Abstract
The human EST1A/SMG6 polypeptide physically interacts with the chromosome end replication enzyme telomerase. In an attempt to better understand hEST1A function, we have started to dissect the molecular interactions between hEST1A and telomerase. Here, we demonstrate that the interaction between hEST1A and telomerase is mediated by protein–RNA and protein–protein contacts. We identify a domain within hEST1A that binds the telomerase RNA moiety hTR while full-length hEST1A establishes in addition RNase-resistant and hTR-independent protein–protein contacts with the human telomerase reverse transcriptase polypeptide (TERT). Conversely, within hTERT, we identify a hEST1A interaction domain, which comprises hTR-binding activity and RNA-independent hEST1A-binding activity. Purified, recombinant hEST1A binds the telomerase RNA moiety (hTR) with high affinity (apparent overall Kd = 25 nM) but low specificity. We propose that hEST1A assembles specifically with telomerase in the context of the hTR–hTERT ribonucleoprotein, through the high affinity of hEST1A for hTR and specific protein–protein contacts with hTERT.
INTRODUCTION
Human EST1A/SMG6 (below referred to as hEST1A), EST1B/SMG5 and EST1C/SMG7 were identified through their sequence similarity with the Saccharomyces cerevisiae Ever Shorter Telomeres 1 (Est1) gene product and the Caenorhabditis elegans and Drosophila melanogaster nonsense-mediated mRNA decay (NMD) factors CeSMG5, CeSMG6 and CeSMG7 and DmSMG5 and DmSMG6 (1–3). NMD leads to rapid decay of mRNAs that carry premature stop codons. Immunoprecipitation experiments using antibodies raised against hEST1A and hEST1B revealed association of these polypeptides with telomerase activity (1,2). We could not detect association between hEST1C/SMG7 and telomerase in co-immunoprecipitation experiments (Nele Hug and J. Lingner, unpublished data). Telomerase association is particularly evident for hEST1A, which appears to associate with ∼70% of active telomerase affinity purified from HeLa cell extracts. Consistent with a direct function of hEST1A at telomeres is its detection at telomeres in vivo by chromatin immunoprecipitation (Claus Azzalin and J. Lingner, manuscript submitted for publication). Over-expression of hEST1A in the human fibrosarcoma-derived cell line HT1080 induces telomere–telomere associations causing chromosome bridges during anaphase and a rapid apoptotic response (1). Telomeric fusions in hEST1A-over-expressing cells may stem from telomere uncapping followed by DNA break repair, reminiscent of the loss of function phenotype of the double strand telomere-binding protein TRF2 (4,5). Thus, we hypothesize that endogenous hEST1A is involved in modulating the telomere structure. Over-expression of full-length hEST1A in kidney 293T cells leads to progressive telomere shortening (2). This effect could be reversed by co-expression of telomerase reverse transcriptase (TERT), suggesting that the shortening depends on modulation of telomerase. Thus, hEST1A may also regulate telomere length. In the yeasts S. cerevisiae and Schizosaccharomyces pombe, the EST1 genes are required for telomerase-mediated telomere elongation in vivo (6,7). S. cerevisiae Est1p recruits telomerase to chromosome 3′ ends through a direct physical interaction with the telomere end-binding protein Cdc13 (8). It is unknown if the human EST1-proteins play a similar essential role for telomerase activation at chromosome ends.
The involvement of human EST1A-C/SMG5-7 in NMD has been substantiated in knock-down experiments, which leads to an increase in abundance of NMD-reporter constructs carrying premature stop codons, and of endogenous mRNAs that are regulated by this pathway (9). Worm SMG5-7 are involved in the PP2A-mediated dephosphorylation of UPF1, a highly conserved 5′ to 3′ helicase playing a central role in NMD (10,11) and genome stability [(12); (see below)]. SiRNA-mediated depletion of human EST1A-C/SMG5-7 leads to impairment of NMD and accumulation of hyperphosphorylated UPF1, over-expression of hEST1A/SMG6 promotes dephosphorylation of UPF1 and hEST1B/SMG5 and hEST1C/SMG7 are found in complexes with phospho-UPF1 (3,10). Thus, the role of SMG5-7 proteins in the phosphorylation cycle of UPF1 appear to be conserved between worms and humans.
The above studies suggest that human EST1-polypeptides may have dual functions in telomere maintenance and RNA surveillance pathways. Indeed, an intimate relation between genome and RNA surveillance pathways is also suggested by the findings that the human phosphoinositide 3-kinase-related protein kinase (PIKK) SMG1 is required for optimal activation of p53 upon genotoxic stress (13). Also UPF1 is required for genome stability (12). Depletion of UPF1 causes human cells to arrest early in S-phase inducing an ATR-dependent DNA damage response. A fraction of UPF1 associates with chromatin in S-phase and upon γ-irradiation, supporting a direct role in genome stability. In contrast, downregulation of the NMD factor UPF2 does not interfere with cell cycle progression and DNA stability, supporting the notion that effects on genome stability are not due to a general loss in NMD and that only some NMD factors play a role in genome stability (14). Towards an attempt to understand the putative roles of hEST1A in telomere/genome stability and RNA surveillance pathways, we identify here functional domains of hEST1A that mediate the interaction with telomerase. We identify a high affinity RNA-binding domain in hEST1A and detect RNA-independent protein–protein interactions that mediate assembly with telomerase. We also identify a segment within the N-terminal part of hTERT, which enables association with hEST1A.
