is distributed throughout the protein sequences (23% identity overall), it is most prominent at the COOH termini (35% identity in the last 60 amino acids). This conserved COOH- terminal domain most likely represents the region of Pex18p/ Pex21p responsible for interaction with Pex7p, since it is present in all the PEX18 and PEX21 library clones isolated with the PEX7 bait, and the shortest interacting clone contains only the last 96 amino acids of Pex21p.
Figure 3.
Bestfit alignment of the protein sequences of Pex18p and Pex21p. The arrowheads indicate the fusion junctions between Gal4p transactivation domain and COOH-terminal regions of Pex18p and Pex21p of the clones isolated from two-hybrid libraries with Pex7p bait.
Pex18p and Pex21p Constitute a Peroxin Family Essential for PTS2 Targeting and Growth on Oleate Medium
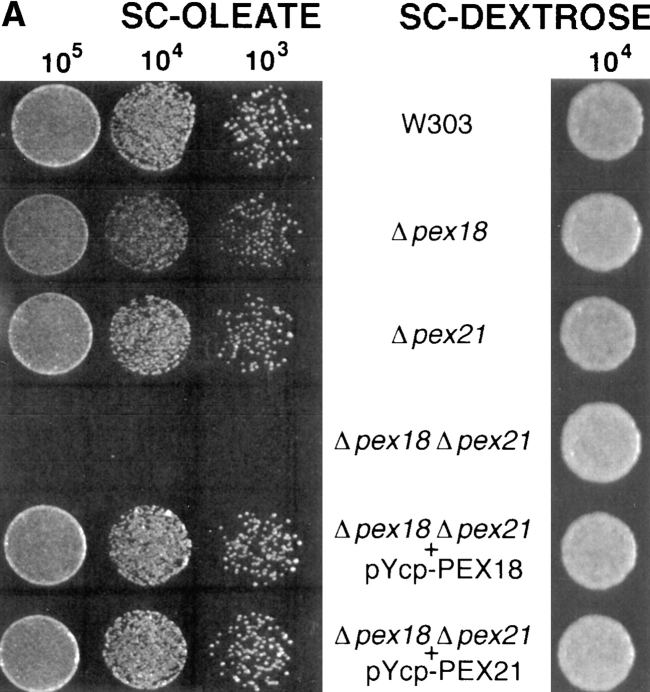
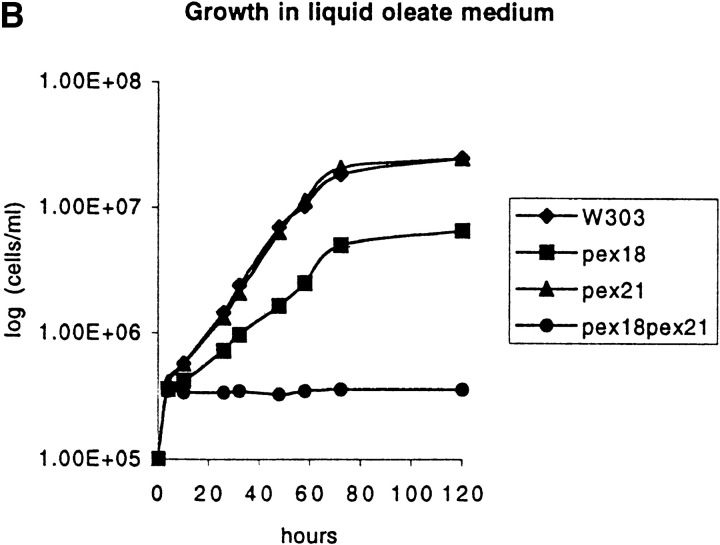
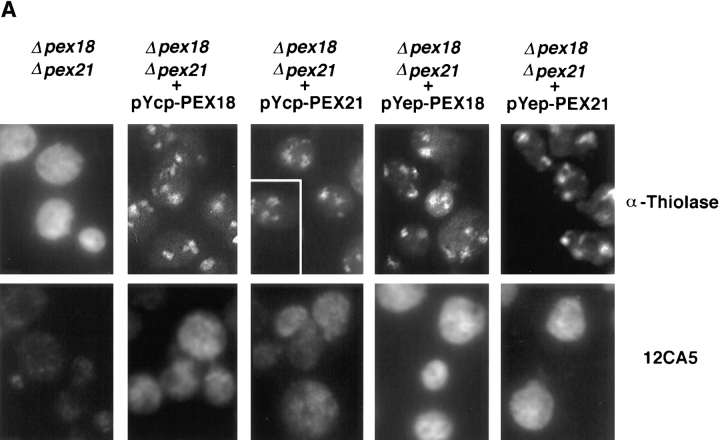
To investigate the roles of Pex18p and Pex21p in peroxisomal biogenesis, the genes encoding these proteins were disrupted, both individually and together, from wild-type strain W303. The disruption strains were initially tested for growth on plates containing oleate as sole carbon source, which will support cell growth only if peroxisomal β-oxidation is functional. Of the single gene disruptions, W303 Δpex21 grew at a rate indistinguishable from wild-type, whereas W303Δpex18 cells grew somewhat more slowly (Fig. 4, A and B). However, the double disruption strain (W303Δpex18Δpex21) showed no growth whatsoever on this medium (Fig. 4, A and B). To investigate the biochemical basis for this growth defect, we next looked at the subcellular distribution of thiolase, since this is a key enzyme of peroxisomal β-oxidation, and is targeted to peroxisomes by the Pex7p-dependent PTS2 targeting pathway (Marzioch et al., 1994; Zhang and Lazarow, 1994). Both by indirect immunofluorescence and subcellular fractionation analyses, peroxisomal packaging of thiolase appeared unperturbed in W303Δpex21 (Fig. 4 C). However, in strain W303Δpex18 ∼50% of thiolase was mistargeted to the cytosol, as assessed by fractionation, and fluorescence was distributed throughout the cytosol (presumably masking the signal from the residual peroxisomal thiolase). Finally, W303Δpex18Δpex21 showed complete redistribution of thiolase to the cytosol (Fig. 4 C). These results strongly indicated that Pex18p and Pex21p were specifically involved in PTS2 targeting. To test this further, we investigated the subcellular distribution in these strains of a plasmid encoded fusion between the PTS2 of S. cerevisiae thiolase (amino acids 1–16) and GFP (PTS2-GFP), under the regulation of the thiolase promoter. The targeting of this fusion protein closely resembled that of thiolase, with predominantly peroxisomal fluorescence seen in wild-type and W303Δpex21, but cytosolic fluorescence in W303Δpex18 and W303Δpex18Δpex21 (Fig. 4 C). Whereas no punctate fluorescence at all was detected in W303Δpex18Δpex21, some W303Δpex21 cells showed a mixture of cytosolic and punctate labeling, consistent with the partial thiolase mistargeting phenotype of this strain.
