Abstract
The path Plasmodium takes across the Anopheles midgut constitutes the major bottleneck during the malaria transmission cycle. In the present study, using a combination of shot-gun cloning and bioinformatic analysis, we have identified 18 miRNAs from Anopheles gambiae including three miRNAs unique to mosquito. Twelve of them are expressed ubiquitously across the body, independently of gender, while the other six exhibited an expression pattern restricted to the digestive system. Strikingly, the expression patterns of four miRNAs, including the three unique to mosquito, are affected by the presence of Plasmodium. We also show that knocking down Dicer1 and Ago1 mRNAs led to an increased sensitivity to Plasmodium infection. Altogether, these data support an involvement of miRNAs as new layers in the regulation of Anopheles defence reaction.
INTRODUCTION
Malaria, caused by the protozoan parasite Plasmodium falciparum, is a major threat to human health, responsible annually for up to 3 million deaths worldwide. This pathogenic parasite has a complex life cycle and is transmitted to humans by the vector insect Anopheles. Interactions between the parasite and the insect vector have been the objective of extensive investigations aimed to identify novel and efficient ways to disrupt or reduce pathogen transmission. Plasmodium invasion of the Anopheles midgut represents a critical step in the parasite-transmission cycle. At this stage, the number of parasites drops drastically, reaching a minimum (1). A key concept that has emerged from recent studies is the existence of a fine balance between mosquito factors that negatively and positively affect the development of the parasite (2–4). Genomic-scale microarrays have been employed to assess transcriptional profiles associated with Anopheles gambiae midgut invasion by Plasmodium berghei, which is the rodent malaria parasite used as a laboratory model (2). Strikingly, defence responses include components not only of systemic humoral immunity (including fat body and mosquito blood cells), but also of intracellular local epithelial reactions from midgut epithelial cells (2,5). The results suggested that actin and microtubule cytoskeleton remodelling is a major response of the epithelium to ookinete penetration (2). Other responses encompass components of innate immunity, cell adhesion, extracellular-matrix remodelling, redox metabolism, detoxification and apoptosis (2).
Recently, an abundant class of tiny non-coding RNAs molecules, sized from 21 to 25 nt, designated as microRNAs (miRNAs), has been shown to play a central role in the regulation of gene expression (6). In metazoa, miRNAs target the 3′ untranslated regions (3′-UTR) of mRNAs, thereby repressing their translation [for review: (6), for mechanism: (7)]. The number of predicted miRNA targets is into the thousands, whereby forming another layer of the regulatory circuitry that exists within cells (8). Thus, miRNAs are the likely candidates for serving as regulators of defence responses to parasites, either by being part of the sensing network detecting the presence of the parasite or by modulating expression levels of defence genes.
In insects, research on miRNAs has been mainly limited to Drosophila melanogaster. In D. melanogaster, experimental methods, both in vitro and in vivo, have confirmed the essential and crucial roles of miRNAs for cellular functions. Indeed, miRNAs have been implicated in numerous biological processes ranging from cell proliferation and apoptosis during development (9,10), cell–cell interactions during development of the peripheral nervous system (10,11), to stress resistance and fat metabolism (12), from cellularization and segmentation on embryos (13) to cardiogenesis (14) and muscle growth (15). Although the roles of miRNAs in non-drosophiloid insects have not yet been established, 38 miRNAs genes in A. gambiae have been previously identified by computational methods [MiRBase, (16)].
MiRNAs, like messenger RNAs, undergo RNA processing events prior to exerting their functional roles within cells. In D. melanogaster, the RNAse III-like nuclear protein drosha processes miRNA precursors from primary transcripts that are synthesized by RNA polymerase II. In the cytoplasm, miRNA maturation from the ∼70 nt long stem-loop precursor is mediated by the RNase III-like protein Dicer1 (17,18). Dicer1 processing of the pre-miRNA results in the production of a ∼22-nt long miRNA/miRNA* duplex intermediate. From this duplex, one strand, the mature miRNA, preferentially enters the protein complex that represses target gene expression, the RNA-induced silencing complex (RISC), whereas the other strand, the miRNA*, is degraded (17,18). In D. melanogaster, the protein Argonaute1 had been shown to be required for miRNA-induced silencing. The miRNA guides the RISC complex to the 3′-UTR of target mRNAs. Animal miRNAs usually base pair with imperfect complementarity to their target. The seed region (nucleotides 2–8) of miRNAs is most important for target recognition and silencing (10). Association of miRNAs with their target mRNAs inhibits translation, but the exact mechanism of translational repression remains unclear.
We used a direct cloning procedure to isolate A. gambiae miRNAs expressed in the midgut of P. berghei infected mosquito. A bioinformatical approach additionally enlarged our initial experimentally identified set of A. gambiae miRNAs, which was followed by expression profiling of miRNAs across the A. gambiae body. As a result, four miRNAs potentially involved in the post-transcriptional regulation of genes implicated in the response of Anopheles midgut epithelium to Plasmodium invasion were selected.
In addition, we investigated whether the miRNA silencing pathways contributes to Anopheles defence reactions against Plasmodium infection. We tested the effects of RNAi-dependent silencing of genes involved in miRNA processing. Indeed, silencing of Dicer1 and Argonaute1 genes pointed to the involvement of miRNAs in parasite survival.