MATERIALS AND METHODS
Protein expression and purification
A GST-tag encoding DNA fragment was amplified by PCR from the pGEX-6P1 vector (GE Healthcare) and subcloned in the pFastBac vector (Invitrogen). A DNA fragment encoding the complete open reading frame (1419 amino acids) of Homo sapiens EST1A (SMG6, KIAA0732) was subcloned in frame downstream of the GST-tag in the modified pFastBac vector. Baculoviruses were prepared following the manual of the Bac-to-Bac Baculovirus expression system (Invitrogen). Hi5 insect cells were transduced with baculoviruses at m.o.i. of 20. Forty-eight hour post-infection, nuclear extracts were prepared as described (15) with slight modifications. Infected Hi5 cell pellets harvested from 135 mm plates (1.5 × 107 cells before infection) were resuspended in 5.75 ml of cold buffer A (50 mM Tris–HCl pH 8.0, 100 mM KCl, 0.1 mM PMSF, 0.1 mM EDTA, 0.1 mM DTT, 0.3% NP40) and kept on ice for 15–20 min for cell lysis. After centrifugation for 5 min at 500g at 4°C, the nuclei containing pellet was washed three times with 1 ml buffer A, repeating in between the centrifugation step. For extraction, the washed nuclei were resuspended in 0.75 ml of cold buffer B (50 mM Tris–HCl pH 8.0, 400 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, protease cocktail inhibitor EDTA-free from Roche), transferred in a microfuge tube and shaken very gently for 30 min at 4°C. The mixture was centrifuged for 5 min with maximal velocity at 4°C. Tween 20 and glycerol were added to final concentrations of 0.2 and 10%, respectively, to the nuclear extract corresponding supernatant. 0.21 ml slurry of glutathione-Sepharose beads (GE Healthcare) were washed with cold 1× PBS and equilibrated with buffer B supplemented with 0.2% Tween 20 and 10% glycerol. The nuclear extract was added to these beads and incubated at 4°C for 1.5 h on a wheel. The flow through was collected upon centrifugation for 1 min at 500g at 4°C. The beads were washed three times with 1.5 ml of cold EB buffer (100 mM Tris–HCl pH 8.0, 400 mM NaCl, 10% glycerol, 0.2% Tween 20, 1 mM DTT, 1 mM EDTA and protease cocktail inhibitor EDTA-free). GST–hEST1A was eluted from the beads with 0.21 ml of cold GEB buffer (buffer EB containing 20 mM GSH) after incubation at 4°C for 45 min on a wheel. The recovered supernatant was applied to a small disposable filter column (Biorad) to remove the remaining beads. The elution step was repeated two more times for 15 min. Upon purification, the GST–hEST1A polypeptide was estimated to be more than 90% pure. As negative control, nuclear extracts of Hi5 cells that had been infected with baculoviruses containing only the GST transgene were prepared and purified following the same procedure.
Plasmids
DNA fragments encoding for the hTR deletion mutants downstream of a T7 promoter were generated by PCR (Pseudo and ΔPseudo) or overlap extension PCR (ΔCR45; see Ref. (16) for the method) using pT7hTER (17) as a template.
hEST1A deletion mutants (1–855), (1–457), (1–237), (606–1419), (783–1419), (606–991) and (606–855) were generated by DNA restriction and ligation of DNA fragments into pcDNA6myc-HisC (Invitrogen) or by ligation into a modified phrGFP-N1 vector (Stratagene) in which a FLAG encoding sequence was used to replace the GFP sequence. In these vectors, expression is driven by the CMV promoter and polyadenylation is triggered by the SV40 poly(A) site. Mutant (243–533) was generated by PCR amplification of the respective hEST1A fragment and subcloning into pcDNA6myc-HisC. Mutant (243–412) was generated by overlap extension PCR (16) and subcloning into pcDNA6myc-HisC.
hTERT deletion mutants (1–832), (1–656) and (1–446) were generated by DNA restriction and ligation in pcDNA6myc-HisC-hTERT (18). All the other hTERT-mutants were generated from pcDNA6myc-HisC-hTERT using a site-directed mutagenesis kit (Stratagene). Further details on plasmid constructions can be obtained upon request.
RNA band shifts
RNAs were transcribed in vitro following the instructions of the Ribomax large-scale RNA production system-T7 kit (Promega). After in vitro transcription, DNA templates were removed by DNase I digestion and the RNA samples were extracted with phenol:chloroform (1:1) and precipitated with ethanol. Analysis of the RNA samples by gel electrophoresis confirmed their correct length and intactness. Full-length hTR was prepared as described (17) and gel purified. Transcribed hTR was dephosphorylated at the 5′ end with CIP (Roche) and extracted with phenol:chloroform (1:1). hTR was 5′-end labeled with [32P]-γ-ATP using T4 polynucleotide kinase and unincorporated nucleotides were removed using mini quick spin RNA columns (Roche). Ribosomal RNA was gel purified from total RNA of HeLa cells prepared with the RNA easy kit (Qiagen). Escherichia coli tRNA was from Roche. RNA–protein complexes were separated on native 3.5% polyacrylamide/0.4% agarose gels that were run in 75 mM Tris–glycine buffer at 4°C (150 V for 30 min followed by 300 V for 3 h). The gels were pre-run for 1 h at 250 V.
For the determination of the overall Kd of GST–hEST1A for hTR, GST–hEST1A (1–100 nM) was incubated with 1 nM 5′ [32P]-labeled hTR (*hTR) in presence of a 250× excess of tRNA as a non-specific competitor in 70 mM Tris–HCl pH 8.0, 80 mM NaCl, 4 mM GSH, 1.2 mM EDTA, 1 mM DTT and 4% glycerol for 30 min at RT. Time-course experiments with 1 nM 5′ [32P]-labeled hTR and 100 nM GST–hEST1A showed that the binding equilibrium was reached already after <1 min of incubation. For the competition experiments, competitor RNAs were added as indicated prior to addition of hEST1A. Prior to loading on the gel, 1 µl of 20% Ficoll was added. Competition experiments were quantified from three independent reactions.
