Abstract
The actin monomer-binding protein, profilin, influences the dynamics of actin filaments in vitro by suppressing nucleation, enhancing nucleotide exchange on actin, and promoting barbed-end assembly. Profilin may also link signaling pathways to actin cytoskeleton organization by binding to the phosphoinositide PIP2 and to polyproline stretches on several proteins. Although activities of profilin have been studied extensively in vitro, the significance of each of these activities in vivo needs to be tested. To study profilin function, we extensively mutagenized the Saccharomyces cerevisiae profilin gene (PFY1) and examined the consequences of specific point mutations on growth and actin organization. The actin-binding region of profilin was shown to be critical in vivo. act1-157, an actin mutant with an increased intrinsic rate of nucleotide exchange, suppressed defects in actin organization, cell growth, and fluid-phase endocytosis of pfy1-4, a profilin mutant defective in actin binding. In reactions containing actin, profilin, and cofilin, profilin was required for fast rates of actin filament turnover. However, Act1-157p circumvented the requirement for profilin. Based on the results of these studies, we conclude that in living cells profilin promotes rapid actin dynamics by regenerating ATP actin from ADP actin–cofilin generated during filament disassembly.
Keywords: yeast, cytoskeleton, cofilin, filament turnover, ATP
Introduction
The ability of the actin cytoskeleton to undergo rapid rearrangements is critical for such cellular processes as cell polarity development, endocytosis, cell motility, and cytokinesis. Actin dynamics are modulated by a host of actin-binding proteins (Pollard 1994). One such protein, profilin, binds to actin monomers and may, through its ability to interact with several proteins and phosphoinositides, link signaling pathways to the actin cytoskeleton (Machesky and Pollard 1993). Profilin can impact actin dynamics by sequestering actin monomers (Carlsson et al. 1977), increasing the rate of exchange of ATP for ADP on actin (Mockrin and Korn 1980; Goldschmidt-Clermont et al. 1991), and promoting barbed-end assembly of actin filaments (Pantaloni and Carlier 1993; Kang et al. 1999). Profilin also increases the rate of actin filament turnover in a synergistic manner with cofilin, a protein that promotes disassembly of actin filaments at their pointed ends (Didry et al. 1998).
In all organisms studied to date, profilin plays a critical role in establishing and maintaining the proper organization of the actin cytoskeleton (Ayscough 1998). We have chosen to study profilin function in the yeast Saccharomyces cerevisiae because it is a genetically tractable organism with a dynamic actin cytoskeleton. The yeast actin cytoskeleton consists of cables, which orient along the mother-bud axis, and cortical patches, which polarize in the developing bud and reorganize depending on the specific stage in the cell cycle (Pruyne and Bretscher 2000). The importance of profilin in the organization of the yeast actin cytoskeleton has been demonstrated by analysis of yeast lacking profilin. These cells grow extremely slowly, become large and rounded, lack detectable actin cables, and no longer polarize their actin patches to the bud (Haarer et al. 1990). In addition to interacting with actin, yeast profilin (Pfy1p) binds to stretches of proline residues in the FH (formin homology) domains of Bni1p and Bnr1p (Kohno et al. 1996; Evangelista et al. 1997; Imamura et al. 1997). Members of the FH protein family bind to proteins of the Rho family of small GTPases and are involved in cytokinesis and the establishment of cell polarity (Wasserman 1998). The interaction between profilin and FH proteins thus provides a link between signaling by small GTPases and the actin cytoskeleton.
Although the interactions of profilin with actin and polyproline have been studied extensively biochemically, the significance of each of these activities in vivo has been much more difficult to test. The importance of the ability of profilin to enhance nucleotide exchange on actin has been controversial. Mammalian and amoeba profilins increase the rate of nucleotide exchange on actin in a concentration-dependent manner, by as much as 1,000-fold (Mockrin and Korn 1980; Goldschmidt-Clermont et al. 1991; Vinson et al. 1998; Selden et al. 1999). However, plant profilins do not affect nucleotide exchange of rabbit muscle actin (Perelroizen et al. 1995, Perelroizen et al. 1996), and yeast profilin only increases the rate of yeast actin nucleotide exchange approximately threefold (Eads et al. 1998). Furthermore, in Xenopus extracts, actin monomers are primarily associated with ATP rather than ADP (Rosenblatt et al. 1995), suggesting that nucleotide exchange might not be a rate-limiting step during actin assembly. However, profilin may be required to desequester and/or promote nucleotide exchange on actin bound by actin monomer-binding proteins, such as cofilin and thymosin β-4 (Pantaloni and Carlier 1993; Blanchoin and Pollard 1998). Data presented here indicate that the ability of profilin to enhance nucleotide exchange on actin is an important function in vivo.
Materials and Methods
Plasmid Construction and Mutagenesis
All mutations were generated using the QuikChange mutagenesis kit (Stratagene). The PFY1 gene encoding yeast profilin was PCR amplified from genomic DNA 301 bp upstream and 159 bp downstream of PFY1 using 5′ and 3′ primers containing engineered SacI and KpnI sites, respectively. The PCR product was cloned into pBluescript to generate pAW1. To mark PFY1 with the LEU2 auxotrophic marker, a NotI site was introduced into pAW1 93 bp upstream of the PFY1 gene to generate pAW2. The LEU2 fragment from pDD802 was ligated to NotI-digested pAW2 to generate pAW3.
All mutations in the PFY1 gene were generated using pAW2 as a template. After mutagenesis and verification of the mutation by restriction enzyme digestion and DNA sequencing, LEU2 was subcloned into the NotI site of each mutated plasmid.
The plasmid for overexpression of yeast profilin in Escherichia coli was pMW172 (Eads et al. 1998). To overexpress mutant profilins, pMW172 was mutagenized.
