Abstract
V(D)J recombination events are initiated by cleavage at gene segments by the RAG1:RAG2 complex, which results in hairpin formation at the coding ends. The hairpins are opened by the Artemis:DNA-PKcs complex, and then joined via the nonhomologous DNA end joining (NHEJ) process. Here we examine the opening of the hairpinned coding ends from all of the 39 functional human VH elements. We find that there is some sequence-dependent variation in the efficiency and even the position of hairpin opening by Artemis:DNA-PKcs. The hairpin opening efficiency varies over a 7-fold range. The hairpin opening position varies over the region from 1 to 4 nt 3′ of the hairpin tip, leading to a 2–8 nt single-stranded 3′ overhang at each coding end. This information provides greater clarity on the extent to which the hairpin opening position contributes to junctional diversification in V(D)J recombination.
INTRODUCTION
The enormous diversity of antigen receptor genes is generated through V(D)J recombination during B and T lymphocyte development. The exons encoding the antigen-binding variable domains of immunoglobulin and antigen receptors (TCR) are assembled by recombination of V (variable), D (diversity) and J (joining) elements or subexons (1–4). The combinatorial diversity of numerous V, D and J elements, as well as nucleotide loss and addition at the recombination junctions (junctional diversification) combine to generate a potential repertoire of 1022 murine antigen receptor molecules (5).
V(D)J recombination is directed by recombination signal sequences (RSS) flanking each coding element. There are two types of RSS: one with a 12 bp spacer between the heptamer and nonamer (12 RSS), and the other with a 23 bp spacer (23 RSS). RAG1/RAG2 proteins, together with HMG1 recognize the RSS and initiate V(D)J recombination by, creating a nick between the 3′ end of the coding segment and the 5′ end of the heptamer. Synapsis of two different RSSs (one 12 and one 23 RSS) results in completion of cleavage, where the 3′-hydroxyl group at the coding end attacks the opposing strand to form a hairpin at the coding end (6). The two blunt signal ends are later joined directly with no nucleolytic processing of the ends. The coding end hairpins are opened by Artemis:DNA-PKcs, and then joined by nonhomologous DNA end joining (NHEJ) (7). Due to the intrinsic error-prone nature of NHEJ, the resulting coding joints usually exhibit nucleotide loss and additions of up to ∼15 bp (8). Nucleotide additions include P (palindromic) nucleotides from asymmetric hairpin opening, as well as N nucleotides from TdT activity. These junctional regions between V, D and J subexons encode the most highly variable regions in the antigen receptor variable domain, called complementarity determining region 3 or CDR3.
The assembly of the antigen receptor repertoire relies on many factors. Coding end and signal sequences can markedly affect the recombination frequency (9–11). The binding of the RAGs to various signal sequences and the varied cleavage efficiency by the RAG complex at different coding segments both contribute to this (12). Previous studies from our group analyzed the 39 functional human VH elements at the human immunoglobulin heavy chain locus for RAG cleavage (13). The preferred cleavage of the RAG complex at different coding segments corresponds well to the observed profile of unselected VH family usage in humans. However, the efficiency of the subsequent NHEJ steps has yet to be determined by direct experiments.
Recombination between the same two coding segments could yield different junctions due to nucleotide loss and addition during NHEJ. But the diversity at the junctions can be restricted when the coding ends bear short regions of homology (14). Homology may account for overrepresentation of certain combinations that are important in immune recognition of common pathogens. For example, the DFL16.1-JH1 maximum homology joint is present in most T15-positive antibodies, which are critical in protection against Streptococcus pneumoniae (15,16). The position of hairpin opening after RAG cleavage may greatly affect the homology between two coding ends (14,17). Here we analyzed the 39 functional VH segments for hairpin opening by Artemis:DNA-PKcs in an in vitro system. The hairpin opening generates 3′ overhangs that vary from 2 to 8 nt, with the dominant length being 4 nt. These biochemical observations provide the essential basis for understanding the extent to which asymmetric hairpin opening contributes to V(D)J recombination junctions formed in vivo.
