Abstract
We find that profilin contributes in several ways to Cdc42-induced nucleation of actin filaments in high speed supernatant of lysed neutrophils. Depletion of profilin inhibited Cdc42-induced nucleation; re-addition of profilin restored much of the activity. Mutant profilins with a decreased affinity for either actin or poly-l-proline were less effective at restoring activity. Whereas Cdc42 must activate Wiskott-Aldrich Syndrome protein (WASP) to stimulate nucleation by the Arp2/3 complex, VCA (verpolin homology, cofilin, and acidic domain contained in the COOH-terminal fragment of N-WASP) constitutively activates the Arp2/3 complex. Nucleation by VCA was not inhibited by profilin depletion. With purified N-WASP and Arp2/3 complex, Cdc42-induced nucleation did not require profilin but was enhanced by profilin, wild-type profilin being more effective than mutant profilin with reduced affinity for poly-l-proline.
Nucleation by the Arp2/3 complex is a function of the free G-actin concentration. Thus, when profilin addition decreased the free G-actin concentration, it inhibited Cdc42- and VCA-induced nucleation. However, when profilin was added with G-actin in a ratio that maintained the initial free G-actin concentration, it increased the rate of both Cdc42- and VCA-induced nucleation. This enhancement, also seen with purified proteins, was greatest when the free G-actin concentration was low. These data suggest that under conditions present in intact cells, profilin enhances nucleation by activated Arp2/3 complex.
Keywords: actin polymerization, nucleation, Cdc42, leukocytes, profilin
Introduction
Neutrophils provide the body's first line of defense against bacterial infection. They are produced in the bone marrow and are carried passively in the circulation throughout the body. At sites of injury or infection, they are activated to move and directed to the site of infection by chemoattractants. This activation is reversible: upon removal of chemoattractant, the cells become round and immobile. Activation involves actin polymerization: 10 s after addition of chemoattractant, the F-actin level doubles (Howard and Oresajo 1985). The F-actin is derived from a reservoir of G-actin bound to thymosin β4 and profilin. This reservoir is large enough that the free G-actin remains fairly constant at ∼0.5 μM during most of the F-actin rise. However, when peak F-actin levels are reached, the free G-actin is calculated to decline to ∼0.15 μM (see Materials and Methods for details).
The chemoattractant-induced F-actin appears to be turning over rapidly, since upon removal of the chemoattractant (or addition of cytochalasin), the concentration of F-actin returns to basal levels, with a half-time between 3 and 10 s (Cassimeris et al. 1990; White et al. 1983). The rapid filament treadmilling is facilitated by both cofilin and profilin. Filament depolymerization is enhanced by cofilin, which increases the pointed-end off-rate, and by profilin, which sequesters G-actin from the pointed end of an actin filament. Profilin also catalyzes the exchange of ADP to ATP–G-actin, which stimulates depolymerization by liberating cofilin (which preferentially binds ADP–G-actin) and polymerization, which is fueled by ATP–G-actin (Blanchoin and Pollard 1998; Didry et al. 1998; Goldschmidt-Clermont et al. 1992; Perelroizen et al. 1996; Pollard and Cooper 1984). Finally, profilin enhances the rate of polymerization by delivering ATP–G-actin to the barbed end of the filament.
However, treadmilling of monomers within a filament probably accounts for a small part of the F-actin dynamics. The increase in F-actin in neutrophils correlates with a rapid increase in filament number (Cano et al. 1991). At steady state, the F-actin turnover probably involves continual nucleation and elongation of new filaments matched by depolymerization and loss of existing filaments. Nucleation of new filaments is much too fast to occur by “spontaneous nucleation” from the free G-actin present. To investigate factors regulating agonist-induced filament nucleation, we have examined Cdc42-induced nucleation in supernatant of lysed neutrophils.
Cdc42, a member of the Rho family of small GTPases, induces actin polymerization in a high speed supernatant of cell extracts (Katanaev and Wymann 1998; Ma et al. 1998a; Moreau and Way 1998; Zigmond et al. 1997). The Cdc42-induced polymerization, like that induced by chemoattractant in intact cells, correlates with an increase in the number of filaments (Zigmond et al. 1998). Each new filament nucleated appears to elongate transiently, the mean filament length, early and late in the time course, being similar. The net increase in filaments results from the rate of filament nucleation exceeding the rate of filament loss.
Cdc42 stimulates actin nucleation through binding to Wiskott-Aldrich Syndrome protein (WASP) or its ubiquitous relative N-WASP (Bear et al. 1998; Machesky and Insall 1998; Machesky et al. 1999; Miki et al. 1998a; Mullins and Pollard 1999; Rohatgi et al. 1999; Winter et al. 1999; Yarar et al. 1999). WASP, in an unstimulated cell, is present in an inactive conformation (Kim et al. 2000; Miki et al. 1998a). Upon binding Cdc42, the conformation of WASP changes and the COOH-terminal VCA (verprolin homology, cofilin homology, and acidic) region is exposed (Kim et al. 2000). The VCA region then activates the Arp2/3 complex to nucleate an actin filament. The Arp2/3 complex–induced nucleation uses F-actin as a cofactor, the new filament at least transiently forming a Y-shaped branch on the existing filament (Machesky et al. 1999).
WASP and N-WASP are also activated by other factors including PIP2, phosphorylation, and clustering (Castellano et al. 1999; Oda et al. 1998; Rohatgi et al. 1999). Finally, WASP contains a proline-rich region that binds a number of SH3 containing proteins including Grb2, several src kinases, and PLCγ (Banin et al. 1996; Bunnell et al. 1996; Finan et al. 1996; Suetsugu et al. 1998). In addition, profilin binds to this proline-rich region. The binding of profilin to proline-rich regions of various proteins including WIP, VASP, and the formin family is thought to contribute to signal transduction to the actin cytoskeleton (Gertler et al. 1996; Lanier et al. 1999; Miki et al. 1998a; Ramesh et al. 1997; Suetsugu et al. 1998; Watanabe et al. 1997). However, the precise role of profilin remains obscure.
In this paper, we focus on the role of profilin in Cdc42–Arp2/3-induced actin nucleation. We show that the presence of profilin is critical for Cdc42-induced nucleation in cell supernatant. We confirm previous studies showing that profilin is not essential for Cdc42-induced nucleation with pure proteins and that addition of profilin can inhibit nucleation (Machesky et al. 1999). We extend these results to show that this inhibition is due to sequestration of free G-actin and that when the free G-actin is not lowered, profilin–actin enhances the rate of nucleation by activated Arp2/3 complex.
Materials and Methods
Reagents
Rabbit pAb, raised against the glutathione S-transferase (GST)–human profilin I or VASP peptide (NH2-CEAFVQELRKRGSP-COOH), were used for immunodepletion of profilin or VASP from supernatants. Recombinant VASP was a gift from Dr. M.-F. Carlier (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). This VASP was able to restore Listeria motility to VASP-depleted supernatants (Laurent et al. 1999). The constructs for the N-WASP, VCA (392–505), and CA (cofilin homology and acidic tail contained in the COOH-terminal fragment of N-WASP) (450–505) fragments were described previously (Miki et al. 1996, Miki et al. 1998a) and the construct of GST–profilin in pGEX2T was a gift from Dr. Y. Takai (Osaka University Medical School, Suita, Japan). We obtained antibodies to WASP from Dr. W. Li (University of Chicago, Chicago, Illinois); to WIP from Dr. N. Ramesh (Harvard Medical School, Boston, Massachusetts); to Arp3 from Dr. L. Machesky (University of Birmingham, Birmingham, UK). Native profilin was isolated from calf spleen by the poly-l-proline (PLP) affinity method (Kaiser et al. 1989; Perelroizen et al. 1994). Thymosin β4 was isolated from calf spleen as described by Cassimeris et al. 1992. The purified Arp2/3 complex isolated from thymus was the gift of Dr. Henry Higgs (Salk Institute for Biological Studies, La Jolla, California). All other reagents were from Sigma–Aldrich unless indicated.
Supernatant of Neutrophil Lysates
High speed supernatant of lysed rabbit peritoneal exudate neutrophils (supernatant) was obtained as described by Zigmond et al. 1997.
Arp2/3 complex was depleted from supernatant using beads coupled to CA (the COOH-terminal fragment of N-WASP containing cofilin and the acidic tail). Supernatant was incubated with GSA–CA beads or control GST beads (1:3, vol/vol) for 1 h on a rocker at 4°C. The beads were removed by centrifugation and fresh beads were added to the supernatant; again at a ratio of 1:3. This was repeated one more time (final of three times).
Profilin was depleted with PLP beads. PLP (10–30 kD; Sigma–Aldrich) was coupled to CNBr–activated Sepharose 4B (Amersham Pharmacia Biotech) according to manufacturer's instructions. After washing with buffer (20 mM Tris-HCl, pH 7.4, 150 mM KCl. and 0.2 mM ATP), PLP beads or control Separose beads were incubated with supernatants (1:3, vol/vol) on a rocker at 4°C for 30 min.
VASP was depleted by immunoprecipitation. Protein A beads were incubated with anti-VASP or 0.1% BSA at 4°C overnight amd then pelleted by centrifugation. After washing with IP buffer (135 mM KCl, 10 mM NaCl, 2 mM MgCl2, 2 mM EGTA, 10 mM Hepes, pH 7.1.), the beads were incubated with supernatants at 4°C for 4 h. The extent of depletion was determined by Western blots with a standard curve derived from serial dilutions of control supernatant run on the same gel and quantified with a PhosphorImager using the ImageQuant program (Molecular Dynamics).
In each case, the concentration of the supernatant was adjusted with IP buffer or concentrated via Centricon 10 (Amicon) to 3 or 4 mg/ml protein for assays of F-actin and nucleation.
Preparation of Profilin Mutants
The site-directed mutation replacing Arg-74 with glutamic acid (R74E mutant) and His-133 with serine (H133S mutant) was performed using QuikChange™ site–directed mutagenesis kit (Stratagene). The mutagenic oligonucleotides (mutated bases are underlined) 5′-GTTCGGTGATCGAGGACTCACTGCTGCAGGATGG-3′ and 5′-GTTATGAAATGGCCTCCTCCCTTCGGCGTTCCC-3′ and their reverse complements were used. The GST–profilin cDNA in pGEX2T (Mammoto et al. 1998) was amplified using Pfu DNA polymerase with these primers for 16 cycles in a DNA thermal cycler (Perkin-Elmer). After digesting parental DNA with DpnI, the amplified mutant DNA was transformed into Escherichia coli (XL 1-Blue strain). The presence of the desired mutations was confirmed by DNA sequencing. The resulting constructs were named R74E and H133S, respectively.