Pull-down experiments of telomerase and quantification of the activity
1 µl of reconstituted and affinity purified telomerase from insect cells (17) or in rabbit reticulocyte lysates (RRL) (Promega) reconstituted telomerase (19) was incubated with 0.5 pmol of purified GST–hEST1A or GST for 1 h at 4°C in 10 µl final volume of 20 mM HEPES pH 7.9, 100 mM KCl, 1 mM DTT, 10% glycerol and 0.2% Tween 20. 9 µl of this mixture was added to 36 µl of 30% slurry of GSH-Sepharose beads (GE Healthcare) in IP buffer (20 mM HEPES pH 7.9, 100 mM KCl, 1 mM DTT, 10% glycerol, 0.2% Tween 20) and incubated 1 h at 4°C on a wheel. The beads were washed three times with 150 µl of IP buffer. In Figure 1, telomerase activity was measured by TRAP using quantitative PCR and the light cycler (Roche) (18).
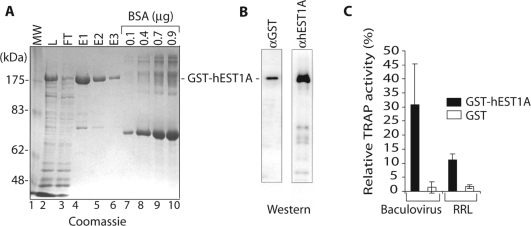
Figure 1.
Expression and purification of recombinant GST–hEST1A protein and interaction with in vitro reconstituted telomerase. (A) SDS–PAGE and Coomassie blue staining of GST–hEST1A fractions derived from Hi5 insect cells. Lane 1: MW: Molecular weight markers. Lane 2: L: GST–hEST1A expressing Hi5 nuclear extract. Lane 3: FT: flow through of GSH-beads. Lanes 4–6: E1, E2 and E3: first, second and third eluates, Lanes 7–10: BSA dilutions. GST–hEST1A is indicated. (B) Western blots with anti-GST and anti-hEST1A antibodies. (C) Relative TRAP activity (%) pulled down with GSH-beads upon incubation of 50 nM GST (white rectangles) or 50 nM GST–hEST1A (fraction E1; black rectangles) with telomerase reconstituted in vitro in insect cell lysates (Baculovirus) or in RRL. Interaction of GST–hEST1A and GST with telomerase in vitro. Bars represent the means ± SD of three independent experiments.
Transfection of human cell lines and immunoprecipitation
The 293T cells were transfected with 4 µg of total plasmid DNA per well of a 6-well plate using Lipofectamine 2000 (Invitrogen). WI38-VA13 cells were transfected in 10 cm dishes with Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Twenty-four hours post-transfection plasmid-containing cells were selected with 4 µg/ml blasticidin for 48 h. Whole cell extracts were prepared by resuspending cells in 600 µl of IPII buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 0.5% TritonX-100), incubating the cell suspension for 30 min at 4°C and centrifugating at 13 000 r.p.m. (16 100g) for 10 min. The supernatant was pre-cleared for 1 h at 4°C with protein A beads (GE Healthcare) and then incubated with 1 µg/ml of either α-Myc (9B11 from NEB) or α-FLAG (M2 from Sigma) antibodies for 1 h at 4°C. 20 µl of a 50% slurry of protein A beads was added and incubated over-night at 4°C. Beads were recovered by spinning 1 min at 6500 r.p.m. (4000g) and washed five times in the IPII buffer. Samples were loaded on 4–20% gradient Tris–HCl PAGE duramide gels (Cambrex). After semi dry transfer and western blotting, the signals were measured using the ChemiGlow (Wytec) substrate on a FluorChem™ 8900 machine (Alpha Innotech).
Immunoprecipitation in rabbit reticulocyte lysate
Myc-hTERT fragments and Flag-hEST1A fragments were in vitro translated in the presence of [35S]-methionine in RRL following the instructions of TnT quick-coupled transcription/translation system (Promega). A total of 110 μl of RRL-mix containing 88 μl of TNT T7 master mix, 4.4 μl [35S]-methionine (1000 Ci/mmol, 10 mCi/ml) and 440 ng plasmid DNA was incubated for 1.5 h at 30°C. Protein A beads were first washed with 1× PBS and then with IPIII buffer [20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 0.2% Tween 20, 0.4 U/μl RNAguard™ (GE Healthcare) and protease cocktail inhibitor (EDTA-free, Roche)] and then blocked for 1 h at 4°C in 1 ml of IPIII buffer supplemented with 100 μg E. coli tRNA and 500 μg BSA (NEB). The pre-blocked beads were resuspended in IPIII buffer to give a 50% slurry. For each immunoprecipitation, 1 μg of either α-Myc (9B11 from NEB) or α-FLAG (M2 from Sigma) antibodies were pre-incubated for 2.5 h at 4°C with 20 μl of 50% slurry of protein A beads (GE Healthcare) in IPIII buffer. For hTR-binding experiments, 1 μl of [32P]-labeled hTR (0.1 pmol/μl), 1 μl of non-labeled hTR (0.4 pmol/μl) and 62.5 µg E. coli tRNA were mixed with 12.5 μl of the RRL-mix to give a final volume of 15.75 µl and incubated 1 h at 30°C. For pre-clearing, each reaction was mixed with 20 μl of 50% slurry of protein A beads (GE Healthcare) in IPIII buffer with 85 μl of IPIII buffer for 1 h at 4°C. The beads were removed by centrifugation at 16 100g for 1.5 min. Then the supernatant was incubated for 2 h at 4°C with 20 μl of the coated protein A beads with the respective antibodies. After immunoprecipitation, beads were recovered by spinning 1 min at 4000g and washed five times in 500 μl IPIII buffer. Samples were loaded on 4–20% gradient Tris–HCl PAGE duramide gels (Cambrex). After fixation following Cambrex instructions, gels were dried between two cellophane sheets (GE Healthcare). [35S]-labeled proteins and [32P]-labeled hTR were detected and quantified on a PhosphoImager. [32P]-labeled hTR could be distinguished from [35S]-labeled proteins by shielding the lower energy [35S]-derived β-rays with a sheet of plastic.