MATERIALS AND METHODS
Oligonucleotides
The oligonucleotides (oligonts) used for the hairpin opening in Figure 1A are shown in Table 1. The oligonts used to generate the 3′ overhang substrate in Figure 1C are YM-149 and YM-68 [sequences were described in (18)]. The sequence for each coding end hairpin substrate is depicted in Figure 3A: 5′-GGGCCC-CE-CE anti-parallel-CCCGGG-3′, and the sequences for CE (coding end) are shown in Table 1. The human VH names in Table 1 are according to the Honjo laboratory (19). For activation of DNA-PKcs, the 60 bp double-strand DNA was annealed form oligonts YM-74 and YM-75 [sequences in (7)]. The pseudo-Y DNA was annealed from oligonts YM-220 and YM-221 [sequences in (20)].
Figure 1.
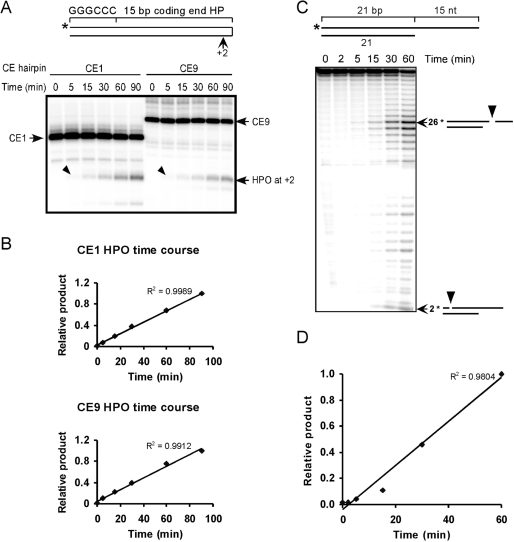
The endonuclease activity of Artemis:DNA-PKcs is linear over a 90-min time course. (A) Two different coding end hairpins 1 and 9 (CE1 and CE9) were assayed for time courses of hairpin opening by Artemis:DNA-PKcs. The arrowheads point to the primary hairpin opening products at the +2 position. (B) The major products at each time point were quantified and normalized to the product at the last time point (90 min). A linear trend line was added and the R-squared value was displayed on the chart. (C) The time course of 3′ DNA overhang cleavage by Artemis:DNA-PKcs. The 3′ DNA overhang substrate was illustrated on the top. The major products are identified using arrows with the nt sizes and corresponding cleavage positions (arrowheads) depicted on the right. (D) The primary cleavage products of 26 nt at each time point were plotted in the same manner as in (B).
Table 1.
Sequences of different human VH coding ends and hairpin opening efficiency by Artemis:DNA-PKcs.
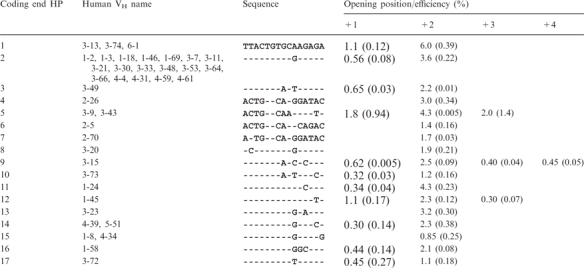 |
The terminal 15 bp of the VH elements were used to create the oligont-based cleavage substrates. The name designations for the human VH elements in the second column are those assigned by the Honjo laboratory (19). The first number represents the family and the second number is the individual VH designation. The full sequence for coding end HP 1 (VH 3-13, 3-74 and 6-1) is shown at the top. Each dash for the other sequences indicates the identical nucleotide, while derivations are indicated by the corresponding nucleotides. Hairpin opening positions +1 to +4 indicate the distance 3′ to the hairpin tip, as illustrated in Figure 4A. Hairpin opening efficiency was calculated based on the percentage of total substrate (hairpin opening product/total signal in each lane is shown in Figure 4). The efficiency data shown in the table are averages of two determinations, and the standard deviations are displayed in parentheses below the efficiency. Any product amount less than 0.30% was regarded as no hairpin opening.
Figure 3.