For expression of the GST fusion proteins, E. coli strain BL21 (DE3) was freshly transformed with the DNA constructs. Expression of the proteins was induced with 0.2 mM IPTG at 30°C for 3 h. Cells were then lysed by sonication in 50 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl, 1 mM DTT, 2 μg/ml aprotinin, 2 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride. After centrifugation at 10,000 g for 10 min at 4°C, the GST–profilin was purified by binding to glutathione-Sepharose beads (Amersham Pharmacia Biotech). Profilin was cleaved from the beads with thrombin. After removal of thrombin by benzamidine beads, profilin was dialyzed against 10 mM Tris-HCl, pH 7.6, and 0.1 mM DTT.
The properties of the profilin mutants were characterized by their ability to bind proteins in neutrophil supernatant. Equal amounts (∼100 μg) of GST–H133S profilin and GST–R74E profilin on 50 μl glutathione-Sepharose beads (Amersham Pharmacia Biotech) were incubated with 400 μl supernatant (7 μg/μl) for 2 h at 4°C. After washing with IP buffer, proteins bound to the beads were separated on SDS-PAGE, blotted onto Immobilon-P Transfer Membrane (Millipore), and probed with various antibodies. The affinities of wild-type and H133S profilin for G-actin were both determined to be 0.1 μM by their ability to sequester actin from the pointed end and decrease the concentration of gelsolin-capped F-actin. The H133S mutant was able to support filament elongation as it was comparable to wild-type profilin in stimulating the rate of Listeria movement in platelet extracts (Egile et al. 1999).
Purification of Recombinant Proteins
Recombinant Cdc42 was expressed in a baculovirus insect cell expression system (Heyworth et al. 1993; Xu et al. 1994) and purified as described previously by Zigmond et al. 1997. The purified Cdc42 was charged with GTPγS as described by Knaus et al. 1992. In this manuscript, Cdc42, unless otherwise stated, means GTPγS-activated Cdc42. The Cdc42 used in these experiments, based on protein concentration (Bio-Rad Laboratories), had three– to tenfold lower activity than that used in previous studies reported from this lab (Zigmond et al. 1997, Zigmond et al. 1998). The Cdc42 appeared as a single band on SDS and between 50 to 100% of the Cdc42 bound GTPγS. Partitioning in Triton X-115 suggested that this Cdc42 was less fully prenylated than that used previously. Experiments comparing nucleation activity and the time course of polymerization in supernatant using two different preparations (i.e., old versus new) gave similar results given the three– to tenfold difference in apparent concentration.
Recombinant N-WASP was expressed in a baculovirus insect cell expression system and purified using FPLC on HiTrap-Heparin column according to the published methods of Miki et al. 1998a. GST–VCA and GST–CA fragments of N-WASP were expressed and purified from E. coli according to Miki et al. 1996.
F-actin Determination
F-actin was quantified from TRITC-phalloidin staining of pelleted material as described originally by Howard and Oresajo 1985 and modified slightly according to Zigmond et al. 1998.
Assays of Nucleation Sites in Supernatant
Pyrenyl-actin assays of nucleation were performed as described previously by Cano et al. 1991. In brief, supernatants with or without agonists (Cdc42 or VCA) were incubated for stated times at 37°C and then diluted 100-fold directly into 1.5 μM pyrenylactin. The initial rate of pyrenylactin polymerization was determined from the rate of change of pyrenyl fluorescence Ex370 /Em410. Because addition of 2 μM cytochalasin B decreased the agonist-induced rate of polymerization by ∼90%, the initial rate of polymerization is primarily due to barbed-end elongation and thus the initial rate was considered proportional to the number of free barbed ends.
Time Course of Polymerization with Purified Proteins
The time course of polymerization induced by N-WASP or VCA in the presence of Arp2/3 complex was followed by using G-actin that contained 10% or less pyrenylactin. Lower enrichments of labeled G-actin (3% or 1%) were used in experiments in which a high concentration of profilin was present. The mixture was placed, without dilution, into a cuvette and the pyrenylactin fluorescence was monitored continuously. In this assay, nucleation activity is detected from the decrease in the lag time before polymerization begins, or the decrease in the time to achieve half-maximal polymerization. Unlike the assay of nucleation sites described above, the rate of polymerization at any given time is the product of the number of barbed ends present and the concentration of G-actin above the critical concentration. With time, the concentration of G-actin decreases to the critical concentration and the fluorescence reaches a plateau.
Calculation of Free G-actin
In supernatants at a concentration of 3 mg/ml protein, there is ∼4 μM profilin, 12 μM G-actin, and 17.5 μM thymosin β4 (Cassimeris et al. 1992). Using the following affinities for G-actin: profilin, 0.1 μM and thymosin β4, 0.6 μM (Cassimeris et al. 1992), we calculate that the concentration of free G-actin in the supernatant is ∼0.52 μM. The concentration of “active” (able to bind G-actin) latrunculin A and vitamin D binding protein (VDBP), was determined from their ability to decrease the initial rate of polymerization of pyrenylactin from spectrin–actin seeds (using a K d for latrunculin A of 0.1 μM, and for VDBP of 1 nM). Using these values, we calculate the concentration of free G-actin in supernatant before or after addition of each protein.
Results
Cdc42 Acts through the Arp2/3 Complex to Induce New Actin Filaments in Neutrophil Supernatant
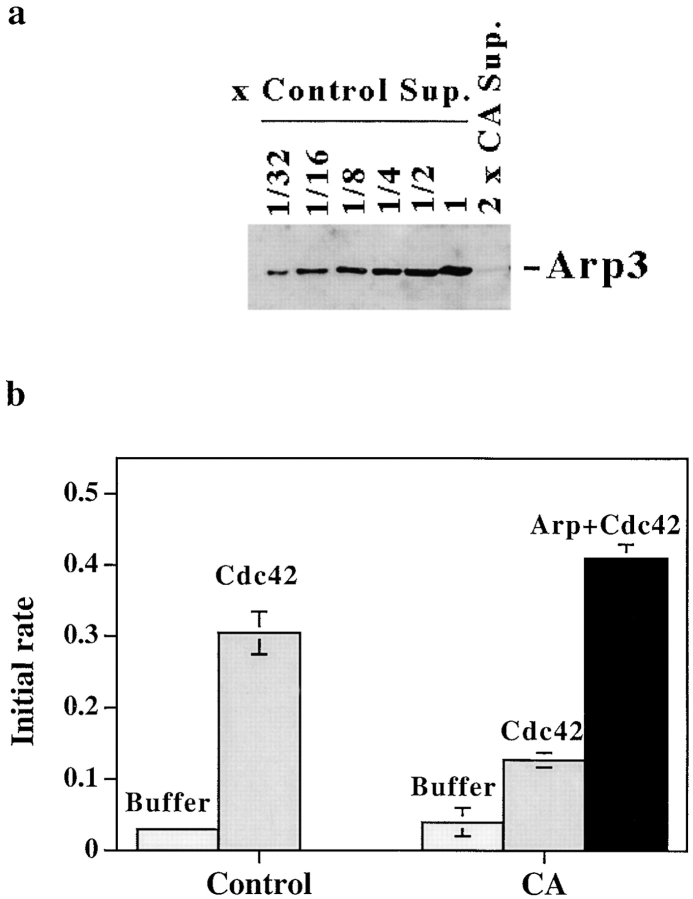
Cdc42, charged with GTPγS (Cdc42) and added to supernatant of lysed neutrophils, increases the number of actin filaments (Zigmond et al. 1998). To determine if the increase in filament number in neutrophil supernatants, like that in extracts of Xenopus oocytes or Acanthamoeba (Ma et al. 1998a; Mullins and Pollard 1999), depended on the Arp2/3 complex, we depleted >99% of the Arp2/3 complex by treating supernatant with beads coupled with CA. This depletion inhibited Cdc42-induced nucleation (Fig. 1, a and b). Addition of 80 nM of purified Arp2/3 complex fully restored the nucleation activity. Addition of Arp2/3 complex to control supernatant did not increase its nucleation activity (not shown). This suggests that in control supernatant, the concentration of Arp2/3 complex does not limit nucleation.
Figure 1.
Depletion of Arp2/3 from supernatant inhibits Cdc42-induced nucleation. (a) Serial dilutions of control (GST bead–treated) supernatant (30, 15, 7.5, 3.75, 1. 8 μg/lane, labeled as control 1, 1:2, 1:4, 1:8, 1:16, and 1:32), or GST–CA bead–treated supernatant (60 μg/lane, 2×CA) were loaded on a 10% gel. The gel was blotted and probed with anti-Arp3 antibody. (b) Control (GST bead–treated) supernatant and CA (CA bead– treated) supernatant were incubated with buffer, 2 μM Cdc42, or Cdc42 plus 80 nM purified Arp2/3 complex (Arp+Cdc42) for 5 min before dilution into 1.5 μM pyrenylactin; the initial rate of pyrenylactin polymerization was determined as a measure of nucleation sites (see Materials and Methods).
