Abstract
unc-94 is one of about 40 genes in C. elegans, that when mutant, displays an abnormal muscle phenotype. Two mutant alleles of unc-94, su177 and sf20, show reduced motility and brood size, and disorganization of muscle structure. In unc-94 mutants, immunofluorescence microscopy shows that a number of known sarcomeric proteins are abnormal, but the most dramatic effect is in the localization of F-actin, with some, abnormally accumulated near muscle cell-to-cell boundaries. Electron microscopy shows that unc-94(sf20) mutants have large accumulations of thin filaments near the boundaries of adjacent muscle cells. Multiple lines of evidence prove that unc-94 encodes a tropomodulin, a conserved protein known from other systems to bind to both actin and tropomyosin at the pointed ends of actin thin filaments. su177 is a splice site mutation in intron 1, which is specific to one of the two unc-94 isoforms, isoform-a; sf20, has a stop codon in exon 5, which is shared by both isoform-a and isoform-b. The use of promoter-GFP constructs in transgenic animals revealed that unc-94a is expressed in body wall, vulval and uterine muscles, whereas unc-94b is expressed in pharyngeal, anal depressor, vulval and uterine muscles, and in spermatheca and intestinal epithelial cells. By western blot, anti-UNC-94 antibodies detect polypeptides of expected size from wild type, wild type-sized proteins of reduced abundance from unc-94(su177), and no detectable unc-94 products from unc-94(sf20). Using these same antibodies, UNC-94 localizes as two closely spaced parallel lines flanking the M-lines, consistent with localization to the pointed ends of thin filaments. In addition, UNC-94 is localized near muscle cell to cell boundaries.
Keywords: striated muscle, myofibrils, thin filaments, tropomodulin, C. elegans
Introduction
Sarcomeres are specialized actin cytoskeletal structures that perform the work of muscle contraction. Sarcomeres are molecular or “nano”-machines consisting of a highly ordered assemblage of many proteins. Despite ever increasing knowledge of the components and functions of sarcomeric proteins, we still do not have a clear picture about how sarcomeres are assembled, and how sarcomeres are maintained in the face of repeated muscle activity. The ability to analyze mutants in the nematode C. elegans is being exploited to obtain insights into these questions.1-3 Molecular genetic experiments using C. elegans complement the typical biochemical analyses that are carried out in mammalian systems. The major striated muscle in the worm lies in the body wall and is used for locomotion. In adults, there are 95 spindle shaped cells that are divided among four quadrants, which lie just underneath a basement membrane, hypodermis and cuticle. Due to the optical transparency of the worm, the myofibrils can be viewed by polarized light, which reveals obvious striations. Bright A-bands alternate with dark I-bands, and each I-band contains a row of dense bodies, which are the analogs of Z-disks of vertebrate striated muscle. Because the striations lie at a slightly oblique angle with respect to the long axis of the worm, this muscle is “obliquely striated.” Rather than filling the entire cell as is the case for vertebrate striated muscle, in C. elegans body wall muscle, the myofibrils are restricted to a narrow zone of ~1.5 microns along one side of the cell. All the M-lines and Z-disks are attached to the muscle cell membrane, making these structures good models for studying muscle costameres4 and focal adhesions of non-muscle cells.3
C. elegans has been used profitably to obtain mutants defective in the formation, function and/or structure of muscle. There are two major classes of muscle-affecting mutations. In one class, the uncoordinated or “Unc” class, the worms develop into adults but are slow moving or paralyzed.5-7 The second class, the “Pat” class of mutants (paralyzed arrested at two-fold) display a characteristic embryonic lethality in which embryos do not move within the eggshell and stop development at the twofold stage.8 A few genes have both hypomorphic Unc and null Pat phenotypes; examples include unc-1129 and unc-4510. To date, nearly all the muscle Unc and many of the Pat genes, have been cloned and studied at the molecular level. The encoded proteins include both previously known and novel components of the thick and thin filaments and their organizing and membrane attachment structures (M-lines and dense bodies). The cloning of M-line and dense body components has revealed a number of familiar components of focal adhesions (perlecan, integrins, vinculin, integrin linked kinase, PINCH), but has also revealed new components of these structures (UNC-112, UNC-98, UNC-96 and UNC-89).3,11 This analysis is consistent with a model in which myofibril assembly is directed by signals first laid down in the ECM and the muscle cell membrane. In addition, most of the components of dense bodies and M-lines are shared, except for the proteins involved in the later stages of assembly; for example, for the dense bodies, vinculin and α-actinin, and for the M-lines, UNC-89.
For thick filaments, the expected genes for the myosins and paramyosin were among the first worm muscle genes identified. New and evolutionarily conserved components of thick filaments were first revealed by this genetic analysis. One example is twitchin,12-14 the founding member of the giant kinases which include mammalian titin and insect projectin. Another example is UNC-45, which is a conserved chaperone for myosin head folding and for the assembly of myosin into thick filaments.10,15 For thin filaments, genes encoding the actins and the troponin-tropomyosin complex have been found. In addition, novel components have been identified (e.g. UNC-8716), and roles in myofibril assembly were first revealed for several previously known proteins. There are many actin binding proteins, many of which regulate actin filament dynamics in many eukaryotic cells. This includes proteins that promote actin polymerization (e.g. profilin), severing (e.g. gelsolins, ADF/cofilin), stability (e.g. tropomyosin), depolymerization (e.g. ADF/cofilin), barbed end capping (e.g. capZ) or pointed end capping (e.g. tropomodulin). Molecular genetic analysis of UNC-60B (an ADF/cofilin protein17,18), tropomyosin19, and UNC-7820,21, have clearly demonstrated the requirement for regulating actin filament dynamics to ensure proper assembly and maintenance of muscle thin filaments.
In 1980, Zengel and Epstein7 reported results of a screen for mutants with altered body wall muscle structure. Their screen involved enrichment for slow moving worms, followed by assessment by polarized light microscopy. Mutants represented new alleles of 10 previously identified genes,5,6 and 7 new genes. Among them was a single mutant allele for a new gene, unc-94. unc-94 (su177) was described as slow moving and by polarized light to have “irregular birefringent areas”. EM showed large collections of thin filaments interspersed with possible intermediate filaments and patches of thick filaments. Here, we report that unc-94 encodes a tropomodulin, an F-actin pointed-end capping protein. Our results show the expected localization of a tropomodulin in the sarcomere of another animal, its in vivo importance, and point to a new role for tropomodulin in regulating F-actin at muscle cell-cell boundaries.
Results
As is the case for most muscle Unc mutants, unc-94(su177) displays a less organized myofilament lattice as compared to wild type (Fig. 1a). There is alternation between normal and increased width of individual birefringent bands. To gain further understanding about the unc-94 mutant phenotype, we sought to identify additional unc-94 mutant alleles. By an F1 non-complementation screen, we recovered a new allele, sf20. As shown in Fig. 1a, sf20 has the same polarized light phenotype as su177. Neither allele shows any obvious defects by polarized light in the organization of pharyngeal muscle (data not shown). There were no marked differences in pharyngeal pumping on plates seeded with bacteria between wild type and either of the two mutant alleles (data not shown).
Figure 1.
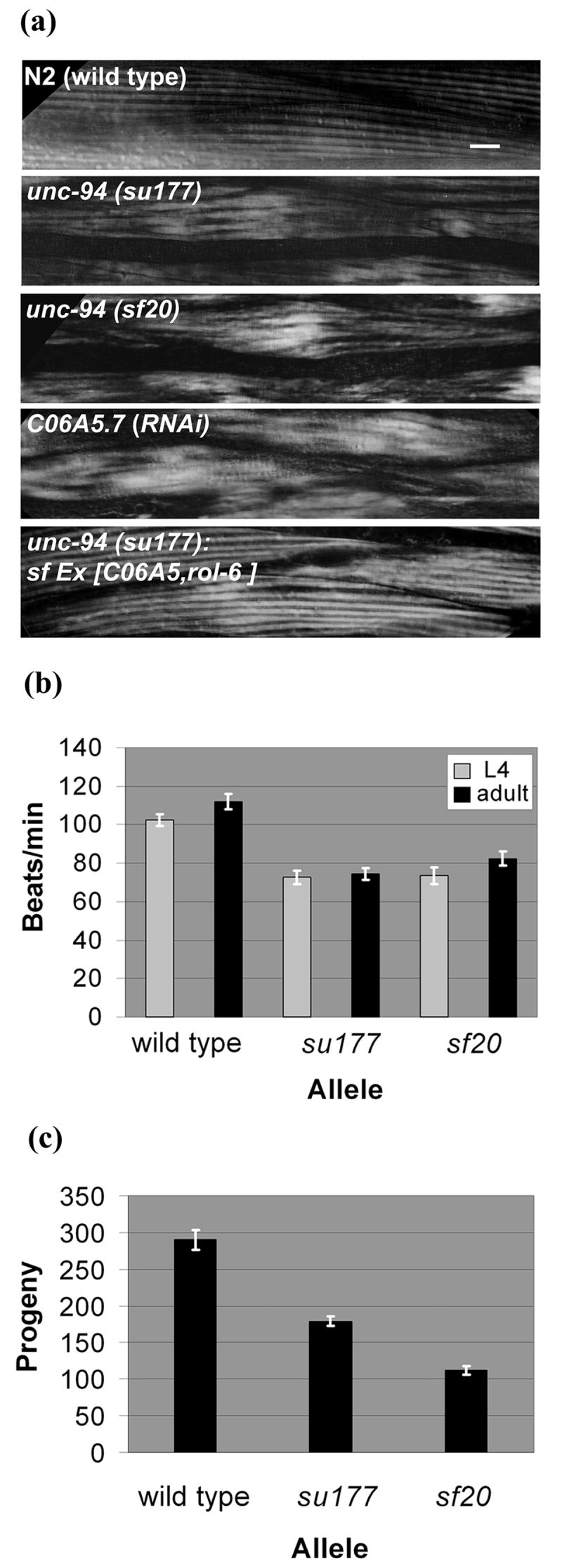
unc-94 mutants show disorganized muscle structure, decreased motility and low brood size. (a) Polarized light microscopy of body wall muscle in adult worms. In wild type muscle there is a normal arrangement of alternating birefringent A-bands with dark I-bands that run parallel to the long axis of the worm. In the muscle of worms expressing the two mutant alleles and C06A5.7 (RNAi) animals, there is reduced organization with alternation between normal and increased width of individual birefringent bands. The mutant phenotype can be rescued in unc-94 transgenic animals that carry an extrachromosomal array of the cosmid C06A5. Scale bar represents 10 μm. (b) Liquid motility assays of wild type and unc-94 animals at the 4th larval (L4) and adult stages of development. Data are shown as means and SEMs, with n=30. Both mutant alleles show reduced motility as compared to wild type. (c) Brood size assay comparing the amount of eggs laid by wild type animals and the unc-94 mutants. As shown, a normal N2 animal can lay between 200-300 eggs. Both unc-94 mutants show a significant decrease in their brood sizes, with su177 laying about 40% fewer eggs and sf20 laying 60% fewer eggs than wild type animals.