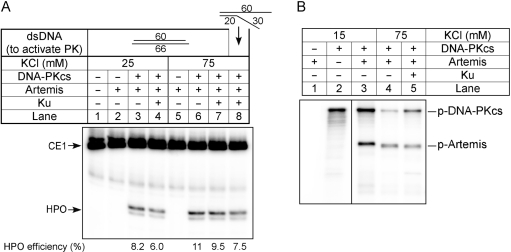
Hairpin opening by Artemis:DNA-PKcs as a function of KCl or Ku. (A) The substrate used here is coding end hairpin 1 (CE1). Proteins in the reactions are indicated above the corresponding lanes. Reactions 1–4 are done in 25 mM KCl, while reactions 5–8 are done in 75 mM KCl. To activate DNA-PKcs, reactions 1–7 included a 60 bp double-strand DNA with short overhangs, whereas reaction 8 instead contains a pseudo-Y structured DNA with a 20 bp double-strand region as depicted. The numbers along the bottom are the quantitation of hairpin opening (product/total). (B) Radioactive kinase assays with DNA-PKcs and Artemis were carried out at the indicated KCl concentrations, and Ku protein was added to reaction 5.
Protein purification
Ku70/Ku86, Artemis-myc-his and DNA-PKcs were purified as described. The purity of the proteins was also demonstrated previously (7,18,20).
In vitro nuclease assay
For the 3′ overhang cleavage assay, the longer oligonucleotide was labeled with T4 polynucleotide kinase (PNK) and [γ-32P] ATP (3000 Ci/mmol, PerkinElmer), and then annealed to the partner unlabeled oligonucleotide at equal molar ratios. The coding end hairpins were labeled at the 5′ end in the same way. Time-course assays were carried out at 37°C in a total volume of 60 μl with the hairpin opening buffer [25 mM Tris (pH 8.0), 10 mM KCl, 10 mM MgCl2, 1 mM DTT, 50 ng/ml of BSA, 0.25 mM of ATP and 0.5 μM of 40 bp DNA as stimulator]. Substrates were added to 25 nM with 100 nM Artemis and 60 nM DNA-PKcs. An aliquot of 10 μl was taken out at each time point; and the reaction was terminated by the addition of 10 μl stop dye (95% Formamide and 10 mM EDTA) and boiling for 5 min. For Figure 3A and B, reactions were done in 10 μl total volume and incubated at 37°C for 1 h. In Figure 3A where Ku was added, the concentration was 40 nM. The reaction products were resolved on denaturing 7 M urea—12% polyacrylamide gels. The gels were then dried and exposed to a PhosphorImager screen, and the image was obtained by using PhosphorImager 445SI (Molecular Dynamics, Sunnyvale, CA, USA) and analyzed with ImageQuant software (v5.0).
In vitro DNA-PKcs kinase assay
The DNA-PK kinase assay was performed in a total volume of 60 μl, which contains 10 mM Tris (pH 7.5), 1 mM EDTA, 10 mM MgCl2, 1 mM DTT, 0.3 μM 35 bp DNA, and 165 nM of [γ-32P] ATP. DNA-PKcs was added to 80 nM, and Artemis-myc-his was added to 100 nM. Reaction mixtures were incubated at 37°C, an aliquot of 10 μl was removed and fractionated on an 8% SDS–PAGE. For the zero time point, a separate reaction with no DNA-PKcs added was prepared. For Figure 3B, lanes 2 and 3, an additional of 60 mM KCl was supplied to make the final KCl concentration 75 mM KCl, and Ku was added to 80 nM. The images were obtained in the same way as for the nuclease assay.
Quantification and plotting
The gel images developed on PhosphoImager were analyzed and quantified with ImageQuant and plotted out using Microsoft Excel. For each assay at least two experiments were done and the quantifications in Figures 1 and 2 were typical of all of the experiments. For the 3′ DNA overhang cleavage assay shown in Figure 1C, quantification of the shorter products (25 and 24 nt) indicated almost identical linear curves (data not shown). For coding end hairpin opening efficiencies shown in Table 1, the averages of two determinations are shown with standard deviations in parentheses.
Figure 2.
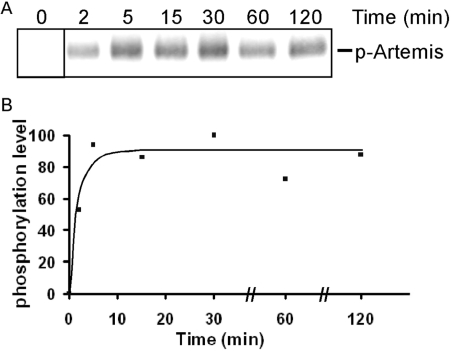
The phosphorylation of Artemis by DNA-PKcs rapidly reaches a plateau. (A) Artemis was incubated with DNA-PKcs in the presence of [γ-32P] ATP at 37°C. A fraction of the incubation mixture was removed at each time point and resolved on SDS–PAGE. For the zero time point, a separate reaction containing all components except DNA-PKcs was used. (B) The level of phosphorylation was quantified and normalized as a percentage of the highest level (100% at 30 min).