Figure 4.
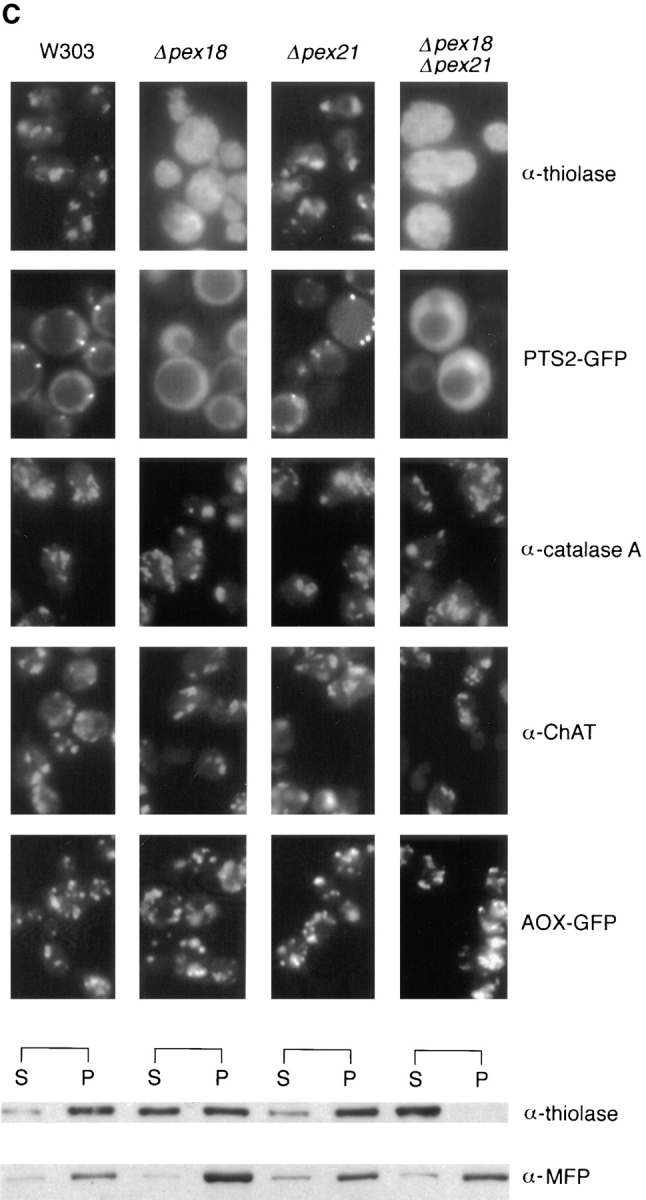
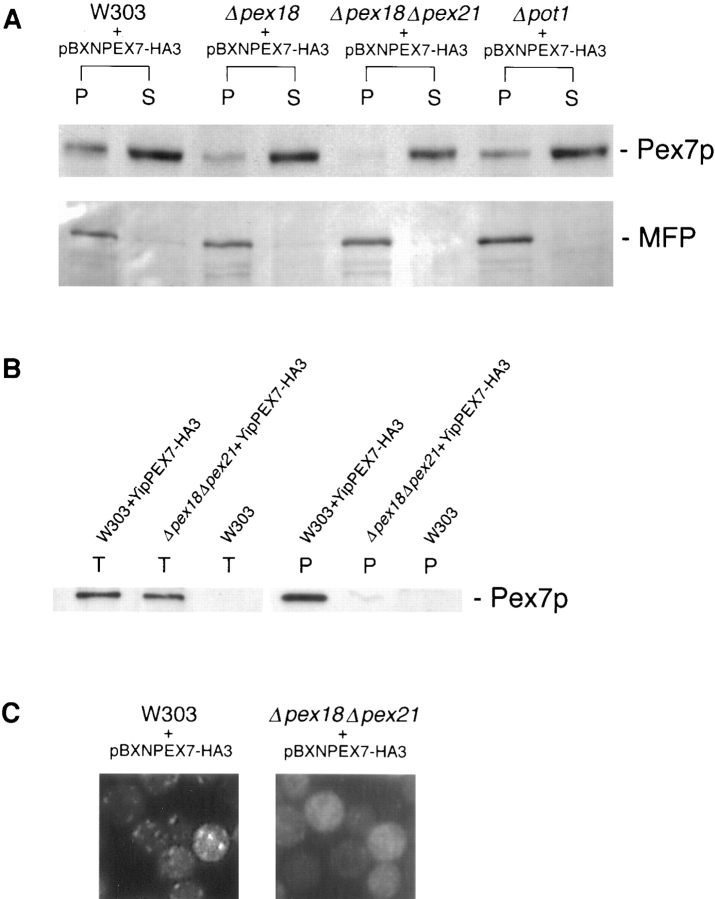
Growth on oleic acid and packaging of thiolase into peroxisomes are abolished in cells lacking both Pex18p and Pex21p, but are supported fully by Pex18p alone, and partially by Pex21p alone. (A) Growth on plates. Cells were grown to mid-log phase in liquid synthetic complete medium, and then applied at various dilutions (105, 104, or 103 cells per spot) to plates containing synthetic complete medium with either oleic acid (SC-OLEATE) or dextrose (SC-DEXTROSE) as sole carbon source, and grown at 30°C. The bottom two panels show the double knockout strain transformed with centromeric plasmids encoding epitope tagged Pex18p (pYcp-PEX18) or Pex21p (pYcp-PEX21). (B) Growth in liquid oleate media. Cells were precultured to mid-log phase in SCD medium and then inoculated at a density of 105 cells/ml into liquid SCO. (C) Fluorescence and cell fractionation. Wild-type cells, and cells lacking Pex18p and/ or Pex21p, were grown for peroxisome induction, and analyzed by immunofluorescence with antibodies against thiolase (top row) and catalase A (third row). These same strains were transformed with plasmids expressing chloramphenicol acetyl transferase (ChAT) appended with the PTS1 −KANL (Purdue and Lazarow, 1996), or a fusion between the thiolase PTS2 and GFP (PTS2-GFP; Huang and Lazarow, 1996), or a fusion between acyl-CoA oxidase and GFP (AOX-GFP), and analyzed with anti-ChAT antibodies (fourth row), or by GFP autofluorescence (second and fifth rows). Induction was with YPGO in all cases except for PTS2-GFP fluorescence, when SCEO was used. For fractionation, cells were grown for peroxisome induction in YPGO, and fractionated into postnuclear pellet (P) and supernatant (S) fractions, which were run in equivalent proportions on SDS-polyacrylamide gels, and immunoblotted to detect thiolase and the β-oxidation multifunctional protein (MFP). All the strains are in a W303 genetic background.
Targeting by PTS1 or the PTS of Acyl-CoA Oxidase Do Not Require Pex18p/Pex21p
To determine whether the involvement of Pex18p/Pex21p in peroxisomal biogenesis extends beyond the PTS2 branch of peroxisomal biogenesis, the disruption strains were assessed for packaging of other peroxisomal proteins. These studies revealed that targeting via the PTS1 pathway, as represented by the multi-functional β-oxidation protein (ends −SKL; Hiltunen et al., 1992), catalase A (ends −SKF; Kragler et al., 1993) and a plasmid-encoded fusion between ChAT and the human catalase PTS1 (−KANL; Purdue and Lazarow, 1996), was not affected by the single or double disruptions (Fig. 4 C). Similarly, targeting of acyl-CoA oxidase (AOX), which is believed to contain a yet-to-be-defined PTS3 signal (since it lacks both PTS1 and PTS2; Dmochowska et al., 1990), was normal in all strains, as assessed by fluorescence analysis of cells expressing a fusion between AOX and GFP which is targeted efficiently to peroxisomes (Skoneczny, M., P.E. Purdue, and P.B. Lazarow, manuscript in preparation).
Pex18p/Pex21p Are Required for Peroxisomal Targeting of Epitope-tagged Pex7p
Reports from several laboratories have concluded that yeast Pex7p is associated, at least in part, with peroxisomes (Marzioch et al., 1994; Zhang and Lazarow, 1994; Elgersma et al., 1998). However, observations conflict over whether this protein is an exclusively intra-peroxisomal import receptor, or has a substantial presence and function in the cytosol. Data from our laboratory indicate that, in S. cerevisiae, Pex7p tagged at the COOH terminus with three repeats of the hemagglutinin nonapeptide recognized by monoclonal antibody 12CA5 (Pex7p-HA3) is, within experimental error, confined to the peroxisomal matrix when expressed at wild-type levels, but also accumulates in the cytosol when overexpressed (Zhang and Lazarow, 1994). Since these results suggest that Pex7p may function within the peroxisomal matrix, we proceeded to investigate the distribution of this same epitope-tagged Pex7p in the W303Δpex18 and W303Δpex18Δpex21 strains, which show deficient PTS2 targeting.
Subcellular fractionation analysis revealed that the peroxisomal localization of Pex7p-HA3 seen in wild-type cells was essentially absent in W303Δpex18Δpex21 cells, with only a trace detectable in the organellar pellet (Fig. 5). This dependence upon Pex18p/Pex21p for the presence of Pex7p in peroxisomes was found not only when Pex7p-HA3 was expressed at wild-type levels (Fig. 5 B), but also when it was markedly overexpressed (Fig. 5, A and C). Consistent with the intermediary PTS2 targeting defect seen in W303Δpex18, this strain showed a level of peroxisomal targeting of Pex7p-HA3 significantly lower than wild-type, but higher than seen in W303Δpex18Δpex21 (Fig. 5 A). As previously reported, absence of thiolase did not affect the intracellular distribution of Pex7p (Fig. 5 A).
Figure 5.