Each unc-94 allele moves more slowly than wild type when viewed by the dissecting microscope. To quantitate this difference, we placed a worm in liquid and counted the number of times the “head” moved away from and returned to an imaginary starting point. This swimming assay was conducted on both L4 larvae and adults because for many muscle Uncs, motility is most compromised in adults. As shown in Fig. 1b, both su177 and sf20 animals have significantly reduced motility as compared to wild type. A worsening in adults was not observed. Because many components of body wall muscle are also expressed in muscles required for egg laying (e.g. vulval and uterine muscles), we hypothesized that egg laying ability might be affected in unc-94 animals. This indeed is the case. Brood size measurements (Fig. 1c) show that both su177 and sf20 lay many fewer eggs as compared to wild type. In fact, sf20 is more severe than su177 by this assay, consistent with the fact that Western blot analysis of sf20 worm extracts failed to detect UNC-94 gene products (Figure 7).
Figure 7.
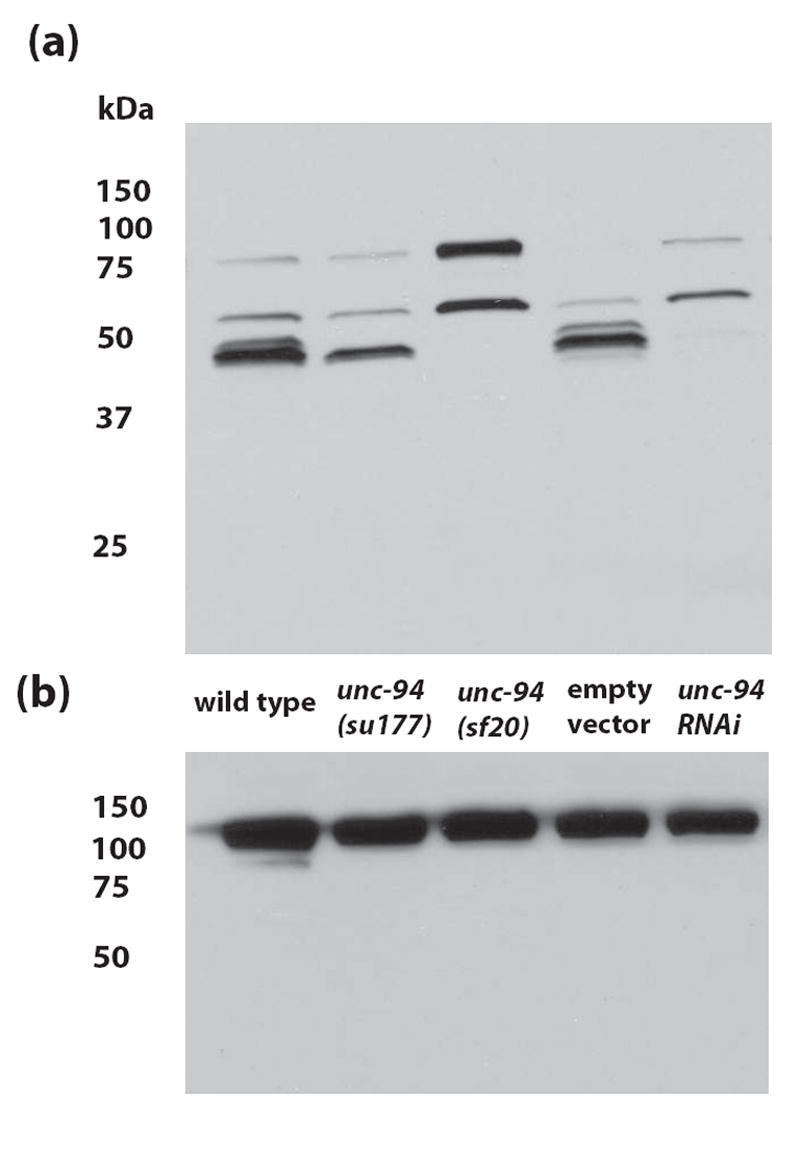
By Western blotting, UNC-94 polypeptides can be detected from wild-type, but are absent from or in reduced amounts in unc-94 mutants or RNAi animals. (a) Affinity-purified anti-UNC-94 antibodies detect proteins of the expected size (~45 kDa) for the products of the unc-94 gene from wild type C. elegans. Note that these bands are absent from unc-94(sf20) and unc-94(RNAi) animals (“Empty vector” refers to the use of the RNAi feeding vector without insert). (b) The same blot was washed and then reacted with anti-paramyosin to demonstrate equal loading of total protein in each lane. The columns of numbers and their positions represent molecular weight markers in kDa.
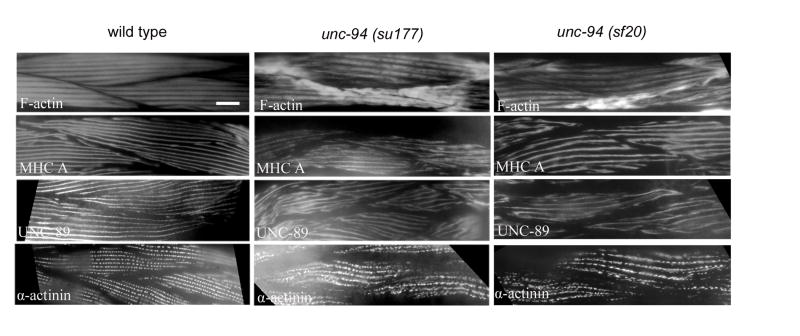
To further characterize the structural defect in adult unc-94 mutant muscle, immunofluorescence microscopy was used to visualize the localization of a number of known sarcomeric proteins. These proteins included F-actin, myosin heavy chain A (MHC A), UNC-89 as a marker for M-lines, and α-actinin as a marker for dense bodies. As shown in Fig. 2, each of these proteins shows some degree of mislocalization. The severity of the mislocalization appears similar in the two mutant alleles, su177 and sf20. As revealed by MHC A staining, the thick filaments appear discontinuous and at places perhaps broken. UNC-89 staining suggests that M-lines are also somewhat disorganized; the M-lines appear rather wavy and of variable width. α-actinin staining shows that the dense bodies are not as well defined and not arranged in as regular a pattern of rows as in wild type. However, the most dramatic effect is on the localization of F-actin as revealed by phalloidin staining. In either unc-94 (su177) or sf20, there is abnormal accumulation of F-actin near muscle cell-to-cell boundaries, and yet in most other areas of the myofilament lattice, F-actin localization and I-band organization appear normal. Significantly, in both unc-94 mutants, including the stronger allele sf20 (which has no detectable UNC-94 proteins by Western blot), there are still gaps (H-zones) between the I bands.
Figure 2.
Immunofluorescent localization of several known sarcomeric proteins in wild type and unc-94 mutant muscle. Antibodies were used to detect MHC A (thick filaments), F-actin (thin filaments), UNC-89 (M-lines), and α-actinin (dense bodies). Each of these proteins shows some degree of mislocalization, in a similar way for both mutant alleles. The most dramatic effect is on the localization of F-actin, with abnormal accumulation near muscle cell / cell boundaries; in most other areas of the cell, I-band organization appears normal. Scale bar represents 10 μm.