Profilin Depletion Inhibited Cdc42–induced Actin Nucleation
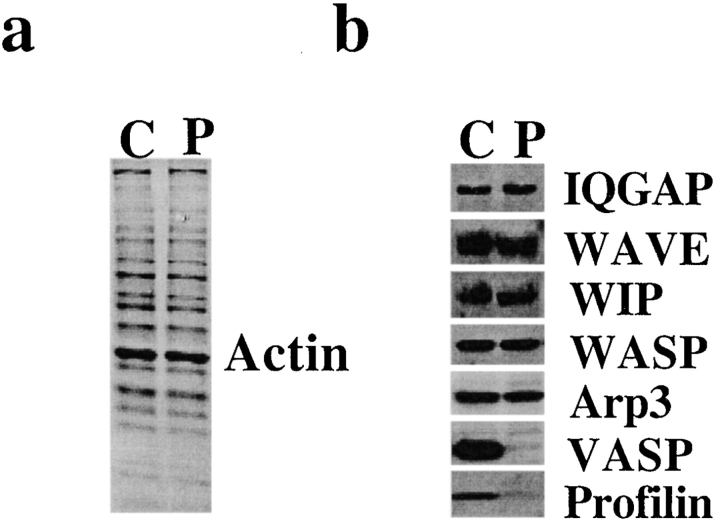
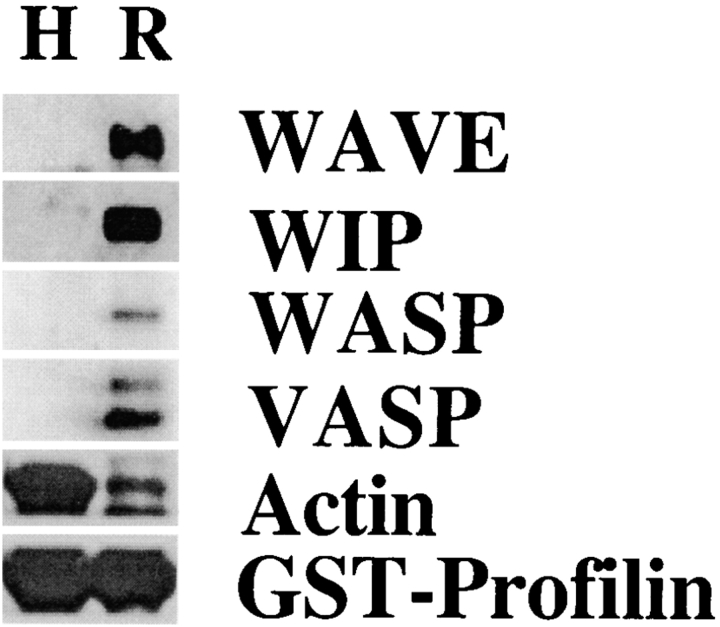
To investigate the role of profilin in Cdc42–induced actin nucleation, we treated supernatant with beads coupled to PLP. The PLP bead treatment depleted >90% of the profilin, >90% of the VASP, and 5–10% of the actin in the supernatant. There was no detectable depletion of WASP, WIP, WAVE, Arp3, or IQGAP (Fig. 2). PLP treatment severely inhibited the ability of Cdc42 to induce actin nucleation (Fig. 3 a). The number of nucleation sites induced by Cdc42 in PLP-treated supernatant was reduced to 10 ± 12% (n = 7) of mock-treated supernatant.
Figure 2.
Treatment of supernatant with PLP depletes profilin and VASP. (a) Coomassie blue–stained SDS-PAGE of PLP bead–treated supernatant (P) and control (Sepharose bead–treated) supernatant (C). Little or no change in protein staining was detected. (b) Western blots of PLP-treated supernatant (P) and control supernatant (C) stained with antibodies to IQGAP, WAVE, WIP, WASP, Arp3, VASP, and profilin. Of these, only VASP and profilin were decreased by PLP treatment. Each gel lane was loaded with 30 μg protein.
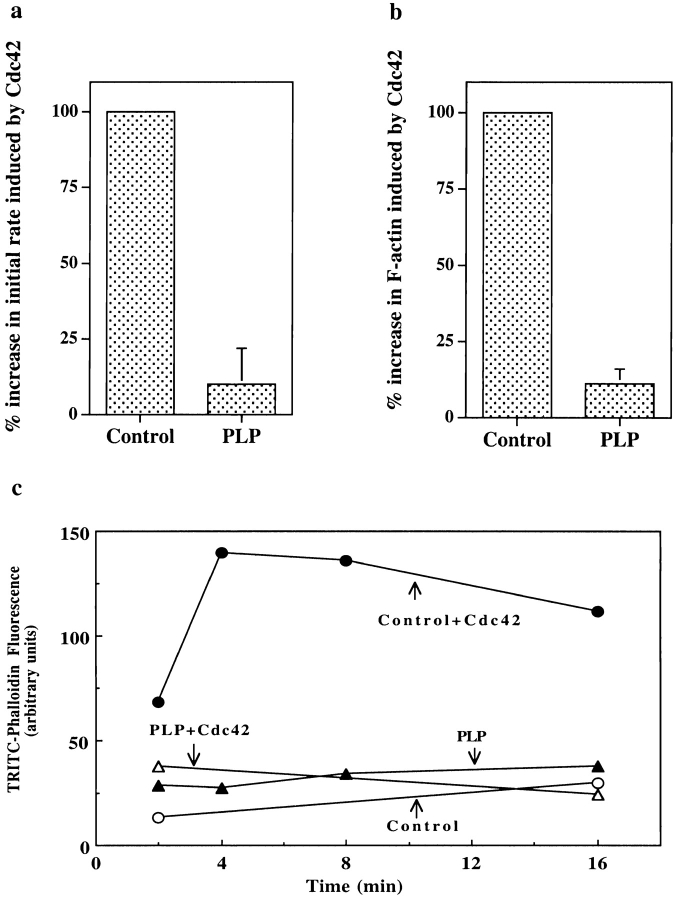
Figure 3.
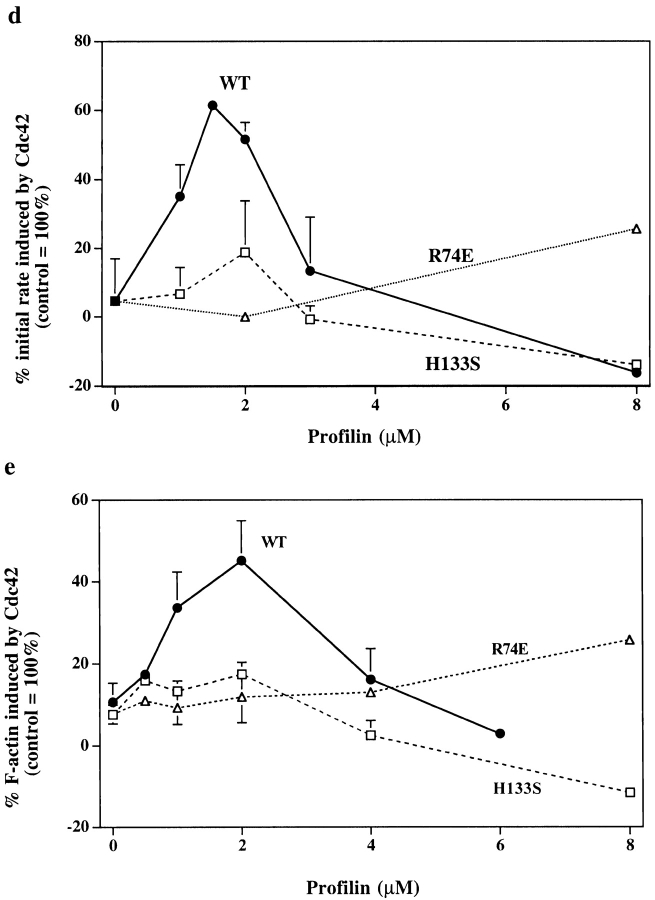
PLP treatment inhibits induction of nucleation sites and actin polymerization by Cdc42. Control (Sepharose bead–treated) and PLP (PLP-bead–treated) supernatants were incubated with buffer or 1 μM Cdc42 for 5 min. (a) To measure filament number, the supernatants were diluted into 1.5 μM pyrenylactin and the initial rate of increase in pyrenylactin fluorescence was determined as a measure of nucleation sites (see Materials and Methods). Data from several experiments were normalized by setting the Cdc42-induced increase in polymerization rate in the control supernatant as 100%. PLP treatment decreased the Cdc42-induced change in initial rate to 10.0 ± 12.0% (mean ± SD, n = 7) of control. (b) To measure F-actin, the supernatants were diluted into TRITC–phalloidin, which stains F-actin. The F-actin was pelleted and the TRITC–phalloidin extracted and its fluorescence used as a measure of F-actin (see Materials and Methods). The data from different experiments were normalized by setting the increase in F-actin induced by Cdc42 in control supernatant as 100%. PLP treatment decreased F-actin induced to 11.2±4.8% of control (mean ± SD, n = 12). (c) Time course of polymerization. PLP-treated (▴) or control (Sepharose bead–treated, ○) supernatants were incubated for 5 min without (open symbols) or with (filled symbols) 1 μM Cdc42 before dilution into TRITC–phalloidin. The amount of TRITC–phalloidin bound was measured as in Fig. 3 b. (d and e) Re-addition of profilin restores ability of Cdc42 to induce actin nucleation sites and F-actin in PLP-treated supernatant. Various concentrations of wild-type (WT) or mutant (H133S and R74E) profilins were added to PLP-treated supernatant, which was then incubated for 5 min with Cdc42 before assaying for nucleation sites (d, as described in Fig. 3 a) or F-actin (e, as described in Fig. 3 b). Data are expressed as the percentage of Cdc42–induced nucleation sites in control (Sepharose bead–treated) supernatant; addition of 1.5–2 μM WT profilin restored the nucleation response to ∼60% of control level (d) and F-actin to ∼50% (e) of control level. Little or no restoration was seen with either H133S or R74E profilin.
If the ability of Cdc42 to increase F-actin levels depends on its ability to create new filaments, profilin depletion should also inhibit Cdc42–induced actin polymerization. Indeed, PLP treatment decreased the Cdc42-induced increase in F-actin to 11.2 ± 4.8% (n = 12) of control level (Fig. 3 b). Polymerization was not merely slowed since even after 16 min of Cdc42 induction, the F-actin level increased only slightly (Fig. 3 c). Supplementing the supernatant with 100 μg/ml brain lipid and/or 0.1 μM F-actin did not increase the response to Cdc42.
To determine if the inhibition was due to profilin and/or VASP depletion, we investigated whether re-addition of these proteins would restore activity. Addition of 1–2 μM recombinant profilin restored ∼60% of Cdc42-induced nucleation and ∼50% actin polymerization (Fig. 3c and Fig. d). Purified spleen profilin and recombinant profilin were similar in their ability to restore activity. Higher concentrations of profilin inhibited both nucleation and polymerization (see below), even though control supernatant contains about 4 μM profilin. Inclusion of 0.5 μM G-actin with 1.5 μM profilin restored Cdc42-induced nucleation to >80% of the control level (data not shown). Higher concentrations of profilin–actin increased nucleation above control levels (see below). Thus, complete restoration of activity can be achieved by replacing both the depleted profilin and the depleted actin.
Addition of VASP (up to 1 μM) had no effect on nucleation or polymerization, with or without profilin. The conclusion that VASP is not required for Cdc42-induced polymerization was supported by experiments in which immunodepletion of >90% VASP, with no detectable depletion of profilin, had no effect on actin polymerization (data not shown).