Peroxisomal targeting of Pex7p is somewhat decreased in cells lacking Pex18p, and almost abolished in cells lacking both Pex18p and Pex21p. (A) Strains overexpressing epitope tagged Pex7p (plasmid pBXNPEB1-HA3 in Zhang and Lazarow [1994], now referred to as pBXNPEX7-HA3) were fractionated into postnuclear pellet (P) and supernatant (S) fractions, which were then analyzed by SDS-polyacrylamide gels and immunoblotting with monoclonal antibody 12CA5 (to detect the tagged Pex7p) and antibodies against the β-oxidation multifunctional protein (MFP). (B) Immunoprecipitation of epitope tagged Pex7p expressed at wild-type levels (plasmid YipPEB1-HA3 in Zhang and Lazarow [1994], now referred to as YipPEX7-HA3). Total cell extracts (T) and organellar pellets (P) from YPGO-induced cultures of W303, W303-expressing tagged Pex7p, and W303Δpex18Δpex21-expressing tagged Pex7p were separated on SDS-polyacrylamide gels, and immunoblotted with monoclonal antibody 12CA5. (C) Strains overexpressing epitope tagged Pex7p were analyzed by immunofluorescence with 12CA5.
Pex7p Can Interact with the Thiolase PTS2 and Pex18p Simultaneously
Since Pex18p and Pex21p participate in the PTS2 peroxisomal targeting pathway, we next tested whether they were capable of two-hybrid interaction with thiolase, which contains an NH2-terminal PTS2. To do this, the PEX18 and PEX21 clones previously isolated from the two-hybrid libraries were each cotransformed into PJ69-4A with plasmids encoding full-length thiolase, or thiolase lacking the NH2-terminal 16 amino acid PTS2, fused to the Gal4p DNA-binding domain, and plated on media lacking adenine or histidine. Interestingly, full-length thiolase exhibited a clear interaction with Pex18p (Ade+, His+, β-gal+), and a somewhat lesser interaction with Pex21p (His+, β-gal+), whereas truncated thiolase showed no interaction (Fig. 1). These results suggested that Pex18p/Pex21p may be capable of interaction with the PTS2 of thiolase. However, an alternative explanation was suggested by the fact that Pex7p has now been observed to interact with both thiolase (Rehling et al., 1996; Zhang and Lazarow, 1996; Elgersma et al., 1998) and Pex18p/Pex21p, raising the possibility that cellular Pex7p is serving as a bridge between the Pex18p/Pex21p and thiolase fusions. Therefore, these transformations were repeated in PJ69-4A from which the PEX7 gene had been disrupted (PJ69-4AΔpex7). In this strain, no interactions were detected, suggesting that the Gal4p activation seen in the full-length thiolase cotransformants probably results from simultaneous interaction of cellular Pex7p with both thiolase and Pex18p (or Pex21p) fusions, although the presence of other bridging proteins can not be ruled out at this stage. Consistent with this, the Pex7p-Pex18p/Pex21p interaction was not affected by disruption of the thiolase gene, and the Pex7p-thiolase interaction was not altered by disruption of the PEX18 and PEX21 genes (Fig. 1).
Pex18p Localizes to the Cytoplasm and Outer Surface of Peroxisomes
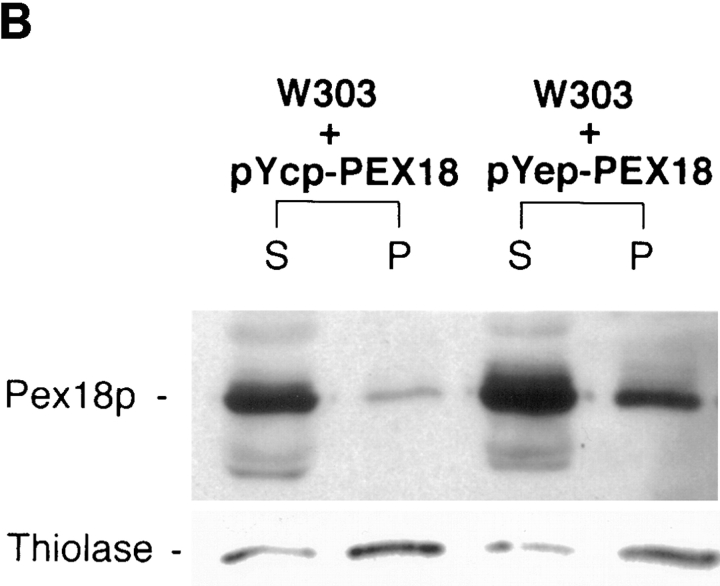
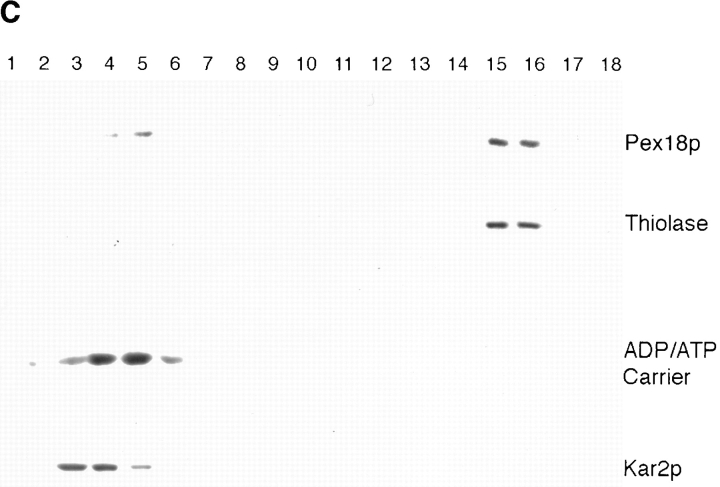
To localize Pex18p and Pex21p within cells, clones encoding these proteins appended at their COOH terminus with three copies of the hemagglutinin nonapeptide epitope were generated and expressed in yeast cells. Both tagged proteins were capable of rescuing oleate growth (Fig. 4 A) and thiolase packaging (Fig. 6 A) in W303Δpex18Δpex21 when expressed from the oleate-inducible POX1 promoter in low-copy plasmids. Fractionation analysis revealed that Pex18p was largely cytosolic, but also present at low levels (<10% of the total) in the organellar pellet (Fig. 6 B). This pattern of distribution was also seen when tagged Pex18p was expressed in cells lacking either thiolase, Pex7p or Pex14p, a putative Pex7p receptor on the external face of the peroxisomal membrane (results not shown). Increasing expression of tagged Pex18p increased the amount of targeting to the organellar pellet, indicating that targeting is not saturated at the lower expression level. Nycodenz gradient fractionation of the organellar pellet further localized the pelletable Pex18p to peroxisomes, although trace amounts also copurified with mitochondria (Fig. 6 C). Protease protection analysis of the organellar pellet (or peroxisomal peak fractions from the gradient) revealed that almost all of the peroxisomal tagged Pex18p is readily proteolytically digested under conditions in which thiolase (a matrix protein) is only digested in the presence of detergent (Fig. 7), although lengthy exposure of the immunoblots did identify a small pool of protected Pex18p. This suggests that peroxisomal Pex18p is primarily located on the external face of the peroxisomal membrane, but that the existence of a small pool of intraperoxisomal Pex18p can not be ruled out. A primarily cytosolic localization of Pex18p was supported by immunofluorescence analysis, which showed fluorescence throughout the cell (Fig. 6 A). A similar immunofluorescence picture was obtained with cells expressing tagged Pex21p (Fig. 6 A), indicating that this protein is also primarily cytosolic, which is not surprising in view of the apparent redundancy of function of Pex18p and Pex21p. However, we were unable to determine whether any Pex21p is organellar, since the tagged Pex21p was highly unstable during cellular fractionation procedures (data not shown).