RESULTS
Time course of Artemis endonuclease activity
The coding end hairpin substrates used for in vitro cleavage are illustrated in Figure 1A. They consist of the last 15 bp of each different human VH coding ends and a 6 bp GC sequence to minimize breathing at the end opposite the hairpin. The 5′ ends of the oligonts were labeled with 32P, as indicated by the asterisk. To assess the sequence effect on coding end hairpin opening efficiency, we first examined the time course of the endonuclease activity to make sure our assay conditions were in the linear range. Two of the coding end hairpin substrates (CE1 and CE9, see Table 1 for sequences) were assayed for hairpin opening by Artemis:DNA-PKcs as a function of time. We find that the primary product of hairpin opening is at 2 nt on the 3′ side of the hairpin tip (Figure 1A). The endonucleolytic activity of Artemis remains constant throughout the 90-min incubation (Figure 1B).
Various DNA structures containing single-/double-strand transitions can be opened by the Artemis:DNA-PKcs complex (20), and a 3′ DNA overhang substrate shows higher cleavage efficiency. We also assayed the time course for 3′ overhang cleavage. Cleavage by Artemis:DNA-PKcs occurs primarily at 4 and 5 nt 3′ of the single-/double-strand transition (Figure 1C). The endonuclease activity also remained linear for over 1 h (Figure 1D). Therefore, we chose to use 60 min incubations for the coding end hairpin opening assay.
DNA-PKcs phosphorylation is rapid and is not the rate-limiting step for hairpin opening
The kinase activity of DNA-PKcs is required to activate the endonucleolytic activity of Artemis (7,21–23). The time course of DNA-PKcs autophosphorylation is very rapid (24). Artemis forms a physical complex with DNA-PKcs and is phosphorylated by DNA-PKcs. We examined the time course of Artemis phosphorylation and found that phosphorylation is also rapid and reaches a plateau within 5 min (Figure 2). The difference in time course between the kinase and the nuclease activity indicate that phosphorylation of Artemis and autophosphoryation of DNA-PKcs are not the rate-limiting steps for the endonucleolytic reaction.
Physiological salt concentrations and addition of Ku did not increase the endonucleolytic activity of Artemis:DNA-PKcs
It has been suggested that under higher salt conditions (75 mM KCl), the endonucleolytic activity is reliant on Ku (25). The same study showed hairpin opening by Artemis at the hairpin tip, which is inconsistent with the hairpin opening pattern observed in the chromosomes of primary thymic T cells and lymphoid cell lines (26) and observed in vitro (7). To ensure that our in vitro hairpin opening assay reflects physiological conditions, we compared activity at 75 and 25 mM KCl [our original condition (7)] for hairpin opening. The cleavage position and efficiency remained the same at both KCl concentrations (Figure 3A, lanes 3 versus 6). Addition of Ku did not change the hairpin opening in 75 mM KCl (Figure 3A, lanes 6 versus 7), and at 25 mM KCl, Ku is slightly inhibitory (Figure 3A, lanes 3 versus 4). Use of a pseudo-Y DNA molecule as the activator for DNA-PKcs did not change the reaction outcome (Figure 3A, lanes 8), consistent with our previous studies (7).
The same paper from Goodarzi et al. (25) also implied that with 75 mM KCl, there is less Artemis phosphorylation by DNA-PKcs than with lower salt conditions, and Ku could increase the level of Artemis phosphorylation. But kinase reactions in Goodarzi et al. were conducted using Artemis purified from E.coli, which is catalytically inactive in vitro (21). Here we carried out a kinase assay with active Artemis purified from 293T cells. As expected, with 75 mM KCl, the amount of overall phosphorylation is reduced by about 3-fold (Figure 3B, lanes 3 versus 4). The addition of Ku slightly reduces Artemis phosphorylation, while it increases DNA-PKcs autophosphorylation (Figure 3B, lanes 4 versus 5). Based on the electrophoretic mobility, the number of phosphorylated residues was also reduced at 75 mM KCl, as indicated by a faster migration on SDS–PAGE gels. Despite the decreased phosphorylation at 75 mM KCl, the nuclease activity was not affected. Therefore, we decided to carry out all of the following hairpin opening assays with our original conditions (25 mM KCl) (7).
