Abstract
Recent data showed that p53 stimulates the expression of genes encoding not only pro- but also antioxidant enzymes. It was suggested that antioxidant genes could be induced under physiologic levels of stress while the prooxidant ones respond to higher level of stress. Results presented in this article illustrate an additional degree of complexity. We show that the expression of Haeme-oxygenase 1 (HO-1), a stress-inducible gene that codes for an enzyme having antioxidant properties, is stimulated in a p53-dependent manner in the thymus and spleen of irradiated mice. We prove that HO-1 is a direct p53 target gene by showing that the p53RE identified within human and mouse genes is specifically bound by p53. The threshold of irradiation dose required to induce a significant response of HO-1 in the lymphoid organs of the irradiated mice is higher than that for Waf1/p21 that encodes an universal inhibitor of cell cycle. Moreover, induction of HO-1 occurs later than that of Waf1/p21. Finally, the higher stimulation of HO-1 is reached when Waf1/p21 stimulation starts to decrease.
INTRODUCTION
The toxicological mechanism of ionizing radiation is generally accepted to be mediated primarily by generation of reactive oxygen species (ROS). Based on a number of observations, it was suggested that ROS are the common mediators of apoptosis (1,2). Several mechanisms by which ROS could induce apoptosis have been proposed. (i) ROS act on mitochondria, causing a disruption of the mitochondrial membrane potential and subsequently the release of cytochrome c; (ii) ROS up-regulate the expression of Fas and FasL and (iii) ROS modify the activity of transcription factors involved in the cell death and survival pathways [references in (2)].
Many of the signalling pathways activated by ionizing radiation converge on p53, a protein encoded by the tumour suppressor gene p53, whose major function is to protect organisms from proliferation of potentially tumourigenic cells. Post-translational modifications of p53 in response to ionizing radiation lead to the activation of its principal property that is to recognize a specific DNA motif leading to the transcriptional stimulation of the genes having such a p53 responsive element (p53RE) within their promoter or intronic sequences (3–5). Global mapping of transcription factor binding sites using chromatin-immunoprecipitation-based techniques placed the number of human genes potentially transactivated by p53 between 542 (6) and 1600 (7). Among them, more than one hundred have been functionally characterized. A number of these genes involved in DNA repair, regulation of cell cycle and apoptosis, could account for the tumour suppression activity of p53 [references in (8,9)]. More recently, p53 target genes involved in the regulation of glucose metabolism and oxidative stress have also been identified [references in (10)]
Earlier data showed that the extent of p53-induced apoptosis can be modulated by decreasing or increasing the extent of oxidative stress (11,12). In 1997, a Sequential Analysis of Gene Expression (SAGE) in the colorectal cancer line DLD-1 forced to express wt-p53 led Polyak et al. to identify, for the first time, a stimulation (>10 fold) of several genes predicted to encode proteins that could generate or respond to oxidative stress (13). At least one, PIG3, appears to be directly regulated by p53 through a p53RE element located in the promoter. The expression of PIG3 is also stimulated in response to γ-irradiation in a number of transformed cell lines expressing wt-p53 (14). PIG6 is another gene whose expression was found to be stimulated in DLD-1 cells infected by recombinant adenovirus expressing wt-p53. This gene encodes a mitochondrial proline oxydase (POX) that mediates the reversible conversion of proline to pyrroline-5-carboxylate (P5C) with the concomitant transfer of electrons to cytochrome c (13,15). Using doxorubicin to initiate p53-dependent apoptosis, Donald et al. (15) have shown that the expression of POX is up-regulated in a time- and dose-dependent manner in another p53-wt human colon cancer cell line (LoVo). The p53-dependent stimulation of POX catalyses the proline-dependent ROS generation suggesting again that ROS could act as a downstream mediator of p53-induced apoptosis.
The ferrodoxine reductase gene (FRDX) is another redox-related gene that was found to be up-regulated in HCT116 carcinoma cells in response to endogenous wt-p53 activation by 5-fluoruracil (16). A p53RE element has been recognized within the promoter sequence identifying FRDX as a direct p53 target gene (16,17). FRDX encodes a mammalian mitochondrial cytochrome P-450 NADPH reductase. It has been shown that its over-expression sensitizes HCT116 cells to apoptosis induced by ROS-producing agents such as hydrogen peroxide (H2O2) or doxorubicin (17).