To Restore Activity, Profilin Must Be Able to Bind Actin and Polyproline
Profilin can simultaneously bind actin and PLP. To determine whether binding to one or both of these sites was required to restore nucleation, we used site-directed mutagenesis to create two profilin mutants. The R74E mutant has a decreased affinity for actin (Korenbaum et al. 1998); the H133S mutant has a decreased affinity for PLP (Bjorkegren-Sjogren et al. 1997), but a normal affinity for actin (Egile et al. 1999). The binding of actin and proline-rich proteins to the mutant profilins was examined by incubating supernatant with beads containing GST–R74E profilin or GST–H133S profilin. Western blots revealed that H133S profilin beads bound much more actin, but much less WAVE, WIP, WASP, and VASP, than did R74E profilin beads (Fig. 4).
Figure 4.
Proline-rich proteins in supernatant bind to R74E profilin but not H133S profilin. Beads containing GST–H133S–profilin (H) or GST–R74E–profilin (R) were incubated for 2 h with supernatant before washing and extracting with SDS. Western blots of the extracts were probed with antibodies to proline-rich proteins, WASP, WIP, WAVE, and VASP. The blots were also probed with antibody to actin. Actin binds to H133S–profilin much better than to R74E–profilin. Anti-profilin was included to monitor the amount of profilin on the beads.
The H133S profilin had a reduced ability to restore Cdc42-induced nucleation or polymerization to PLP-treated supernatant (Fig. 3 d). At concentrations >2 μM, H133S profilin, like wild-type profilin, inhibited nucleation. No restoration (or inhibition) of nucleation was seen with up to 32 μM R74E profilin. Thus, both the PLP- and actin-binding sites of profilin are needed to enhance Cdc42-induced nucleation. Profilin might function at two independent sites, one binding to the proline-rich domain, the other to actin, however, this seems not to be the case since simultaneous addition of H133S and R74E to PLP-depleted supernatant did not restore activity (data not shown). Thus, the same molecule of profilin must simultaneously bind a proline-rich domain and actin.
Profilin Was Not Required for Nucleation in Supernatants Induced by VCA
Cdc42 stimulates nucleation by binding to a member of the WASP family which then activates nucleation by the Arp2/3 complex (Kim et al. 2000; Ma et al. 1998b; Machesky et al. 1999; Mullins and Pollard 1999; Rohatgi et al. 1999; Yarar et al. 1999). Since both WASP and Arp2/3 bind profilin (Mullins et al. 1998; Suetsugu et al. 1998), we sought to determine which interaction was needed for profilin function. GST–VCA, a constitutively active fragment of N-WASP (containing the verpolin homology, cofilin homology, and acidic tail domains but not the profilin binding–proline-rich domain) is a potent stimulator of actin nucleation and polymerization in neutrophil supernatant. The ability of GST–VCA to induce nucleation sites was similar in PLP-depleted and control supernatant (Fig. 5). Thus, GST–VCA-induced nucleation did not require profilin. This suggested that the role of profilin in Cdc42 induced-nucleation was upstream of the activated Arp2/3, probably in the activation of WASP.
Figure 5.
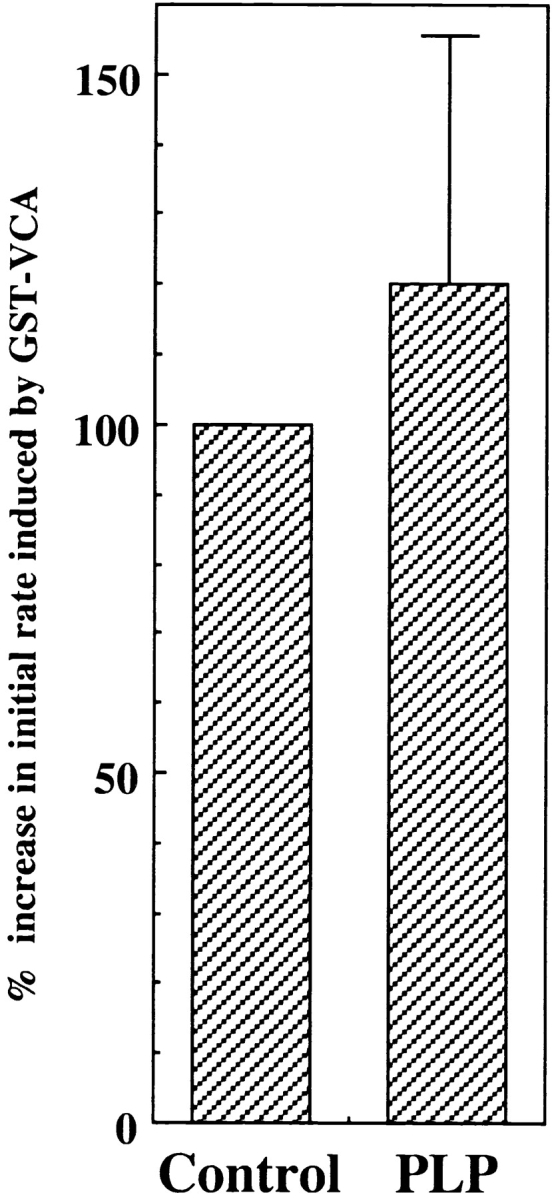
PLP treatment did not inhibit actin nucleation induced by GST–VCA. PLP (PLP-bead treated) and control (Sepharose bead–treated) supernatants were incubated for 5 min with 100 nM of GST–VCA before dilution into pyrenylactin, and the initial rates of polymerization was determined (see Fig. 3 and Methods). The data are expressed as the percentage (mean ± SD, n = 4) of control supernatant set as 100%.
Profilin Modulation of Nucleation Induced by Cdc42, Lipids, N-WASP, and the Arp2/3 Complex
To characterize how profilin enhanced the ability of Cdc42 to activate WASP, we examined its effect on nucleation induced by purified proteins. Cdc42 in combination with phosphotidylinositol lipids stimulate N-WASP to activate the Arp2/3 complex in vitro (Rohatgi et al. 1999). We found that profilin, though not required, could enhance Cdc42/lipid-induced nucleation by N-WASP (Fig. 6 a). The enhancement by profilin was small (less than or equal to twofold) and most effective at early times after Cdc42 addition. As in the cell supernatant, both the actin-binding and polyproline-binding sites of profilin were critical for its activity. H133S profilin only slightly enhanced nucleation activity, and R74E increased nucleation only at a concentration 10–20 times higher than wild-type profilin. Inhibition by higher concentrations of wild-type and H133S profilin was due to their binding G-actin, since when equal concentrations of free G-actin were added with the profilin there was no inhibition (Fig. 6 a).
Figure 6.
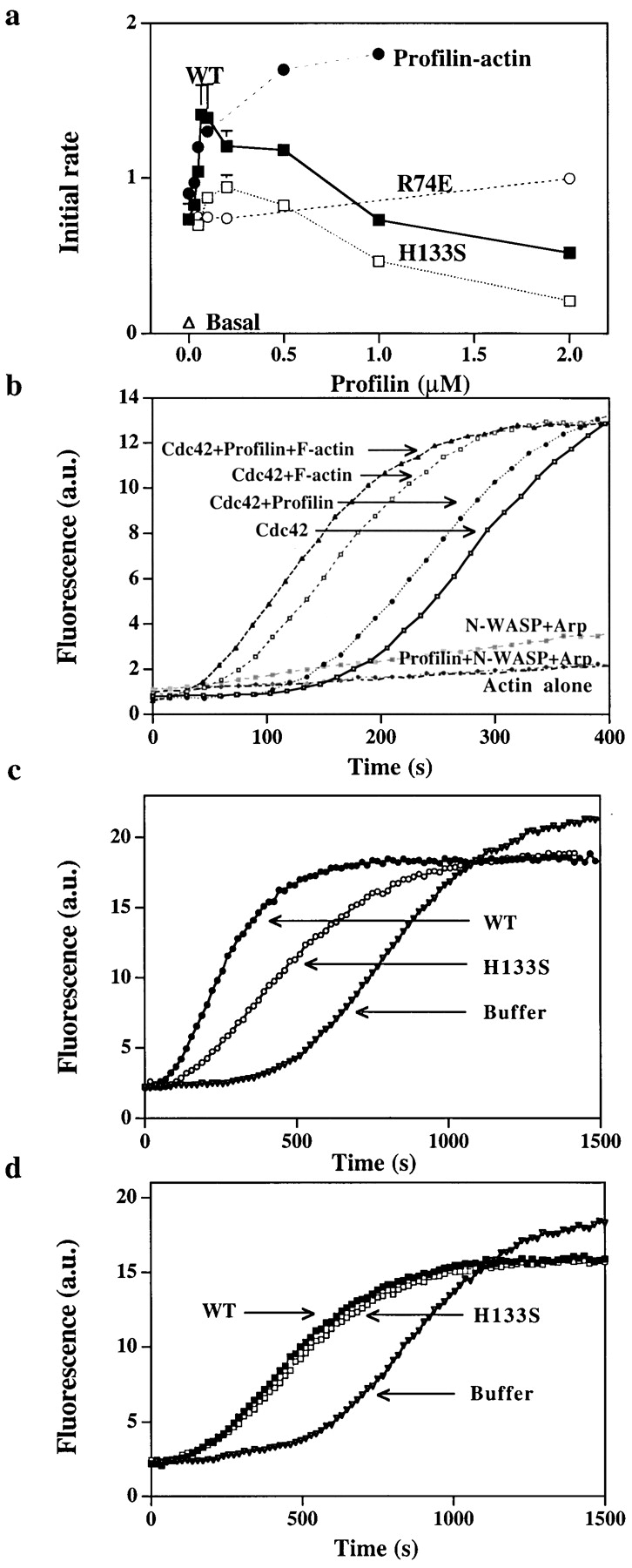
Effect of profilin on nucleation by purified N-WASP and Arp2/3. (a) Purified Arp2/3 complex (5 nM) was incubated with N-WASP (100 nM), Cdc42 (100 nM), lipid (15 ng/μl), G-actin (2 μM), and various concentrations of wild-type profilin (WT, ▪), or mutant profilin (H133S, □ or R74E, ○) for 2 min at 37°C before the samples were diluted 100-fold into 1.5 μM pyrenylactin and the initial rate of polymerization was determined. In one sample, wild-type profilin was added with enough G-actin (profilin–actin, •) to keep the initial free G-actin concentration at 2 μM. (b) Effect of profilin on the lag before polymerization. G-actin (2 μM containing 10% pyrenylactin) was incubated (1) in a cuvette alone (Actin alone), (2) purified Arp2/3 complex (5 nM) and N-WASP (100 nM) (N-WASP Arp), (3) Arp2/3 complex, N-WASP, and profilin (100 nM) (Profilin+ N-WASP+Arp), (4) Arp2/3 complex, N-WASP, Cdc42 (30 nM), and lipid (15 ng/μl) (Cdc42), (5) Arp2/3 complex, N-WASP, Cdc42, lipid with profilin (100 nM), (Cdc42+profilin), (6) Arp2/3 complex, N-WASP, Cdc42, lipid with F-actin (100 nM) (Cdc42+F-actin), or (7) Arp2/3 complex, N-WASP, Cdc42, lipid, profilin with F-actin (Cdc42+profilin+F-actin). Pyrenyl fluorescence was monitored over time. In this assay, unlike those shown previously, the pyrenylactin is included with the reactants and its polymerization is followed continuously in the cuvette. Thus, nucleation in this assay is reflected by the decrease in the lag before polymerization. (c) Effect of profilin on N-WASP activated nucleation in the presence of thmyosin β4. Thmyosin β4 (10 μM) and G-actin (4 μM, 4% labeled, i.e., pyrenylactin) were incubated in a cuvette with purified Arp2/3 complex (30 nM), N-WASP (200 nM), Cdc42 (30 nM), and lipid (15 ng/μl) without (buffer) or with 0.5 μM profilin, either wild-type (WT) or H133S. The pyrenyl fluorescence (arbitrary units, a.u.) was monitored over time as in Fig. 6 b. (d) Effect of profilin on GST–VCA activated nucleation in the presence of thmyosin β4. Thmyosin β4 (10 μM) and G-actin (4 μM, 4% labeled) were incubated in a cuvette with purified Arp2/3 complex (30 nM) and GST–VCA (5 nM) without (buffer) or with 0.5 μM profilin, either wild-type (WT) or H133S. The pyrenyl fluorescence was monitored over time.