Figure 6.
Pex18p and Pex21p are functional when COOH terminally tagged with HA epitopes, and localize primarily to the cytosol. Pex18p is partially peroxisomal. (A) Cells lacking both Pex18p and Pex21p (Δpex18Δpex21)before and after transformation with low copy (pYcp-PEX18, pYcp-PEX21) or high copy (pYep-PEX18, pYep-PEX21) plasmids encoding HA epitope-tagged Pex18p or Pex21p were analyzed by immunofluorescence with anti-thiolase (top panels) and 12CA5 (detects the HA-tagged proteins; bottom panels). (B) Wild-type cells expressing epitope-tagged Pex18p from low copy (pYcp-PEX18) and high copy (pYep-PEX18) plasmids were fractionated into organellar pellet (P) and supernatant (S) fractions, which were run in equivalent proportions on SDS-polyacrylamide gels, and immunoblotted with monoclonal 12CA5 to detect Pex18p (top), and anti-thiolase (bottom). (C) The organellar pellet from wild-type W303 cells transformed with plasmid pYep-PEX18 was further fractionated by Nycodenz gradient centrifugation. Fractions were run on a polyacrylamide gel and immunoblotted to detect Pex18p (monoclonal 12CA5), and to detect thiolase, ADP/ATP carrier and Kar2p, which are markers for the peroxisomes, mitochondria and endoplasmic reticulum, respectively. The fractions are labeled from top to bottom of the gradient.
Figure 7.
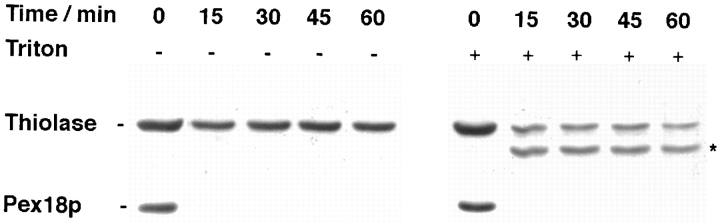
Protease protection analysis shows that peroxisomal Pex18p is primarily located on the external face of the peroxisomal membrane. An organellar fraction of wild-type cells overexpressing HA-tagged Pex18p was digested with a final concentration of 5 μg/ml Proteinase K-agarose in the presence and absence of 0.1% Triton X-100. Reactions were stopped with 1 mM PMSF after various times, and the samples were separated by SDS-PAGE, and immunoblotted to detect thiolase, a known matrix protein, and the tagged Pex18p. A proteolytic thiolase digestion product is indicated by an asterisk.
Discussion
Our current model of peroxisome biogenesis describes a branched pathway of matrix protein import, with each branch representing an import receptor specific for one of several distinct classes of PTS, followed by a shared membrane translocation process. At least three branches exist, two representing the well-characterized COOH-terminal PTS1 and NH2-terminal PTS2 signals, and the other(s) representing proteins lacking either of these, most notably acyl-CoA oxidase. In recent years, much has been learned about PTS2 targeting through the cloning and characterization from several species of Pex7p, an import receptor which interacts with PTS2 sequences and is required for peroxisomal targeting of proteins bearing these signals (Marzioch et al., 1994; Zhang and Lazarow, 1994, 1996; Rehling et al., 1996; Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997; Elgersma et al., 1998). In this paper, we report the identification and initial characterization of Pex18p and Pex21p, two novel S. cerevisiae peroxins required for PTS2 targeting.