The Artemis:DNA-PKcs complex opens coding end hairpins consistently but with minor variations
Coding end hairpin substrates were designed based on the 39 functional human VH elements sequences. Artemis:DNA-PKcs recognizes the single-/double-strand transition region on different substrate structures. Therefore the sequences that would affect hairpin opening efficiency probably lie close to the hairpin tip. The terminal 15 bp of the coding end sequences from the 39 VH elements is highly homologous. Sequence alignment revealed 17 different sequences. Accordingly, we created 17 different coding end hairpin opening substrates (CE1 to CE17). To minimize breathing at the blunt end, we incorporated a 6 bp GC clamp to stabilize the hairpin structure (Figure 4A).
Figure 4.
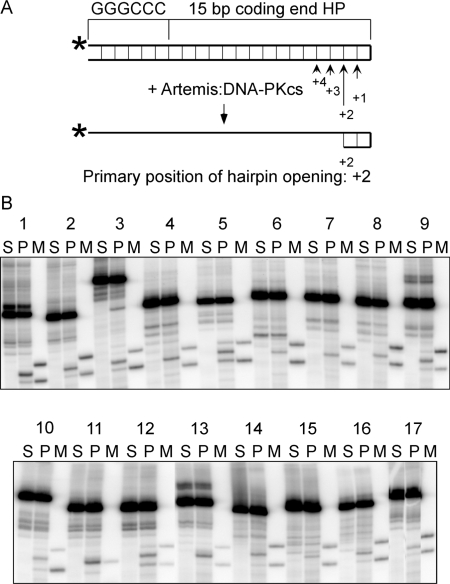
The hairpin opening pattern by Artemis:DNA-PKcs for the 17 different coding end hairpins derived from 39 human VH elements. (A) The illustration shows the configuration of coding end hairpin substrates and the cleavage positions by Artemis:DNA-PKcs. The base pairings are indicated as thin lines between the top and bottom strands. The length of the arrows reflects different cleavage efficiency at the four positions (+1 to +4). (B) The hairpin opening products were resolved by denaturing PAGE. The numbers 1–17 correspond to the sequence numbers in Table 1. For each coding end hairpin (HP), there is a set of three lanes. The hairpin opening products (P) were in the middle, with uncut substrate on the left (S), and the hairpin opening markers (M) on the right. For most substrates, the markers correspond to hairpin opening at +1 and +3 positions, except for substrate 11, with only one marker at the +2 position. CE3 substrate appears to show one minor hairpin opening product at greater than the +4 position.
The coding end hairpin substrates were individually incubated with Artemis and DNA-PKcs at 37°C for 1 h, and the products were resolved on denaturing polyacrylamide gels (Figure 4B, ‘P’ lanes). Because the substrates have hairpin structures, they cannot be fully denatured in the gel and often run as multiple bands. The substrates (Figure 4B, ‘S’ lanes) were loaded next to the products to help identify the cleavage products. For every substrate, two different markers with sequences and lengths of the expected hairpin opening products were prepared and loaded onto the same gel adjacent to the product lanes (Figure 4B, ‘M’ lanes). The exact position of hairpin opening was determined by comparing products with the markers. All of the hairpins were opened primarily at the +2 position (Figure 4B), consistent with our previous observations (7). However, about two-thirds of the hairpin substrates (11 out of 17) were also opened at the +1 position, and three substrates were also opened at +3 and one substrate was opened at +4 position (Table 1, Figure 4). The amount of major cleavage products (opened hairpin) at the +2 position varied over a 7-fold range among this set of natural human VH substrates, with 1–4% of most hairpin opening substrates converted to product. We compared the sequences of the coding end hairpins and found no distinct pattern for preferred cleavage. It is surprising, however, that a one base-pair difference located 5 bp away from the hairpin tip can change the hairpin opening efficiency considerably (compare CE 1, 2 and 17). This study shows that the sequence differences between human VH coding ends must affect hairpin structure in a manner to which Artemis is highly sensitive.