More recent evidence suggests that p53 could also play a role in antioxidant metabolism by inducing the expression of proteins that function to lower ROS level. ROS levels are normally controlled by the antioxidant defence system including low and high molecular weight components. Among them, superoxide dismutase (SOD), glutathione peroxidase (GPx) and catalase play a central role (18). The GPX1 gene that encodes glutathione peroxidase-1 has been characterized as a direct p53 target gene (19). But the SOD2 gene that encodes the mitochondrial MnSOD enzyme has been found to be either up- or down-regulated by p53, depending on the context (20–22). The aldehyde deshydrogenase 4 (ALDH4), another gene encoding an antioxidant enzyme has also been identified as a p53 target gene (23). It encodes a mitochondrial-matrix NAD+-dependent enzyme. Inactivation of ALDH4 enhances p53-dependent apoptosis, and its forced expression reduces intracellular ROS levels (23).
In the same vein, expression of two mammalian sestrin homologs, SESN1 and SESN2 that encode PA26 and Hi95, is stimulated by p53 (24). These products have been reported to be essential for regeneration of overoxidized peroxiredoxins, the enzymes involved in the decomposition of H2O2 (25). Finally, TIGAR (TP53-induced glycolysis and apoptosis regulator), a novel p53 target gene, selectively inhibits ROS-sensitive apoptosis (26).
The fact that p53 can stimulate the expression of pro- and antioxidant genes strongly suggests that p53 could have opposing roles in the regulation of ROS depending on the nature and the intensity of the stress, and on the cellular context. As most of the published data refer to experiments performed with tumour cell lines, we wanted to analyse the p53-dependent regulation of stress-related genes in a physiological cell environment in response to a genotoxic stress that induces apoptosis specifically in p53 positive cells. It is well known that p53 is absolutely required for irradiation-induced apoptosis in thymus and spleen, both in vitro and in vivo (27–29). We therefore used thymus and spleen obtained from whole body irradiated p53+/+ and p53−/− mice as a source of differentially expressed mRNA. We report that the stimulation Haeme-oxygenase 1 (HO-1) in response to γ-irradiation is directly under the control of the p53 gene.
MATERIALS AND METHODS
Cells and their treatment
U-2OS and MCF-7 cells are derived from a human osteosarcoma and a human breast carcinoma, respectively. SKNSH-DDp53 and SKNSH-CMVNeo cell lines, generously provided by Dr S. Lain, have been described previously (30). They are derived from SK-N-SH neuroblastoma cells transfected with plasmids pCMVNeop53DD and pCMV-Neo, respectively. pCMVNeop53DD encodes the p53DD truncated protein that includes the mouse p53 amino acid residues 1–14 and 302–390 (31). MCF-7/R-A1 cells, a gift from Dr S. Chouaib, was obtained from MCF-7 by continuous exposure to increasing doses of TNF-α (32). This cell line expresses a p53 mutated at amino acid residue 280 (R→K). Cells were maintained at 37°C in DMEM (U-2OS, MCF-7, SKNSH-DDp53, SKNSH-CMVNeo) or RPMI (MCF-7/R-A1) supplemented with 10% fetal calf serum, and 100 U/ml of penicillin and 100 mg/ml streptomycin. The SKNSH-DDp53 and SKNSH-CMVNeo cells were maintained at 0.5 mg/ml G418.
Plasmids
pGL3-E1bTATA (pluc-min) firefly luciferase reporter, which contains a E1B minimal promoter upstream of the luciferase coding sequence, is the reporter plasmid used to measure the basal luciferase activity (33). The pGL3-HO1 plasmid was obtained by inserting between the HindIII and NcoI sites of pluc.min, a fragment of the mouse HO-1 5′non-translated sequence, obtained by PCR amplifcation using the oligonucleotide pair [CTGAAGCTTACGGTCTCCAGTCGCCTCCAG]/[CGTACCATGGTCATCACCGGACTGGGCTAGTTC]. The recognition sequences of HindIII and NcoI restriction enzymes are underlined. The amplified sequence of 151 bp encompasses the HO-1 5′non-translated sequence from nt −131 to nt +2 (+1 accounting for the A of the ATG initiation codon) including the putative p53-responsive element (Table 1). The cloned amplified fragment was sequenced to confirm the absence of mutation. The pGL3-HO1mut plasmid was obtained from pGL3-HO1 by PCR-directed mutagenesis by introducing three point mutations within the second decamer of the p53-responsive element (Figure 3). The sequence of the mutated fragment was verified. The pDDm-TO was obtained by inserting the sequence that encodes the mouse p53DD truncated protein into the Invitrogen vector pcDNA4/TO/B.