Yeast Strain Construction
Standard techniques were used to grow, manipulate, and transform yeast strains (Rose et al. 1989). Complete disruption of the PFY1 gene was performed using PCR-mediated gene replacement as described (Goode et al. 1998), using the HIS3 plasmid pRS303 (Sikorski and Hieter 1989). The disruption was generated in yeast strain DDY1102 to create strain DDY2001 (MAT a/α ade2-1/+ his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 +/lys2-801 +/pfy1Δ::HIS3 ura3-52/ura3-52). DDY2001 was sporulated to generate the PFY1 deletion strain DDY2034 (MAT a his3Δ200 leu2-3,112 lys2-801 pfy1Δ::HIS3 ura3-52).
Plasmids carrying either PFY1 or the mutant pfy1 alleles marked with LEU2 were digested with BssHII and transformed into DDY2001 for integration at the pfy1Δ::HIS3 site. His−, Leu+ colonies were selected, and the presence of the mutation was verified by PCR and restriction enzyme digestion. Diploids were sporulated to generate haploid strains carrying PFY1 or mutant pfy1 alleles. The genotype of each strain was identical to that of the pfy1Δ strain DDY2034, except pfy1 alleles were marked with LEU2. The DDY strain numbers for the strains used in this study are listed in Table .
Table 1.
Integrated Alleles of PFY1
| Growth | |||||
|---|---|---|---|---|---|
| Allele | Mutation | Feature | YPD | YPD ± formamide | DDY strain number |
| PFY1 | None | Wild-type | Wild-type | Wild-type | 2044 |
| pfy1-2 | R75A | Charged | Wild-type | Wild-type | 2011 |
| pfy1-3 | R90A, K92A | Charged | Wild-type | Wild-type | 2030 |
| pfy1-15 | R23A, R75A | Charged | Wild-type | Wild-type | 2027 |
| pfy1-17 | R23A, K92A | Charged | Wild-type | Wild-type | 2029 |
| pfy1-4 | K66A | Actin binding | Conditional (25–34°C) | Lethal (37°C) | 2008 |
| pfy1-5 | R71A | Actin binding | Wild-type | Lethal (37°C) | 2014 |
| pfy1-6 | R80A | Actin binding | Wild-type | Lethal (37°C) | 2025 |
| pfy1-16 | K66A, R80A | Actin binding | Conditional (25–34°C) | Not determined | 2038 |
| pfy1-7 | H81A | Actin binding | Wild-type | Wild-type | 2017 |
| pfy1-8 | D73A, D74A | Charged | Wild-type | Wild-type | 2012 |
| pfy1-9 | D7A | Polyproline binding | Wild-type | Wild-type | 2019 |
| pfy1-10 | Q3A | Polyproline binding | Wild-type | Wild-type | 2020 |
| pfy1-11 | S1A | Polyproline binding | Wild-type | Wild-type | 2023 |
| pfy1-12 | W29A | Polyproline binding | Lethal | Not determined | 2003 |
| pfy1-13 | W2A | Polyproline binding | Conditional (25–34°C) | Not determined | 2036 |
| pfy1-14 | Y119A, Y125A | Polyproline binding | Conditional (25–34°C) | Lethal (37°C) | 2033 |
| pfy1-18 | Y119A | Polyproline binding | Wild-type | Lethal (37°C) | 2041 |
| pfy1Δ | Deletion | Profilin null | Conditional (25–30°C) | Lethal (37°C) | 2034 |
Phenotypic Analysis and Microscopy
For phenotypic analysis of each pfy1 allele, strains derived from two independent transformants were examined. Growth on formamide was examined on YPD plates supplemented with 2.5% formamide at 25 and 37°C. Actin organization was examined by fixing and staining cells grown at 23°C, or shifted to 37°C for 4 h, with rhodamine-phalloidin (Cope et al. 1999). Uptake of Lucifer yellow by fluid phase endocytosis was performed as described (Belmont et al. 1999b), except that the wash buffer was of 50 mM potassium phosphate, pH 7.5, 10 mM NaN3, and 10 mM NaF.
Protein Purification
Recombinant yeast profilin was purified from E. coli as described for wild-type cofilin expressed as a nonfusion protein (Lappalainen et al. 1997), except that buffers contained 20 mM Tris, pH 8.0. Profilin concentration was determined spectrophotometrically as described (Eads et al. 1998).
Yeast cofilin was purified as a GST fusion protein and cleaved from GST by thrombin digestion (Lappalainen et al. 1997). Yeast actin was purified using DNAse I affinity chromatography as described (Rodal et al. 1999).
In Vitro Assays
Binding of profilin to monomeric actin was measured by determining the amount of actin that copelleted with profilin bound to poly-l-proline (PLP) Sepharose. PLP–Sepharose was prepared as described (Rozycki et al. 1991). Variable concentrations of profilin were incubated with excess PLP–Sepharose beads in G-actin buffer (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.5 mM ATP, 0.2 mM DTT) at 4°C for 1 h. The beads were then washed three times with G-actin buffer, and G-actin was added to a final concentration of 5 μM. Samples were incubated at 4°C for 1 h. Beads were washed once with G-actin buffer, twice with G-actin buffer plus 100 mM NaCl, and were then resuspended in SDS sample buffer and subjected to SDS-PAGE. Gels were stained with Coomassie brilliant blue and quantified using the Alpha Imager 2000 system (Alpha Innotech Corp.).
Binding of profilin to monomeric actin was also measured indirectly using an actin polymerization assay. Various concentrations of profilin were mixed with 5 μM G-actin, and polymerization was initiated by the addition of 2 mM MgCl2, 100 mM KCl at room temperature. After 45 min, the reactions were centrifuged at 90,000 rpm for 20 min in a TLA100 rotor at 25°C. Protein concentration in supernatants and pellets was analyzed by SDS-PAGE as above.
Urea denaturation of profilin was measured by incubating 0.5 μM profilin in PBS buffer containing various concentrations of urea overnight at 4°C. The fluorescence of each protein was monitored at 370-nm emission and 280-nm excitation wavelengths. Two independent data sets for each profilin were normalized and averaged. Data was collected on a PTI spectrofluorometer.
The rate of actin treadmilling in the presence of profilin and cofilin was measured by monitoring the release of inorganic phosphate as described (Belmont et al. 1999a).