MATERIALS AND METHODS
Bioinformatics
Database searches used online submission of BLAST jobs and were performed accessing the ENSEMBL server (Ensembl v45-Jun 2007) (http://www.ensembl.org), the NCBI (6 December 2005: BLAST 2.2.13 released) (http://www.ncbi.nlm.nih.gov/BLAST/), the Broad Institute (http://www.broad.mit.edu/cgi-bin/annotation/disease_vector/aedes_aegypti/blast_page.cgi), the plasmoDB (http://plasmodb.org/) and the FlyBase (http://flybase.bio.indiana.edu/blast/). Sequences 100 nt apart of the putative miRNAs were extracted from the various genomes and fold online (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) (19). Sub-regions that folded into a stem-loop with at least 20 bp were then folded independently to ensure that they maintain stem-loop structure. MiRBase (Release 9.2: May 2007; http://microrna.sanger.ac.uk/cgi-bin/sequences/browse.pl) is the new home of microRNA data on the web, providing data previously accessible from the miRNA Registry (20). All miRNA gene sequences were deposited to the MiRBase Registry prior to publication of results.
To distinguish miRNAs from mRNA degradation products, we evaluated the ability of the RNAs corresponding to the genomic sequences surrounding our cloned small RNAs to fold into potential hairpin miRNA precursors. In addition, as conservation is being used as a valid criterion for the classification of small RNAs as bona fide miRNAs (21), we searched for sequence conservation among insects such as D. melanogaster and Aedes aegypti.
We enlarged our set of Anopheles miRNAs to those encoded close to the miRNAs cloned in this study. We used a 50 kb cutoff on the basis of experiments performed by Baskerville and Bartel (22). Bioinformatic searches were based on clusters reported previously in D. melanogaster (23,24) and orthologues were looked for in the A. gambiae genome.
Biological material and infections
Anopheles gambiae strain G3 rearing and maintenance was carried out as described previously (25). Mosquitoes were fed on anaesthetized IRC mice that had been infected with the P. berghei GFP-con259cl2 (26). Infected mosquitoes were kept at 21°C until dissection. For each experiment, batches of females of the same age and from the same cage were fed in parallel on an infected mouse and on a regular mouse.
Mosquito dissections and total RNA extractions
Dissections were performed on ice in PBS (25 mM HEPES–NaOH pH7, 0.6 mM MgCl2, 4 mM KCl, 1.8 mM NaHCO3, 150 mM NaCl, 1.7 mM CaCl2) and kept on ice. Dissections of body were performed on young adults between 1 and 2 days old. Anopheles thorax and head were separated from the abdomen. Then, midgut was dissected from the abdomen. The remaining part of the abdomen, which includes the fat body, the ovaries, muscles and abdominal cuticle, was labelled as leftover.
Dissection of midguts and leftovers on Anopheles female blood-fed on naive or infected mouse were performed 24–48 h post-blood feeding. In this case, the midguts also contained the undigested blood bolus.
Total RNAs were extracted from dissected tissue using the TRIzol Reagent (Invitrogen) according to the supplier's instructions.
Small RNA cloning
Short RNAs were cloned using a protocol inspired by (27). Two hundred fifty micrograms of total RNAs from infected midguts were fractioned on a denaturing 8% polyacrylamide gel (7 M urea, 1 × TBE buffer). RNAs in the size range between 15 and 27 nt were excised from the gel, eluted and ethanol precipitated. Gel purified 15–27 nt RNAs were ligated to a 3′ adapter oligonucleotide (p UUUaaccgcgaattccagx: uppercase, RNA; lowercase, DNA; p, phosphate; x, 4-hydroxymethylbenzyl) using T4 RNA ligase (Roche) (20 µl reaction, 37°C, 30 min, 5 µM 3′ adapter, 50 mM Tris–HCl pH 7.6, 10 mM MgCl2, 0.2 mM ATP, 0.1 mg/ml acetylated BSA, 15% DMSO, 20 U T4 RNA ligase). The ligation reaction was stopped by the addition of an equal volume of stopmix (8 M urea, 50 mM EDTA) and directly loaded on a 15% denaturing gel. After electrophoresis, the ligation product was recovered from the gel. Next, the 5′ adapter oligonucleotide (acggaattcctcactAAA: uppercase, RNA; lowercase, DNA) was ligated to the phosphorylated ligation product as described above. The new ligation product was gel purified and eluted from the gel slice in the presence of reverse transcription primer (Gactagctggaattcgcggttaaa), used as carrier. Reverse transcription (15 µl reaction, 42°C, 30 min, 150 U Superscript II reverse transcriptase (Life Technologies) in the buffer provided by the manufacturer) was followed by PCR using a 5′ primer (Gagccaacggaattcctcactaaa) and the 3′ RT primer. The PCR product was purified on a native 8% polyacrylamide gel and ligated into the pCR2.1-TOPO® vector using the TOPO-TA® cloning kit (Invitrogen). Clones were submitted for custom sequencing (MWG or in house sequencing facility).