RESULTS
Recombinant hEST1A associates with active telomerase
To study the biochemical properties of hEST1A, we expressed recombinant hEST1A as an N-terminal glutathione (GST)-fusion protein from a baculoviral vector in Hi5 insect cells. Nuclear extracts were prepared and GST–hEST1A was purified on Sepharose-glutathione beads (GSH-beads) (Figure 1). Analysis of column eluates on Coomassie-brilliant blue stained polyacrylamide gels revealed one major band in the purified fraction (Figure 1A), which was recognized on western blots by GST-antibodies and hEST1A antibodies raised against the full-length hEST1A (Figure 1B). This indicates the presence of nearly homogeneously pure full-length GST–hEST1A in the eluted fractions. To test the ability of recombinant GST–hEST1A to associate with telomerase, the purified protein was incubated with telomerase, expressed and reconstituted in insect cells (17) or in RRL (20). Upon binding of GST–hEST1A via its tag to GSH-beads, the amount of associated telomerase activity was determined in TRAP telomerase activity assays, in which the products were quantified by real-time PCR (Figure 1C). Ten to thirty percent of telomerase activity associated with the beads in a GST–hEST1A-dependent manner indicating efficient association of recombinant EST1A with telomerase. GST alone did not bind substantial amounts of telomerase.
Recombinant hEST1A binds telomerase RNA with high affinity but low specificity
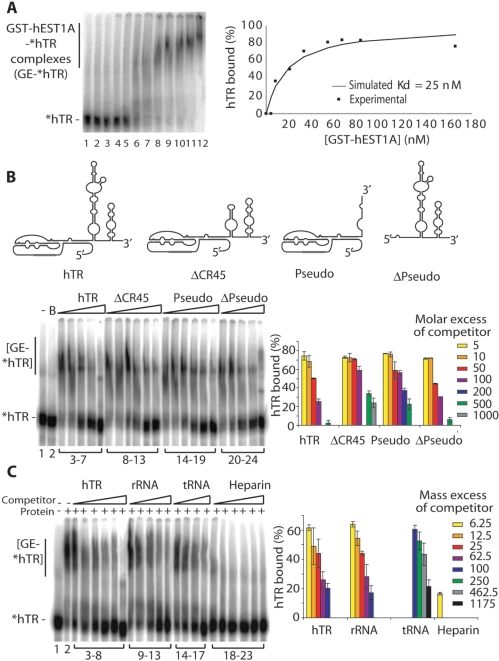
Association of hEST1A with telomerase may be mediated by protein–protein and/or protein–RNA interactions. In order to test the ability of hEST1A to bind the telomerase RNA moiety hTR, RNA band shift experiments were carried out (Figure 2). Increasing amounts of GST–hEST1A were incubated with [32P]-5′-end labeled in vitro transcribed hTR. Separation of the fractions on non-denaturing polyacrylamide gels indicated formation of slower migrating GST–hEST1A–hTR complexes (Figure 2A). The experimental data for overall hEST1A–hTR-binding fitted best a theoretical curve with a Kd of 25 nM (Figure 2A). The spread-out appearance of the GST–hEST1A–hTR complexes may be due to conformational flexibility, complex heterogeneity and concomitant binding of several hEST1A polypeptides per hTR molecule (see below). Thus this analysis gives an estimate for the overall Kd of hEST1A–hTR complexes without distinguishing individual subtypes. Other gel recipes, removal of the GST-tag from hEST1A or the use of hTR fragments did not reduce the heterogeneity. To test binding affinity for different domains of hTR, different fragments of hTR were transcribed in vitro and tested for their ability to inhibit the formation of the full-length GST–hEST1A–hTR complex (Figure 2B). The Δ-Pseudo fragment had a similar capacity as unlabeled full-length hTR, to compete for the formation of the [32P]-labeled full-length GST–hEST1A–hTR complex (Figure 2B). However, two other hTR fragments (Pseudo and ΔCR45) also competed well with hTR for hEST1A-binding at slightly higher concentrations. Since the three tested hTR fragments contained distinct non-overlapping domains, this indicated several hEST1A–hTR interaction sites. To further address specificity of RNA binding, other non-related RNAs were tested in a competition experiment (Figure 2C). This experiment indicated that the affinity of hEST1A for rRNA is similar as for hTR, while tRNA competes only at ∼20-fold higher concentration, indicating very low affinity of hEST1A for this RNA. Heparin efficiently inhibited hTR binding.
Figure 2.
Recombinant GST–hEST1A interacts with hTR with high affinity but low specificity. (A) EMSA analysis of [32P] 5′-end labeled in vitro transcribed hTR (*hTR, 1 nM) with increasing amounts of purified GST–hEST1A (fraction E1). Lane 1: *hTR alone. Lane 2: *hTR incubated in EMSA buffer. Lanes 3–12: *hTR incubated with 0.83, 4.15, 8.3, 20.75, 33.2, 41.5, 53.95, 66.4, 83.0 and 166 nM of GST–hEST1A. GST–hEST1A–*hTR complexes are indicated. Right panel: Percentage of *hTR bound to GST–hEST1A plotted as a function of the concentration of GST–hEST1A. The experimental points fitted well with a curve with a Kd of 25 nM. (B) EMSA competition experiment to identify GST–hEST1A-binding specificity for different hTR domains. The secondary structure models are schematically depicted. Lane 1: *hTR alone. Lane 2: B: *hTR incubated with EMSA-buffer. Lanes 3–7: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing molar equivalent of unlabeled full-length hTR (5-, 10-, 50-, 100- and 500-fold excess). Lanes 8–13: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing molar equivalents of cold DCR45 (5-, 10-, 50-, 100-, 500- and 1000-fold excess). Lanes 14–19: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing molar equivalent of cold Pseudo (5-, 10-, 50-, 100-, 200- and 500-fold excess). Lanes 20–24: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing molar equivalent of cold ΔPseudo (5-, 10-, 50-, 100- and 500-fold excess). Right panel: Quantification of competition experiments. (C) EMSA competition experiment with additional RNA molecules. Lane 1: *hTR incubated with EMSA buffer. Lane 2: *hTR incubated with 50 nM GST–hEST1A (fraction E1). Lanes 3–8: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing mass equivalents of cold full-length hTR (6.25-, 12.5-, 25-, 62.5-, 100- and 250-fold mass excess over hTR). Lanes 9–13: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing quantity of cold gel purified ribosomal RNA (rRNA) (6.25-, 12.5-, 25-, 62.5- and 100-fold mass excess over hTR). Lanes 14–17: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing quantity of cold E. coli tRNA (100-, 250-, 450- and 1800-fold mass excess over hTR). Lanes 18–23: 50 nM GST–hEST1A (fraction E1) incubated with *hTR and increasing quantity of heparin (6.25-, 12.25-, 25-, 62.5-, 100- and 250-fold mass excess over hTR). Right panel: quantification of competition experiments.