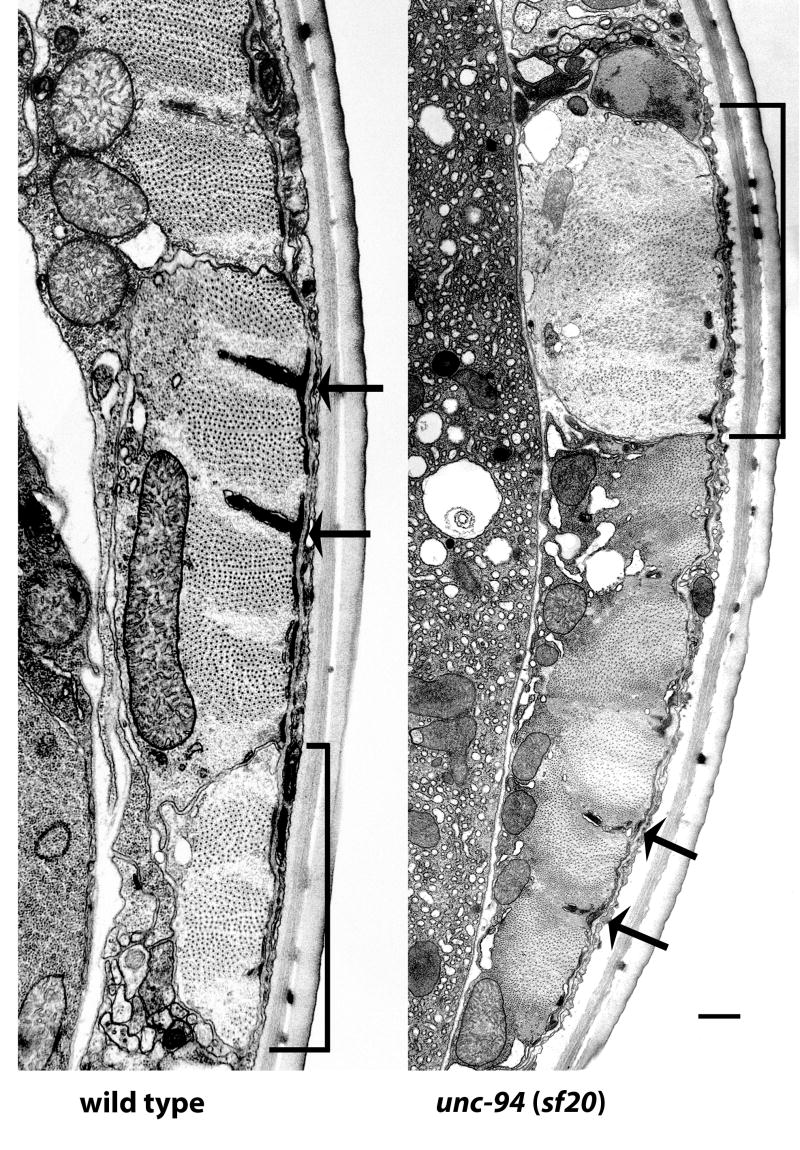
As an additional approach for understanding the role of unc-94 in myofibril organization, we examined the new unc-94 mutant allele sf20 by electron microscopy. Figure 3 shows cross sections of wild type and unc-94(sf20) body wall muscle, in each case, showing the broad part of one muscle cell, and an adjacent thin process of a neighboring cell (indicated with brackets). In wild type, the individual sarcomeres have well organized A-bands of thick filaments centered about M-lines, and I-bands of thin filaments centered about dense bodies. In unc-94(sf20), the sarcomeres are clearly disorganized, especially having irregular (short, thin and jagged) dense bodies and a lack of M-lines; the muscle cell process is dilated and contains a higher ratio of thin to thick filaments than wild type. In other sections of sf20 muscle, near cell-cell boundaries, there are large accumulations of what appear to be thin filaments without associated thick filaments (see Supplementary Figure 1).
Figure 3.
Electron micrographs of body wall muscle from wild type and from unc-94(sf20). These low power electron micrographs show most of one muscle cell next to a muscle cell process (indicated with a bracket), cut in transverse section. Thick filaments appear as large dots; thin filaments appear as barely discernible thin dots, most prominent around dense bodies. Dense bodies are indicated with arrows. In wild type highly ordered sarcomeres are present, with thin filaments in normal positions, either in I-bands, surrounding dense bodies, or in A-bands associated with thick filaments. Note that in sf20, the muscle cell process is dilated and contains a higher ratio of thin to thick filaments than wild type; also, a small amount of dense body material is found close to the cell membrane. In the adjacent muscle cell, sarcomere organization is better, but dense bodies are irregular. Scale bar, 500 nm.
After having characterized the phenotype of unc-94 mutants, a combination of deficiency and SNP mapping was used to determine where in the C. elegans genome unc-94 resides (Fig. 4). Initial studies7 of unc-94 mapped the gene to a location on chromosome I, between dpy-5 and unc-13 (2.03 map units and 2MB apart). Therefore, this region of the genetic map was inspected for deficiencies, but only one deficiency was available, qDf16. When animals containing this deficiency were mated with su177 animals, all of the out-crossed F1 progeny showed wild-type organization of body wall muscle by polarized light. From this result, it was concluded that the area which is deleted in this particular deficiency does not uncover the location of unc-94. SNP mapping was employed to limit unc-94’s position to a 500kb region, between (and including) cosmids F26B1 and C06A5, which contains a set of twelve overlapping cosmids. The left and right boundaries of this region agree with the results of the deficiency mapping, because this 500kb region was not deleted in qDf16. After scanning the 500 kb region for muscle expressed genes, a gene encoding a tropomodulin was identified, tmd-1(C06A5.7). Inspection of WormBase revealed that there are two predicted isoforms for C06A5.7 called a and b, each one containing 10 coding exons. Sequencing of cDNA clones confirms these predicted splicing patterns. Conceptual translation indicates that TMD-1a is 392 residues (calculated molecular weight of 44,399) and TMD-1b is 401 residues (molecular weight of 45,536). Isoforms a and b differ in their first and third exons: exon 1 of tmd-1a encodes 7 amino acids, and exon 1 of tmd-1b encodes 19 amino acids, with no obvious sequence similarity. Exon 3 of isoform a encodes an extra 3 residues at its 5’ end, which is not present in isoform b. The only notable feature of the N-terminal 19 residues of TMD-1b is that it contains a segment of 4 consecutive prolines.
Figure 4.
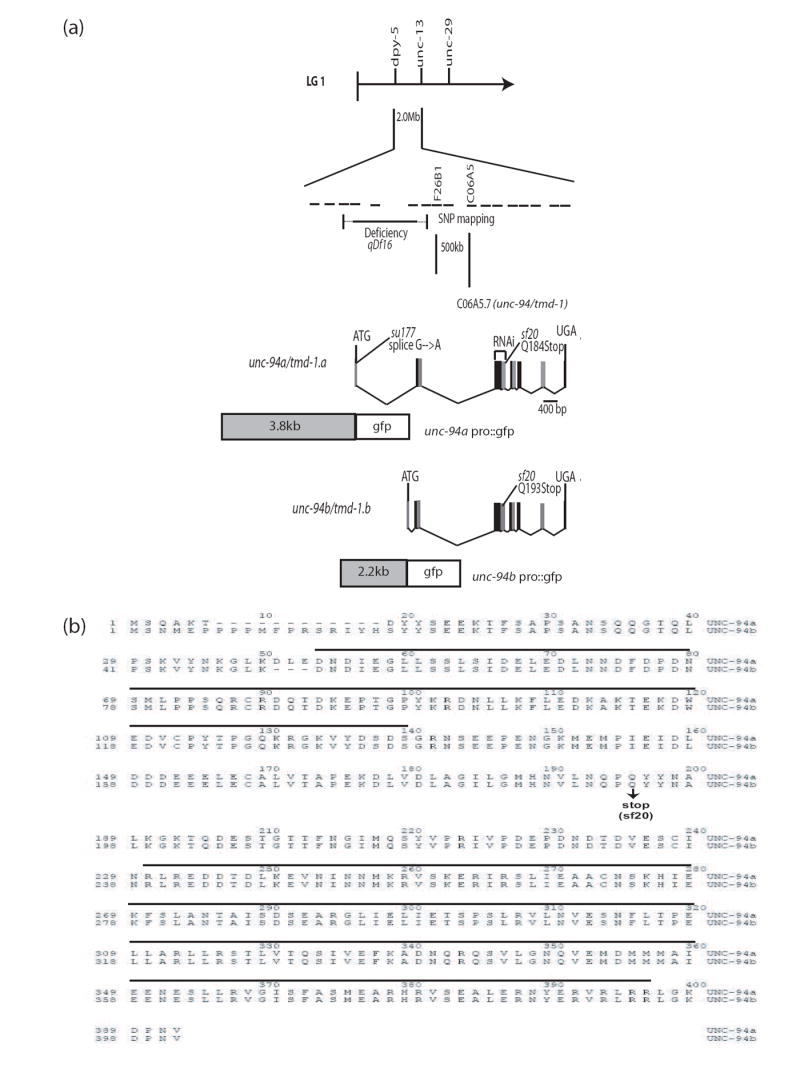
Genetic and physical mapping of unc-94, location of mutation sites for su177 and sf20, and the sequences of UNC-94a and b. (a) By using a combination of deficiency and SNP mapping, unc-94 was placed within a 500 kb region between/within cosmids F26B1 and C06A5. (Each dashed line represents the span of one cosmid insert.) After scanning the region for candidate genes, cosmid C06A5 was injected into unc-94 (su177) animals and was able to rescue the Unc-94 mutant phenotype. When rrf-3 animals were fed dsRNA for C06A5.7, their progeny showed a polarized light phenotype similar to that of unc-94 (su177 and sf20). As shown, WormBase predicts two isoforms for C06A5.7, a and b. In the figure, for clarity consecutive exons are represented as black or grey boxes. After having sequenced both unc-94 alleles, it was noted that su177 contains a splice site mutation in the first intron for isoform a, while sf20 contains a nonsense mutation in the fifth exon, which is shared among the two isoforms. Putative promoter regions plus the first exon of each isoform were used to create promoter-gfp fusions in order to analyze expression patterns. (b) Alignment of UNC-94 a and b protein sequences. The UNC-94 proteins are identical except in two places: (1) at their N-termini because of different first exons, and (2) beginning at residue 39 of UNC-94a which has an additional 3 residues (DLE) not found in UNC-94b because the 5’ end of exon 3 in UNC-94a is 9 bp earlier than in UNC-94b. Indicated is the mutation site of sf20: it is a C-to-T transition which converts glutamine 184 of isoform a (and glutamine 193 of isoform b) to a stop codon (CAA to TAA). The horizontal black lines indicate a putative tropomyosin-binding domain in the amino-terminal half, and a putative actin-binding domain in the carboxy-terminal half of each protein.