MiRNA profiling using primer extension
Experiments were performed on 2 µg of total RNA using the Omniscript RT kit (Qiagen) according to the manufacturer's instructions. Oligonucleotides complementary to the 3′ end of the miRNAs (Table S2) were end-labelled with gamma [32P]-ATP and T4 polynucleotide kinase (28). Reverse transcription products were separated on a 10% denaturing polyacrylamide gel. RNA or DNA dideoxynucleotide sequencing reactions were run in parallel to allow identification of the stop positions (28). Experiments were conducted at least twice on each sample and repeated on total RNAs extracted from two distinct mosquito populations.
Radioactivity was measured with a BAS 2000 Bio-Imager. Intensity of the reverse transcriptase stop counted for each sample was corrected by the total count of radioactivity in the sample. We run in parallel reverse transcription assays on 5S ribosomal RNA as internal control. The calculated mean of variation for the intensity of the 5S rRNA stop was ±19% and the standard variation was 7% (n = 5). The estimated confidence interval was calculated according to ± (mean + 2 standard variation). All the intensities of the reverse transcriptase stops were normalized to that measured in the female midgut, chosen as the internal standard for each tested miRNA. Ratios were accepted as significantly different from the value observed in the female gut if located outside of the interval [0.67; 1.33].
Double-stranded RNA preparation and silencing
Anopheles Dicer1 gene (ENSANGG00000014308) fragment was amplified by PCR reaction from genomic DNA using dcr1-A and dcr1-B primers (Table S1) and cloned into pGEM®-T vector (Promega) in both orientations. Anopheles Argonaute1 gene (ENSANGG00000006700) fragment and drosha gene (ENSANGG00000009133) fragment were amplified by PCR reaction from genomic DNA using the ago1-A and ago1-B primers and drosha-A and drosha-B primers, respectively (Table S1) and cloned into pCR®2.1-TOPO® vector (Invitrogen) in both orientations. Both sense and antisense RNAs were synthesized and purified as described in (28). Annealing was performed by mixing equimolar amounts of both strands in water, denaturing them at 95°C for 5 min and slow cooling for 30 min. A nano-injector (Nanoject, Drummond) was used to introduce 69 nl of double-stranded RNAs (dsRNAs) (3 mg/ml) in the thorax of CO2-anaesthetized mosquito females. The lacZ dsRNA was produced from the pLLlacZ plasmid, a pLL100 derivative containing two T7 promoters, a kind gift from Levashina (29).
Effect of silencing on mRNA level was analysed 4 days after dsRNA injection by quantitative real-time PCR. Total RNA from at least 10 females was extracted with TRIzol reagent (Invitrogen) and 2 µg of total RNA were reverse transcribed using random primers and the SuperscriptIII Platinum enzyme (Invitrogen). Quantitative PCR reactions were assembled using specific primers (Table S1) and the Syber-green Q-PCR mix (Invitrogen) and run on the Mx4000 (Stratagene) following the manufacturer's instructions. The gene (ENSANGG00000014460) encoding the ribosomal S7 protein was used as an internal control (2).
Phenotypic analysis of dsRNA-injected mosquitoes
Mosquitoes were blood-fed on a mouse infected with Plasmodium. Mosquito midguts were dissected 7–10 days after infection and fixed in 4% formaldehyde. Fluorescence microscopy was done with a confocal microscope 100 M with software package LSM 510 version 3.2 (Zeiss, Jena, Germany) in multitrack mode. Excitation/emission wavelengths were 488/505 to 545 nm for GFP. Transmitted light reference images were captured using differential interference contrast optics and argon laser illumination at 488 nm. Image processing occurred in ImageJ (1.37v, Wayne Rasband, National Institutes of Health, USA, http://rsb.info.nih.gov/ij/) and Photoshop 6.0 (final image assembly; Adobe Systems, San Jose, CA, USA). Statistical analysis of differences between control and silenced groups was evaluated by Mann–Whitney test using graphPad Prism version 4.0 for MacOSX (GraphPad Software).
RESULTS
Construction of the small RNA library and bioinformatic analysis of sequences
In order to identify A. gambiae miRNAs that could potentially be involved in a midgut defence mechanism against invasion by Plasmodium, we generated a specialized cDNA library encoding small non-coding RNAs from total RNA of Anopheles midgut samples 24–48 h after being infected with P. berghei (see Materials and Methods section).
From the cDNA library, ∼300 cDNA clones were sequenced. Our first screen was based on sequence quality (less than three undetermined nucleotides) and length (between 20 and 25 nt) criteria. Subsequently, 152 clones (corresponding to 110 different sequences) were analysed. Most sequences (90%) were observed only once, indicating that the dataset probably does not represent the complete pool of short RNAs present in infected midgut epithelium.
Our functional annotation was based on queries from all available sequences of NCBI Genebank including prokaryotic genomes as well as sequences from insects and mammals (see Materials and Methods section). Annotation was refined using already assembled genomes from Mus musculus and A. gambiae and P. berghei (see Materials and Methods section). Our choice to examine diverse databases was directed by the complexity of the studied sample. We found that 61% of the clones could clearly be assigned to the A. gambiae genome, 14% to Mus musculus and 14% to P. berghei (Table 1). In addition, 5% of the clones (7 sequences) were assigned to different bacteria. Six percent of the clones (8 sequences) exhibited no hit to any of the database and do not map to any of the genomes (Anopheles, mouse, Plasmodium or bacteria). A detailed analysis of sequences that do not belong to the A. gambiae genome is given in Supplementary Data.