Recombinant hEST1A has also been reported to bind telomeric DNA oligonucleotides (2). However, the RNA-binding domain of hEST1A appears distinct from the DNA-binding domain. First, telomeric and non-telomeric DNA oligonucleotides were not able to compete with formation of the GST–hEST1A–hTR complex even when present at a 1000-fold molar excess (Supplementary Data, Figure 1). Second, the DNA-binding domain of hEST1A has been mapped by Lea Harrington's laboratory (Toronto, personal communication) to a fragment encompassing amino acids 1–208 of hEST1A. This domain does not overlap with the hTR-binding domain of hEST1A (TRID), which has been mapped to amino acids 243–412 in the next section.
Identification of the hTR-binding domain of hEST1A
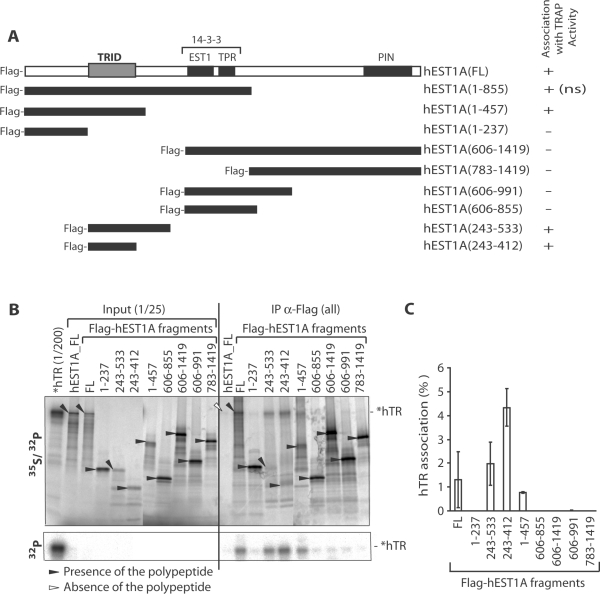
To map the hEST1A domain that mediates hTR binding, FLAG-hEST1A fragments (Figure 3A) were expressed in RRL in presence of [35S]-methionine and incubated with [32P]-labeled hTR. After immunoprecipitation of FLAG-hEST1A with α-FLAG antibodies, samples were separated by SDS–PAGE. [35S]-labeled proteins and [32P]-labeled hTR were detected upon analysis on a PhosphoImager (Figure 3B). [32P]-labeled hTR could be unequivocally distinguished from [35S]-labeled proteins by shielding the lower energy [35S]-derived β-rays with a sheet of plastic (Figure 3B, lower part). In the absence of the FLAG-epitope (lane hEST1A_FL), neither hEST1A nor hTR was detected in the immunoprecipitate, demonstrating the specificity of the assay for FLAG-tagged hEST1A. A FLAG-hEST1A fragment encompassing amino acids 243–412 immunoprecipitated hTR even more efficiently than full-length FLAG-hEST1A, while FLAG-hEST1A fragments lacking this domain did not reveal efficient hTR-binding activity (Figure 3C). We therefore refer to this domain as hTR-interaction domain (TRID).
Figure 3.
Identification of the hEST1A hTR-binding domain (TRID). (A) Schematic representation of previously assigned hEST1A domains and hEST1A truncations. EST1, EST1 homology domain; TPR, tetratricopeptide repeats; PIN, PilT amino-terminal domain (1). The EST1 and TPR domains of the related SMG7/hEST1C form a 14-3-3 domain with a classic phosphoserine-binding pocket (31). For each polypeptide the association with TRAP activity is indicated [determined in Figure 4; for hEST1A (1–855) association with TRAP activity is not shown (ns)]. TRID stands for hTR Interacting Domain identified in B and C. (B) [32P] 5′-end labeled in vitro transcribed hTR (*hTR) was incubated with in RRL translated [35S] methionine-labeled full-length hEST1A, full-length Flag-hEST1A (FL) and Flag-hEST1A fragments (amino acid fragments are indicated). After immunoprecipitation with α-FLAG antibodies and five washes, samples were separated by 4–20% gradient SDS–PAGE. [35S] and [32P] signals were revealed by analysis on a PhosphoImager. Upon placing a plastic film between the gel and the PhosphoImager screen, only the [32P] signal was detected (lower part). Filled black arrows indicate the presence of the polypeptide in the immunoprecipitated fractions and empty arrows indicate the absence of the polypeptide. (C) Quantification of hTR associated with Flag-hEST1A fragments based on the gel in panel B and at least two additional experiments. The values were corrected by the immunoprecipitation efficiency of the individual polypeptides.