Results
Actin and Polyproline Binding Functions of Profilin In Vivo
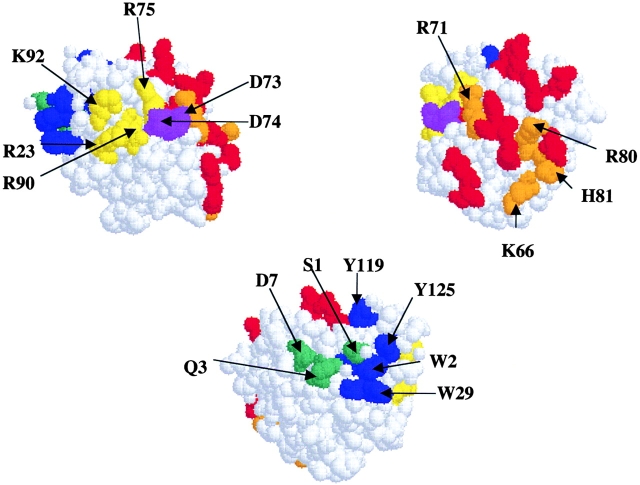
To identify the regions of profilin critical for its function in vivo, we made specific point mutations in conserved amino acid residues located on the yeast profilin surface (Schutt et al. 1993; Eads et al. 1998). Residues predicted to be involved in actin binding or polyproline binding, as well as charged amino acids clustered on the surface of the protein, were replaced with alanine (Fig. 1). Mutant profilin (pfy1) alleles were integrated into the endogeneous profilin locus in the yeast genome, and haploid strains were analyzed for growth at various temperatures (Table ). Immunoblotting showed that mutant profilins were expressed at levels indistinguishable from wild-type profilin at both 25 and 37°C (data not shown). We conclude that amino acids in the actin- and polyproline-binding regions of the protein are important for profilin function, whereas charged amino acids located on other surfaces of the protein are not. The phenotypes of all of the mutations were less severe than the profilin deletion, except in the case of pfy1-12, which appeared to be lethal.
Figure 1.
Space filling model of yeast profilin, showing three rotations. Residues mutated in this study are indicated by arrows. Residues are color coded to illustrate function. Residues predicted to contact actin are red and orange. Orange residues were mutated to alanine. Residues forming the polyproline binding site are blue and green. Blue residues are hydrophobic and are predicted to have stronger effects on polyproline binding than green residues. Conserved basic residues and acidic residues are colored yellow and purple, respectively.
Actin organization in all mutant strains was examined. Strains with growth defects also displayed disordered actin, the severity of which was proportional to the severity of the growth phenotype. We focused on two mutant profilin alleles with severe growth phenotypes, pfy1-4 and pfy1-14, with mutations located in the actin- and polyproline-binding regions, respectively. Actin organization in a pfy1-4 strain is shown in Fig. 2. The actin phenotype for pfy1-14 was indistinguishable from that of pfy1-4 (data not shown). At the permissive temperature of 25°C, strains expressing either pfy1-4 or pfy1-14 had a slight defect in localizing actin patches to the bud, and cables were not detectable. These mutant cells were also more round and slightly larger than wild-type cells, and 6% of cells from a log phase culture of pfy1-4 or pfy1-14 mutants had more than one bud. When pfy1-4 or pfy1-14 cells were shifted to the nonpermissive temperature of 37°C, the cells became more swollen and actin patches became completely nonpolarized, more brightly stained with phalloidin, and more clumpy. The occurrence of multibudded cells increased to as much as 20%, and in some instances, a thick cable-like structure was observed crossing the length of the cell or curving within the mother cell. The disordered actin cytoskeleton and abnormal morphology of pfy1-4 and pfy1-14 are similar to the phenotype of the profilin null (Haarer et al. 1990). Therefore, it appears that the actin- and polyproline-binding regions of profilin are critical for both growth and actin organization in vivo.
Figure 2.
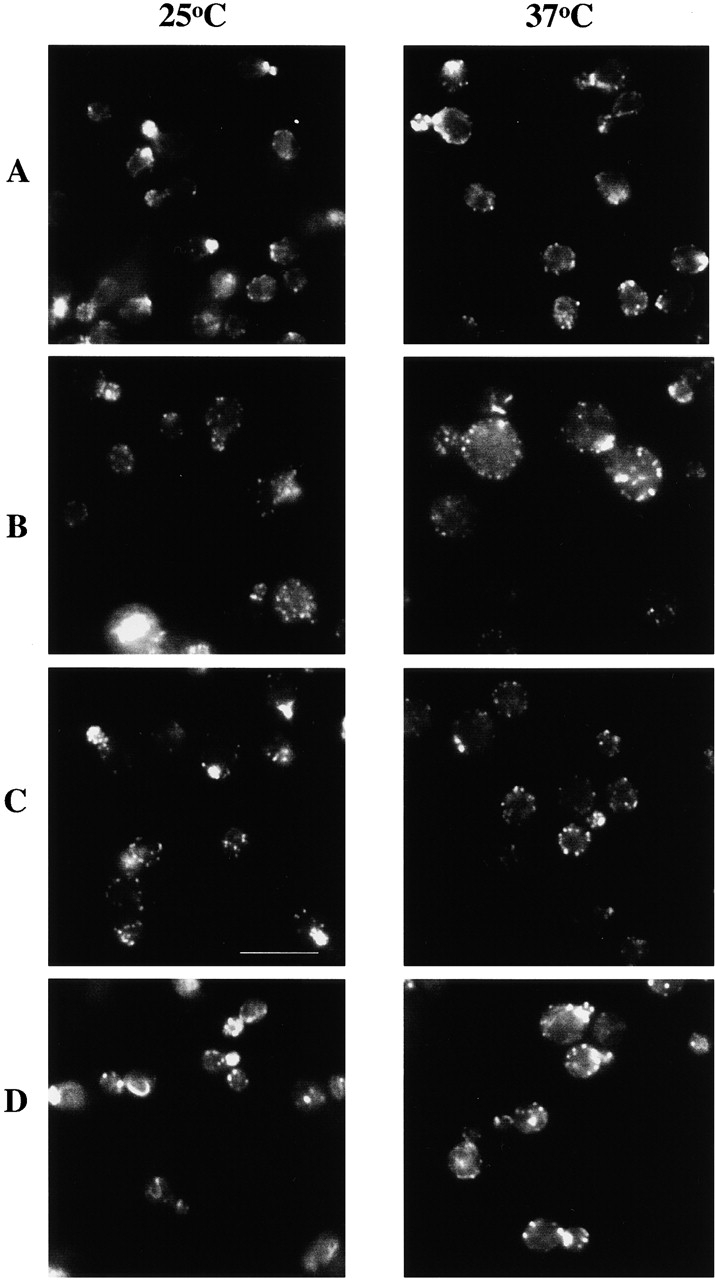
Rhodamine-phalloidin staining of yeast expressing pfy1-4 and act1-157. Yeast cells were grown to log phase in rich media at 25°C then fixed with formaldehyde, or were shifted to 37°C for 3 h, and then fixed. The F-actin was stained with rhodamine phalloidin. Yeast expressing PFY1 (A), pfy1-4 (B), pfy1-4 and act1-157 (C), and act1-157 (D). Bar, 10 μm.