Substantial evidence indicates that Pex18p and Pex21p are specifically involved in the PTS2, and no other, branch of peroxisomal protein biogenesis. Most importantly, peroxisomal import of PTS2-targeted thiolase, or of a fusion protein consisting of the yeast thiolase PTS2 (amino acids 1–16) fused to GFP, is completely abolished in cells lacking Pex18p and Pex21p, whereas targeting of PTS1 proteins, and of acyl-CoA oxidase (which has neither PTS2 nor PTS1) fused to GFP, are normal. In addition, Pex18p and Pex21p each interact with Pex7p both in two-hybrid assays and in vitro. Pex7p has been shown to function as a PTS2 receptor, not only in yeast, but also in humans, where its deficiency causes rhizomelic chondrodysplasia punctata (RCDP; Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997). Although thiolase is the only currently known yeast PTS2 protein, other human PTS2 proteins have been identified and shown to be deficient in RCDP (De Vet et al., 1997; Mihalik et al., 1997; Jansen et al., 1997), indicating that Pex7p (and, by extension, Pex18p and Pex21p) mediate targeting not just of thiolase, but of PTS2s in general. It is considerable interest to note that whereas the molecular basis of PTS2 targeting pathway is conserved between S. cerevisiae and humans, current evidence indicates that this may not be the case for Yarrowia lipolytica. In this latter yeast, no orthologue for Pex7p has yet been identified, but a recent report describes a novel peroxin, Pex20p, required for thiolase import (Titorenko et al., 1998). YlPex20p shows no obvious similarity in molecular function to either Pex7p or to the Pex18p/Pex21p peroxins described in this paper. Instead, it functions by binding cytosolic thiolase in a PTS2-independent manner, and mediating the subsequent oligomerization and biogenesis of thiolase. Proteome-wide searches with the Y. lipolytica Pex20p sequence fail to identify any significant candidates for a S. cerevisiae Pex20p orthologue and, while it is too early to exclude the possible existence of ScPex20p, the weight of evidence presently available tends to suggest that the molecular machinery for PTS2/thiolase targeting in S. cerevisiae (and humans) is substantially different from that found in Y. lipolytica.
A number of observations indicate that Pex18p and Pex21p constitute a two-member family of peroxins of related and redundant function. First, as mentioned above, both of these proteins were identified by interaction with the same bait, Pex7p, in a two-hybrid screen, and they both bind epitope-tagged Pex7p in vitro. Second, they are similar in size and display significant sequence homology to each other, but show no striking similarity to any other S. cerevisiae proteins. This homology is distributed throughout their entire lengths, but is particularly noteworthy in the COOH-terminal domain common to all the clones isolated by virtue of interaction with Pex7p. Third, presence of either Pex18p or Pex21p, but not the other, is sufficient to support substantial levels of PTS2 targeting, with abolition of this pathway only occurring when both these proteins are absent.
Although Pex18p and Pex21p share functional redundancy, they appear to make quantitatively different contributions to PTS2 targeting. For example, peroxisomal targeting of thiolase is indistinguishable from wild-type in cells lacking Pex21p, whereas cell lacking Pex18p mislocalize ∼50% of thiolase to the cytosol. Although the respective contributions of Pex18p and Pex21p to PTS2 targeting in wild-type cells have not been established, these data indicate that Pex18p may bear most of the burden. The level of thiolase targeting in Δpex18 cells (and Δpex21 cells) is sufficient to support growth on oleic acid as sole carbon source, which requires functional peroxisomal β-oxidation. This provides an explanation as to why pex18 and pex21 mutants have not been identified by genetic screens for pex mutants unable to use oleic acid. It is, however, interesting to note that Δpex18 cells grow more slowly than wild-type (or Δpex21) cells on oleic acid, suggesting that levels of peroxisomal thiolase may be rate limiting for β-oxidation, although the existence of yet-to-be-identified additional PTS2-targeted proteins required for peroxisomal β-oxidation can not be ruled out (Elgersma et al., 1998).
What might the function(s) of Pex18p and Pex21p be? Data presented in this paper show that peroxisomal targeting of Pex7p is virtually abolished in a strain lacking these peroxins, suggesting that their primary function may be associated with the biogenesis of Pex7p. However, interpretation of this data is complicated by the controversy that surrounds the localization and function of Pex7p. We have previously reported that in S. cerevisiae, Pex7p COOH terminally tagged with HA epitopes is both functional and exclusively peroxisomal (within experimental error) when at wild-type PEX7 expression levels (Zhang and Lazarow, 1994). In contrast, a predominantly cytosolic localization of the S. cerevisiae protein was reported by one group, based on overexpression of Pex7p appended NH2-terminally with a c-myc tag (Marzioch et al., 1994). In a different yeast, P. pastoris, Pex7p was localized to both the peroxisomal matrix and the cytosol (Elgersma et al., 1998). In human cells, we have found that overexpressed Pex7p is partly peroxisomal and partly cytosolic (Purdue, P.E., and Paul B. Lazarow, manuscript in preparation), whereas others report an exclusively cytosolic localization (Braverman et al., 1997). This range of localization results has spawned a variety of hypotheses regarding Pex7p function. Whereas we have proposed that a peroxisomal matrix localization suggests that Pex7p may function as an intraperoxisomal receptor (Zhang and Lazarow, 1994), others have proposed models in which Pex7p may act as a mobile receptor, moving (with PTS2 cargo) from cytosol to either the peroxisomal outer surface, or the peroxisomal matrix, delivering cargo, and recycling for additional rounds of import (Marzioch et al., 1994; Erdmann et al., 1997; Elgersma et al., 1998). Interestingly, in spite of the fundamental differences between these various models, they all stipulate a requirement for Pex7p to move from cytosol to peroxisome, and can therefore all accommodate in principle Pex18p/Pex21p as factors required for this targeting event. Since Pex18p and Pex21p both interact with Pex7p, we can now envisage this pair of peroxins acting as a receptor(s) for targeting of Pex7p from the cytosol to peroxisomes, and that loss of PTS2 targeting in Δpex18 Δpex21 cells results from Pex7p failing to reach its site of action, the peroxisomal matrix (according to the intraperoxisomal receptor model), or failing to cycle between cytosol and peroxisome (according to the mobile receptor model). Localization of Pex18p to the cytosol and peroxisomal surface is consistent with a role for this protein in ferrying Pex7p to the peroxisomal membrane translocation machinery. Pex14p and Pex13p have been identified as peroxisomal membrane proteins capable of interacting with Pex7p (Albertini et al., 1997; Brocard et al., 1997; Erdmann et al., 1997; Girzalsky et al., 1998). These peroxins are not directly involved in the Pex18p-Pex7p and Pex21p-Pex7p interactions, as evidenced by the maintenance of these interactions in two-hybrid strains lacking Pex13p or Pex14p, but they are required for PTS2 targeting, raising the possibility that Pex7p is transferred from Pex18p/ Pex21p to Pex14p and/or Pex13p at the outer face of the membrane. The mobile receptor model suggests that interaction of Pex7p with Pex14p/Pex13p at the external face of the peroxisomal membrane may correspond to a docking event, before recycling of Pex7p to the cytosol, but it seems equally likely that Pex14p/Pex13p may be part of the translocation machinery required for import of Pex7p. It remains to be seen whether yet more peroxins are involved in the PTS2 pathway and the biogenesis of Pex7p. Although all three previously known Pex7p-interacting proteins (thiolase, Pex13p and Pex14p) are dispensable for the Pex7p-Pex18p and Pex7p-Pex21p interactions, it remains a possibility that other, as yet unidentified, peroxins are involved in mediating these interactions.