With the pure proteins, the time course of actin polymerization could be directly monitored in the cuvette containing G-actin spiked with pyrenylactin. The combination of Cdc42 and lipid added to N-WASP, Arp2/3 complex, and G-actin increased nucleation as detected by a decrease in the lag time before polymerization, as described previously by Rohatgi et al. 1999. Addition of 100 nM profilin further decreased the lag time (Fig. 6 b). In the absence of Cdc2 and lipid, profilin slightly increased the lag time. F-actin decreased the lag period for polymerization induced by the Arp2/3 complex, as described previously by Machesky et al. 1999. The combination of profilin and F-actin further decreased the lag period (Fig. 6 b). The combined data with supernatants and with pure proteins suggest that profilin enhances activation of N-WASP by Cdc42 and lipid.
The effects of profilin on Cdc42 activation of purified N-WASP were small compared with the effects in the cell supernatant. In the supernatant, a reservoir of G-actin buffered with thymosin β4 allows addition of a low concentration of profilin to form profilin–actin without significantly decreasing the free G-actin. To determine if a G-actin reservoir would enhance the effects of profilin, we examined the effect of profilin in the presence of 10 μM thymosin β4 and 6 μM G-actin (K d = 1 μM). Under these conditions, addition of 0.5 μM profilin will form 0.45 μM profilin–actin, and only decreasing the free G-actin from 1–0.88 μM. Indeed, addition of profilin decreased the long lag (∼900 s to half maximal polymerization) by 64% (±6% range in two experiments) and increased the maximal rate of polymerization (Fig. 6 c). H133S profilin was less effective than wild-type profilin, decreasing the lag by 55 ± 3% and having no effect on the maximal rate. In the presence of the thymosin-actin buffer, profilin also decreased the lag for polymerization induced by VCA, but in this case the decrease by wild-type or H133S profilin was similar (47 ± 3%) (Fig. 6 d). Thus, the presence of a G-actin reservoir accounts for some of the differences between the effects of profilin in supernatant and with pure proteins.
Inhibition of Nucleation by Elevated Concentration of Profilin Is Due to G-actin Sequestration
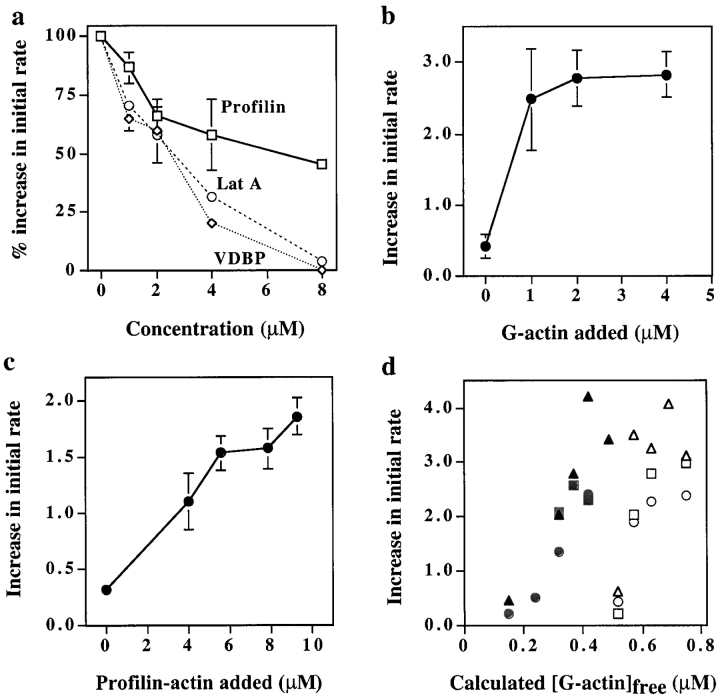
In the experiments above, we have examined the effects of depleting profilin from cell supernatant. Since supernatant preparation involves a ∼tenfold dilution of cytoplasm, the concentration of profilin in the intact cell is higher than in the supernatant. Thus, we investigated the effects of increasing the profilin concentration. Adding even 1 μM profilin to cell supernatant or purified proteins decreased the rate of nucleation (see Fig. 3 d and 6 a). Since high concentrations of a profilin mutant, which does not bind G-actin (R74E), did not inhibit nucleation, it seemed likely that the inhibition was due to G-actin sequestration. Indeed, addition of latrunculin A or VDBP, both of which sequester G-actin, also inhibited nucleation (Fig. 7 a). Addition of thymosin β4 also inhibited nucleation (not shown), although, due to its lower affinity, higher concentrations were required to sequester a comparable amount free G-actin. Cdc42-induced nucleation could not be detected when the calculated free G-actin was decreased to 0.1 μM (see Materials and Methods). This concentration of G-actin appears to limit nucleation with pure proteins as well. Addition of 5 nM Arp2/3 complex and 50 nM VCA to 0.5 μM actin filaments at steady state (G-actin is at its critical concentration of 0.1 μM) resulted in little or no increase in the number of filament ends, even after a two-hour incubation (data not shown).
Figure 7.
Effects of altering the concentration of G-actin. (a) Addition of G-actin sequestering factors inhibits nucleation. Various concentrations of latrunculin A (○), VDBP (⋄), or profilin (□) were added to supernatant before warming for 5 min in the presence of 2 μM Cdc42. The supernatant was then diluted into pyrenylactin and the initial rate of polymerization determined. The data from several experiments were normalized by setting the initial rate of polymerization in the absence of added sequestering agent as 100%; error bars represent the SD. (b) Addition of G-actin enhanced Cdc42-induced nucleation. Various concentrations of G-actin were added to supernatant and warmed at 37°C for 5 min in the absence or presence of 2 μM Cdc42 before dilution into pyrenylactin. The initial rate of polymerization was determined. The difference in the initial rate of polymerization with and without Cdc42 was plotted versus the concentration of G-actin added. (c) Addition of profilin–actin to supernatant enhances Cdc42-induced nucleation. Profilin and G-actin (P/A = 5:3.3, 7:4.6, 10:7, 12:7.9 μM) were added to the supernatant in a ratio that, combined with the 4 μM profilin, 17.5 μM thymosin β4, and 12 μM G-actin already in the supernatant (see Materials and Methods), gave an initial free G-actin concentration ∼0.42 μM. After incubation with 2 μM Cdc42 for 2 min at 37°C, the samples were diluted into pyrenylactin and the initial rate of pyrenylactin polymerization was measured. Data shown are from a representative experiment; error bars represent duplicate samples. (d) Profilin–actin allows nucleation at lowered concentrations of G-actin. Profilin (10 μM), with different concentrations of G-actin (3, 4, 5, 6, or 7 μM), was added to supernatant to give various initial concentrations of the free G-actin (filled symbols; free G-actin calculated as described in Methods). In parallel samples, different concentrations of G-actin (0, 0.5, 1, 1.5, and 2 μM) were added to supernatant to give different concentrations initial free G-actin (open symbols). Samples were incubated for 5 min at 37°C with buffer or 2 μM Cdc42 before dilution into pyrenylactin and the initial rate of polymerization determined. The data plotted for each sample are the difference in rate with and without Cdc42 as a function of the calculated initial free G-actin concentration. Data from three experiments using different supernatant preparations are plotted using different symbols (circles, triangles, or squares).
Increasing the free G-actin concentration in the supernatant by addition of exogenous G-actin increased Cdc42-induced nucleation. Up to 2 μM G-actin could be added without increasing basal nucleation (nucleation in the absence of Cdc42); addition of higher concentrations caused a parallel increase in basal and agonist-induced nucleation (Fig. 7 b). Combined, these data suggest that Cdc42-induced nucleation in neutrophil supernatant is a function of the free G-actin calculated to be between 0.1–0.8 μM (see Materials and Methods). G-actin bound to profilin or thymosin β4 did not effectively substitute for free G-actin in this nucleation reaction.
Profilin–Actin Enhanced Cdc42-induced Nucleation
With a better understanding of the G-actin requirement for nucleation, we next examined the effect of profilin added to supernatant in combination with enough G-actin to maintain a constant concentration of free G-actin. Under these conditions, the presence of profilin–actin enhanced nucleation by Cdc42 without increasing basal nucleation. The effects of profilin–actin were not due merely to an increased pool of G-actin, since in a parallel experiment, addition of the same amount of G-actin in combination with enough thymosin β4 to maintain the same concentration of free G-actin did not enhance nucleation (not shown). The Cdc42-induced nucleation sites increased with increasing concentrations of profilin–actin (Fig. 7 c). To induce a comparable number of nucleation sites, profilin–actin was required at ∼10 times the concentration of free G-actin. Since the concentration of profilin–actin in the neutrophil cytoplasm is about 20-fold higher than that of free G-actin (20 μM versus 0.5 μM; see Materials and Methods), the contribution by profilin–actin is likely to be physiologically meaningful.