Table 1.
Information on the Anopheles miRNAs cloned in this study
| Name | Sequence | Chroa | Location | Strand | Conservationb | Referencesc | |
|---|---|---|---|---|---|---|---|
| aga let-7 | TGAGGTAGTTGGTTGTATAGTAA | 3R | 10270762 | 10270783 | − | Metazoa | 1,2,3 |
| aga mir-306 | TCAGGTACTGGATGACTCTCAG | 3R | 5888646 | 5888667 | − | Arthropoda | 1 |
| aga mir- 996 | TGACTAGATTACATGCTCGTCT | 2R | 55572844 | 55572865 | − | Dm, Aae | 1 |
| aga mir-275 | TCAGGTACCTGAAGTAGCGCGC | 3R | 38377402 | 38377423 | + | Arthropoda | 1,2,3 |
| aga mir- 989 | TGTGATGTGACGTAGTGGTAC | 3L | 2905485 | 2905505 | − | Culicidae | 1 |
| aga mir-12 | TGAGTATTACATCAGGTACTGGT | 2R | 37888125 | 37888147 | + | Arthropoda | 1 |
| aga mir-281 | CTGTCATGGAATTGCTCTCTT | 2L | 17362447 | 17362467 | + | Arthropoda | 1,2,3 |
| aga mir-34 | TGGCAGTGTGGTTAGCTGGT | 2R | 28232721 | 28232740 | − | Metazoa | 1,2 |
| aga mir- 1175 | TGAGATTCTACTTCTCCGACTTAA | 2R | 9129834 | 9129857 | − | Culicidae 1 | 1 |
| aga mir-1174 | TCAGATCTACTTCATACCCATG | 2R | 9129644 | 9129665 | − | Culicidae 1 | 1 |
aChro stands for chromosome.
bDm stands for Drosophila melanogaster, Aae for Aedes aegypti
c1, this work; 2, Wang et al., 2005; 3, MiRBase.
Anopheles small non-coding RNAs
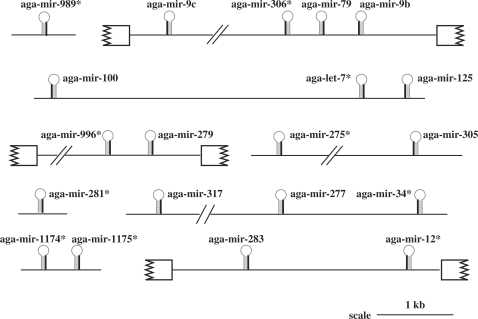
Ten cloned sequences met all the criteria to be bona fide miRNAs (Table 1). We identified eigth additional A. gambiae miRNAs employing the synteny of miRNA gene clustering (Figure 1 and Supplementary Data, Table S2).
Figure 1.
Cluster organization. The symbol (*, asterisk) indicates the miRNA cloned in this study. Exons are symbolized using broken boxes. Position of the mature miRNA within its stem-loop precursor is indicated using a darker line. Additional information is gathered in Supplementary Data (Table S2).
Four miRNAs were unrelated to any previously described miRNAs in the MiRBase namely aga-miR-996, aga-miR-989, aga-miR-1174 and aga-miR-1175. Orthologues of mature aga-miR-996 and its hairpin precursor were identified in the genome of D. melanogaster and A. aegypti. Dme-miR-996 was located 1.58 kb away from the already described dme-miR-279 gene. This cluster [miR-279, miR-996] was conserved in A. gambiae (distance: 0.47 kb), as well as in A. aegypti (distance: 4.7 kb). Strikingly, the seed sequences of miR-279 and of miR-996 are identical, suggesting that miR-996 and miR-279 might have arisen from recent gene duplication. A putative orthologue of aga-miR-989 (including conservation of its precursor) is found in A. aegypti whereas it is absent in D. melanogaster. Orthologues of aga-miR-1174 and aga-miR-1175 (and their precursors) were not found in D. melanogaster but were conserved in A. aegypti. In addition, the genes of these two mosquito-specific miRNAs were encoded 168 nt apart in A. gambiae and genomic clustering was conserved in A. aegypti (139 nt apart).
Aga-miR-275, aga-miR-281 and aga-let-7 were already described in the MiRBase. Aga-miR-12, aga-miR-34 and aga-miR-306 are new Anopheles miRNAs and are the orthologues of previously described D. melanogaster miRNAs (MiRBase). Aga-let-7 displayed one mutation compared with its Drosophila counterpart (MiRBase) whereas aga-miR-306 exhibited three sequence changes compared with its Drosophila orthologue (Table 1). Importantly, in each case, sequence variations were located outside of the seed region of the miRNA.