TRID binds active telomerase
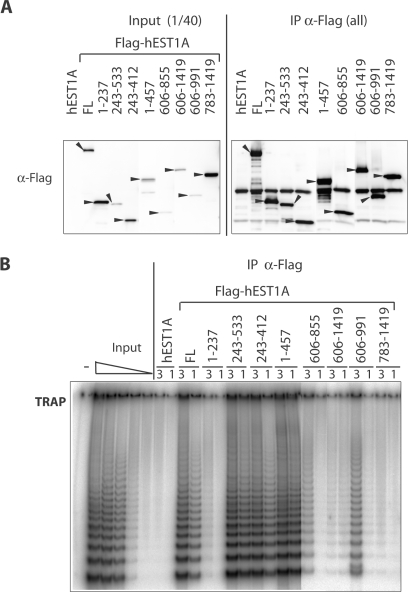
To map the hEST1A domain, which mediates association with telomerase activity, FLAG-hEST1A fragments were transiently expressed in 293T cells and FLAG-hEST1A was immunoprecipitated with α-FLAG antibodies. Expression of FLAG-hEST1A fragments and their enrichment in the immunoprecipitate was tested in western blots with α-FLAG antibodies (Figure 4A). Presence of active telomerase was assessed in TRAP-assays (Figure 4B). As expected, immunoprecipitation of telomerase activity depended on presence of the FLAG epitope on hEST1A [Figure 4B, compare untagged hEST1A (lanes 7–8) with FLAG-hEST1A (lanes 9–10)]. Again, the FLAG-hEST1A fragment encompassing amino acids 243–412 was sufficient to immunoprecipitate telomerase activity while FLAG-hEST1A fragments lacking this domain immunoprecipitated considerably less (fragment 606–991) or no telomerase activity (Figure 4B). Thus, TRID is able to associate with active telomerase probably via its association with hTR.
Figure 4.
Identification of a hEST1A fragment that binds active telomerase. The 293T cells were transiently transfected with expression vectors for full-length EST1A, full-length Flag-hEST1A (FL) and Flag-hEST1A fragments (amino acid fragments are indicated). (A) Upon immunoprecipitation with α-FLAG antibodies and five washes, samples were loaded on 4–20% gradient SDS–polyacrylamide gels and FLAG-tagged EST1A fragments were revealed by western blotting with α-FLAG antibodies. Input refers to extracts of the transiently transfected 293T cells before immunoprecipitation. (B) TRAP assay before and after immunoprecipitation with α-FLAG antibodies. 3 and 1 refer to relative amounts of fraction used in the assay.
Identification of a hTERT fragment (EID) that binds hEST1A and hTR
To identify a domain within hTERT that mediates interaction with hEST1A, we transiently co-expressed Myc-hTERT fragments with FLAG-hEST1A in 293T cells (Figure 5). Extracts were prepared and FLAG-hEST1A was immunoprecitated with α-FLAG antibodies. Presence of FLAG-hEST1A in the immunoprecipitate was checked in western blots with α-FLAG antibodies and co-immunoprecipitated hTERT was detected with α-Myc antibodies (Figure 5B). An hTERT fragment encompassing amino acids 147–311 efficiently associated with hEST1A and other hTERT fragments that contained this domain also immunoprecipitated FLAG-hEST1A. However, N-terminal fragments of hTERT (1–144 or 1–78) did not immunoprecipitate FLAG-hEST1A. We refer to the fragment encompassing amino acids 147–311, which binds hEST1A as EID (for hEST1A-interaction domain).
Figure 5.
Identification of an hTERT fragment (EID) that binds hEST1A. (A) Schematic representation of full-length hTERT and hTERT fragments. The different hTERT domains are indicated (32). For each polypeptide the molecular weight is given in kDa and the binding of hEST1A as determined in B is indicated (+, binding; −, no binding and ?, not known). (B) Transient co-expression in 293T cells of FLAG-hEST1A and full-length hTERT, full length Myc-hTERT (FL) or Myc-hTERT fragments (amino acid numbers are indicated). FLAG-hEST1A was immunoprecipitated using α-FLAG antibodies. Upon immunoprecipitation and five washes, samples were loaded on 4–20% gradient SDS–polyacrylamide gels. The presence of Myc-hTERT fragments was determined by western blotting with α-Myc antibodies and the immunoprecipitation efficiency was determined by western blotting with α-FLAG antibodies. The presence of the polypeptides is indicated by black arrows whereas white arrows indicate absence of the polypeptides.
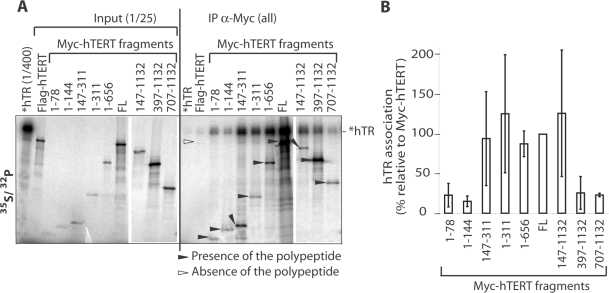
To test if hEST1A binding by hTERT involves also hTR, we measured hTR-binding activity of different hTERT fragments. hTERT fragments were expressed in presence of [35S]-methionine in RRL and incubated with [32P]-labeled in vitro transcribed hTR (Figure 6). After immunoprecipitation of Myc-hTERT with α-Myc antibodies, samples were separated by SDS–PAGE. [35S]-labeled proteins and [32P]-labeled hTR were detected upon analysis on a PhosphoImager as in Figure 3. Weak hTR-binding activities were detected with Myc-hTERT-fragments containing amino acids 1–78, 1–144, 397–1132 and 707–1132. Most efficient hTR-binding activity was present in the EID-fragment (amino acids 147–311), which also bound hEST1A in Figure 5.
Figure 6.
RNA-binding activity of hTERT fragments (see Figure 5A for schematic representation of the constructs). (A) [32P] 5′-end labeled in vitro transcribed hTR (*hTR) was incubated with in RRL in presence of [35S] methionine translated full-length FLAG-hTERT, full-length Myc-hTERT (FL) or Myc-hTERT fragments (amino acid numbers are indicated). Upon immunoprecipitation with α-Myc antibodies and five washes, samples were loaded on 4–20% gradient SDS–polyacrylamide gels. The analysis was done as in Figure 3. (B) Quantification of hTR associated (%) with the Myc-hTERT fragments based on the gel in panel A and at least two additional experiments. The values were corrected by the immunoprecipitation efficiency of the individual polypeptides.