Table 1.
Real-time PCR analysis: sequence of the oligonucleotide pairs, annealing temperature, and product size
| Genes | Species | Acc number | Primer pairs | Ta(°C)a | Size(bp)b |
|---|---|---|---|---|---|
| GAPDH | Murine | BC083079 | F CATGGCCTTCCGTGTTCCTA R TGCCTGCTTCACCACCTTCT | 60 | 105 |
| Human | NM_002046 | F AGCTCACTGGCATGGCCTTC R ACGCCTGCTTCACCACCTTC | 60 | 117 | |
| HO-1 | Murine | NM_010442 | F GGAGATAGAGCGCAACAAGC R CCATACCAGAAGGCCATGTC | 60 | 105 |
| Human | NM_002133 | F CCAGTGCCACCAAGTTCAAG R CAGCTCCTGCAACTCCTCAA | 60 | 143 | |
| Waf1/p21 | Murine | NM_007669 | F TTGTCGCTGTCTTGCACTCT R ATCTGCGCTTGGAGTGATAG | 60 | 124 |
aTa: annealing temperature; bSize of the amplified fragment.
Figure 3.
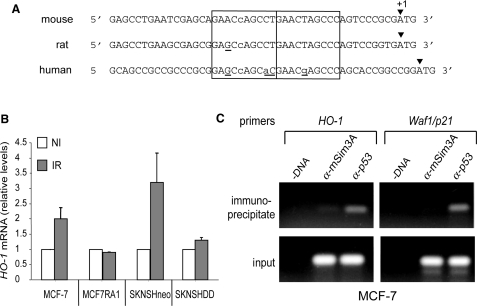
The human HO-1 gene is directly transactivated by p53. (A) Alignment of the 5′ non-coding sequence of mouse, rat and human HO-1 gene. In open boxes, the two adjacent decamers that respond to the definition of El Deiry et al. (41) are shown. The nucleotide variants as compared to the consensus sequence (see legend of Figure 2) were indicated by lower case letters and nucleotide variations in rat and human p53 decamers as compared to mouse are underlined. (B) Stimulation of HO-1 gene expression in irradiated human transformed cells expressing wt-p53 (MCF-7 and SKNSH Neo) but not in cells expressing mutant-p53 (MCF-7/R-A1 and SKNSH DD). Cells were non-irradiated (NI) or irradiated (IR) at a dose of 10 Gy. Total RNA was extracted 3 h post-irradiation. Expression of HO-1 was analysed by real-time quantitative RT-PCR, normalized relative to GAPDH mRNA levels and expressed taking as 1 the HO-1 mRNA level of the non-irradiated cells. (C) ChIP assay of HO-1 and Waf1/p21 in MCF-7 cells. Cross-linked p53 protein/DNA complexes were immunoprecipitated with either anti-p53 or anti-mSim3A (negative control) before PCR amplification as described in Materials and Methods section. Input (positive control) indicates a portion of the sonicated chromatin that has been amplified after immunoprecipitation.
Mice and irradiation
Animal procedures were carried out in accordance with French Government regulations. The p53−/− and p53+/+ C57BL/6 mice studied were progeny of p53 heterozygote animals obtained from IFFA-CREDO (Orléans, France). The genotype background was determined by PCR amplification of genomic DNA extracted from tail biopsies as described previously (34). Six- to eight-week-old mice were given whole-body γ-irradiation at the doses indicated from a 137Caesium source at a dose rate of 0.2 Gy/min (IBL 637, CisBio International). Control and irradiated animals were sacrificed at the time intervals indicated. Tissues were harvested and snap frozen in liquid nitrogen and kept at −80°C until used.