Furthermore, increasing the concentration of profilin–actin decreased the concentration of free G-actin needed for nucleation. To vary the concentration of free G-actin in the presence of profilin–actin, 10 μM profilin was added to supernatant along with increasing concentrations of G-actin. In parallel samples, increasing concentrations of G-actin alone were added. The nucleation induced by Cdc42 was then measured. The number of nucleation sites versus the calculated free G-actin concentration is shown in Fig. 7 d.
G-actin and Profilin–Actin Also Stimulate Nucleation Induced by GST–VCA
To determine if stimulation by increased concentrations of profilin–actin in supernatant was unique to Cdc42-induced nucleation, we examined nucleation induced by GST–VCA. Profilin–actin enhanced nucleation and decreased the requirement for free G-actin, similar to GST–VCA- and Cdc42-induced nucleation (not shown). In summary, in cell supernatant, profilin–actin is less efficient than free G-actin in supporting Arp2/3-mediated nucleation activated by either Cdc42 or GST–VCA, but profilin–actin, in the presence of the same initial free G-actin concentration, enhances nucleation. To examine the mechanism of this enhancement we switched to a pure protein system.
Profilin–Actin Enhances Nucleation by GST–VCA-activated Arp2/3 Complex
The activation of nucleation by GST–VCA and Arp2/3 complex depends on the concentration of free G-actin, even in the presence of F-actin. Therefore, to determine if profilin–actin could enhance the rate of nucleation with pure proteins, we examined the time course of polymerization of samples with profilin/G-actin ratios calculated to give the same initial free concentration of G-actin as control samples in the absence of profilin. The rationale for this strategy is as follows. At any particular concentration of barbed ends, free G-actin will elongate the filaments at a rate proportional to its concentration, independent of the presence or absence of profilin. If profilin–actin is also present it will contribute to filament elongation with a rate constant ∼70% of that of free G-actin (Gutsche-Perelroizen et al. 1999), releasing profilin. Thus, both G-actin and profilin–actin will be consumed by elongation in approximate proportion to their quantity present. However, some of the profilin released will sequester G-actin, and so, whereas the amount of free G-actin is initially the same, it will fall more rapidly in the presence of profilin. Therefore, under the null hypothesis that profilin–actin makes no contribution to nucleation, we would predict that in the presence of profilin, compared with a control with the same initial free G-actin: (a) the concentration of free G-actin will begin to fall more rapidly and follow a lower trajectory over time; therefore (b) the concentration of barbed ends will increase more slowly and also follow a lower trajectory; and so (c) the fractional completion of the conversion of G-actin to F-actin will be slowed.
Fig. 8 a shows a test of prediction b above. Profilin/G-actin ratios giving a free G-actin concentration of ∼0.1 μM (which on its own did not increase nucleation above background) enhanced nucleation by VCA and the Arp2/3 complex, contrary to the prediction. As in the supernatant, a much higher concentration of profilin–actin than free G-actin was required to achieve a comparable number of nucleation sites. H133S profilin was as effective as wild-type profilin in this respect (not shown), consistent with there being no proline-rich region in VCA.
Figure 8.
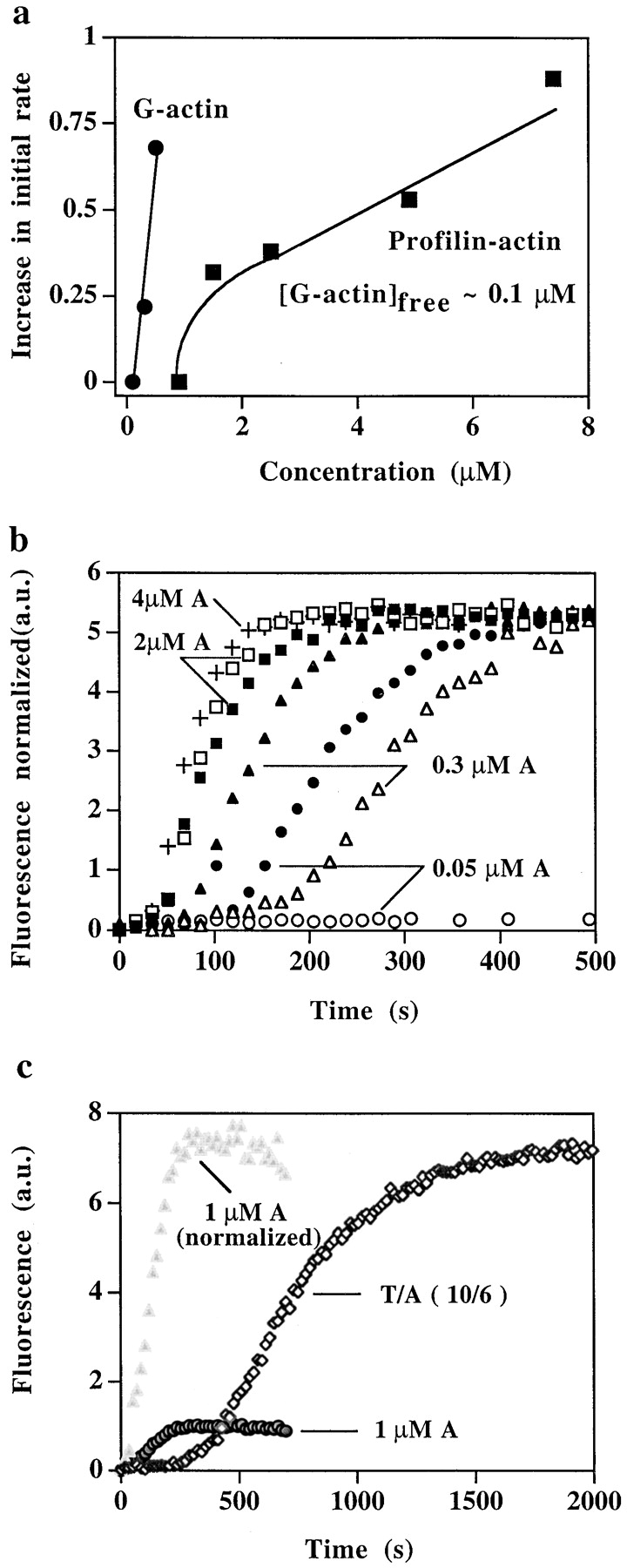
Effect of profilin–actin on nucleation by VCA and Arp2/3 complex. (a) Effect of G-actin and profilin–actin on the number of nucleation sites induced by VCA and Arp2/3 complex. The Arp2/3 complex (100 nM), GST–VCA (200 nM), and 0.2 μM F-actin (phalloidin stabilized) were incubated with different concentrations of G-actin (0.1, 0.3, and 0.5 μM, •), or with profilin and G-actin added in a ratio to give an initial free G-actin = 0.1 μM (P/A = 15:7.5, 10:5, 5:2.6, 3:1.6, and 2:1 μM, ▪). After incubation for 2 min at 37°C, the samples were diluted into pyrenylactin and the initial rate of polymerization determined. Data shown are the increase in rate of polymerization over basal (without Arp2/3) in a representative experiment. (b) Profilin–actin enhances nucleation by VCA activated Arp2/3 complex. Arp2/3 complex (5 nM) was incubated with GST–VCA (50 nM), 0.1 μM F-actin, and either with different concentrations of G-actin (0.05 μM, ○, 0.3 μM, ▵, 2 μM, □, or 4 μM, +) or with 4 μM G-actin (1% labeled) and different concentrations of profilin, to give an initial free G-actin concentration = 0.05 μM (profilin = 16 μM, •), 0.3 μM (profilin = 5 μM, ▴), or 2 μM (profilin = 3.3 μM, ▪). The pyrenyl fluorescence was followed continuously (as in Fig. 6 b). The fluorescence of each sample (except the sample with 0.05 μM G-actin alone, which did not polymerize) was normalized to the same final level. (c) Time course of nucleation and polymerization in the presence of thymosin β4. The Arp2/3 complex (5 nM) was incubated with GST–VCA (50 nM) in the presence of 1 μM G-actin (3% labeled) (gray circles, 1 μM A) or in the presence of a ratio of thymosin β4/actin (T/A = 10:6) to give a free G-actin of 1 μM (⋄). The polymerization was followed continuously in the cuvette. The increased pool of G-actin present in the T/A samples allowed more polymerization, however the lag before polymerization was increased. To better compare the lag period between samples, fluorescence of the G-actin sample is also shown normalized (gray triangles, 1 μM A [normalized]) to the same final fluorescence as the thymosin β4/actin sample.
Fig. 8 b shows a test of prediction c above. The time course of the conversion of G-actin to F-actin was followed by using partially pyrenylated G-actin with the same initial concentration of G-actin (0.05, 0.3, or 2 μM) in the presence and absence of profilin and normalized to the same final fluorescence. In the presence of profilin, a low percentage of pyrenyl-actin was used, since profilin preferentially binds unlabeled actin, a fact that does not qualitatively alter the predictions made under the null hypothesis. Again, the outcome was contrary to the prediction: when the G-actin concentration was low, the presence of profilin accelerated nucleation. These results suggest that profilin–actin either participates directly in Arp2/3-induced nucleation or enhances it in some less direct way, for example by stabilizing nascent nuclei or by increasing the concentration of a cofactor. The ability of profilin–actin to stimulate Arp2/3 complex nucleation depended on the concentration of free G-actin. When the concentration of G-actin was ≥1 μM, the presence of profilin–actin did not accelerate nucleation, presumably because any effect was masked by the more efficient G-actin.
Increasing the total G-actin pool can, if it releases an appropriate concentration of free G-actin, increase the amount of actin polymerized and the number of nucleation sites produced. To determine whether increasing the total G-actin pool might, in some unexplained way, affect the rate of nucleation, we increased the reservoir of G-actin by adding G-actin together with thymosin β4. Since under our conditions, thymosin β4–actin does not contribute to filament elongation, we would expect its influence to be expressed via its effect of buffering the free G-actin as it is consumed. Therefore, under the null hypothesis that neither thymosin β4 nor the total G-actin pool affect nucleation, we would predict that in the presence of thymosin β4, compared with a control with the same initial concentration of free G-actin: (a) the concentration of free G-actin will fall more slowly and follow a higher trajectory over time; therefore (b) the concentration of barbed ends will increase more rapidly and also follow a higher trajectory; and so (c) the rate of the conversion of G-actin to F-actin will be increased.