Protein–RNA and protein–protein interactions mediate association of human EST1A with hTERT
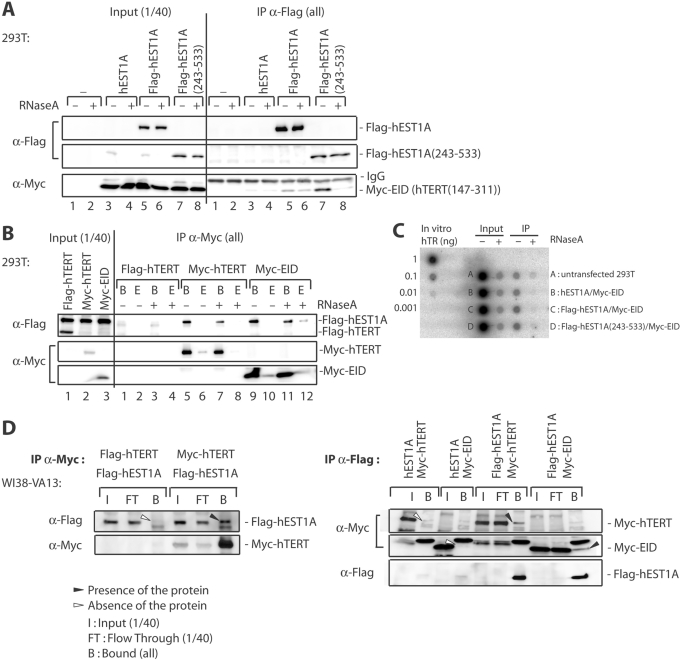
The above experiments delineated hEST1A and hTERT domains that at the same time mediated the interaction between hEST1A and hTERT as well as with hTR. To test the hypothesis that the interaction between hEST1A and hTERT was mediated by hTR, we transiently co-expressed hEST1A and hTERT fragments in 293T cells and tested their interaction by co-immunoprecipitation upon treatment of the extracts with RNase A (Figure 7). The extent of hTR destruction by RNase A treatment was determined by assessing presence of hTR upon Northern hybridization of dotblots with a randomly labeled full-length hTR probe (Figure 7C). Approximately 99% of the hTR signal disappeared in extracts and more than 95% of the signal disappeared in the immunoprecipitates upon RNase A treatment. Immunoprecipitated full-length FLAG-hEST1A co-immunoprecipitated Myc-EID (hTERT-fragment 147–311) and this interaction was not alleviated upon RNase A treatment (Figure 7A, compare lanes 5 and 6 on the right panel). Thus, while EID is able to bind hTR it establishes also protein–protein contacts with hEST1A. In contrast, when a TRID-containing FLAG-hEST1A fragment [hEST1A-fragment (243–533)] was immunoprecipitated, the interaction with EID was RNase A sensitive (Figure 7A, compare lanes 7 and 8 on the right panel). Thus, EID cannot establish stable protein–protein contacts with TRID as observed with full-length hEST1A. Consistent with these results, when either full-length Myc-hTERT or Myc-EID were co-expressed with full-length FLAG-hEST1A and immunoprecipitated, FLAG-hEST1A remained associated with Myc-hTERT as well as with Myc-EID even upon RNase A treatment of the beads after the immunoprecipitation (Figure 7B, compare in the right panel lanes 5 with 7 and lanes 9 with 11).
Figure 7.
Protein–RNA and protein–protein interactions mediate association of human EST1A with hTERT. The indicated polypeptides were transiently co-expressed in 293T (Figure 7A and B) or WI38-VA13 (Figure 7D) cells and immunoprecipitated (IP) with the indicated antibodies. Polypeptides were revealed by western blotting with antibodies indicated on the left. (A) 293T cell extracts (600 µl) were either treated (+) or not treated (−) with 3 µg RNase A for 10 min at 25°C before starting the immunoprecipitation with α-FLAG antibody. After five washes, samples were loaded on 4–20% gradient SDS–polyacrylamide gels. The presence of Myc-EID was determined by western blotting with α-Myc antibodies and the immunoprecipitation efficiency was determined by western blotting with α-FLAG antibodies. (B) Transient co-transfection in 293T cells of Flag-hEST1A and full-length FLAG-hTERT, full-length Myc-hTERT (FL) and Myc-EID. Immunoprecipitations were carried out with α-Myc antibodies. After five washes, the beads were either treated (+) or not treated (−) or with 0.5 µg RNase A/12 µl for 10 min at 25°C. The eluate (E) was also analyzed to check for the release of Flag-hEST1A. The beads were washed one time before being loaded on 4–20% gradient SDS–polyacrylamide gels. The presence of Flag-hEST1A was determined by western blotting with α-FLAG antibodies and the immunoprecipitation efficiency was determined by western blotting with α-Myc antibodies. (C) Dot blot analysis of total RNA extracted from 293T cell extracts before and after RNase A treatment, revealed with a randomly labeled full-length hTR probe. Extracts (Input) were treated with 0.005 µg RNase A/µl of extract and beads (IP) were treated with 0.04 µg of RNase A/µl of bead suspension as in B. (D) Transient co-transfection in WI38-VA13 cells of Flag-hEST1A and full-length FLAG-hTERT, full-length Myc-hTERT (FL) and Myc-EID. Immunoprecipitations were carried out with α-Myc antibodies (left panel) or α-Flag antibodies (right panel) as in A and B.
To further corroborate hTR-independent interactions between hTERT and hEST1A, we assessed the interaction between both polypeptides upon transient expression in WI38-VA13, an immortal human fibroblast cell line that does not express hTR (Figure 7D). Again, Myc-hTERT co-immunoprecipitated Flag-hEST1A (Figure 7D, left panel) and Flag-hEST1A co-immunoprecipitated Myc-hTERT and Myc-EID. This therefore confirms that full-length Myc-hTERT and Myc-EID interact with hEST1A also in the absence of hTR via protein–protein contacts.