Microarray analysis
We used cDNA arrays of mouse stress/toxicology genes (140 genes) spotted on nylon membrane (Atlas™ filter arrays, Clontech, Palo Alto, CA, USA). The filters include also nine housekeeping control cDNA and negative controls (M13 mp18(+) strand DNA, lambda DNA, pUC18 DNA). The complete list of the cDNA and control as well as their accession number is available on the web (http://www.clonteh.com/atlas/genelists/index.html). Total RNA was isolated using RNA-plus (kit RNA-plus; Appligene) and potentially contaminating DNA was removed by treating with RNAse-free DNAse I (RQ1, RNAse-free DNAse Promega 1 U/µl). Labelled cDNA probe synthesis, microarray hybridization and wash steps were performed according to the manufacturer's recommendations. Briefly, 32P-labelled cDNA probes were generated by reverse transcription of 2–5 μg of each DNAse I-treated total RNA sample in the presence of [α32P] dATP (3000 Ci/mmol, 10 mCi/ml; NEN) and purified by spin column chromatography (Nucleospin® Extraction Kit; Clontech). Microarray hybridizations were carried out on microarray filters with 32P-labelled probe (1–3 × 106 c.p.m.) in 6 ml ExpressHyb™ hybridization solutions (Clontech) supplemented with heat-denatured sheared salmon testes DNA (100 μg/ml), in roller bottles, in a hybridization incubator, at 68°C overnight. After extensive washing (three times with 2 × SSC, 1% SDS, and once with 0.1× SSC, 0.5% SDS each for 30 min at 68°C, once with 2 × SSC for 5 min at room temperature), damp membranes were sealed in a plastic wrap and autoradiographed in phosphorimager cassettes for 5 days. The cDNA microarray autoradiograms were scanned with ImageQuant (Molecular Dynamics, Sunnyvale, CA, USA) and the images were analysed using the AtlasImage 2.0 software (Clontech, USA). The background was calculated using default external background calculation, which takes into consideration the background signals at blank space between the different panels of the arrays. Signal threshold was set as background-based signal threshold. The signal intensities were normalized globally using the sum method (AtlasImage, Clontech, USA).
Quantitative RT-PCR
Total RNA was prepared using the RNA-plus solution (Bioprobe) as previously described (34). RNA (30 µg) was incubated 30 min at 37°C with 1 U DNAse–RNAse-free (Boehringer) and 40 U RNAsine (Promega) in a final volume of 30 µl. Following this treatment, RNA was heated 2 min at 95°C, and then ice cooled. Reverse transcription was performed on 3 µg total RNA in the presence of 12 µl buffer RT X5 (GibcoBRL), 60 U RNAsine (Promega), 3 µl of a mixture of the four dNTPs (20 mM each), 150 ng of random primers (Invitrogen) and 15 U of AMV reverse transcriptase (Promega) in a final volume of 60 µl. After an incubation of 30 min at 42°C, additional 15 U of AMV reverse transcriptase (Promega) were added and incubation was prolonged for 30 min at 42°C. At the end of the reaction, AMV was heat inactivated 2 min at 95°C. Real-time PCR was done using FastStart DNA Master SYBR Green I kit and the LightCycler apparatus according to the protocol provided by the manufacturer (Roche Molecular Biochemicals). For each gene the sequence of primer pairs and the annealing temperature are given in Table 1.
Luciferase assay
Cells (2 × 105) were plated in 6-well plates (Falcon) and transfected with 200 µl of DNA-calcium phosphate precipitate containing plasmids as indicated in figure legends. Total DNA concentration was balanced to 30 µg/ml with pBS (a minimal vector, Stratagene). Each transfection was done in duplicate and repeated four to five times. Six hours after adding the DNA precipitate, the cells were washed twice, re-fed with fresh medium and incubated at 37°C for 18 h. The cells were then lysed with 200 µl/well of passive lysis buffer provided with the ‘luciferase assay kit’ (Promega) and proceeded as previously described using the Microlumat LB96P luminometer (Berthold EG&G Instrument) (35).
