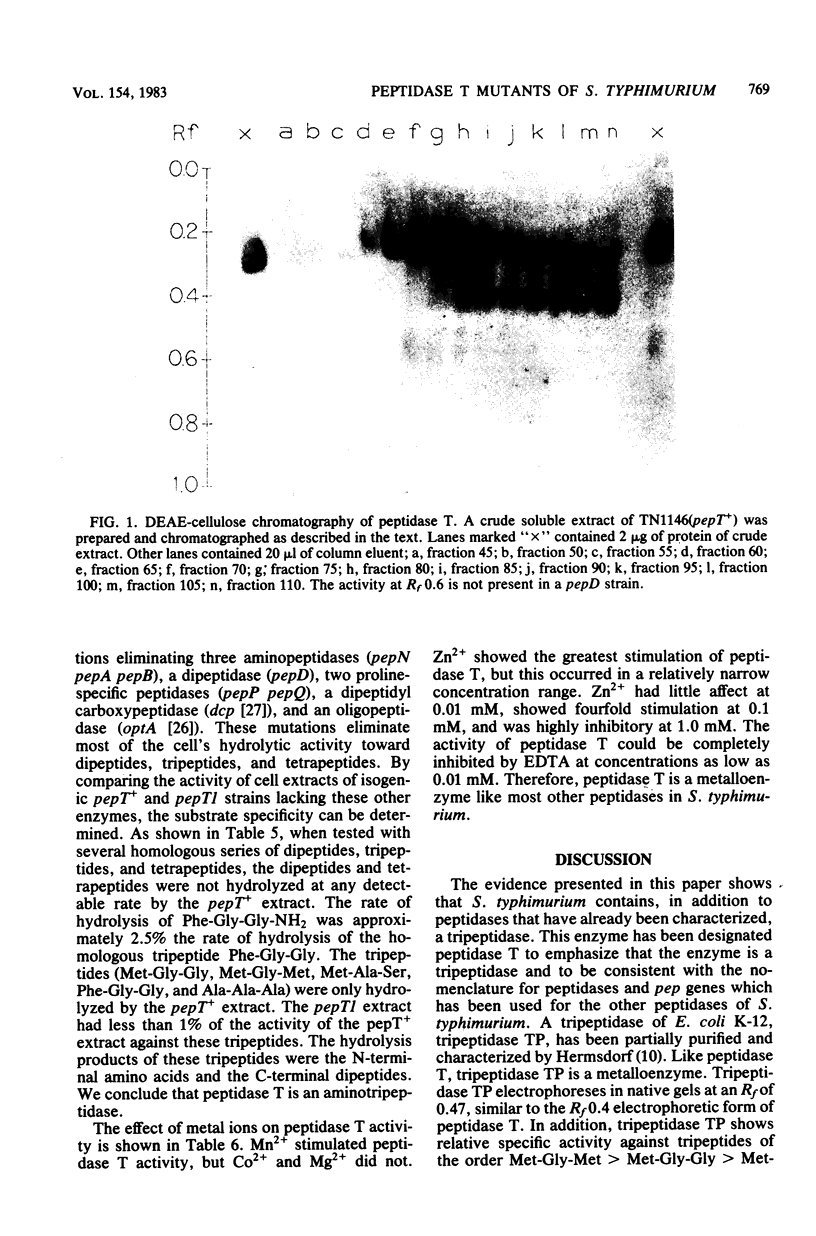
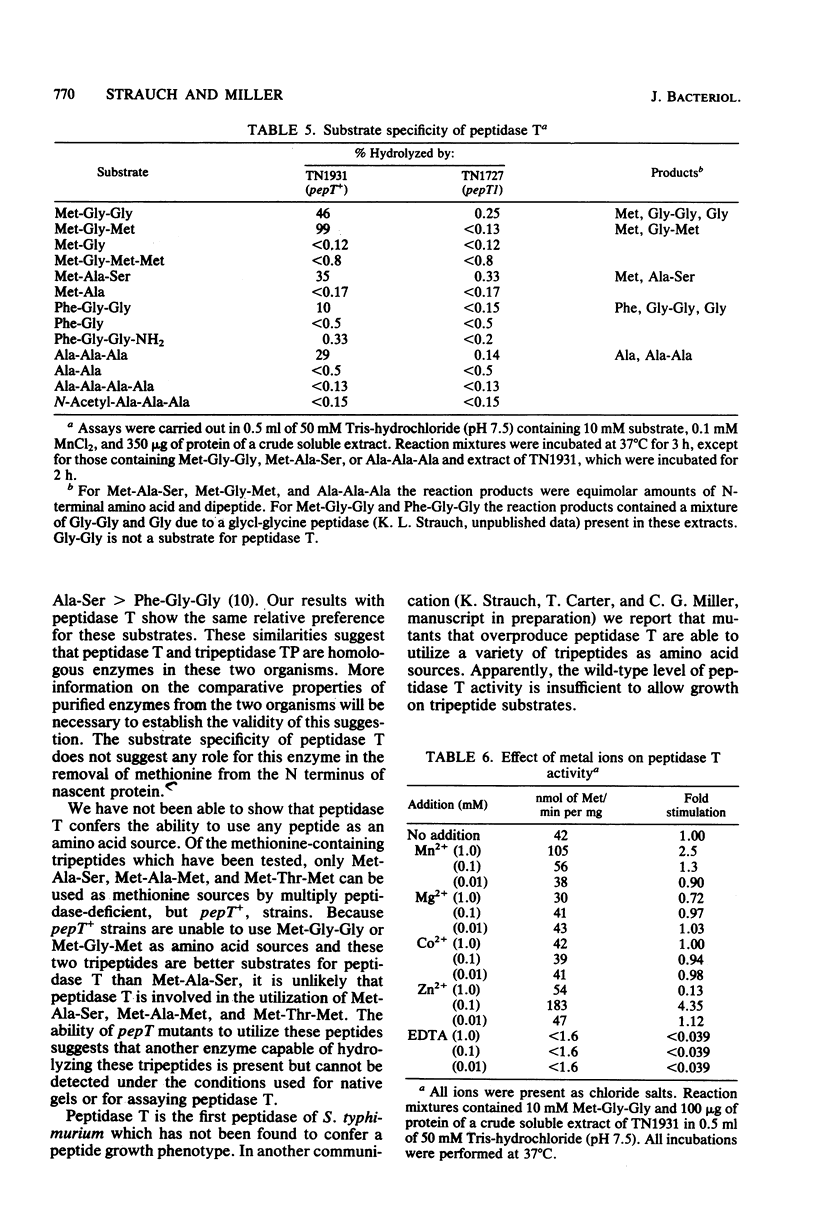
Abstract
Salmonella typhimurium contains an enzyme, peptidase T, that hydrolyzes a variety of tripeptides. Specificity studies with a peptidase activity stain after gel electrophoresis of crude cell extracts showed that peptidase T hydrolyzes tripeptides containing N-terminal methionine, leucine, or phenylalanine. Little or no activity could be detected against dipeptides, N-blocked or C-blocked tripeptides, and tetrapeptides. Analysis of reaction products by high-pressure liquid chromatography showed that peptidase T removes the N-terminal amino acid from tripeptides. Mutants lacking peptidase T were isolated by screening microcultures grown in the wells of plastic microtitration plates for hydrolysis of Met-Ala-Ser or Met-Gly-Gly. Mutations (pepT) that eliminate this enzyme were found to be phage P22 cotransducible with purB at approximately 25 map units on the S. typhimurium map. Comparison of the growth properties of mutant and wild-type strains suggests that peptidase T does not function in utilization of tripeptides to provide amino acids during growth.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N., Ames G. F., Young J. D., Tsuchiya D., Lecocq J. Illicit transport: the oligopeptide permease. Proc Natl Acad Sci U S A. 1973 Feb;70(2):456–458. doi: 10.1073/pnas.70.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L. A., Kaesberg P. Cleavage of the N-terminal formylmethionine residue from a bacteriophage coat protein in vitro. J Mol Biol. 1973 Sep 25;79(3):531–537. doi: 10.1016/0022-2836(73)90404-x. [DOI] [PubMed] [Google Scholar]
