Abstract
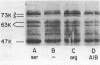
The involvement of a protein methyl transfer system in the chemotaxis of Pseudomonas aeruginosa was investigated. When a methionine auxotroph of P. aeruginosa was starved for methionine, chemotaxis toward serine, measured by a quantitative capillary assay, was reduced 80%, whereas background motility was unaffected or increased. When unstarved bacteria were labeled with L-[methyl-3H]methionine, a labeled species of 73,000 molecular weight which was methylated in response to stimulation by L-serine was identified. Under appropriate electrophoretic conditions, the 73,000 molecular weight species was resolved into two bands, both of which responded to stimulation by L-serine, L-arginine, and alpha-aminoisobutyrate (AIB) with an increased incorporation of methyl label. Arginine, which elicited the strongest chemotactic response in the capillary assay, also stimulated the greatest methylation response. Methylation of the 73,000 molecular weight species reached a maximum 10 min after stimulation by AIB and returned to the unstimulated level upon removal of the AIB. In vitro labeling of cell extracts with S-adenosyl[methyl-3H]methionine indicated that the 73,000 molecular weight species are methylated by an S-adenosylmethionine-mediated reaction. These results indicate that chemotaxis of P. aeruginosa toward amino acids is mediated by dynamic methylation and demethylation of methyl-accepting chemotaxis proteins analogous to those of the enteric bacteria.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973 Jan;74(1):77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Ames G. F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci U S A. 1980 May;77(5):2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci U S A. 1980 May;77(5):2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Failure of sensory adaptation in bacterial mutants that are defective in a protein methylation reaction. Cell. 1978 Dec;15(4):1231–1240. doi: 10.1016/0092-8674(78)90049-1. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P., Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981 Jan;145(1):43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L. Maltose chemoreceptor of Escherichia coli. J Bacteriol. 1975 Apr;122(1):206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedblom M. L., Adler J. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J Bacteriol. 1980 Dec;144(3):1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene S. J., Toews M. L., Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977 May 25;252(10):3214–3218. [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Mesibov R., Ordal G. W., Adler J. The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. Weber law and related phenomena. J Gen Physiol. 1973 Aug;62(2):203–223. doi: 10.1085/jgp.62.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton R. C., Montie T. C. Chemotaxis by Pseudomonas aeruginosa. J Bacteriol. 1979 Jan;137(1):274–280. doi: 10.1128/jb.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins C., Dahlquist F. W. The methyl-accepting chemotaxis proteins of E. coli: a repellent-stimulated, covalent modification, distinct from methylation. Cell. 1981 Aug;25(2):333–340. doi: 10.1016/0092-8674(81)90051-9. [DOI] [PubMed] [Google Scholar]
- Sherris D., Parkinson J. S. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: a requirement for methionine in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Jan;74(1):183–187. doi: 10.1073/pnas.74.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson M. W., Cohen M. A., Merrick J. M. Purification and properties of the periplasmic glucose-binding protein of Pseudomonas aeruginosa. J Bacteriol. 1977 Aug;131(2):672–681. doi: 10.1128/jb.131.2.672-681.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A. H., Ordal G. W. In vivo and in vitro chemotactic methylation in Bacillus subtilis. J Bacteriol. 1981 Feb;145(2):958–965. doi: 10.1128/jb.145.2.958-965.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]