To determine if unc-94 was truly tmd-1, several experiments were performed. First, RNAi by feeding was used to knock down tmd-1 (C06A5.7) in wild type animals. As shown in Figure 1, C06A5.7 RNAi phenocopies Unc-94 by polarized light. Next, we injected su177 animals with C06A5 cosmid DNA to test whether the cosmid could rescue the Unc-94 phenotype when it is carried by the animals as an extrachromosomal array. The last panel of Figure 1a shows that CO6A5 rescues the unorganized patterning of A and I-bands to that of wild type. To gain additional evidence that unc-94 is tmd-1, we sequenced tmd-1 protein coding regions from genomic DNA of su177 and sf20. The su177 allele is a G-to-A transition in the splice donor site of the first intron, specific for isoform a (Figure 4). In contrast, the sf20 allele is a nonsense mutation in exon 5 which is shared by both tmd-1 isoforms; it is a C-to-T transition which converts glutamine 184 of isoform a and glutamine 193 of isoform b to the stop codon UAA. Since unc-94 mutants were isolated before the sequence-based identification of tmd-1, we refer to the gene, henceforth, as unc-94.
Given that there are two predicted isoforms for unc-94, which differ primarily in their first exons, we sought to determine whether their expression patterns were different. Promoter-GFP plasmids were created that contained the putative promoter regions and first exons fused in-frame to GFP (Figure 5). Animals carrying transgenic arrays of these plasmids were examined to determine where the promoters were expressed by the presence of GFP signal. As shown in Figure 5, unc-94a is expressed in body wall muscle (Figure 5 a and c), but not in pharyngeal muscle (Figure 5b), and is also expressed in vulval and uterine muscles (Figure 5d). unc-94b is not expressed in body wall muscle (Figure 5e), but is expressed in pharyngeal muscle (Figure 5f) and the vulval and uterine muscles (Figure 5g; arrowhead). In addition, unc-94b is expressed in a number of other tissues including spermatheca (Figure 5g, arrows), gut epithelial cells (Figure 5g, asterisks), and anal depressor muscle (Figure 5h; arrow). In summary, unc-94a but not unc-94b is expressed in body wall muscle, whereas b but not a is expressed in pharyngeal and anal depressor muscles. Both isoforms are expressed in vulval and uterine muscles.
Figure 5.
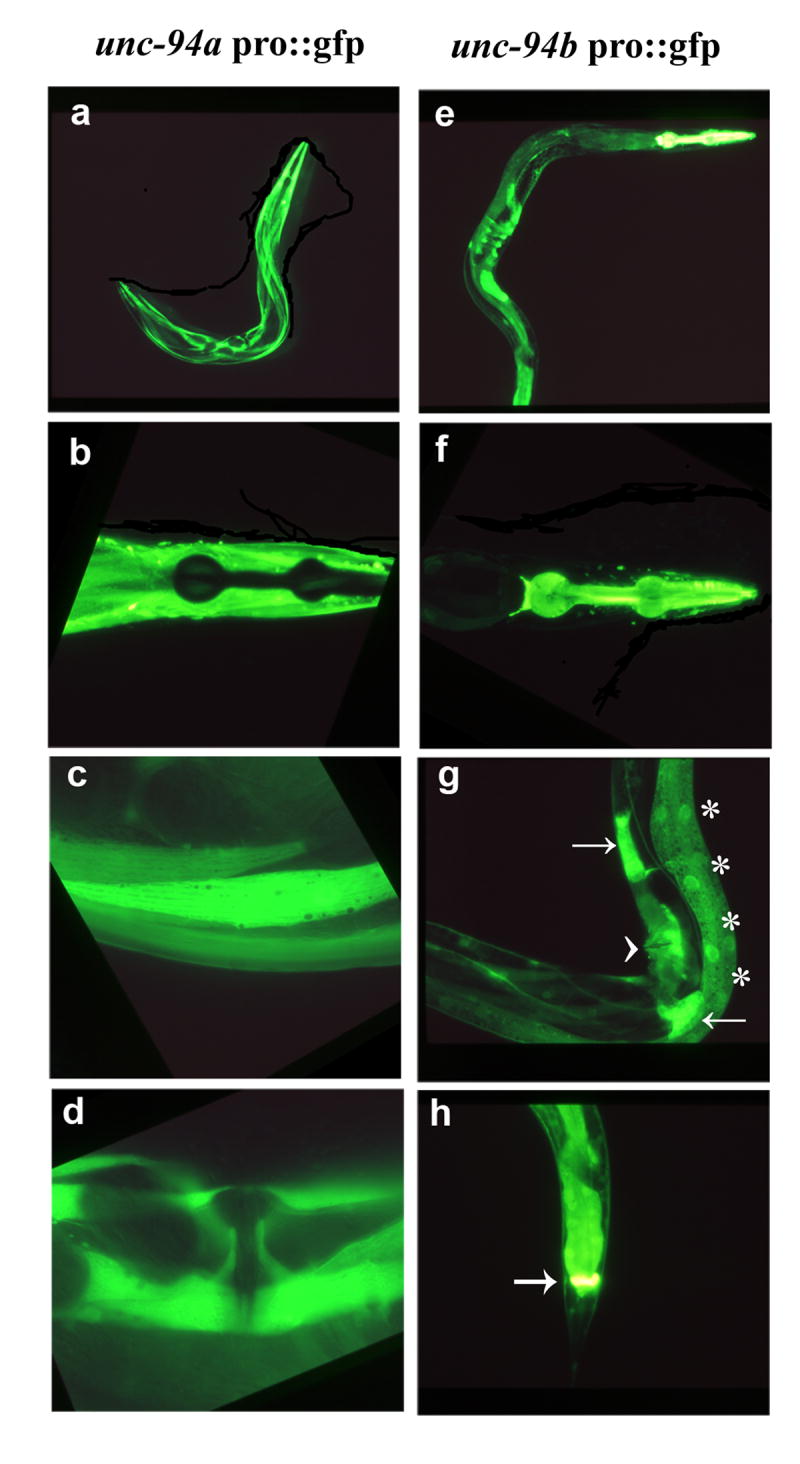
Fluorescent images of GFP expression in transgenic animals that carry unc-94 promoter elements. (a-d) Isoform a is primarily expressed in body wall (a and c), uterine and vulva muscle (d). As shown in (b), isoform a is not expressed in pharyngeal muscle. (e-h) Isoform b is highly expressed in the pharynx (e and f) and anal depressor muscles (h) and can also be detected in the muscle of the vulva and uterus (arrowhead), spermatheca (arrows), and intestinal epithelial cells (asterisks) (g).
To gain further understanding into how the mutations in su177 and sf20 result in phenotypes, we performed Northern analysis. Equal amounts of total RNA from wild type and the two unc-94 mutants were separated on a gel, transferred to a membrane and hybridized with: (1) a probe consisting of most of exons 2 through 4 and expected to detect both unc-94a and unc-94b transcripts; (2) a probe consisting of the unc-94a-specific first exon (mostly 5’ UTR); and (3) a probe for detecting the unc-15 (paramyosin) mRNA to verify that equal amounts of RNA were indeed loaded. (The short unc-94b-specific sequence (mostly 5’UTR of 90 bp) precluded preparation of a convenient isoform b-specific probe.) As shown in Figure 6, each unc-94 probe detects from wild type and the mutants a fairly broad band measured to be approximately 2 kb. cDNA analysis indicates that unc-94a transcript is 2, 135 nucleotides, and that the unc-94b transcript is 1,927 nucleotides. Since the sizes of these mRNAs may be too close to be resolvable on a gel, the observed 2 kb broad band is close to what was expected. Significantly, both unc-94 mutant alleles show decreased levels of unc-94a transcripts. The decreased level of unc-94a mRNA in su177 might result from reduced efficiency of splicing of the entire unc-94a transcript, since the su177 mutation lies in the splice donor site of the first intron. In sf20, the decreased level of unc-94a and possibly unc-94b transcripts likely results from the premature stop codon in sf20 targeting the messages for degradation by the nonsense mediated decay system.22
Figure 6.