- Barak Z., Gilvarg C. Triornithine-resistant strains of Escherichia coli. Isolation, definition, and genetic studies. J Biol Chem. 1974 Jan 10;249(1):143–148. [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I. Bacteriophage mu as a transposition element. Annu Rev Genet. 1976;10:389–412. doi: 10.1146/annurev.ge.10.120176.002133. [DOI] [PubMed] [Google Scholar]
- Capecchi M. R. Initiation of E. coli proteins. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1517–1524. doi: 10.1073/pnas.55.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N., Howe M. M., Ingraham J. L., Pierson L. S., 3rd, Turnbough C. L., Jr Infection of Salmonella typhimurium with coliphage Mu d1 (Apr lac): construction of pyr::lac gene fusions. J Bacteriol. 1981 Jan;145(1):299–305. doi: 10.1128/jb.145.1.299-305.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsdorf C. L. Tripeptide-specific aminopeptidase from Escherichia coli AJ005. Biochemistry. 1978 Aug 8;17(16):3370–3376. doi: 10.1021/bi00609a030. [DOI] [PubMed] [Google Scholar]
- Hong J. S., Ames B. N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housman D., Gillespie D., Lodish H. F. Removal of formyl-methionine residue from nascent bacteriophage f2 protein. J Mol Biol. 1972 Mar 14;65(1):163–166. doi: 10.1016/0022-2836(72)90498-6. [DOI] [PubMed] [Google Scholar]
- Kirsh M., Dembinski D. R., Hartman P. E., Miller C. G. Salmonella typhimurium peptidase active on carnosine. J Bacteriol. 1978 May;134(2):361–374. doi: 10.1128/jb.134.2.361-374.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lenny A. B., Margolin P. Locations of the opp and supX genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1980 Aug;143(2):747–752. doi: 10.1128/jb.143.2.747-752.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney W. C., Hermodson M. A. Separation of large denatured peptides by reverse phase high performance liquid chromatography. Trifluoroacetic acid as a peptide solvent. J Biol Chem. 1980 Dec 10;255(23):11199–11203. [PubMed] [Google Scholar]