Genetic Interactions Provide Evidence for Profilin's In Vivo Function
Although profilin mutagenesis revealed residues critical for function, no phenotypic differences between mutants in the actin- and polyproline-binding regions were observed. To elucidate specific roles for these regions in vivo, yeast strains carrying mutant pfy1 alleles were crossed to yeast containing mutant actin (act1) alleles, and mutant alleles of various cytoskeletal proteins (Table ). The actin mutants chosen for this analysis carry mutations in the nucleotide-binding pocket and differ in their effects on rates of nucleotide exchange and on filament dynamics (Belmont et al. 1999a,Belmont et al. 1999b). Nucleotide exchange rates were increased (act1-157) or decreased (act1-158), or filament dynamics were greatly reduced (act1-159). The genetic interactions revealed two aspects of profilin function. First, actin-binding (pfy1-4) and polyproline-binding (pfy1-14) mutants of profilin can be distinguished genetically. Crossing pfy1-4 with act1-157 resulted in the allele-specific suppression of the temperature sensitivity of pfy1-4, whereas pfy1-14 and act1-157 were synthetic lethal. Second, because Act1-157p exhibits a higher intrinsic rate of nucleotide exchange in vitro compared with wild-type yeast actin (Belmont et al. 1999b), the suppression of the temperature sensitivity of pfy1-4 by act1-157 is consistent with the possibility that profilin's ability to catalyze nucleotide exchange on actin is important in vivo. Further evidence supporting a role for profilin in actin nucleotide exchange is provided by the observed synthetic lethal interaction between pfy1-4 and act1-158, an actin mutant that displays intrinsically slower rates of nucleotide exchange.
Table 2.
Crosses between pfy1 Alleles and Mutant Alleles of Cytoskeletal Proteins
| pfy1-4 | pfy1-14 | |
|---|---|---|
| act1-157 | ts at 37°C suppressed | SL |
| act1-158 | SL | SL |
| act1-159 | SL | SL |
| twf1Δ | SL | SL |
| srv2Δ | SL | ND |
| las17ts | NE | ND |
| abp1Δ | SS | ND |
| bnr1Δ | SS | ND |
| arp2-1 | NE | ND |
| pfy1-4 | pfy1-18 | |
| cof1-4 | NE | NE |
| cof1-5 | SL | SS |
| cof1-6 | SS | NE |
| cof1-19 | SL | NE |
| cof1-22 | SL | NE |
The result of each cross is listed: ts, temperature sensitivity; SL, synthetic lethal; SS, synthetic sick; NE, no effect; ND, not determined. Crosses were chosen based on the specific properties of these cytoskeletal proteins. Twf1p and Srv2p function as actin monomer sequestering proteins, Las17p is the yeast WASP homologue, and Arp2-1p is the yeast Arp2 homologue in the Arp2/3 complex. All strains listed are available as DDY strains, except bnr1Δ and arp2-1, which were kindly provided by C. Boone and B. Winsor, respectively.
act1-157 Suppresses Actin Cytoskeleton and Fluid Phase Endocytic Defects of pfy1-4 at 37°C
We investigated the nature of the suppression of the temperature sensitivity of pfy1-4 by act1-157 by examining actin cytoskeleton organization in these mutant cells. Cells expressing pfy1-4 at 37°C became very large with depolarized actin patches (Fig. 2 B). In contrast, cells expressing pfy1-4 and act1-157 at 37°C (Fig. 2 C) were normal in size and better able to polarize their actin patches to the bud. However, the double mutants still lacked detectable actin cables and were more round than wild-type cells. These results suggest that act1-157 can suppress some, but not all, of the actin defects of pfy1-4 at 37°C.
Previously, we have shown that mutations that decrease actin filament turnover also inhibit fluid phase endocytosis (Lappalainen and Drubin 1997; Belmont and Drubin 1998). Therefore, we examined fluid phase endocytosis in pfy1-4 and act1-157 mutants using Lucifer yellow as a marker. As shown in Fig. 3, yeast expressing pfy1-4 were defective in Lucifer yellow uptake. At 37°C, only ∼18% of cells expressing pfy1-4 showed detectable Lucifer yellow staining in the vacuole. In contrast, 77% of cells expressing both pfy1-4 and act1-157 showed Lucifer yellow staining, a level comparable to staining in wild-type cells (79%) and act1-157 cells (63%). In addition, of those 18% of pfy1-4 cells showing Lucifer yellow staining in the vacuole, the staining was consistently weaker in intensity than in double mutant cells (pfy1-4 and act1-157). Interestingly, suppression of the endocytic defects of pfy1-4 by act1-157 was not observed at 25°C (Fig. 3). The suppression of actin and endocytic defects of pfy1-4 by act1-157 at 37°C suggests that the ability of profilin to enhance nucleotide exchange on actin is critical for fluid phase endocytosis at high temperatures.