Western blot analysis
Mice tissues were squashed between two glass slides, dispersed in PBS and centrifuged for 5 min at 5000 r.p.m. at 4°C. The pellets were resuspended, at 4°C, in 500 µl of lysis buffer [20 mM HEPES pH 7.6; 2 mM EGTA; 5 mM EDTA; 30 mM sodium fluoride; 10 mM sodium pyrophosphate; 2 mM sodium orthovanadate; protease inhibitor cocktail (Roche); 5 µM pepstatin A; 0.5% nonidet P-40). After sonication, the extracts were centrifuged for 30 min at 68 000 r.p.m. at 4°C (Beckman TL-100 rotor). Total protein concentration of the supernatants were determined by the Bradford assay (Biorad Protein Assay). A volume of supernatant corresponding to 80 µg of total protein was separated on a 12% SDS–PAGE. After electrophoresis, the proteins were electrotransferred to nitrocellulose filters and the filters were probed with either the polyclonal anti-Haeme-oxygenase 1 (M-19) produced in goat (Santa Cruz Biotechnology, sc-1797; 1:250 dilution) to reveal mouse HO-1 or with the anti-p21 antibody clone SX118 produced in mice (BD Pharmingen, ref. 556 430; 1:500 dilution) or with the anti-actin (20–33) antibody produced in rabbit (Sigma Aldrich; ref. A5060; 1:500 dilution). After incubation with the secondary antibody conjugated with horseradish peroxidase diluted 1:10 000 [anti-goat peroxidase (Vector) to reveal HO-1; anti-mouse peroxidase and anti-rabbit peroxidase to reveal p21/Waf1 and actin, respectively (Santa Cruz Biotechnology)], detection was performed with Chemiluminescence Reagent Plus (NEN).
Electrophoretic mobility shift assays (EMSA)
Wild-type human p53 protein was overexpressed in Sf21 cells using a baculovirus expression vector (36). Oligomers [TCGAGGAACCAGCCTGAACTAGCCCG/CTAGCGGGCTAGTTCAGGCTGGTTCC], containing the putative HO-1 p53RE were annealed and labelled with α32P dCTP using Klenow fragment of the DNA Polymerase and then used as probe. A mix containing 0.5 µl p53-overexpressing Sf9 cellular extract, 3.3 µl of 3× buffer [60 mM HEPES pH 7.6; 300 mM NaCl; 4.5 mM MgCl2; 30 mM DTT; 0.3% Triton 100X; 3 mM Pefabloc (Boehringer); 60% glycerol); 30 ng of purified anti-p53 antibody PAb 421 (Calbiochem); 100 ng of sonicated, non-denatured, salmon sperm DNA and 0.5 ng of radiolabel probe in a final volume of 10 µl was incubated for 30 min at room temperature. For competition experiments, 100- and 500-fold molar excess of unlabelled double-stranded oligonucleotides, corresponding to the consensus (specific = TCGACGGACATGCCCGGGCATGTCCGTCGA annealed to itself) or non-consensus (non-specific = GCGGAATTCAAAAGTTAGAGATAATCCTACGGGTCGGCCTCGAGGCG annealed to CGCCTCGAGGCCGACCCGTAGGATTATCTCTAACTTTTGAATTCCGC) sequence of p53–DNA-binding site, were added to the binding reaction before the addition of the labelled probe. The protein–32P-DNA complexes were separated by electrophoresis at 4°C on 4–15% PhastGel (Amersham Pharmacia Biotech) as previously described (33).
Chromatin immunoprecipitation assays
Fifteen centimetre dishes of subconfluent MCF-7 cells were subjected to ChIP assay as previously described (37). Briefly, cells were treated with 1% formaldehyde to cross-link genomic DNA and proteins. Cells were lysed with SDS-lysis buffer containing a protease-inhibitor cocktail and sonicated to generate DNA fragments 300–800 bp long. Chromatin was immunoprecipitated with the monoclonal antibody DO-7, directed against p53 (a gift from Dr D. Lane) for 16 h. Immunoprecipitates produced by anti-Sin3A(AK-11) (Santa Cruz Biotechnology Inc., sc-767) served as control. After immunoprecipitation, DNA–protein cross-links were reversed by incubation at 65°C for 4 h and genomic DNA was extracted. The immunoprecipitated genomic DNA were amplified with the oligonucleotide pair GTCAACGCCTGCCTCCTCTC/GGTTGCGGACGCTCCATC flanking the suspected binding site of p53 in HO-1, and, as a positive control, with the oligonucleotide pair GTGGCTCTGATTGGCTTTCTG/CTGAAAACAGGCAGCCCAAG that flanks the Waf1/p21 p53RE (38).