It is curious to note that, despite screening sufficient library clones with Pex7p bait to cover the entire genome several times over, we did not isolate any interacting library clones for Pex14p, Pex13p, or for any PTS2 proteins, including thiolase. To some extent, this can be accounted for by the intrinsic underrepresentation or absence in two-hybrid libraries of some 5′ coding regions, due to the presence of upstream in-frame termination codons close to translational initiation codons (James et al., 1996). Thus, whereas a transactivation domain plasmid designed to express the extreme NH2-terminal thiolase PTS2 does interact with the Pex7p bait, the existence of such a clone in the two-hybrid libraries is legislated against by the presence of a TAA triplet 15–18 bases upstream of the thiolase (and PTS2) initiation codon (Igual et al., 1991). This explains why thiolase was not identified in our screens, and suggests that the failure to detect other proteins with NH2-terminal PTS2s in our library screens does not necessarily imply that such proteins do not exist, a conclusion consistent with a recent suggestion that Pex11p may be PTS2 targeted (Passreiter et al., 1998). Since the regions of Pex14p and Pex13p which interact with Pex7p have not been mapped, it is unknown whether the failure to detect these proteins can be accounted for in a similar way. Another interesting observation arising from our two-hybrid analysis is that thiolase, fused to the Gal4p DNA-binding domain, interacts with Pex18p (or Pex21p) fused to the transactivation domain, but that this is dependent upon the presence of cellular Pex7p. Disruption of the PEX7 gene from the two-hybrid host strain abolishes this interaction. The most likely explanation of this is that cellular Pex7p, despite its low abundance, can simultaneously bind the thiolase and Pex18p fusions, and thus create a functional Gal4p hybrid, which in turn suggests that the thiolase and Pex18p/ Pex21p binding sites of Pex7p are distinct. A similar scenario has recently been proposed for yeast peroxins Pex5p, Pex7p, Pex14p and Pex17p (Huhse et al., 1998; Girzalsky et al., 1998), where the Pex5p-Pex17p and Pex5p- Pex7p interactions are abolished when PEX14 is disrupted from the two-hybrid host strain. Such findings emphasize that the two-hybrid system can detect indirect, as well as direct, interactions, and while this extends the potential of this type of analysis, it also counsels caution in the interpretation of its results.
Acknowledgments
We would like to thank Dr. Philip James, University of Wisconsin, for supplying the two-hybrid libraries and PJ69-4A strain, and Dr. Marek Skoneczny for construction of the acyl CoA oxidase-GFP fusion expression plasmid.
This work was supported by National Institutes of Health Grant DK19394.
Abbreviations used in this paper
- AOX
acyl CoA oxidase
- GFP
green fluorescent protein
- HA
hemagglutinin
- pex
peroxin
- PTS
peroxisomal targeting sequence
- RCDP
rhizomelic chondrodysplasia punctata
References
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JAKW, Veenhuis M, Kunau WH. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Curr. Prot. Mol. Biol. John Wiley and Sons, Inc., New York. 13.6.1–13.6.4.
- Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D. Human PEX7encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nature Genet. 1997;15:369–376. doi: 10.1038/ng0497-369. [DOI] [PubMed] [Google Scholar]
- Brocard C, Lametschwandtner G, Koudelka R, Hartig A. Pex14p is a member of the protein linkage map of Pex5p. EMBO (Eur Mol Biol Organ) J. 1997;16:5491–5500. doi: 10.1093/emboj/16.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. “Marker swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- De Vet ECJM, Van den Broek BTE, Van den Bosch H. Nucleotide sequence of human alkyl-dihydroxyacetonephosphate synthase cDNA reveals the presence of a peroxisomal targeting signal 2. Biochim Biophys Acta Lipids Lipid Metab. 1997;1346:25–29. doi: 10.1016/s0005-2760(97)00014-3. [DOI] [PubMed] [Google Scholar]
- Dmochowska A, Dignard D, Maleszka R, Thomas DY. Structure and transcriptional control of the Saccharomyces cerevisiae POX1gene encoding acyl-coenzyme A oxidase. Gene. 1990;88:247–252. doi: 10.1016/0378-1119(90)90038-s. [DOI] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet. 1995;9:115–125. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- Elgersma Y, Elgersma-Hooisma M, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. . J Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. The peroxisomal targeting signal of 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. . Yeast. 1994;10:935–944. doi: 10.1002/yea.320100708. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Kunau WH. Peroxisomes: organelles at the crossroads. Trends Cell Biol. 1997;7:400–407. doi: 10.1016/S0962-8924(97)01126-4. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- Girzalsky, W., P. Rehling, J. Kipper, L. Blank, W.-H. Kunau, and R. Erdmann. 1998. Involvement of Pex13p in PTS2-dependent peroxisomal protein import. International Symposium on Peroxisomes: Biogenesis, Function and Disease, Fukuoka, Japan. P19.