Figure 3.
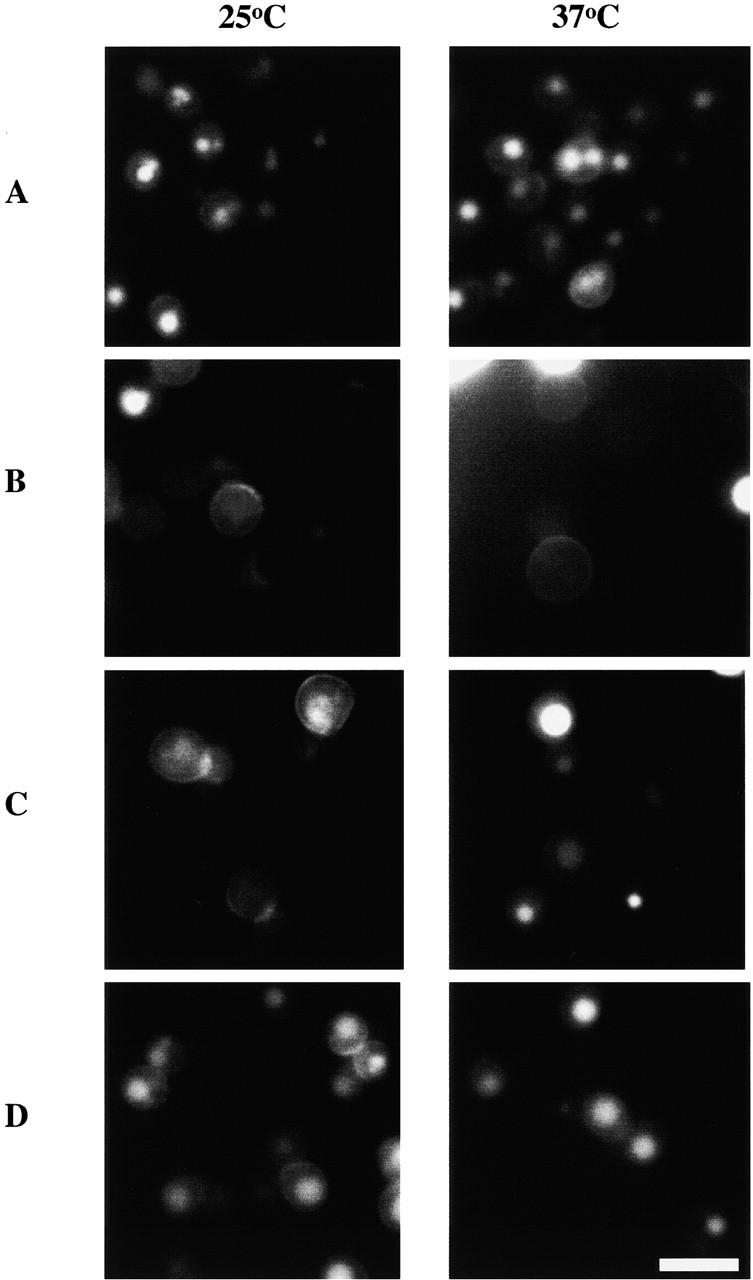
Uptake of Lucifer yellow by fluid-phase endocytosis in yeast expressing pfy1-4 and act1-157. Yeast cells were grown to early log phase at 25°C and some were shifted to 37°C. After 2 h, uptake of Lucifer yellow into the vacuole was monitored by fluorescence microscopy. Yeast expressing PFY1 (A), pfy1-4 (B), pfy1-4 and act1-157 (C), and act1-157 (D). Note that in the periphery of B at 37°C, very bright Lucifer yellow staining of dead cells expressing pfy1-4 is evident. Bar, 10 μm.
The Mutation in pfy1-4 Disrupts Binding to Actin Monomers
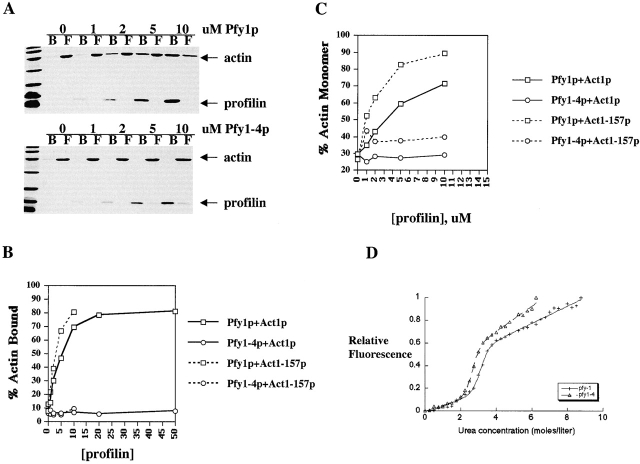
To better understand the underlying molecular basis for these observed genetic interactions, we performed a series of biochemical experiments using purified Pfy1-4p and Act1-157p. Three methods were employed. First, we measured the binding of actin to profilin using a PLP cosedimentation assay (polyproline binds to profilin, which in turn binds to actin monomers). As illustrated in Fig. 4 A, wild-type Pfy1p bound to Act1p in a concentration-dependent manner. In contrast, Pfy1-4p was unable to bind Act1p even at 50 μM Pfy1-4p, the highest concentration tested. Next, we measured actin monomer sequestration during actin assembly (Fig. 4 C). Pfy1p sequestered Act1p monomers in a concentration-dependent manner, whereas Pfy1-4p failed to sequester monomers, consistent with its inability to bind to Act1p. The results from these two experiments indicate that Pfy1p binds to Act1p with a K d of 4–5 μM, consistent with previous reports (Eads et al. 1998). Finally, we examined ATP exchange on actin. Pfy1p was able to increase the rate of Act1p ATP exchange by about threefold, whereas Pfy1-4p had no effect (data not shown). Taken together, these results indicate that Pfy1-4p is unable to bind to Act1p.
Figure 4.