DISCUSSION
It has been known that asymmetric opening of hairpinned coding ends contributes to junctional addition. However, the variability in the position of hairpin opening and efficiency of opening has not been examined previously. The study here shows that different hairpin sequences are opened by the native DNA-PKcs and full-length Artemis in variable ways and with varying efficiency. The variation in position is important from the standpoint of immunologic diversity. The variation in efficiency of this hairpin opening step will be important in future considerations of the rate-limiting steps of V(D)J recombination.
Correlation between hairpin opening by the Artemis:DNA-PKcs complex and the phosphorylation by DNA-PKcs
The endonucleolytic activity of Artemis:DNA-PKcs remains linear for at least 90 min in vitro, while the phosphorylation of Artemis and DNA-PKcs itself is relatively rapid, requiring <10 min. Phosphorylation of DNA-PKcs and Artemis is probably the ‘priming’ stage of the cleavage reaction. The phosphorylated protein complex then changes conformation and cleaves DNA at the single-/double-strand transition of the DNA substrate. This is based on the fact that Artemis continues to cleave endonucleotically for over 1 h after the maximal phosphorylation of Artemis and DNA-PKcs. Autophosphorylation is known to facilitate the release of DNA-PKcs from DNA ends (27), but DNA-PKcs may retain the ability to bind DNA and assist the action of Artemis on other DNA molecules.
Despite a higher salt condition (75 mM KCl), which reduces the kinase activity of DNA-PKcs, this higher salt had no effect on the Artemis endonuclease activity. Different clusters of autophosphorylation sites exist in DNA-PKcs, and they are implicated in its various functions (28–30). Possibly the weaker phosphorylation observed at 75 mM KCl includes the activating cluster and is sufficient for kinase activation. Similar reasoning could be applied to the phosphorylation of Artemis. Fourteen phosphorylation sites have been identified on Artemis using an in vitro kinase assay (21). Among these 14 sites, certain fractions or combinations of residues may have more critical regulatory roles than the others.
The activity of Artemis:DNA-PKcs was not affected by Ku in vitro. A previous study by Goodarzi et al. suggested that Ku dependence is only detectable at higher salt conditions (75 mM KCl) (25). However, in our in vitro assay, addition of Ku did not affect the kinase activity of the DNA-PKcs or the endonuclease activity of Artemis:DNA-PKcs at 75 mM relative to lower KCl concentrations. Ku increases the affinity of DNA-PKcs to DNA ends, thereby helping to activate the kinase (31). Possibly, in vivo, Ku is essential because the immediate repair of the DNA breaks is critical for cell survival, whereas in vitro, a high concentration of DNA ends in the test tube and long reaction times permit adequate binding of DNA-PKcs to the DNA ends.
Hairpin opening at the coding ends of the functional human VH segments
The coding ends were opened in a generally similar pattern despite substantial sequence differences. This further confirms that Artemis:DNA-PKcs is a structure-specific rather than a sequence-specific endonuclease and cleaves all single-/double-strand transitions. The hairpin opening positions all fall within 1–4 nt to the 3′ side of the hairpin tip, with the 2 nt position (+2) being the primary hairpin opening site. The resulting coding ends contain 3′ overhangs of inverted repeats, 2–8 nt in length, among which, 4 nt overhangs are the dominant species. Such inverted repeats, termed P (palindromic) nucleotides, were identified in V(D)J recombination junctions in all vertebrates. Statistical analysis on the human IgM sequences revealed potential P nucleotides of 1–4 nt at the V–D–J junctions of the IgH gene (32), which coincides with our observation of hairpin opening at the +1 to +4 positions.
There are examples where the P nucleotides are a key determinant of antibody or T-cell receptor (TCR) specificity. First, T15 antibodies are optimally protective against Streptococcus pneumoniae and other phosphocholine-bearing pathogens. The DJ junction in the T15 antibodies have a predominant, identical sequence, which is driven by alignment of microhomologies that include P nucleotides (33). Second, the nonclassical murine MHC class I molecules T10 and T22 have been found to be the natural ligands for a sizable population (0.2–2%) of murine gamma-delta T cells. The specific recognition of T22 by the gamma-delta TCRs relies on the use of a 2 nt P-nucleotide during D–J recombination, which encodes a leucine at a key position in this particular TCR (34). Third, neonatal murine Ig VDJ segments lack N regions at their junctions. Their junctions are often directed by terminal microhomology, and P nucleotides are an important part of this terminal homology (35).