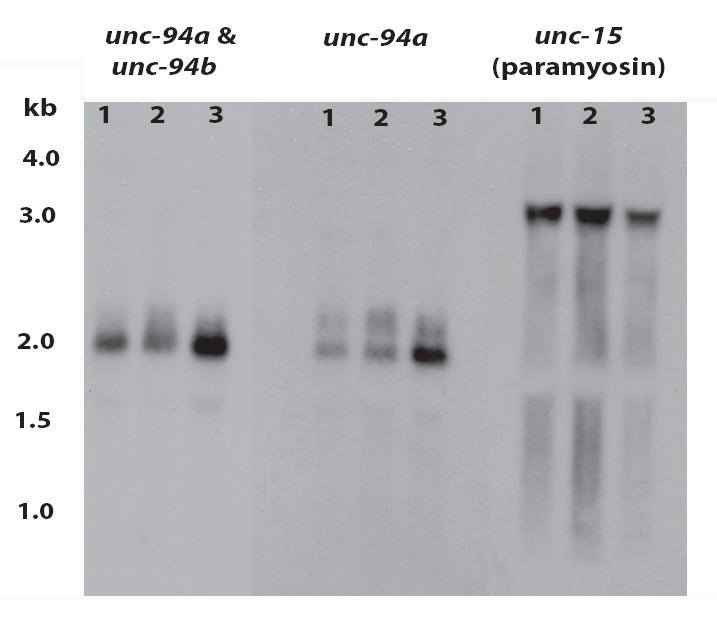
By Northern blot, unc-94 mutations result in decreased levels of unc-94 mRNAs. Total RNA from wild type (“3”), unc-94(sf20) (“2”) , and unc-94(su177) (“1”) mutant worms were separated on a gel, transferred to a membrane and hybridized with probes that were expected to detect both unc-94a and unc-94b, unc-94a alone, or unc-15 transcripts (as loading control). Each unc-94 probe detects a broad band of approximately 2 kb, close to the size expected for unc-94a (2,135 nucleotides) and unc-94b (1,927 nucleotides) by cDNA analysis. Note that the unc-94a mRNA is decreased in both su177 and sf20. The numbers denote the sizes, in kb, of the RNA markers.
Affinity-purified rabbit antibodies generated to residues 144-401 of UNC-94b/TMD-1b23 were used in immunoblot and immunofluorescent experiments. As shown in Figure 7, these antibodies react primarily to a band of ~45 kD and two faint bands just slightly above and below it; there are also two moderate intensity bands of ~60 and ~75 kD. Based on the absence of these ~45 kD bands in unc-94(sf20) and unc-94(RNAi) animals, it is very likely that these proteins are the products of unc-94. These bands are also consistent with the predicted sizes of UNC-94 isoforms from sequence analysis. Even after an overnight exposure of the Western blot, no detectable ~45 kD bands could be detected from sf20. unc-94(su177) shows decreased intensity of the 45 kD bands: the faint upper band (presumably isoform b, predicted to be 45,536 Da) is missing, and the main band (presumably isoform a, predicted to be 44,399 Da) is reduced. This result is unexpected given the fact that the mutation in su177 lies in an isoform a-specific exon. Perhaps the a and b isoforms run anomalously on a gel, or isoform a is posttranslationally modified, causing it to migrate at a higher than expected position. At present, the identity of the 60 and 75 kD bands are unknown. The C. elegans genome has a second tropomodulin encoding gene called tmd-2. The region of TMD-1b used as immunogen does have some similarity (32% identity, 55% similarity) to both TMD-2a (35.5 kD) and TMD-2b (73 kD) so there is the potential for cross-reactivity. Thus, the band detected on the Western at ~75 kD is possibly TMD-2b. We performed RNAi for tmd-2 using the Ahringer library feeding clone, but we did not detect a phenotype or a change in the Western blot pattern using this anti-UNC-94 antibody as compared to wild type (data not shown).
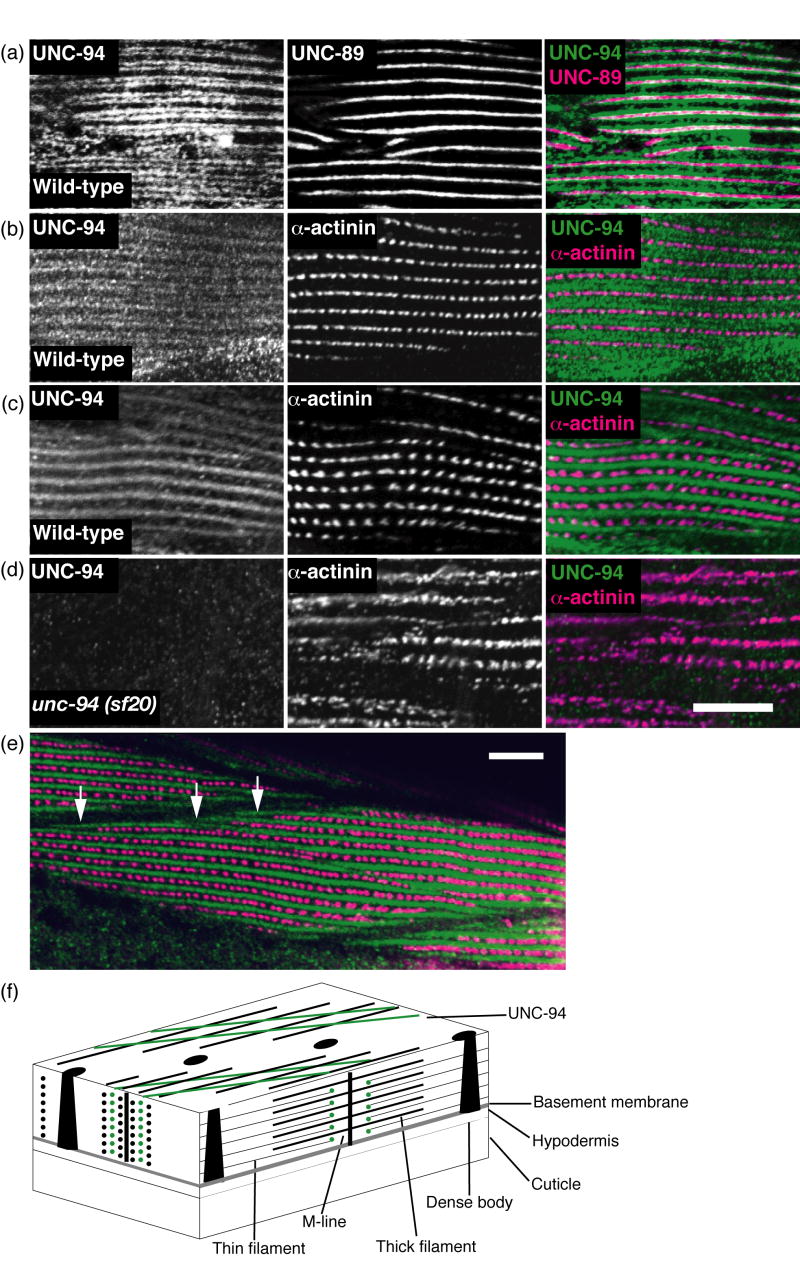
The same antibodies were used to localize UNC-94 in worm body wall muscle. Separate batches of adult wild type worms were fixed using either the Nonet method24 (Figure 8a, b) or the Finney and Ruvkun method25 (Figure 8c,d, e). Anti-UNC-94 was co-incubated with either marker antibodies to dense bodies (α-actinin) or M-lines (UNC-89). As shown in Figure 8a, using the Nonet method of fixation, UNC-94 localizes as two closely spaced parallel lines flanking the M-lines. This localization is in the I-bands, but not within the row of dense bodies where the barbed ends of the thin filaments are located (Figure 8b). Thus, our localization is consistent with the known localization of tropomodulins in striated muscle of other animals, in which tropomodulins are located at the pointed or minus ends of thin filaments (see model of worm body wall muscle and UNC-94 localization in Figure 8f). Figure 8c shows anti-UNC-94 staining of worms fixed by the most commonly used method of fixation.25 These images reveal broad I-band localization with no hint of the two closely spaced bands as seen in Figure 8a and b. A possible explanation is that the structures to which UNC-94 is associated (pointed ends of thin filaments) are fragile and may require rapid fixation not achievable by the Finney and Ruvkun procedure.
Figure 8.
By immunofluorescence, UNC-94 localizes to the pointed ends of thin filaments and to muscle cell boundaries. (a and b) Adult wild type worms were fixed by the Nonet method24 and either co-incubated with anti-UNC-94 and anti-UNC-89 (M-line marker) as shown in (a), or anti-UNC-94 and anti-α-actinin (dense body marker), as shown in (b). UNC-94 is localized to two closely spaced parallel lines closely flanking the M-lines. (c) Adult wild type worms were fixed by the “constant spring” method25 and co-incubated with anti-UNC-94 and anti-α-actinin. By this method, UNC-94 appears as a broad band, probably due to incomplete fixation. (d) Adult unc-94(sf20) worms (also fixed by the constant spring method) were co-incubated with anti-UNC-94 and anti-α-actinin. Note the absence of staining with anti-UNC-94: this suggests that the staining observed in wild type is due to reaction to UNC-94 and not cross reaction to the related protein, TMD-2. (e) The same animals as shown in (c), but at lower magnification. In this view, UNC-94 extends beyond the rows of dense bodies, likely at muscle cell/cell boundaries (white open arrows). (f) Drawing of C. elegans obliquely striated body wall muscle with the localization of UNC-94 (green) to the pointed ends of thin filaments as indicated by this study. Scale bars, 10μm.
Because of the possible cross reactivity between the anti-UNC-94 antibodies and the related protein, TMD-2 (see Figure 7), we wondered if some of the staining of these antibodies in body wall muscle was due to the presence of TMD-2. However, as shown in Figure 8d, this is likely not the case since unc-94 (sf20), which shows no detectable UNC-94 proteins by Western blot, also does not show any detectable staining with the antibody. Thus, this suggests that all the protein detected in body wall muscle is due to UNC-94, and given results of our promoter analysis, specifically, UNC-94a. A lower magnification view (Figure 8e) of UNC-94 and α-actinin staining reveals that UNC-94 extends beyond the rows of dense bodies (white arrows), probably at muscle cell to cell boundaries. This enrichment of UNC-94 near cell to cell boundaries might explain the accumulation of actin filaments near cell boundaries in unc-94 loss of function mutants as revealed by phalloidin staining and EM (Figures 2 and 3, respectively).