Fig. 8 c shows an experiment in which the thymosin β4 to actin ratio was adjusted to give an initial concentration of free G-actin of ∼1 μM (T/A = 10 μM:6 μM), compared with a control in the absence of thymosin β4 with an initial concentration of free G-actin of 1 μM. Again, the prediction under the null hypothesis was not born out: in the presence of thymosin β4, the conversion of G-actin to F-actin was in fact slowed (best seen when the data are normalized to the same final fluorescence). Therefore, not only could we detect no effect of an increased total G-actin pool in enhancing the rate of nucleation, but also we conclude that either thymosin β4 or thymosin β4–actin acts in some way to inhibit nucleation as compared with the control with the same initial concentration of free G-actin. The number of nucleation sites at plateau was, however, increased (data not shown). Thus the reservoir of G-actin did allow continued slow nucleation by the free G-actin resulting in an increase in the final level of nucleation sites.
Discussion
Profilin Has Multiple Effects on Nucleation
These studies illuminate the multiple effects, both positive and negative, that profilin has on Arp2/3 complex–mediated nucleation. In cell supernatant, profilin depletion profoundly inhibited nucleation induced by Cdc42, but not by VCA. Cdc42-induced nucleation was restored by addition of wild-type profilin, but not by mutant profilins unable to bind PLP or actin. PLP binding also contributed to profilin's ability to enhance Cdc42-nucleation via purified N-WASP.
Profilin binds with submicromolar affinity to the proline-rich domain of N-WASP (Suetsugu et al. 1998). Perhaps this binding, in combination with Cdc42 and lipid, induces a conformation of N-WASP that is better able to activate Arp2/3 complex. The proline-rich region is believed to serve as a hinge, and by binding to this region, profilin could modulate the thermodynamic balance in favor of activation (Abdul-Manan et al. 1999; Kim et al. 2000). The adaptor Grb2, which binds via its SH3 domain to the proline-rich region, also enhances Cdc42 activation of N-WASP (Carlier et al. 2000). Because only profilin able to bind actin stimulated nucleation, the profilin–actin bound to WASP may deliver its actin to the Arp2/3 complex nucleation site or to the barbed end of a growing filament (Fig. 9 a).
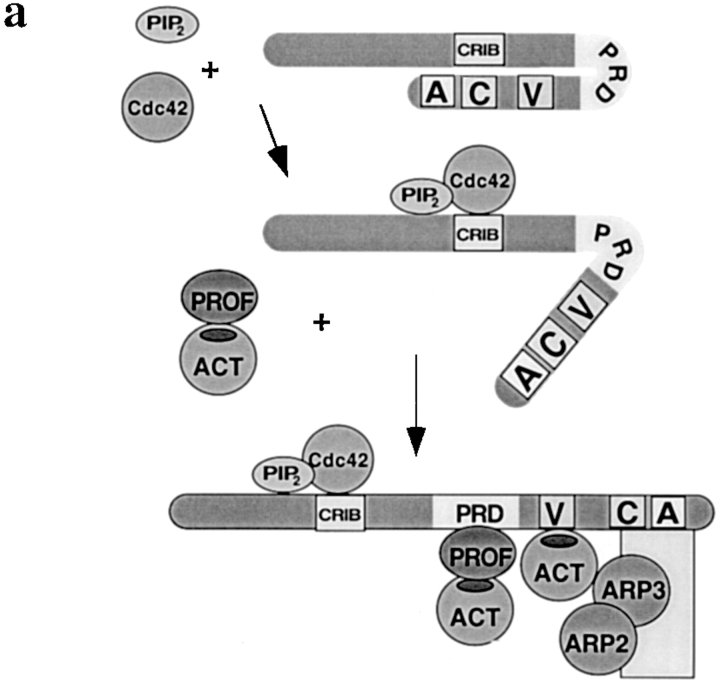
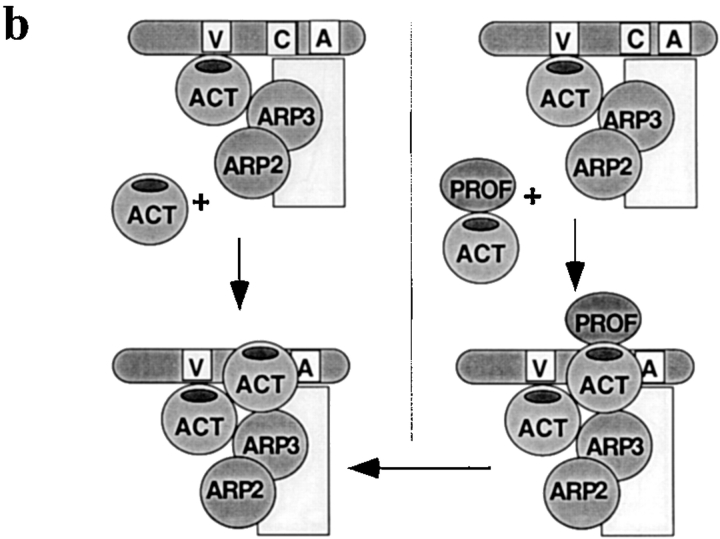
Figure 9.
Models of activation of nucleation by profilin. (a) Activation of WASP by profilin. In the inactive conformation, WASP is folded back on itself with the acidic COOH-terminal domain binding near the CRIB domain (Kim et al. 2000). Since the ability of profilin to increase Cdc42-induced nucleation sites in profilin-depleted supernatant or with pure proteins was optimal with profilin able to bind both actin and PLP, profilin–actin presumably binds to the proline-rich domain (PRD) of WASP. This binding could extend the conformational change induced by Cdc42 and lipid. In the fully “active” conformation, the CA-terminus of WASP is able to bind the Arp2/3 complex, with G-actin bound to the verprolin domain, thereby activating nucleation. Though not illustrated, the three-dimensional structure of this complex may be such that the actin of the profilin–actin complex contributes to the nascent filament. (b) Stimulation of VCA–Arp2/3 nucleation by profilin–actin. In this model, we propose that profilin–actin can bind to a site on the Arp2/3 complex (right side) that binds G-actin (left side) possibly with higher affinity. As proposed above, G-actin binds preferentially to the V domain. Upon dissociation of profilin, the actin from the profilin–actin complex contributes to nucleation of a new filament.
Profilin Effects on Supernatant Are Not Fully Explained by Results with Purified Proteins
Though studies with the purified proteins help define the molecular interactions, they do not fully reflect the situation in the supernatant. Addition of profilin increased Cdc42-induced nucleation in profilin-depleted supernatant more than sixfold, but with pure reagents only about twofold. Most likely, other components in the supernatant, including thymosin β4 and cofilin, increase the importance of profilin. Other profilin-binding proteins, such as WIP, may also play a role in the supernatant.
Inhibitory Effects of Profilin on Nucleation Are Due to G-actin Sequestration
The inhibitory effects of profilin on nucleation are due in part to its sequestration of G-actin. Nucleation by Arp2/3 complex in supernatant or with pure proteins is a function of the free G-actin concentration (Fig. 7; Higgs et al. 1999). When profilin addition resulted in lowered free G-actin concentration, it inhibited the rate of nucleation (Fig. 7; Higgs et al. 1999; Machesky et al. 1999). Thus, profilin–actin is less effective than free G-actin in supporting Arp2/3 complex-mediated nucleation. This decreased activity is probably not due to profilin–actin's slightly (30%) slower barbed-end elongation rate, nor does it seem to be due merely to an inhibition of spontaneous nucleation, since it was observed in the presence of F-actin.
Rather, it appears that profilin–actin is less effective than free G-actin in some aspect of the nucleation reaction. G-actin binds to the verprolin domain of WASP proteins. This domain, present in all members of the family, is needed for activation of Arp2/3 by COOH-terminal fragments of WASP family proteins (Machesky et al. 1999; Rohatgi et al. 1999). WASP family members with deletion in the verprolin domain are defective in vivo (Miki et al. 1998a,Miki et al. 1998b). Since the same surface of G-actin (domains one and/or three) is involved in binding to the verprolin domain and to profilin (Egile et al. 1999; Higgs et al. 1999; Machesky and Insall 1998), G-actin can not bind both simultaneously. The inability of profilin–actin to bind the verprolin domain may explain why it is less effective than free G-actin (Fig. 9 b).
Profilin–Actin Can Enhance Nucleation Independent of Polyproline Binding
Profilin–actin can enhance the rate of nucleation under conditions where the free G-actin concentration was maintained, i.e., when profilin was added as profilin–actin or added to a reservoir of G-actin buffered with thymosin β4. This enhancement does not require that profilin be able to bind PLP and may result from several different actions of profilin. When F-actin, a cofactor in Arp2/3 complex-mediated nucleation, is limiting, profilin–actin could enhance nucleation by its ability to elongate at barbed ends thereby increasing F-actin. Indeed, the enhancement by profilin was greater in the absence than in the presence of F-actin, suggesting that it might function by increasing F-actin or some other cofactor.
The enhancement by profilin–actin was greatest, in the presence or absence of F-actin, when the free G-actin concentration was low. The presence of profilin–actin allowed nucleation when the free G-actin concentration was too low to support nucleation on its own (>0.1 μM). Consistent with this result, branched filaments formed in the presence of Arp2/3 complex, VCA, and profilin/actin at a ratio expected to give a low concentration of free G-actin (Blanchoin et al. 2000). The fact that nucleation can occur when the free G-actin is below its critical concentration need not imply that G-actin is not required. Since the K d of binding of G-actin to verprolin is ∼0.6 μM (Egile et al. 1999; Higgs et al. 1999), 0.06 μM G-actin is sufficient to bind ∼10% of the verprolin domains. When G-actin is limiting, profilin–actin could contribute by binding directly to the Arp2/3 complex to form a new filament and/or by barbed-end elongation to stabilize a nascent filament.
Profilin has long been known to enhance actin treadmilling. These studies show that profilin also enhances Arp2/3 complex-mediated actin nucleation. Profilin contributes both to the activation of WASP by Cdc42 and to nucleation by activated Arp2/3 complex. Profilin–actin allows nucleation at decreased free G-actin concentrations. Because neutrophil cytoplasm contains about 40 μM profilin, the pool of profilin–actin in a resting cell (assuming the concentration of free G-actin = 0.5 μM) is ∼34 μM. After chemoattractant stimulation, when the F-actin level doubles, the free G-actin level may decrease to 0.1 μM, but the profilin–actin would still be present at ∼24 μM. This profilin–actin can allow rapid nucleation of new actin filaments to continue.