Profilin binding to monomeric actin. Purified yeast actin and recombinant yeast profilin were incubated together to examine actin monomer binding. A, PLP–Sepharose cosedimentation assay. Various concentrations of Pfy1p and Pfy1-4p were bound to PLP–Sepharose and then incubated with 5 μM actin. Actin bound to the beads (B) was separated from the free actin (F) by centrifugation and analyzed by SDS-PAGE and Coomassie staining. B, Quantitation of results from the PLP–Sepharose cosedimentation experiment using various combinations of Act1p, Act1-157p, Pfy1p, and Pfy1-4p. Shown is a plot of the percent of actin bound versus profilin concentration. C, Actin assembly assay. Increasing concentrations of Pfy1p or Pfy1-4p were incubated with 5 μM Act1p or Act1-157p. Steady state reactions were centrifuged, analyzed by SDS-PAGE, and quantitated. D, Urea denaturation of Pfy1p and Pfy1-4p.
To insure that Pfy1-4p is properly folded, we examined its stability in an urea denaturation assay (Fig. 4 D). Pfy1-4p shows a similar denaturation profile to wild-type Pfy1p. In addition, Pfy1-4p binds to PLP with the same affinity as Pfy1p (Fig. 4 A, and data not shown). These results indicate that Pfy1-4p is stable and properly folded, but does not bind to monomeric actin.
Yeast Profilin and Cofilin Cooperate to Increase Turnover of Yeast Actin Filaments
The ability of actin to rapidly polymerize and depolymerize is critical to its functions in vivo. Cofilin promotes depolymerization from the pointed end of actin filaments, generating ADP-actin monomers, and it inhibits exchange of ATP for ADP. Thus, the ability of profilin to recharge actin monomers with ATP, or alternatively to promote assembly at barbed ends of actin filaments (Pantaloni and Carlier 1993), may be important for the speed of actin filament turnover when cofilin is present. Indeed, vertebrate profilin and cofilin have been shown to synergize to increase the rate of actin turnover, or treadmilling, in vitro (Didry et al. 1998). We examined the turnover rate of yeast actin filaments in vitro by measuring phosphate released from actin filaments after polymerization in the presence of yeast profilin and cofilin. The combined action of Pfy1p and Cof1p increased the rate of actin filament turnover by approximately fivefold compared with actin alone (Fig. 5 A). In contrast, the rate of actin filament turnover in the presence of Pfy1-4p and Cof1p was similar to a reaction that contained only Cof1p (two- to threefold stimulation). This result is consistent with the observation that Pfy1-4p does not bind to actin. Thus, yeast profilin and cofilin synergize to increase the rate of actin filament turnover in vitro, consistent with published results on vertebrate profilins and cofilins (Didry et al. 1998).
Figure 5.
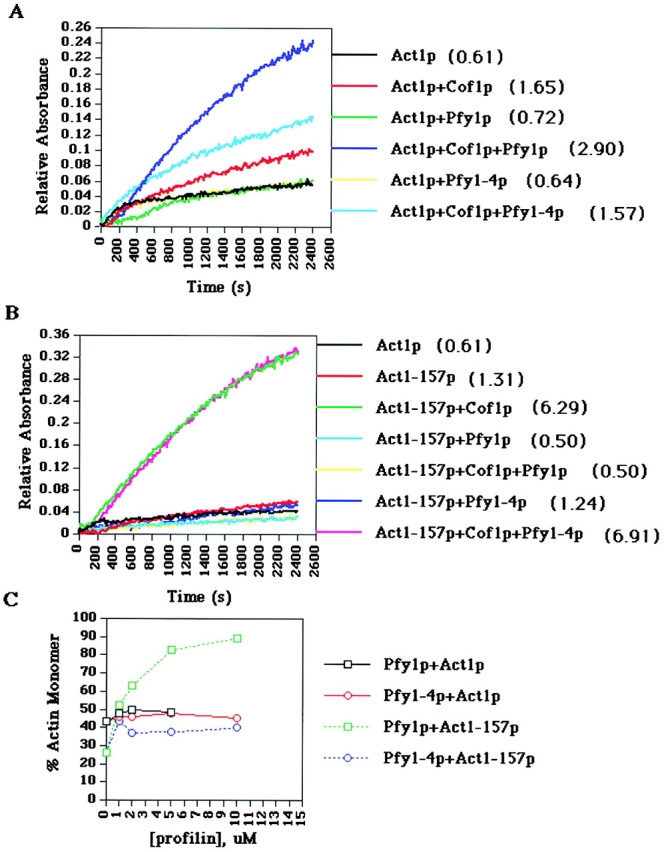
Actin filament turnover assays and steady state assembly. In A and B, purified Act1p, Act1-157p, Pfy1p, Pfy1-4p, and Cof1p were incubated in various combinations at a concentration of 25 μM for each protein. Actin assembly was initiated by the addition of polymerization salts, and phosphate released from actin filaments was measured. Rates of actin treadmilling are measured by comparing the slopes of the linear portions of the graph for each reaction, after steady state levels of assembly have been reached. The numbers in parentheses represent average rates of treadmilling from several experiments, expressed in pmol phosphate/s. A, Actin treadmilling using Act1p. B, Actin treadmilling using Act1-157p. C, Actin assembly in the presence of Pfy1p and Cof1p. Reactions were performed as in Fig. 4 C, except Cof1p was included in each reaction at a concentration of 5 μM.
If Pfy1p and Cof1p also act synergistically in vivo to control actin dynamics, then crosses of pfy1 and cof1 mutants would be predicted to be synthetic lethal. As shown in Table , synthetic lethal interactions were observed between pfy1-4 and several cof1 alleles. The combined results of these experiments show that synergy between profilin and cofilin on actin turnover are important in vitro and in vivo.