DISCUSSION
Human EST1A was discovered through its sequence similarity with S. cerevisiae Est1p but its precise function at telomeres has remained unclear. We show here that hEST1A associates with telomerase through dual interactions with the telomerase RNA moiety and the TERT. The TRID is contained within a 168 amino acid fragment in the N-terminal part of hEST1A, a region that does not contain any recognizable structural motifs or sequence homologies with other proteins in the database. TRID binds to hTR with high overall affinity (25 nM) but low specificity. Distinct non-overlapping fragments of hTR as well as rRNA competed for binding of full-length hTR with similar efficiency while tRNA was a poor ligand for hEST1A. TRID is also distinct from the DNA-binding domain of hEST1A (2), which resides more N-terminal [amino acids 1–208, Lea Harrington's laboratory (Toronto, personal communication)]. Low specificity of RNA binding was also observed in vitro with yeast Est1p, which displayed a similar affinity (50 nM) for the yeast telomerase RNA TLC1 and other RNA substrates (21). However, in vivo a bulged stem of TLC1, which has no obvious counterpart in human telomerase RNA, mediates association with Est1p (22). Thus, it is conceivable that similarly to yeast Est1p, hEST1A also interacts with a specific hTR structure in vivo in the context of the assembled hTR-hTERT complex. This would be analogous to some ribosomal proteins, which achieve RNA sequence-specific binding only in the context of a partially assembled ribonucleoprotein particle (23,24).
Consistent with this notion, we also identify protein-mediated interactions between hEST1A and TERT. TRID interacts with EID in a strictly RNA-dependent manner whereas full-length hEST1A remains associated with hTERT or with EID even upon digestion of hTR with RNases, or when expressed in a cell line that lacks hTR. Consistent with our results in cell culture, telomerase RNA-independent association of hEST1A and hTERT was observed in RRL (2). It remains to be determined which domains outside of TRID establish protein-mediated interactions with TERT. Interestingly, the EST1/TPR domain of the hEST1A/SMG6-related SMG7/hEST1C polypeptide has a 14-3-3 fold with a classic phosphoserine-binding pocket (25). However, other regions of the protein could also be involved.
We also identified a TERT domain that associates with hEST1A. The EID is comprised between amino acids 147–311. EID contains hTR-binding activity and at the same time it establishes RNase-resistant contacts with hEST1A. Previous studies with hTERT have identified two regions responsible for interactions with TR, termed RNA interaction domain (RID) 1 and 2 (26). The C-terminal part of RID1 (amino acids 1–250), which associates with the hTR template-pseudoknot domain (27) overlaps with the N-terminal part of EID (amino acids 147–311). A hTERT fragment containing the entire RID1 and EID regions did not pull-down more hTR than EID on its own (Figure 6) suggesting that EID and RID1 contain one and the same RNA-binding domain. However, deletion of residues 1–24 in RID1 abolished binding of the template-pseudoknot domain (27), suggesting that either the RNA-binding activity in EID is distinct from RID1 or that the 1–25Δ RID1-mutant displayed a folding problem that abolished RNA-binding activity. Interestingly, the recently solved structure of a Tetrahymena TERT region named TEN, which corresponds to human RID1, shows a positively charged flexible C-terminal tail. This tail, which is present at the N-terminus of EID has been proposed to be involved in telomerase protein–RNA interactions (28). No functions have been assigned before to the C-terminal part of EID. Scanning mutagenesis with short linkers did not identify hTERT-mutants in this region that abolished telomerase function in vivo (29), but protein–protein interactions may involve larger surfaces that were not fully inactivated with this approach. Furthermore, hEST1A–hTR interactions may partially compensate for the loss of direct hTERT–hEST1A contacts.
The regulation of human telomerase at telomeres and the putative role of hEST1A in this process are poorly understood at the molecular level. The functional analysis of hEST1A has been hampered by its involvement in NMD (3) and essential roles of hEST1A for S-phase progression presumably in conjunction UPF1 ((12) and unpublished data). S. cerevisiae Est1p is thought to mediate telomerase recruitment through its direct interaction with Cdc13p, which binds to the telomeric 3′ overhang (8). POT1, the putative human ortholog of Cdc13p inhibits telomerase activity in vitro when bound to the 3′end of a telomeric oligonucleotide substrate (19,30). Trials to detect a physical interaction between POT1 and hEST1A have been unsuccessful so far and addition of recombinant hEST1A to in vitro assays could also not alleviate the inhibitory effects of POT1 on telomerase activity (our unpublished data). However, telomerase recruitment by S. cerevisiae Cdc13p and Est1p has also not yet been reproduced in vitro. The here reported identification and characterization of functional domains of hEST1A that mediate interaction with telomerase may allow to specifically disrupt these interactions to understand the possible roles of hEST1A in telomere length control and telomere capping.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Isabel Kurth for affinity-purified telomerase from insect cells and for technical advice, Claus Azzalin for comments on the manuscript and Lea Harrington for communicating unpublished results. Work in the laboratory was supported by grants from the Swiss National Science Foundation, the Swiss Cancer League, the Human Frontier Science Program and the EU 6th Framework Programme. Funding to pay the Open Access publication charges for this article was provided by HFSP.
Conflict of interest statement. None declared.
REFERENCES
- 1.Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 2.Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 3.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 5.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 6.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 7.Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 2003;13:575–580. doi: 10.1016/s0960-9822(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 8.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 9.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–3534. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 21.Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 22.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 24.Brodersen DE, Clemons W.M., Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 25.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Moriarty TJ, Huard S, Dupuis S, Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 29.Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 31.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelleher C, Teixeira MT, Forstemann K, Lingner J. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 2002;27:572–579. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.