- Glover JR, Andrews DW, Subramani S, Rachubinski RA. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. . J Biol Chem. 1994;269:7558–7563. [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. colishuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Wenzel B, Beyer A, Erdmann R, Fossa A, Kunau W-H. Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the FOX2gene and gene product. J Biol Chem. 1992;267:6646–6653. [PubMed] [Google Scholar]
- Huang K, Lazarow PB. Targeting of green fluorescent protein to peroxisomes and peroxisome membranes in S. cerevisiae. Mol Biol Cell. 1996;7(Suppl.):494a. [Google Scholar]
- Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau WH. Pex17p of Saccharomyces cerevisiaeis a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual JC, Matallana E, Gonzalez-Bosch C, Franco L, Perez-Ortin JE. A new glucose-repressible gene identified from the analysis of chromatin structure in deletion mutants of yeast SUC2locus. Yeast. 1991;7:379–389. doi: 10.1002/yea.320070408. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen GA, Ferdinandusse S, Ijlst L, Muijsers AO, Skjeldal OH, Stokke O, Jakobs C, Besley GTN, Wraith JE, Wanders RJA. Refsum disease is caused by mutations in the phytanoyl-CoA hydroxylase gene. Nat Genet. 1997;17:190–193. doi: 10.1038/ng1097-190. [DOI] [PubMed] [Google Scholar]
- Kragler F, Langeder A, Raupachova J, Binder M, Hartig A. Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. . J Cell Biol. 1993;120:665–673. doi: 10.1083/jcb.120.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau W-H. PAS7encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO (Eur Mol Biol Organ) J. 1994;13:4908–4917. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalik SJ, Morrell JC, Kim D, Sacksteder KA, Watkins PA, Gould SJ. Identification of PAHX, a Refsum disease gene. Nature Genet. 1997;17:185–189. doi: 10.1038/ng1097-185. [DOI] [PubMed] [Google Scholar]
- Motley AM, Hettema EH, Hogenhout EM, Brites P, Ten ALMA, Asbroek, Wijburg FA, Baas F, Heijmans HS, Tabak HF, Wanders RJA, Distel B. Rhizomelic chondrodysplasia punctata is a peroxisomal protein targeting disease caused by a non-functional PTS2 receptor. Nat Genet. 1997;15:377–380. doi: 10.1038/ng0497-377. [DOI] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Passreiter M, Anton M, Lay D, Frank R, Harter C, Wieland FT, Gorgas K, Just WW. Peroxisome biogenesis: Involvement of ARF and coatomer. J Cell Biol. 1998;141:373–383. doi: 10.1083/jcb.141.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Peroxisomal biogenesis: multiple pathways of protein import. J Biol Chem. 1994;269:30065–30068. [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J Cell Biol. 1996;134:849–862. doi: 10.1083/jcb.134.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Zhang JW, Skoneczny M, Lazarow PB. Rhizomelic chondrodysplasia punctata is caused by deficiency of human PEX7, a homologue of the yeast PTS2 receptor. Nat Genet. 1997;15:381–384. doi: 10.1038/ng0497-381. [DOI] [PubMed] [Google Scholar]
- Rehling P, Marzioch M, Niesen F, Wittke E, Veenhuis M, Kunau W-H. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7gene. EMBO (Eur Mol Biol Organ) J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 1–186.
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. PEXgenes on the rise. Nat Genet. 1997;15:331–333. doi: 10.1038/ng0497-331. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO (Eur Mol Biol Organ) J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Titorenko VI, Veenhuis M, Rachubinski RA. Pay32p of the yeast Yarrowia lipolyticais an intraperoxisomal component of the matrix protein translocation machinery. J Cell Biol. 1995;131:1453–1469. doi: 10.1083/jcb.131.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlecky SR, Nuttley WM, McCollum D, Sock E, Subramani S. The Pichia pastorisperoxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO (Eur Mol Biol Organ) J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer R, Shio H, Han Y, Cohen G, Lazarow PB. Peroxisomes in Saccharomyces cerevisiae: immunofluorescence analysis and import of catalase A into isolated peroxisomes. Mol Cell Biol. 1991;11:510–522. doi: 10.1128/mcb.11.1.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolyticais required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei IJ, Hilbrands RE, Swaving GJ, Waterham HR, Vrieling EG, Titorenko VI, Cregg JM, Harder W, Veenhuis M. The Hansenula polymorpha PER3gene is essential for the import of PTS1 proteins into the peroxisomal matrix. J Biol Chem. 1995;270:17229–17236. doi: 10.1074/jbc.270.29.17229. [DOI] [PubMed] [Google Scholar]
- Van Der Leij I, Franse MM, Elgersma Y, Distel B, Tabak HF. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1993;90:11782–11786. doi: 10.1073/pnas.90.24.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JW, Lazarow PB. PEB1 (PAS7) in Saccharomyces cerevisiaeencodes a hydrophilic, intraperoxisomal protein which is a member of the WD repeat family and is essential for the import of thiolase into peroxisomes. J Cell Biol. 1994;129:65–80. doi: 10.1083/jcb.129.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JW, Lazarow PB. Peb1p (Pas7p) is an intraperoxisomal receptor for the NH2-terminal, type 2, peroxisomal targeting sequence of thiolase: Peb1p itself is targeted to peroxisomes by an NH2-terminal peptide. J Cell Biol. 1996;132:325–334. doi: 10.1083/jcb.132.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]