- McHugh G. L., Miller C. G. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):364–371. doi: 10.1128/jb.120.1.364-371.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G., Green L. Degradation of proline peptides in peptidase-deficient strains of Salmonella typhimurium. J Bacteriol. 1983 Jan;153(1):350–356. doi: 10.1128/jb.153.1.350-356.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G., Mackinnon K. Peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G., Schwartz G. Peptidase-deficient mutants of Escherichia coli. J Bacteriol. 1978 Aug;135(2):603–611. doi: 10.1128/jb.135.2.603-611.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., Green L., Miller C. G. Oligopeptidase-deficient mutants of Salmonella typhimurium. J Bacteriol. 1983 Mar;153(3):1259–1265. doi: 10.1128/jb.153.3.1259-1265.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., Miller C. G. Dipeptidyl carboxypeptidase-deficient mutants of Salmonella typhimurium. J Bacteriol. 1983 Mar;153(3):1252–1258. doi: 10.1128/jb.153.3.1252-1258.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALLER J. P. THE NH2-TERMINAL RESIDUES OF THE PROTEINS FROM CELL-FREE EXTRACTS OF E. COLI. J Mol Biol. 1963 Nov;7:483–496. doi: 10.1016/s0022-2836(63)80096-0. [DOI] [PubMed] [Google Scholar]
- Yen C., Green L., Miller C. G. Degradation of intracellular protein in Salmonella typhimurium peptidase mutants. J Mol Biol. 1980 Oct 15;143(1):21–33. doi: 10.1016/0022-2836(80)90122-9. [DOI] [PubMed] [Google Scholar]