Acknowledgments
We are very grateful to Henry Higgs for helpful comments on a draft of the manuscript.
This work was supported by National Institutes of Health grant AI-19883 to S.H. Zigmond.
Footnotes
Abbreviations used in this paper: CA, cofilin homology and acidic tail contained in the COOH-terminal fragment of N-WASP; GST, glutathione S-transferase; PLP, poly-l-proline; VCA, verpolin homology, cofilin, and acidic domain contained in the COOH-terminal fragment of N-WASP; VDBP, vitamin D binding protein; WASP, Wiskott-Aldrich Syndrome protein.
References
- Abdul-Manan N., Aghazadeh B., Liu G.A., Majumdar A., Ouerfelli O., Siminowitch K.A., Rosen M.K. Structure of Cdc42 in complex with the GTPase-binding domain of the Wiskott-Aldrich syndrome protein. Nature. 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- Banin S., Truong O., Katz D.R., Waterfield M.D., Brickell P.M., Gout I. Wiskott-Aldrich syndrome protein (WASp) is a binding partner for c-Src family protein-tyrosine kinases. Curr. Biol. 1996;6:981–988. doi: 10.1016/s0960-9822(02)00642-5. [DOI] [PubMed] [Google Scholar]
- Bear J.E., Rawls J.F., Saxe C.L., III. SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 1998;142:1325–1335. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkegren-Sjogren C., Korenbaum E., Nordberg P., Lindberg U., Karlsson R. Isolation and charactarization of two mutants of human profilin I that do not bind poly-l-proline. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1997;418:258–264. doi: 10.1016/s0014-5793(97)01376-8. [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K.J., Higgs H.N., Marchand J.B., Kaiser D.A., Pollard T.D. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Henry P.A., Kolluri R., Kirchhausen T., Rickles R.J., Berg L.J. Identification of Itk/Tsk Src homology 3 domain ligands. J. Biol. Chem. 1996;171:25464–25656. doi: 10.1074/jbc.271.41.25646. [DOI] [PubMed] [Google Scholar]
- Cano M., Lauffenburger D.A., Zigmond S.H. Kinetic analysis of F-actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulations increases actin filament number without altering filament length distribution. J. Cell Biol. 1991;115:677–687. doi: 10.1083/jcb.115.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., Nioche P., Broutin-L'Hermite I., Boujemaa R., Le Clainche C., Egile C., Garbay C., Ducruix A., Sansonetti P., Pantaloni D. GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich Syndrome Protein (N-WASp) with actin-related protein (ARP2/3) complex. J. Biol. Chem. 2000;275:21946–21952. doi: 10.1074/jbc.M000687200. [DOI] [PubMed] [Google Scholar]
- Cassimeris L., McNeill H., Zigmond S.H. Chemoattractant stimulated neutrophils contain two populations of actin filaments that differ in their spatial distributions and relative stabilities. J. Cell Biol. 1990;110:1067–1075. doi: 10.1083/jcb.110.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Safer D., Nachmias V.T., Zigmond S.H. Thymosin β4 sequesters the majority of G-actin in resting human polymorphonuclear leukocytes. J. Cell Biol. 1992;119:1261–1270. doi: 10.1083/jcb.119.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano F., Montcourrier P., Guillemot J.-C., Gouin E., Machesky L., Cossart P., Chavrier P. Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr. Biol. 1999;9:351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- Didry D., Carlier M.-F., Pantaloni D. Synergy between actin depolymerization factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 1998;1998:25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Egile C., Loisel T.P., Laurent V., Li R., Pantaloni D., Sansonetti P.J., Carlier M.-F. Activation of the Cdc42 effector N-WASP by the Shigella flexneri IscA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan P.M., Soames C.J., Wilson L., Nelson D.L., Stewart D.M., Truong O., Hsuan J.J., Kellie S. Identification of regions of the Wiskott-Aldrich Syndrome Protein responsible for association with selected Src homology 3 domains. J. Biol. Chem. 1996;271:26291–26295. doi: 10.1074/jbc.271.42.26291. [DOI] [PubMed] [Google Scholar]
- Gertler F.B., Niebuhr K., Reinhard M., Wehland J., Soriano P. Mena, a relative of VASP and Drosophila enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont P.J., Furman M.I., Wachsstock D.H., Safer D., Nachmias V.T., Pollard T.D. Regulation of actin nucleotide exchange by thymosin β4 and profilin. Mol. Biol. Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche-Perelroizen I., Lepault J., Ott A., Carlier M.F. Filament assembly from profilin–actin. J. Biol. Chem. 1999;274:6234–6243. doi: 10.1074/jbc.274.10.6234. [DOI] [PubMed] [Google Scholar]
- Heyworth P.G., Knaus U.G., Xu X., Uhlinger D.J., Conroy L., Bokoch G.M., Curnutte J.T. Requirement for posttranslational processing of Rac GTP-binding proteins for activation of human neutrophil NADPH oxidase. Mol. Biol. Cell. 1993;4:261–269. doi: 10.1091/mbc.4.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H.N., Blanchoin L., Pollard T.D. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Howard T.H., Oresajo C.O. A method for quantifying F-actin in chemotactic peptide activated neutrophilsstudy of the effects of tBOC peptide. Cell Motil. 1985;5:545–557. doi: 10.1002/cm.970050609. [DOI] [PubMed] [Google Scholar]
- Kaiser D.A., Goldschmidt-Clermont P.J., Levine B.A., Pollard T.D. Characterization of renatured profilin purified by urea elution from poly-L-proline agarose columns. Cell Motil. Cytoskelet. 1989;14:252–262. doi: 10.1002/cm.970140211. [DOI] [PubMed] [Google Scholar]
- Katanaev V.L., Wymann M.P. GTPgS-induced actin polymerization in vitroATP- and Phosphoinositide-independent signalling via Rho-family proteins and a plasma membrane-associated guanine nucleotide exchange factor. J. Cell Sci. 1998;111:1583–1594. doi: 10.1242/jcs.111.11.1583. [DOI] [PubMed] [Google Scholar]
- Kim A.S., Kakalis L.T., Abdul-Manan N., Liu G.A., Rosen M.K. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Knaus U.G., Heyworth P.G., Kinsella B.T., Curnutte J.T., Bokoch G.M. Purification and characterization of Rac 2a cytosolic GTP-binding protein that regulates human neutrophil NADPH oxidase. J. Biol. Chem. 1992;267:23575–23582. [PubMed] [Google Scholar]
- Korenbaum E., Norberg P., Bjorkegren-Sjogren C., Schutt C.E., Linberg U., Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–9283. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- Lanier L.M., Gates M.A., Witki W., Menzies A.S., Wehman A.M., Macklis J.D., Kwiatkowski D., Soriano P., Gertler F.B. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Laurent V., Loisel T.P., Harbeck B., Wehman A., Grobe L., Jockusch B.M., Wehland J., Gertler F.B., Carlier M.-F. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes . J. Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Cantley L.C., Janmey P.A., Kirschner M.W. Corequirement of specific phosphoinositide and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts J. Cell Biol. 140 1998. 1125 1136a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Rhatgi R., Kirschner M.W. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42 Proc. Natl. Acad. Sci. USA 95 1998. 15362 15367b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M., Insall R.H. Scar1 and the related Wiskott-Aldrich Syndrome protein WASP regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky L.M., Mullins R.D., Higgs H.N., Kaiser D.A., Blanchoin L., May R.C., Hall M.E., Pollard T.D. Scar, a WASp-related protein, activates dendritic nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A., Sasaki T., Asakura T., Hotta I., Imamura H., Takahashi K., Matsuura Y., Shirao T., Takai Y. Interactions of drebrin and gephyrin with profilin. Biochem. Biophys. Res. Commun. 1998;243:86–89. doi: 10.1006/bbrc.1997.8068. [DOI] [PubMed] [Google Scholar]
- Miki H., Miura K., Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in PIP2-dependent manner downstream of tyrosine kinases. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki H., Sasaki T., Takai Y., Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP Nature 391 1998. 93 96a [DOI] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac EMBO (Eur. Mol. Biol. Organ.) J 17 1998. 6932 6941b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V., Way M. Cdc42 is required for membrane dependent actin polymerization in vitro. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1998;427:353–356. doi: 10.1016/s0014-5793(98)00443-8. [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Kelleher J.F., Xu J., Pollard T.D. Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments. Mol. Biol. Cell. 1998;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.D., Pollard T.D. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr. Biol. 1999;9:405–415. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- Oda A., Ochs H.D., Druker B.J., Ozaki K., Watanabe C., Handa M., Miyakawa Y., Ikeda Y. Collagen induces tyrosine phosphorylation of Wiskott-Aldrich syndrome protein in human platelets. Blood. 1998;92:1852–1858. [PubMed] [Google Scholar]
- Perelroizen I., Didry D., Christensen H., Chua N.H., Carlier M.F. Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J. Biol. Chem. 1996;271:12302–12309. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- Perelroizen I., Marchand J.-B., Blanchoin L., Didry D., Carlier M.-F. Interaction of profilin with G-actin and poly(l-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Anton I.M., Hartwig J.H., Geha R.S. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc. Natl. Acad. Sci. USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M.W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Miki H., Takanawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B.M., Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target for Rho small GTPase and is a ligand for profilin. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.R., Naccache P.H., Sha'afi R.I. Stimulation by chemotactic factors of actin association with cytoskeleton in rabbit neutrophils (role of calcium and cytochalasin) J. Biol. Chem. 1983;258:14041–14047. [PubMed] [Google Scholar]
- Winter D., Lechler T., Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Xu X., Barry D.C., Settleman J., Schwartz M.A., Bokoch G.M. Differing structural requirements for GTPase-activating protein responsiveness and NADPH oxidase activation by Rac. J. Biol. Chem. 1994;269:23569–23574. [PubMed] [Google Scholar]
- Yarar D., To W., Abo A., Welch M.D. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr. Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Zigmond S.H., Joyce M., Borleis J., Bokoch G.M., Devreotes P.N. Regulation of actin polymerization in cell-free systems by GTPγS and Cdc42. J. Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S.H., Joyce M., Yang C., Brown K., Huang M.Z., Pring M. Mechanism of Cdc42-induced actin polymerization in neutrophil extracts. J. Cell Biol. 1998;142:1001–1012. doi: 10.1083/jcb.142.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]