Discussion
We have shown here that the mutationally identified gene unc-94 corresponds with the sequence-predicted gene tmd-1, which encodes a tropomodulin. This discovery demonstrates yet another level of conservation in sarcomere components between C. elegans and mammals. Here we show that mutation in or RNAi against unc-94 results in reduced organization of myofibrils, abnormal accumulation of F-actin near muscle cell-to-cell boundaries, reduced motility and brood size. In cultured chick cardiomyocytes, immunodepletion of tropomodulin results in abnormally long actin filament bundles and a reduction in the number of beating cells,26 while overexpression results in shortened actin filaments and myofibril degeneration.27 These complementary results are believed to be due to the fact that tropomodulin is a pointed end F-actin capping protein.28 Our immunofluorescent localization of UNC-94 to two closely spaced parallel lines closely flanking the M-line (Figure 8a), is consistent with UNC-94 being located, as expected, at the pointed ends of thin filaments in the middle of the sarcomere (Figure 8f). Consistent with our immunolocalization results, C. elegans TMD-1 has recently been shown to cap the pointed ends of tropomyosin coated F-actin in vitro.23 While our sequencing of UNC-94 reveals only 36% sequence identity to the C-terminal actin binding domain of chicken E-Tmod, an overlay of the two structures reveals striking conservation of three dimensional structure29 with a least squares fit in LSQMAN revealing a root-mean squared deviation between Ca atoms of 1.24 Å. Furthermore, a docking model of UNC-94 (TMD-1) to actin29 (http://sgce.cbse.uab.edu/sgce/Structures/79-D4/dock.wrl), reveals that the 5 charged residues involved in charge-charge interaction between chicken E-Tmod and actin30 are not only conserved in C. elegans, but are also predicted to be involved in charge-charge interaction with actin.
In sum, the genetic, phenotypic, sequencing, protein localization and structural data suggest that UNC-94 acts as a pointed end capping protein in the striated muscles of C. elegans. Thus, we were surprised to observe that in unc-94 mutants, including sf20 which has no detectable UNC-94 proteins by immunoblot (Figure 7), there are still H zones, as revealed by phalloidin staining (Figure 2). This indicates that the thin filaments are not abnormally long in unc-94 mutants, and thus although UNC-94 is a tropomodulin located at the pointed ends of thin filaments, it may not be required to regulate thin filament lengths. However, sf20 may not be a null allele, and it may still produce sufficient UNC-94 proteins to regulate thin filament length. Several alternative explanations are also possible: (1) UNC-94 may be redundant to the second tropomodulin-like protein in C. elegans encoded by the separate gene, tmd-2. Consistent with this idea, tmd-2 is expressed in body wall muscle, based on both an mRNA tagging strategy31 and SAGE data32 in WormBase (http://www.wormbase.org). (2) Even in mammals, whether tropomodulins function to prevent abnormal elongation of thin filaments in vivo, is not certain. For example, Tmod1 is the only tropomodulin isoform expressed in heart muscle cells, and cardiomyocytes developing from Tmod1 null embryonic stem cells, have thin filaments of normal length.33 Moreover, in mammalian muscle, the giant protein nebulin which interacts with tropomodulin34 (McElhinny et al., 2001) also has a role in regulating thin filament lengths.35 Interestingly, C. elegans does not appear to have a full length nebulin protein, but rather has one small protein with several nebulin-like repeats closely related to LASP, called Ce LASP (K. Wang, K. Mercer, G. Benian, unpub. data).
In addition to identifying UNC-94 as a C. elegans tropomodulin located at the pointed ends of muscle thin filaments, our data raise a new question about tropomodulin function. We have made two observations that suggest that tropomodulins may have a role in attaching sarcomeres to the muscle cell surface: (1) in unc-94 mutant animals, there is an abnormal accumulation of actin filaments near muscle cell-to-cell boundaries (Figure 2 and Supplementary Figure 1), and (2) in wild type muscle, there is localization not only to the pointed ends of thin filaments, but also near muscle cell to cell boundaries (Figure 8e). In fact, using the same anti-UNC-94/TMD-1 antibodies, the protein has been shown recently to localize to the cell borders of nematode hypodermal cells, acting functionally with α-catenin.23 A role for tropomodulin at cell-cell contacts has previously been proposed for lens fiber cells (a type of epithelial cell),36 and overexpression of Tmod1 in cardiac tissue causes dilated cardiomyopathy and disrupted intercalated disks (a type of intercellular junction) in TOT transgenic mice.37 Tropomodulin may be involved in binding to a muscle attachment complex. A possible mechanism is suggested by the recent finding that tropomyosin is involved in erythrocyte membrane mechanical stability.38 Since, in the erythrocyte cytoskeleton, tropomyosin is bound to actin and tropomodulin, perhaps UNC-94 links actin to the cell membrane via tropomyosin in C. elegans muscle. Alternatively, it could be that at muscle cell-to-cell boundaries, in addition to usual roles in binding actin and tropomyosin, UNC-94 interacts with a different protein with a low affinity. For example, Tmod4 has been shown to bind an intermediate filament protein in lens fiber cells, and this does not block its actin capping activity.39
By use of promoter-GFP experiments, we have shown that unc-94 utilizes two promoters that express two very similar proteins in different sets of cells and tissues. Thus, as shown in Figure 5, unc-94a is expressed in body wall, vulval and uterine muscle, while unc-94b is expressed in pharyngeal, vulval and anal depressor muscles, and the spermatheca and intestinal epithelial cells. Confirmation that native unc-94a is expressed in body wall muscle and is functionally important in that tissue was provided by finding that unc-94(su177), which has a body wall muscle phenotype and no UNC-94 immunostaining (data not shown), is a mutation in intron 1 that is specific for unc-94a. Although the differences between the two isoforms at the protein level are minimal, the use of two alternative promoters might have evolved to permit timing and levels of expression finely tuned for the requirements of particular sets of cells or tissues.
In addition to affecting muscle structure and motility, mutations in unc-94 also affect the total number of progeny laid, (i.e., “brood size”). Both mutant alleles affect brood size. The mutation in su177 affects only the unc-94a isoform, and because this isoform is expressed in vulval and uterine muscles, which are required for egg laying, the reduced brood size is expected. The mutation in sf20 affects the expression of both isoforms and shows an even further reduction in brood size. This more severe brood size defect could be explained by noting that in addition to being expressed in vulval and uterine muscles, unc-94b is also expressed in the spermatheca, the site of fertilization and expulsion of the fertilized egg into the uterus. Alternatively, sf20 might give a more severe reduction in brood size because sf20 is a more severe mutation molecularly than su177 (premature stop vs. splice site mutation).
Interestingly, the alternative splicing pattern for unc-94 is similar to that of the Drosophila melanogaster tropomodulin gene,40 which encodes two isoforms through two different promoters and use of two different first exons. Similar to nematode unc-94, the fly tropomodulin gene uses an upstream promoter to express a smaller isoform (367 residues; first exon encodes 2 residues), and a downstream promoter to express a larger isoform (402 residues; first exon encodes 36 residues). In contrast, all vertebrate Tmod genes studied to date do not show alternative splicing41,42 (C. Conley, unpublished data). A lack of alternative splicing has also been found for a newly-identified tropomodulin homologue from the hemichordate Ciona intestinalis (D. Hoffman and C. Conley, unpublished data).
Alignment of the UNC-94a protein sequence with sequences from the homologous proteins of Drosophila and humans (TMOD1) demonstrates that C. elegans UNC-94a is 35.2% identical to the Drosophila protein and 34.7 % identical to the human protein along the entire alignment (Supplementary Figure 2). The three proteins display two large regions of similarity that correspond to the amino-terminal tropomyosin-binding domain and the carboxy-terminal actin-binding domain of the vertebrate tropomodulins, respectively.41 The second C. elegans tropomodulin gene, tmd-2, uses alternative splicing of its 3’-most exons (confirmed by sequencing cDNAs), to encode two isoforms which differ only at their C-termini. The TMD-2a isoform is predicted to be 308 residues long with a 25 residue C-terminal tail, whereas TMD-2b is 639 residues with a 354 residue C-terminal tail. UNC-94a is only 18.9% identical to TMD-2 (Supplementary Figure 2). TMD-2 seems to lack the amino-terminal tropomyosin binding domain and its amino terminal 35 residues are only weakly similar to the amino-terminal regions of the other tropomodulins. One possible reason for the lack of an obvious tropomyosin binding domain is that the N-terminal region of each TMD-2 isoform has not yet been defined. Alternatively, TMD-2 might indeed lack a tropomyosin binding region because it has a different function and localization in the sarcomere. This is suggested by the unusual sequence of the unique C-terminal 354 residues of TMD-2b: (1) It is enriched for the amino acids P, E, V, K and A (total of 55.4%). (2) PFAM predicts a PPAK motif (342-366), which is the 28 residue repeat that comprises the main elastic region of vertebrate titin called the “PEVK region”.43-45 (3) The computer program Radar predicts two copies of another repeat (305-331 and 424-443). Thus, the amino acid composition and presence of short repeating elements, one of which is similar to vertebrate titin, suggest that this region of TMD-2b might be elastic.