Expression profile: focus on the parasite effect
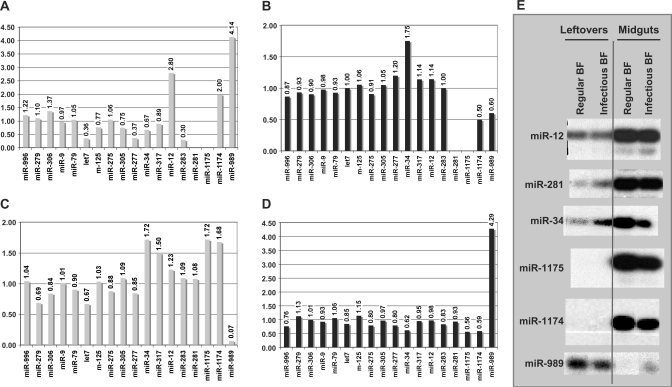
The miRNA expression profiles were determined during the course of Plasmodium invasion of the midgut, using reverse transcription assays (Figure 2). We have chosen to consider Plasmodium invasion to have an effect if changes in the level of the reverse transcription stops between two samples differed by more than 33% (see Materials and Methods section). Three types of expression profiles were encountered.
Figure 2.
Incidence of regular and infectious blood feeding on miRNA expression profiles. The intensity of the reverse transcriptase stops was quantified (see Materials and Methods section). (A) Effect of blood feeding on miRNA expression in leftovers samples. The female leftovers signal was set as the internal reference. (B) Effect of P. berghei presence on miRNA expression in leftovers samples. The blood-fed female leftovers signal was set as the internal reference. (C) Effect of blood feeding on miRNA expression in midguts samples. The female midguts signal was set as the internal reference. (D) Effect of P. berghei presence on miRNA expression in midguts samples. The blood-fed female midguts signal was set as the internal reference. (E) Representative examples of autoradiographies of the primer-extension assays. Total RNAs were extracted from leftovers or from midguts, 24–48 h after blood feeding on a regular mouse or after blood feeding (BF) on P. berghei-infected mouse.
Aga-miR-34, aga-miR-1174 and aga-miR-1175 exhibited a decrease in the expression level in infected compared with blood-fed midgut samples (Figure 2). The ratios (expression with/expression without Plasmodium) are 0.62 for aga-miR-34, 0.56 for aga-miR-1175 and 0.59 for aga-miR-1174. Strikingly, only aga-miR-989 displayed an enhanced expression level in the presence of the parasite: it is four times stronger in midgut samples infected with Plasmodium than in uninfected midgut samples (Figure 2). In addition, aga-miR-989 exhibits a strong decrease of expression (ratio 0.6) in infected leftover samples compared to uninfected ones.
In contrast, the other tested miRNAs (14 over 18) did not exhibit any changes in their expression signal: they remained constant in RNA samples extracted from both infected and uninfected mosquitoes (Figure 2).
Expression profile: focus on the blood effect
We determined the effect of blood feeding on the leftovers and the midguts miRNA contents (Figure 2). Reverse transcription assays were performed and the ratio of the intensity between the reverse transcriptase stop for blood-fed females over sugar-fed females was calculated (Figure 2).
Changes in leftovers were observed for aga-miR-277 (ratio 0.36) and aga-miR-12 (ratio 2.80). In midguts, changes in expression levels were observed for aga-miR-317 (ratio 1.50), aga-miR-1175 (ratio 1.72) and aga-miR-1174 (ratio 1.68). Expression of aga-let7 decreased in leftovers (ratio 0.36) as well as in midguts (ratio 0.67). Concomitant changes were observed for aga-miR-34 (ratio of 0.67 and of 1.72 in leftovers and midguts, respectively) and for aga-miR-989 (ratio of 4.14 and of 0.07 in leftovers and midguts, respectively).
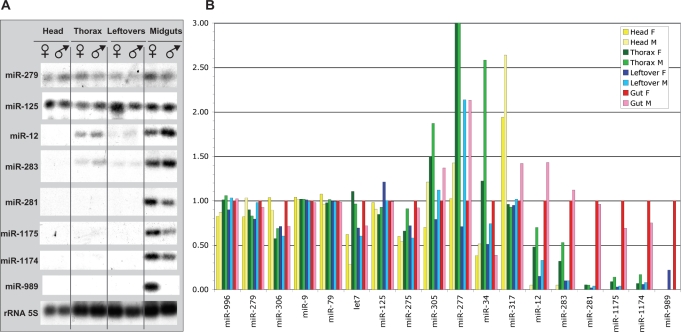
General expression patterns along the mosquito body
The expression profiles of the miRNAs were determined using reverse transcription assays (Figure 3). Experiments were run on total RNAs extracted from different body parts of male or female Anopheles: head, thorax, gut and leftover. Based on the observations, we could define three classes: (i) miRNAs or clusters of miRNAs expressed only in one body part; (ii) clusters for which the miRNAs have distinct patterns with preferential expression in one body part and (iii) clusters for which all the miRNAs encoded are expressed evenly and ubiquitously throughout the body.
Figure 3.
Expression profiling of Anopheles miRNAs across gender and body parts. (A) Representative examples of the autoradiographies of primer-extension assays performed on total RNA samples from Anopheles. (B) Profiling of the miRNA expression in Anopheles body parts using primer-extension assays. The intensities of the reverse transcriptase stops were quantified (see Materials and Methods section). The stop intensities obtained using total RNAs extracted from the female gut was set as the internal reference. F is used for female and M for male. As stated in Materials and Methods section, ratios are required to be outside of the estimated confidence interval [0.67; 1.33] to be deemed as significant.