Most coding joints in V(D)J recombination do not retain P nucleotides at the coding ends, but rather show nucleolytic resection. Artemis has a 5′→3′ exonuclease activity but no 3′→5′ exonuclease activity (7). Hence, it would not further process the 3′ overhang unless this end were to breath. Breathing of the ends may expose the 5′ end to the exonuclease activity of Artemis. The resulting longer 3′ overhang may then be cleaved by Artemis. The varying extent of nucleotide loss during coding joint formation observed at different loci may be a reflection of the level of microhomology inherent to the sequences.
The efficiency of hairpin opening at the 17 different coding end hairpins varies over a 7-fold range. We examined the differences in coding end sequence carefully and could not identify a preferred cutting sequence. Previous studies revealed marked differences between the nicking efficiencies by the RAG complex at the various human VH elements (13). The hairpin opening efficiency by Artemis:DNA-PKcs complex displays no correlation with the VH element utilization preferences in vivo. Given that the other NHEJ proteins (Ku, pol mu and lambda, and XLF:DNA ligase IV:XRCC4) are structure-specific enzymes or binding proteins, our results further support the dominating role of RAG nicking efficiencies for VH element utilization.
ACKNOWLEDGEMENTS
This work was supported by NIH grants to M.R.L. Funding to pay the Open Access publication charges for this article was provided by M.R.L.
Conflict of interest statement. None declared.
REFERENCES
- 1.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol. Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 2.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Ann. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 3.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 4.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 5.Lieber MR. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- 6.McBlane JF, Gent DC, v, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis:DNA-PKcs complex in V(D)J recombination and in nonhomologous end joining. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 8.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell. Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 10.Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- 11.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1067. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 12.Yu K, Lieber MR. The mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol. Cell. Biol. 1999;19:8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu K, Taghva A, Lieber MR. The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex. J. Biol. Chem. 2002;277:5040–5046. doi: 10.1074/jbc.M109772200. [DOI] [PubMed] [Google Scholar]
- 14.Gerstein RM, Lieber MR. Extent to which homology can constrain coding exon junctional diversity in V(D)J recombination. Nature. 1993;363:625–627. doi: 10.1038/363625a0. [DOI] [PubMed] [Google Scholar]
- 15.Feeney AJ, Clarke SH, Mosier DE. Specific H chain junctional diversity may be required for non-T15 antibodies to bind phosphorylcholine. J. Immunol. 1988;141:1267–1272. [PubMed] [Google Scholar]
- 16.Claflin JL, Berry J. Genetics of the phosphocholine-specific antibody response to Streptococcus pneumoniae. Germ-line but not mutated T15 antibodies are dominantly selected. J. Immunol. 1988;141:4012–4019. [PubMed] [Google Scholar]
- 17.Roth DB, Menetski JP, Nakajima P, Bosma MJ, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in SCID mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh C-L, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease can cleave gaps, flaps, and loops. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-PKcs phosphorylation sites of human artemis. J. Biol. Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 22.Niewolik D, Pannicke U, Lu H, Ma Y, Wang LC, Kulesza P, Zandi E, Lieber MR, Schwarz K. DNA-PKcs dependence of artemis endonucleolytic activity: differences between hairpins and 5′ or 3′ overhangs. J. Biol. Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- 23.Pannicke U, Ma Y, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease Artemis. EMBO J. 2004;23:1987–1997. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlissel MS. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol. Cell. Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 28.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson KJ, Gaeta B, Sewell W, Collins AM. Exonuclease activity and P nucleotide addition in the generation of the expressed immunoglobulin repertoire. BMC Immunol. 2004;5:19. doi: 10.1186/1471-2172-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feeney AJ. Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. J. Immunol. 1991;147:4343–4350. [PubMed] [Google Scholar]
- 34.Chien YH, Konigshofer Y. Antigen recognition by gammadelta T cells. Immunol. Rev. 2007;215:46–58. doi: 10.1111/j.1600-065X.2006.00470.x. [DOI] [PubMed] [Google Scholar]
- 35.Feeney AJ. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J. Immunol. 1992;149:222–229. [PubMed] [Google Scholar]