Materials and Methods
Strains and Genetics
Two alleles of unc-94 were employed in this study. The first allele, su177, was isolated and described by Zengel and Epstein7 using a motility and polarized light screen. When we obtained unc-94 (su177) from the Caenorhabditis Genetics Center, it had only been outcrossed three times. These 3x outcrossed animals were used for polarized light, motility, and immunofluorescence experiments. We noticed the same polarized light phenotype in these animals as was reported by Zengel and Epstein. The motility of the animals was about 50% less than the motility of wild type animals and by immunofluorescence we noticed that the structure of all myofibrillar components tested for were compromised. After outcrossing these animals an additional two times (now 5x outcrossed), we repeated the motility assay, and noticed that the motility increased about 20%. The polarized light phenotype did not change; therefore immunofluorescence experiments were not repeated. We recovered the second unc-94 allele, sf20, by using EMS mutagenesis and a polarized light F1 non-complementation screen. This allele was also outcrossed five times and previously mentioned experiments, in addition to brood size measurements, were performed using these animals. unc-94 (RNAi) animals were created by feeding46 rrf-3 animals47 (which are hypersensitive to RNAi) bacteria expressing double stranded RNA (dsRNA) for C06A5.7.
Polarized light microscopy, motility, brood size measurements, and electron microscopy
The procedures for polarized light microscopy and motility assays were performed as described in Mercer et al.11 Brood size was determined by taking 10 L4 hermaphrodites (P0) of each allele (wt, su177 and sf20) and putting these animals, individually, onto seeded agar plates. These animals were allowed 24 hours to mature and lay fertilized eggs. After this period, P0 animals were singly transferred to a new plate and were allowed 24 hours to lay more eggs. Laying and transferring were repeated until the P0 animals laid only unfertilized eggs (oocytes). After the P0 animals had been transferred from a plate, that plate, containing F1 larvae and eggs, was kept and the number of F1 progeny was scored by picking/counting individual worms as they were removed from the plate. Electron microscopy of wild type and unc-94(sf20) was performed using general methods described by Hall,48 specifically the “conventional two step fixation” method described at http://www.wormatlas.org/anatmeth/anatmeth.htm.
During the first aldehyde fixation step, the worms were cut in half using a razor blade, to allow better penetration of fixative.
Immunofluorescent localization of antibodies to known myofibril components
The procedure used for immunofluorescent localization in adult muscle was described in Mercer et al.49 The antibodies and the dilutions that were used for each were as follows: anti-MHC A, 1:40050; anti-UNC-89, 1:20051; and anti-α-actinin, 1:200.52 Phalloidin staining of thin filaments was carried out as described by Ono.20 Images were obtained with a Zeiss Axioskop microscope using Fuji Sensia 100 slide film and scanned and processed with Adobe Photoshop.
Genetic and physical mapping of unc-94, and determination of mutation sites
Zengel and Epstein7 used three-factor mapping to place unc-94 between dpy-5 and unc-13 on chromosome I. To narrow down this 2MB region to a 500kb region (representing 12 overlapping cosmids), we used a combination of deficiency mapping and single nucleotide polymorphism (SNP) mapping.53 The Hawaian strain of C. elegans was mated with the triple mutant, dpy-5 unc-94 unc-13, producing a number of recombinant animals. Sequencing SNPs of Dpy-5, Unc-94, non-Unc-13 individuals revealed that unc-94 lies either within or to the left of cosmid F26B1, creating a left breakpoint. After sequencing SNPs of non-Dpy-5 Unc-94 Unc-13 individuals, we were able to create a right breakpoint that included the cosmid C06A5. After scanning the 500kb region for candidate predicted genes on WormBase, we thought a likely candidate for unc-94 was C06A5.7, which encodes a tropomodulin, a known actin regulatory protein. Thus, the cosmid C06A5 was tested for its ability to rescue the Unc-94 phenotype in transgenic animals. (Cosmid DNA was prepared using a QIAGEN Plasmid Midiprep kit; QIAGEN, Valencia,CA). We were able to detect rescue of the mutant phenotype in five different lines. We performed RNAi, by feeding, in rrf-3 animals using C06A5.7 (tmd-1) clones from the Ahringer library (available through GeneService, Cambridge, United Kingdom). Progeny from worms that were fed double stranded RNA- producing bacteria for C06A5.7 showed an Unc-94 phenotype by polarized light.
To determine mutation sites for both mutant alleles, we prepared genomic DNA from the mutant animals by means of phenol/chloroform extraction. Primers were then designed to amplify genomic regions of all C06A5.7 exons (for both isoforms) and ~100 base pairs of their flanking intronic sequences. For each exon/intron, the primers that were used for PCR amplification were also used for sequencing. Sequences were obtained from both strands.
Analysis of unc-94/tmd-1 and tmd-2 coding sequences
Plasmids containing cDNAs for the predicted transcripts unc-94a/tmd-1a, unc-94b/tmd-1b, tmd-2a, and tmd-2b were obtained from the Kohara lab54 : yk1262e07, yk786f09, and yk618b4 for unc-94a/tmd-1a, yk1191a05, yk1009c10 and yk1056g10 for unc-94b/tmd-1b, yk724h2 for tmd-2a, and yk569e4 and yk416c2 for tmd-2b. Plasmid DNA was then prepared using a QIAGEN Plasmid Miniprep kit (QIAGEN, Valencia, CA) and sequenced (Certigen, Lubbock, Texas) using forward primer pME18F2: TCAGTGGATGTTGCCTTTAC and reverse primer ME-1250RV: TGTGGGAGGTTTTTTCTCTA. To sequence all cDNAs except y724h2 and yk1191a1 Certigen designed internal primers against sequenced portions of the cDNA; the sequence of these primers is available upon request from Certigen. Sequencing confirmed the predicted structure of unc-94a/tmd-1a and unc-94b/tmd-1b, but indicated that exons 5 and 6 of tmd-2a and tmd-2 b were shorter than initially predicted. The revised coding sequences of the tmd-2 transcripts were submitted to www.wormbase.org and also confirmed by the C. elegans orfeome project.55 yk262e07 (unc-94a/tmd-1a) was sent to the Southeast Collaboratory for Structural Genomics, which independently confirmed the sequence and confirmed the protein sequence of the C-terminal, actin binding domain of TMD-1/UNC-94 by solving the crystal structure.56,29 Amino acid sequences were multiply-aligned using the ClustalW algorithm implemented on Lasergene. In addition, TMD-2 sequences were analyzed for protein domains by PFAM57 (version 21.0; www.sanger.ac.uk/Software/Pfam), and for repeating motifs using the program Radar58 (www.ebi.ac.uk/Radar).
Generation of transgenic lines carrying unc-94 promoter constructs
To obtain promoter sequences for unc-94 isoforms a and b, we designed forward and reverse primers CATTCTGCAGATTTTTCAGGTGCCGAGAGTAACATTTTCAAAC and CATTGGATCCAGTTTTAGCCTGACTCATCGCTGATGG (respectively) for isoform a and designed forward and reverse primers CATTCTGCAGCTTATCTCTCACTGGTTCCAGAA-CAGGTGAC and CATTGGATCCATGATAAATTCGTGATCTAGGAAACATGGGTGG (respectively) for isoform b. These primers were used for PCR amplification using genomic DNA as template. In the case of isoform a, 3.8 kb of sequence upstream of the predicted start methionine plus the a-specific first exon were fused in-frame to GFP. For isoform b, 2.2 kb of sequence upstream plus b-specific first exon were fused to GFP. Both PCR products were then digested with PstI and BamHI. These fragments were ligated into the promotorless gfp vector pPD95.77 (provided by Andy Fire, Stanford University, Stanford, CA), which had been previously cut with the same two restriction enzymes. The ligation reactions were then used to transform E. coli strain XL1 Blue for plasmid amplification. For each isoform, two clones were pooled and injected (25ng/μl) along with the reporter gene, rol-6 (80ng/μl) into gravid N2 hermaphrodites. These injections resulted in the production of two transgenic lines for each isoform. GFP fluorescent images of different adult muscle structures were obtained as described in Mercer et al.11
Northern blot
Total RNA from mixed stage populations of wild type and the two unc-94 mutant alleles was prepared using the TRIzol Reagent and a protocol provided by Invitrogen, Inc. A northern blot was prepared and hybridized using materials and methods described in the NorthernMax kit from Ambion, Inc. A 1.3% agarose formaldehyde MOPS buffer gel was used to separate the RNAs (18 μg per lane) and transferred to a nylon membrane. The 0.5—10 kb RNA Ladder from Invitrogen was used as a size marker. DNA probes were labeled with 32P using the DECAprime II random primed labeling kit from Ambion, Inc. Three probes, produced by PCR from cDNA and gel-purified were used; (1) an unc-94a-specific probe containing the entire unc-94a-specific first exon generated by primers ATTTCGTCGTGGAAAGCCTGAG and CAGTTTTAGCCTGACTCATCGCTG; (2) a probe that recognizes both unc-94a and unc-94b transcripts containing most of exons 2 and 4 generated by primers CCTTCTCAGCACCGTCAGCG and CAGGGGCAGTTACAAGAGCAC; and (3) a probe that recognizes the 3’ end of the unc-15 (paramyosin) gene using primers CGCGGATCCGAGGAACAAGAACACTCGATG AND GCGGTCGACTTAATAATCGTCTTCCGTGAC.