Cofilin Induces Rapid Steady State Turnover of Act1-157 Filaments in the Absence of Profilin
To study the mechanism by which act1-157 suppresses the temperature sensitivity of pfy1-4, we repeated the phosphate release assays using purified Act1-157p. We found that Act1-157p filaments in the absence of profilin or cofilin release phosphate approximately twice as fast as Act1p, suggesting that at steady state nucleotide exchange might be rate-limiting for actin assembly (Fig. 5 B). In the presence of Cof1p, the rate of turnover of Act1-157p filaments is increased to approximately tenfold higher than that of Act1p alone. Pfy1-4p does not alter this increased rate of turnover, consistent with the observation that Pfy1-4p does not bind to Act1-157p (Fig. 4 B). Thus, Act1-157p turns over rapidly in the absence of Pfy1p. This result may explain why act1-157 is able to suppress the phenotypes of pfy1-4.
Two models that might explain our results and the role of profilin in promoting rapid actin filament turnover are: that profilin is required to increase the rate of nucleotide exchange under conditions of rapid turnover (i.e., in the presence of cofilin); or that profilin is required to dissociate ADP-bound actin monomers from cofilin. Although it is difficult to rigorously distinguish between these two models, we favor the first one for the following reasons. First, Act1-157 exhibits a higher intrinsic rate of nucleotide exchange, but no significant differences in polymerization or depolymerization kinetics compared with wild-type actin (Belmont et al. 1999b). If the second model were correct, we would predict that act1-157 suppresses pfy1-4 via a mechanism in which ADP-bound Act1-157p has a lower affinity for cofilin, and thus circumvents the need for profilin to dissociate the ADP-actin/cofilin complex. Using the polyproline cosedimentation assay, we indirectly examined the affinity of Cof1p for Act1-157p by testing for the ability of Cof1p to compete with Pfy1p in binding to actin. We found that Cof1p has a similar affinity for ADP-bound wild-type actin as compared with ADP-bound Act1-157p (data not shown). An additional prediction of the second model is that cof1-19, which binds less tightly to actin monomers, but has normal disassembly promoting activity on filaments, would at least partially suppress pfy1-4 (Lappalainen, P., personal communication). However, cof1-19 is synthetically lethal with pfy1-4 (Table ). Therefore, we favor a model in which profilin increases actin filament turnover in the presence of cofilin by increasing the exchange of ATP for ADP-bound actin monomers that are released as a result of the actin depolymerizing activity of cofilin.
One initially surprising observation was that, although Act1-157p filaments turn over rapidly in the presence of Cof1p, turnover is extremely slow in the presence of both Cof1p and Pfy1p. This was unexpected because act1-157 exhibits minor actin defects in vivo. We further investigated the assembly of Act1-157 filaments in the presence of Pfy1p and Cof1p and discovered that Pfy1p tightly sequesters Act1-157p in the presence of Cof1p and prevents its assembly (Fig. 5 C). In addition, based on actin assembly and polyproline cosedimentation assays (Fig. 4B and Fig. C), Pfy1p binds with a higher affinity to Act1-157p (1–2 μM, compared with 4–5 μM for Act1p). The lack of strong actin defects in act1-157 cells remains a mystery, but may indicate that the monomer-sequestering activity of profilin is regulated by additional proteins in vivo.
Discussion
In this study, we have demonstrated that the actin- and polyproline-binding regions of yeast profilin are critical for its function in vivo. The phenotypes of yeast carrying mutations in these two regions of profilin appeared identical, preventing us from identifying functional differences. Only by performing genetic analysis of double mutants in profilin and actin, and correlating these results with biochemical data on purified mutant proteins, can we conclude that one role for profilin in S. cerevisiae is to increase the rate of actin filament turnover. Our studies support the hypothesis that nucleotide exchange on actin can be a rate-limiting step for actin assembly during rapid actin rearrangements in vivo (Mockrin and Korn 1980; Goldschmidt-Clermont et al. 1991; Vinson et al. 1998; Selden et al. 1999). One function of profilin may be to provide a pool of ATP actin competent to assemble quickly onto filament barbed ends in response to internal or external signals. Indeed, profilin is important for achieving maximal rates of actin-based motility of Listeria bacteria (Loisel et al. 1999). Yeast require rapid actin filament turnover for efficient fluid phase endocytosis (Lappalainen and Drubin 1997; Belmont and Drubin 1998). In this study, we observed that endocytosis is defective in yeast carrying a mutant profilin that cannot bind to actin, and is rescued at high temperature by an actin mutant that displays high rates of nucleotide exchange and filament turnover independent of profilin. These results strongly suggest that, through its ability to catalyze nucleotide exchange on actin, profilin promotes actin filament dynamics in vivo.
An important aspect of the experiments presented here is the use of both profilin and cofilin in actin filament turnover assays. Cofilin increases the rate of actin turnover by promoting depolymerization, yet it actually decreases the rate of nucleotide exchange on actin (Blanchoin and Pollard 1998). The combination of cofilin's ability to depolymerize actin filaments and decrease the rate of actin nucleotide exchange would be predicted to generate a large pool of ADP actin. Profilin's ability to enhance exchange on actin might therefore be necessary to maximize actin turnover in the presence of cofilin. Indeed, profilin has been shown to overcome the inhibition of nucleotide exchange by the cofilin-related protein, actophorin (Blanchoin and Pollard 1998), and we have obtained similar results with yeast profilin, cofilin, and actin (Wolven, A.K., and D.G. Drubin, unpublished results). Including both profilin and cofilin in the actin filament turnover assays more accurately reflects the in vivo situation, and strengthens the conclusion that actin nucleotide exchange can be a rate-limiting step that requires profilin in vivo.
An alternative hypothesis to explain the results obtained here is that the role of profilin is to dissociate cofilin from ADP actin, rather than promote nucleotide exchange. While our results would suggest that this is not the case, we cannot rule out a model in which profilin dissociates ADP actin from cofilin by catalyzing ATP exchange on actin. It is possible that dissociation of ADP actin-cofilin and nucleotide exchange occur in a concerted manner and are mechanistically coupled.