Strikingly, four of the tested miRNAs are expressed only in midguts. It included the three mosquito-specific miRNAs, aga-miR-1175, aga-miR-1174 and aga-miR-989 as well as aga-miR-281 (Figure 3). In addition, aga-miR-989 expression is even more restricted as it can only be observed in females.
MiRNAs encoded in the cluster [aga-miR-34, aga-miR-277 and aga-miR-317] exhibited complex expression patterns (Figure 3). Noteworthy, aga-miR-34 expression was more pronounced in thorax of males (ratio 2.58) and females (ratio 1.22) and in midguts of females (ratio 1) compared to the other body parts (Figure 3). Aga-miR-277 was expressed predominantly in the thorax and a clear sexual dimorphism is observed. Aga-miR-317 is equally expressed in males and females, at a constant level in thorax, leftovers and midguts, and twice as much in heads. MiRNAs from the cluster [aga-miR-12; aga-miR-283] are predominantly expressed in midguts and thorax (Figure 3).
Clusters [aga-miR-996, aga-miR-279], [aga-miR-306; aga-miR-79; aga-miR-9b], [aga-miR-275; aga-miR-305] and [aga-let-7; aga-miR-125] are expressed ubiquitously. With few exceptions, their levels were identical in the different body parts as well as between males and females (Figure 3).
Silencing of the Dicer1 and Ago1 mRNAs
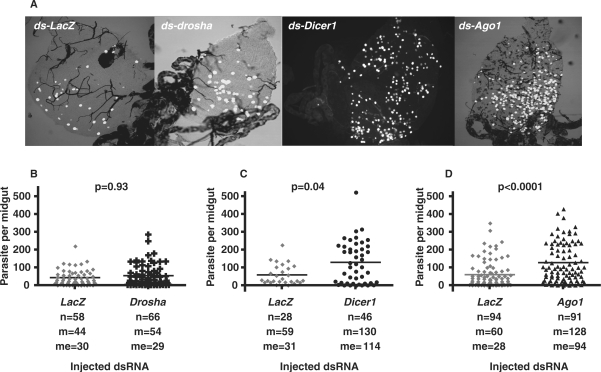
No evidence has been published yet establishing that mosquito genes of the silencing pathway would affect parasite development in the midgut. We conducted a functional screen using the recently established gene silencing technique for adult A. gambiae (30). To achieve gene silencing, drosha, Argonaute1, Dicer1 or lacZ double-stranded RNAs were injected in the body cavity of newly emerged female mosquitoes of the G3 strain that are susceptible to the model rodent parasite P. berghei. Injection of lacZ dsRNA was used as a control to ensure that the observed effects did not simply reflect the dsRNA treatment. The reduction in drosha, Argonaute1 or Dicer1 transcript levels was determined 4 days later by real-time PCR. After injection of drosha dsRNA, the transcript level was unchanged. Transcript levels were reduced to 63% (±9%) after injection of Dicer1 dsRNA and to 50% (±12%) after injection of Argonaute1 dsRNA compared to transcript levels in the lacZ dsRNA control. At the same time point, 4 days after injection, the mosquitoes were allowed to feed on a mouse infected by a carrying-GFP parasite (26). We counted the number of live GFP-expressing oocysts on the dissected midguts 10 days post-infection. Correlating with the inefficiency of the silencing of drosha mRNA, no effect was observed on the parasite count after injection of drosha dsRNA. Depletion of Dicer1 and Argonaute1 led to a substantial increase in oocysts numbers: a significant 2-fold increase in the parasite number was observed (Figure 4).
Figure 4.
Survival of P. berghei in drosha, Dicer1 and Argonaute1 depleted mosquitoes. Females were infected 4 days after injection, and midguts were dissected 10 days later and fixed. (A) Representative composite image between the green fluorescent channel and a differential interference contrast (DIC) image of dissected midguts showing parasite development in the dsRNA-injected mosquitoes. (B–D) Survival of P. berghei parasites in control ds-lacZ and in ds-drosha-, ds-Dicer1- or ds-Ago1-injected mosquitoes. Data were collected from three independent experiments. The mean of parasite number observed in each experiment was similar so that the data were pooled. Statistically significant differences between samples were evaluated by Mann–Whitney test and P-values are indicated on the graph. Black bars represent mean parasite numbers per group. Here, n represents the number of guts, m the average number of parasites per gut and me the median number of parasites per gut.
Thus, both Argonaute1 and Dicer1 play an important role in the mosquito resistance to P. berghei. The simplest model of action for Argonaute1 and Dicer1 would be through miRNA-mediated post-transcriptional regulation of the genes involved in defence reactions.
DISCUSSION
In this study, we used a direct cloning procedure to isolate A. gambiae miRNAs expressed in the P. berghei infected midguts. We included in our assays miRNAs genes identified bioinformatically based on their vicinity to miRNAs identified experimentally in our screen. The expression profiles of these A. gambiae miRNAs within the different body segments were determined for both males and females. Assays were also carried out to determine the effect of Plasmodium presence within the blood meal on the miRNAs expression level.