Western blot and immunofluorescent localization of UNC-94/TMD-1
Extracts of Laemmli-soluble proteins from wild type, su177, sf20, worms fed E. coli harboring an empty RNAi vector, or unc-94(RNAi) worms were prepared by the method of Hannak et al.59 The protein concentrations of these extracts were determined by the filter paper dye-binding method of Minamide and Bamburg.60 After separation of 8 μgs of each extract on a 12% SDS-PAGE and transfer to nitrocellulose, the immunoblot was reacted with affinity-purified rabbit anti-TMD-1b (residues 144-401), the generation of which is described in Cox et al.,23 at a 1:400 dilution and visualized by enhanced chemiluminescence (ECL) (Pierce, Inc.). To verify that equal amounts of total protein were loaded in each lane, the blot was washed and then incubated with anti-paramyosin (monoclonal 5-23),50 and visualized by ECL. The same affinity-purified anti-TMD-1b antibodies were used to localize UNC-94 in wild type and sf20 adult muscle. Figure 8a and b show the results using the picric acid fixation method described in Nonet et al.24 Anti-TMD-1 was used at 1:50 dilution, and anti-α-actinin (MH35) and anti-UNC-89 (MH42) were used at 1:200 dilutions. Figure 8 c, d and e show results using the standard paraformaldehyde/methanol fixation method described by Finney and Ruvkun25 and modified by Benian et al.51 Anti-TMD-1 was used at 1:100 dilution, and anti-α-actinin (MH35) and anti-UNC-89 (MH42) were used at 1:200 dilutions. Rabbit antibodies were visualized by anti-rabbit antibodies conjugated with Alexa 488 (Molecular Probes, Inc.), and mouse antibodies were visualized by anti-mouse antibodies conjugated with Cy3 (Jackson Immunochemicals). Images were captured with a Carl Zeiss LSM 510 confocal microscopy system.
Supplementary Material
Acknowledgments
We thank Hiroshi Qadota for valuable advice on the project and help with the microscopy, Jeannette Taylor for EM images, Andy Fire for the promoterless GFP vector, Alan Coulson for cosmid clones, and Yuji Kohara for cDNA clones. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the National Institutes of Health. These studies were supported by grant AR052133 from the National Institutes of Health to G.M.B., by grant GM58038 from the National Institutes of Health and grant 4218 from the Muscular Dystrophy Association to J.H.
Abbreviations used
- GFP
green fluorescent protein
- RNAi
RNA mediated interference
- MHC A
myosin heavy chain A
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waterston RH. Muscle. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1988. pp. 281–335. [Google Scholar]
- 2.Moerman DG, Fire A. Muscle: structure, function and development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1997. pp. 417–470. [PubMed] [Google Scholar]
- 3.Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. Wormbook, ed. The C. elegans Research Community, WormBook. 2006 January 16; doi: 10.1895/wormbook.1.81.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 4.Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. UNC-98 links an integrin-associated complex to thick filaments in C. elegans muscle. J Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner S. The Genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure in C. elegans. Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- 7.Zengel JM, Epstein HF. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]
- 8.Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG. The unc-112 gene in Caenhorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J Cell Biol. 2000;150:253–264. doi: 10.1083/jcb.150.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barral JM, Bauer CC, Ortiz I, Epstein HF. unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol Biol Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moerman DG, Benian GM, Barstead RJ, Schreifer L, Waterston RH. Identification and intracellular localization of the unc-22 gene product of C. elegans. Genes Devel. 1988;2:93–105. doi: 10.1101/gad.2.1.93. [DOI] [PubMed] [Google Scholar]
- 13.Benian GM, Kiff JE, Neckelmann N, Moreman DG, Waterston RH. The sequence of twitchin: an unusually large protein implicated in regulation of myosin activity in C. elegans. Nature. 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- 14.Benian GM, L’Hernault SW, Morris ME. Additional sequence complexity in the muscle gene unc-22, and its encoded protein, twitchin, of C. elegans. Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- 16.Goetinck S, Waterston RH. The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J Cell Biol. 1994;127:79–93. doi: 10.1083/jcb.127.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKim KS, Matheson C, Marra MA, Wakarchuk MF, Baillie DL. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- 18.Ono S, Baillie DL, Benian GM. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohri K, Ono S. Actin filament disassemblying activity of Caenorhabditis elegans actin-interacting protein1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- 22.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 23.Cox EA, Yamashiro S, Ono S, Hardin J. TMD-1/tropomodulin acts with HMP-1/α-catenin to reinforce adherens junctions under stress during morphogenesis. 2007 Submitted. [Google Scholar]
- 24.Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- 25.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 26.Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- 27.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 28.Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu S, Symersky J, Li S, Carson M, Chen L, Meehan E, Luo M. Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain. Proteins. 2004;56:384–386. doi: 10.1002/prot.10597. [DOI] [PubMed] [Google Scholar]
- 30.Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophys J. 2002;83:2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal sluctering of muscle-espressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 32.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 33.Ono Y, Schwach C, Antin PB, Gregorio CC. Disruption in the tropomodulin1 (Tmod1) gene compromises cardiomyocyte development in murine embryonic stem cells by arresting myofibril maturation. Devel Biol. 2005;282:336–348. doi: 10.1016/j.ydbio.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 34.McElhinny AS, Kolmerer B, Fowler VM, Labeit S, Gregorio CC. The N-terminal end of nebulin interacts with tropomodulin at the pointed ends of the thin filaments. J Biol Chem. 2001;276:583–592. doi: 10.1074/jbc.M005693200. [DOI] [PubMed] [Google Scholar]
- 35.McElhinny AS, Schwach C, Valichnac M, Mount-Patrick S, Gregorio CC. Nebulin regulates the assembly and lengths of the thin filaments in striated muscle. J Cell Biol. 2005;170:947–957. doi: 10.1083/jcb.200502158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee A, Fischer RS, Fowler VM. Stabilization and remodeling of the membrane skeleton during lens fiber cell differentiation and maturation. Dev Dyn. 2000;217:257–270. doi: 10.1002/(SICI)1097-0177(200003)217:3<257::AID-DVDY4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Ehler E, Horowits R, Zuppinger C, Price RL, Perriard E, Leu M, Caroni P, Sussman M, Eppenberger HM, Perriard J-C. Alterations at the intercalated disk associated with the absence of muscle LIM protein. J Cell Biol. 2001;153:763–772. doi: 10.1083/jcb.153.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An X, Salomao M, Guo X, Gratzer W, Mohandas N. Tropomyosin modulates erythrocyte membrane stability. Blood. 2007;109:1284–1288. doi: 10.1182/blood-2006-07-036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer RS, Quinlan RA, Fowler VM. Tropomodulin binds to filensin intermediate filaments. FEBS Lett. 2003;547:228–232. doi: 10.1016/s0014-5793(03)00711-7. [DOI] [PubMed] [Google Scholar]
- 40.Mardahl-Dumesnil M, Fowler VM. Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscle. J Cell Biol. 2001;155:1043–1053. doi: 10.1083/jcb.200108026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:27–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 42.Conley CA, Fowler VM. Tropomodulin genes in Gallus domesticus and in mammals: gene structure, protein homologies and tissue distribution. Cytogen & Gen Res. 2005;109:457–459. [Google Scholar]
- 43.Greaser M. Identification of new repeating motifs in titin. Proteins: Struct Funct Genet. 2001;43:145–149. doi: 10.1002/1097-0134(20010501)43:2<145::aid-prot1026>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 44.Bang M-L, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 45.Gutierrez-Cruz G, Van Heerden A, Wang K. Modular motif, structural folds and affinity profiles of PEVK segment of human fetal skeletal muscle titin. J Biol Chem. 2001;276:7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- 46.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 47.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RHA. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 48.Hall DH. Electron microscopy and three-dimensional image reconstruction. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: modern biological analysis of an organism. Academic Press; San Diego: 1995. pp. 396–436. [Google Scholar]
- 49.Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller DM, Ortiz Il, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- 51.Benian GM, Tinley TL, Tang X, Borodovsky M. The C. elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of immunoglobulin and signal transduction domains. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill K, Harfe BD, Dobbins CA, L’Hernault SW. dpy-18 encodes an alpha-subunit of prolyl-4-hydroxylase in C. elegans. Genetics. 2000;155:1139–1148. doi: 10.1093/genetics/155.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohara Y. Large scale analysis of C. elegans cDNA. Tanpakushitsu Kakusan Koso. 1996;41:715–720. [PubMed] [Google Scholar]
- 55.Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, Moore T, Hudson JR, Jr, Hartley JL, Brasch MA, Vandenhaute J, Boulton S, Endress GA, Jenna S, Chevet E, Papasotiropoulos V, Tolias PP, Ptacek J, Snyder M, Huang R, Chance MR, Lee H, Doucette-Stamm L, Hill DE, Vidal M. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 56.Ding H, Qiu S, Bunzel RJ, Luo D, Arabashi A, Lu S, Symersky J, Nagy LA, DeLucas LJ, Li S, Luo M. Purification, nanocrystallization and preliminary X-ray analysis of a C-terminal part of tropomodulin protein 1, isoform A, from Caenorhabditis elegans. Acta Crystallogr D Biol Crystallogr. 2003;59:1106–1108. doi: 10.1107/s0907444903008217. [DOI] [PubMed] [Google Scholar]
- 57.Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer ELL. Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucl Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heger A, Holm L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins: Struct Funct Genet. 2000;41:224–237. doi: 10.1002/1097-0134(20001101)41:2<224::aid-prot70>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 59.Hannak E, Oegema K, Kirkham M, Gonczy P, Habermann B, Hyman AA. The kinetically dominant assembly pathway for centrosomal asters in C. elegans is γ-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minamide LS, Bamburg JR. A filter paper dye-binding assay for quantitative determination of protein without interference from reducing agents or detergents. Anal Biochem. 1990;190:66–70. doi: 10.1016/0003-2697(90)90134-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.