In contrast to vertebrate and amoeba profilins, plant profilins do not appear to enhance the rate of nucleotide exchange on actin, although rabbit muscle actin was used in the studies of plant profilin (Perelroizen et al. 1996). In yeast, profilin only modestly increases the rate of ATP exchange on actin (Eads et al. 1998). Such results have led to speculation that in plants and yeast, profilin may play other roles in actin dynamics. Our results indicate that the ability of profilin to promote nucleotide exchange on actin is important in S. cerevisiae. However, profilin has additional activities in vitro and our results also suggest that other actin related and unrelated functions of profilin must be important in vivo. Evidence for this conclusion comes from the observations that act1-157 does not completely alleviate the slow growth and disrupted actin organization in yeast expressing pfy1-4, and that pfy1-4 yeast grow better than pfy1Δ yeast. Other important activities of profilin might include promoting barbed-end assembly, preventing pointed-end assembly, sequestering actin monomers to inhibit spontaneous nucleation, and modulating lipid chemistry (Goldschmidt-Clermont et al. 1991; Pantaloni and Carlier 1993; Kang et al. 1999). The context under which actin is assembling in vivo may determine which actin-related activity of profilin is used. Evidence that profilin may have other activities is also provided by the synthetic lethal interactions observed between pfy1-4 and srv2Δ, and between pfy1-4 and twf1Δ (Table ). Srv2p and Twfp can both sequester actin monomers (Freeman et al. 1995; Goode et al. 1998). These double mutants may have problems regulating actin monomer pools.
Acknowledgments
The authors would like to thank Bruce Goode, Avital Rodal, and Daria Siekhaus for helpful discussions and critical reading of this manuscript.
A.K. Wolven was supported by a fellowship from the National Institutes of Health (GM19969). D.G. Drubin is supported by grant GM42759 from the National Institutes of Health.
Footnotes
Amy K. Wolven's present address is Incyte Genomics, Palo Alto, CA 94304.
Lisa D. Belmont's present address is Enogen, Inc., Salinas, CA 93901.
Abbreviation used in this paper: PLP, poly-l-proline.
References
- Ayscough K.R. In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol. 1998;10:102–111. doi: 10.1016/s0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Belmont L.D., Drubin D.G. The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Orlova A., Drubin D.G., Egelman E.H. A change in actin conformation associated with filament instability after Pi release Proc. Natl. Acad. Sci. USA 96 1999. 29 34a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Patterson G.M., Drubin D.G. New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites J. Cell Sci 112 1999. 1325 1336b [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Carlsson L., Nystrom L.-E., Sundkvist I., Markey F., Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Cope M.J., Yang S., Shang C., Drubin D.G. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didry D., Carlier M.F., Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 1998;273:25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Eads J., Mahoney N.M., Vorobiev S., Wen K.-K., Rubenstien P.A., Haarer B.K., Almo S.C. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M.S., Chow C.J., Adames N., Pringle J.R., Peter M., Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Freeman N.L., Chen Z., Horenstein J., Weber A., Field J. An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 1995;270:5680–5685. doi: 10.1074/jbc.270.10.5680. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont P.J., Machesky L.M., Doberstein S.K., Pollard T.D. Mechanism of the interaction of human platelet profilin with actin. J. Cell Biol. 1991;113:1081–1089. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Drubin D.G., Lappalainen P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer sequestering protein. J. Cell Biol. 1998;142:723–733. doi: 10.1083/jcb.142.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B.K., Lillie S.H., Adams A.E.M., Magdolen V., Bandlow W., Brown S.S. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J. Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H., Tanaka K., Hihara T., Umikawa M., Kamei T., Takahashi K., Sasaki T., Takai Y. Bni1P and Bnr1Pdownstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F., Purich D.L., Southwick F.S. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kohno H., Tanaka K., Mino A., Umikawa M., Imamura H., Fujiwara T., Fujita Y., Hotta K., Qadota H., Watanabe T. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D.G. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Fedorov E.V., Fedorov A.A., Almo S.C., Drubin D.G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel T.P., Boujemaa R., Pantaloni D., Carlier M.F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Machesky L.M., Pollard T.D. Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 1993;3:381–385. doi: 10.1016/0962-8924(93)90087-h. [DOI] [PubMed] [Google Scholar]
- Mockrin S., Korn E. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- Pantaloni D., Carlier M.-F. How profilin promotes actin filament assembly in the presence of thymosin β-4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Perelroizen I., Carlier M.F., Pantaloni D. Binding of divalent cation and nucleotide to G-actin in the presence of profilin. J. Biol. Chem. 1995;270:1501–1508. doi: 10.1074/jbc.270.4.1501. [DOI] [PubMed] [Google Scholar]
- Perelroizen I., Didry D., Christensen H., Chua N.H., Carlier M.F. Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J. Biol. Chem. 1996;271:12302–12309. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. Structure of actin binding proteinsinsights about function at atomic resolution. In: Spudich J.A., editor. Annual Review of Cell Biology, Vol. 10. Annual Reviews Inc; Palo Alto, CA: 1994. pp. 207–249. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. J. Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Rodal A.A., Tetreault J.W., Lappalainen P., Drubin D.G., Amberg D.C. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston F.M., Hieter P. Methods In Yeast GeneticsA Laboratory Course Manual 1989. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: pp. 198 [Google Scholar]
- Rosenblatt J., Peluso P., Mitchison T. The bulk of unpolymerized actin in Xenopus egg extracts is ATP-bound. Mol. Biol. Cell. 1995;6:227–236. doi: 10.1091/mbc.6.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycki M., Schutt C., Lindberg U. Affinity chromatography-based purification of profilinactin. Methods Enzymol. 1991;196:100–118. doi: 10.1016/0076-6879(91)96012-g. [DOI] [PubMed] [Google Scholar]
- Schutt C.E., Myslik J.C., Rozycki M.D., Goonesekere N.C.W., Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Selden L.A., Kinosian H.J., Estes J.E., Gershman L.C. Impact of profilin on actin-bound nucleotide exchange and actin polymerization dynamics. Biochemistry. 1999;38:2769–2778. doi: 10.1021/bi981543c. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson V.K., De La Cruz E.M., Higgs H.N., Pollard T.D. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]