It is assumed that miRNA genes located in a gene cluster are first transcribed as a single primary transcript, which is subsequently processed to generate the individual miRNAs. Expression profiling of those miRNAs and of ‘encoded near-by’ miRNAs allowed us to gather information on a total of 18 miRNAs (7 clusters). Indeed, miRNAs from two out of the seven tested clusters, aga-miR-279/aga-miR-996, aga-miR-306/aga-miR-9b/aga-miR-79, have similar expression patterns. In agreement with the accumulating evidence, this observation suggests that they are transcribed as polycistrons (22,31). In most of the other clusters, including aga-let-7/aga-miR-125, aga-miR12/aga-miR-283, limited differences in the expression level along body parts (or dependent on sex) were observed. Two models could explain these observations. First, the generalized notion that clustered miRNA genes are co-expressed may not always be true. Alternatively, transcription of the cluster as a polycistron would be used, followed by a differentially regulated maturation. Differentially regulated maturation may take place either during pri-miRNA processing, the export to the cytoplasm or the maturation of the pre-miRNAs into the mature miRNA.
Recently, a global analysis of miRNA target gene expression was reported and showed that miRNAs more frequently target ubiquitously expressed genes than tissue-restrictively expressed genes (32). On the other hand, some miRNAs play important roles in driving tissue terminal differentiation and maintaining tissue identity (32). Indeed, we found that most of the tested miRNAs are expressed throughout the mosquito body at an almost steady level. However, in a few cases, expression was tissue-restricted. As an example, the cluster [miR-12; miR-283] is expressed predominantly in the gut and the thorax. In D. melanogaster embryos, these same miRNAs are expressed in the foregut, posterior midgut, hindgut and salivary gland (33). These observations support the model predicting that [miR-12; miR-283] play a role in driving digestive tissue differentiation and maintenance of tissue identity in the adult insect.
The gonotrophic cycle (including host seeking, blood feeding, egg development and oviposition) involves a complex series of biological events, including peritrophic matrix formation, blood digestion, oocyte development, vitellogenesis and excretion. Almost half of the expressed genes showed significant variation in transcript accumulation during the first, second and third day after a blood meal (34). Multiple hormones interact to alter tissue states and to activate genes involved in these processes. Two hormones, the juvenile hormone and the 20-hydroxyecdysone, are most fundamental to ovarian development and competence of the fat body (35,36). Interestingly, in D. melanogaster and in S2 cells, Ecd and the juvenile hormone analogue methoprene exert effects on the expression of these miRNAs, indicating the participation of both of them in the temporal regulation of dme-let-7 expression in vivo (37). Alike, aga-let-7 expression is sensitive to blood feeding, indicating that some aspect of the regulation of aga-let-7 expression in leftover and midgut might be mediated by hormonal signals.
Only four miRNAs, aga-miR-34, aga-miR-1175, aga-miR-1174 and aga-miR-989, displayed changes in the expression levels during Plasmodium infection. Comparing directly naive and Plasmodium infected blood feeding is the major limit of our approach. As a matter of fact, the ingested malaria-infected blood differs from non-infected blood in a number of ways, including the presence of blood stage and gametocyte stage of Plasmodium and their metabolites, and of vertebrate infection-responsive molecules, as well as a higher free-radical concentration (38,39). In addition, we observed changes in gene expression in the leftover tissues, at a time point when the ookinetes are in the midgut epithelium. While the midgut is the primary site of response to the invading ookinete, these changes most likely reflect inter-tissues signalling from the midgut epithelium to haemocytes and fat body cells. Alternatively, parasite-derived molecules that diffuse into the haemolymph may affect A. gambiae biological process in the leftover tissues (40).
However, our second approach provides a more general answer about the miRNA pathway involvement in vectorial competence of the Anopheles. We used gene silencing of drosha, Dicer1 and Argonaute1 mRNAs. We took advantage of the fact that in D. melanogaster as well as in dipterans, a dichotomy characterizes the miRNA and siRNA pathways: whereas, miRNA processing relies on drosha, Dicer1 and Argonaute1 that of siRNA involves instead Dicer2 and Argonaute2. Clear orthologues of these enzymes are found in the genome of A. gambiae (data not shown). Indeed, when Dicer1 or Argonaute1 mRNAs level is decreased, we observed a better survival of the parasite. The simplest explanation would be that as miRNA maturation is impaired, translational regulation of a variety of target genes is altered. In turn, this would led to the modification of the fine tuning of mosquito factors that negatively and positively affect the development of the parasite. Yet, we have yet to establish whether the four miRNAs, aga-miR-34, aga-miR-1175, aga-miR-1174 and aga-miR-989, which exhibit an expression profile dependent on the Plasmodium, are players in vector competence of A. gambiae. In particular, in vivo specific strategies need to be designed to test the phenotypes of gain and loss of function of the four miRNAs regarding parasite development.
Numerous studies were aimed at predicting D. melanogaster miRNA targets. In particular, the study performed by (8) established a list of experimentally validated targets of Drosophila miRNAs and the study performed by Ref. (10) included predictions of the miRNA-target couples using the Anopheles database as a validation criterion for Drosophila couples. As a preliminary analysis, we combined both studies to define a set of experimentally validated targets in Drosophila with putative target sites in Anopheles. We next checked which of these Anopheles genes were tested in the functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion using microarray assays performed by Vlachou and collaborators (2). Only, three genes, ENSANGG00000018286 (CathepsinB), ENSANGG00000021087 (betaTub60D) and ENSANGG00000009998 (dgl1) meet all these criteria. Based on gene ontology of biological processes, Cathepsin would be involved in proteolysis and cell adhesion, betaTub60D in microtubule-based movements and microtubule polymerization and dgl1 in signal transduction and morphogenesis of a polarized epithelium. Strikingly, the expression level of the three mRNAs increases specifically during infection (2). The up-regulation correlates well with the decreased aga-miR-34 expression level we observed.
A good correlation was found between the target mRNA and the miRNA-expression profiles, thereby providing a first glimpse of what could be the miRNA targets and their biological functions. Further investigations, including target predictions for the three mosquito-specific miRNAs, are currently being undertaken. Functional analyses of these miRNAs will be of particular importance. Indeed, exogenous modification of the expression of the miRNAs involved in the regulation of the defence reactions of the mosquitoes could become a powerful tool for influencing the vector competence.
In conclusion, this study provides the first experimental data on Anopheles miRNA expression. Several of the miRNAs studied (let-7, miR-125 and miR-34) are conserved across all species, from mammals, birds, fishes to worms and insects. Twelve others, including miR-F are specific to insects and are found in D. melanogaster, Apis mellifera, Bombix mori, A. gambiae and A. aegypti. However, the description of three new mosquito-specific miRNAs supports the emerging concept that some miRNAs might have a specialized function in a particular organism (41). Indeed, divergence between Aedes and Anopheles arose only 95 million years ago, whereas the Culicidae and Drosophilidae split happened at least 250 million years ago. In addition, mosquitoes are blood-sucking arthropods, a feature that has made them suitable vectors for several human diseases but has involved specific biological adaptations to blood digestion and gonotrophic cycles of reproduction (35).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank C. Isel, T. Geissmann, P. Romby, A. Krol and E. Westhof for helpful discussions and comments on the manuscript, and E. Levashina and C. Frolet for sharing mosquito facilities and for valuable advices on mosquito rearing. This work is supported by subsidies from CNRS and from Strasbourg Louis Pasteur University. S.E. was supported by a CNRS Ph.D. fellowship. Confocal imaging was performed using the RIO imaging facility, Plateforme Inter Unités de Microscopie et d’Imagerie Cellulaire Strasbourg Esplanade. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sinden RE. Molecular interactions between Plasmodium and its insect vectors. Cell. Microbiol. 2002;4:713–724. doi: 10.1046/j.1462-5822.2002.00229.x. [DOI] [PubMed] [Google Scholar]
- 2.Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr. Biol. 2005;15:1185–1195. doi: 10.1016/j.cub.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Meister S, Kanzok SM, Zheng X, Luna C, Li T.-R, Hoa NT, Clayton JR, White KP, Kafatos FC, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito. Anopheles gambiae. Proc. Natl Acad. Sci. USA. 2005;102:11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osta M, Christophides G, Kafatos F. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Dong Y, Abraham EG, Kocan A, Srinivasan P, Ghosh AK, Sinden RE, Ribeiro JMC, Jacobs-Lorena M, et al. Transcriptome analysis of Anopheles stephensi–Plasmodium berghei interactions. Mol. Biochem. Parasitol. 2005;142:76–87. doi: 10.1016/j.molbiopara.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP, Chen C.-Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microARNs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 9.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 10.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 12.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, et al. Antisense-mediated depletion reveals essential and specific functions of MicroRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl Acad. Sci. USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- 17.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 18.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 19.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [doi 10.1261/rna.2183803] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA Profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 24.Lai E, Tomancak P, Williams R, Rubin G. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-[kappa]B-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25:677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Franke-Fayard B, Trueman H, Ramesar J, Mendoza J, van der Keur M, van der Linden R, Sinden RE, Waters AP, Janse CJ. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol. Biochem. Parasitol. 2004;137:23–33. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunel C, Ehresmann C. Secondary structure of the 3' UTR of bicoid mRNA. Biochimie. 2004;86:91–104. doi: 10.1016/j.biochi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 30.Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Z, Jian Z, Shen S.-H, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl Acad. Sci. USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinotti O, Calvo E, Nguyen QK, Dissanayake S, Riberio JMC, James AA. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol. Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- 35.Dana A, Hong Y, Kern M, Hillenmeyer M, Harker B, Lobo N, Hogan J, Romans P, Collins F. Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genomics. 2005;6:5. doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raikhel AS, Kokoza VA, Zhu J, Martin D, Wang S.-F, Li C, Sun G, Ahmed A, Dittmer N, et al. Molecular biology of mosquito vitellogenesis: from basic studies to genetic engineering of antipathogen immunity. Insect Biochem. Mol. Biol. 2002;32:1275–1286. doi: 10.1016/s0965-1748(02)00090-5. [DOI] [PubMed] [Google Scholar]
- 37.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity. Dev. Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 38.Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, Ginsburg H. Oxidative stress in malaria parasite-infected erythrocytes: host–parasite interactions. Int. J. Parasitol. 2004;34:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Lim J, Gowda DC, Krishnegowda G, Luckhart S. Induction of nitric oxide synthase in Anopheles stephensi by Plasmodium falciparum: mechanism of signaling and the role of parasite glycosylphosphatidylinositols. Infect. Immun. 2005;73:2778–2789. doi: 10.1128/IAI.73.5.2778-2789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoSPathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.