RESULTS
In vivo identification of HO-1 as a novel p53-inducible gene
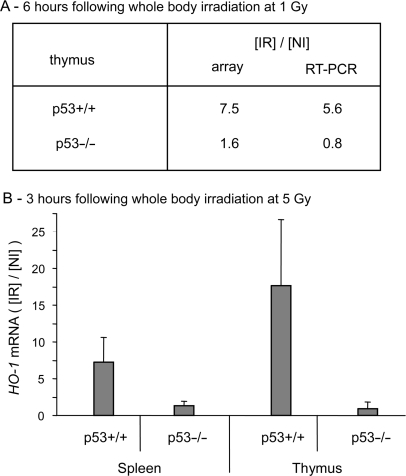
Microarray analysis was used to identify genes involved in oxidative metabolism whose stimulation depends on p53 activation, in vivo. Recently, we showed that 1 Gy is the lowest dose of irradiation to give a significant p53-dependent stimulation of expression of the principal proapoptotic genes, in the thymus and spleen of whole body irradiated mice (39). We decided therefore to use this relative low dose of irradiation to search for oxidant genes whose expression is regulated in response to p53 activation. Wt-p53 (p53+/+) and null (p53−/−) mice were irradiated at a dose of 1 Gy and sacrificed 6 h after irradiation. Radioactively labelled cDNA was synthesized from total RNA extracted from the thymus and hybridized to Altas™ mouse stress/toxicology array membranes as described in the Materials and Methods section. Among the 140 genes present in the membrane, HO-1 (still referred as Hmox1) is the only one that was found to be significantly up-regulated in response to irradiation in p53+/+ but not p53−/− mice. According to the microarray analysis, expression of HO-1 is stimulated by a factor of 7.5 in the thymus of irradiated p53+/+ mice compared to non-irradiated mice. Such stimulation is not observed in the thymus of p53−/− mice (Figure 1A). Microarray analysis was validated by quantitative real-time RT-PCR analysis using the same RNA samples (Figure 1A). The γ-ray induction of HO-1 in p53+/+ but not in p53−/− mice was confirmed in independent experiments, both in thymus and spleen of mice subjected to 5 Gy of whole body irradiation (Figure 1B). These results strongly suggest that the up-regulation of HO-1 in response to γ-irradiation requires a functional p53, at least in the lymphoid organs.
Figure 1.
Gamma-irradiation induced the expression of HO-1 in p53+/+ but not in p53−/− mice. (A) Total RNA was extracted from thymus of p53+/+ and p53−/− mice either non-irradiated or whole body irradiated at a dose of 1 Gy. The thymus was taken 6 h post-irradiation. Hybridization to cDNA arrays of mouse stress/toxicology genes (AtlasTM filter arrays, Clontech, Palo Alto, CA, USA) was performed as described in Materials and Methods section. Signal intensity was normalized as described in Materials and Methods section. The normalized signal calculated for the thymus of irradiated mice was expressed relative to the thymus of non-irradiated mice. Results from microarray analysis (array) were confirmed by real-time quantitative RT-PCR as described in Materials and Methods section. The relative level of HO-1 mRNA measured by RT-PCR was normalized relative to GAPDH mRNA and the level of stimulation was calculated relative to non-irradiated animals. (B) HO-1 mRNA induction in thymus and spleen of mice at 3 h following whole body irradiation at a dose of 5 Gy. The level of HO-1 mRNA estimated by real-time quantitative RT-PCR was normalized as described below. The graphs represent the mean of four independent experiments with their respective standard deviation. NI, non-irradiated mice; IR, irradiated mice.
Identification of a putative p53RE within the 5′non-coding region of the mouse HO-1 gene
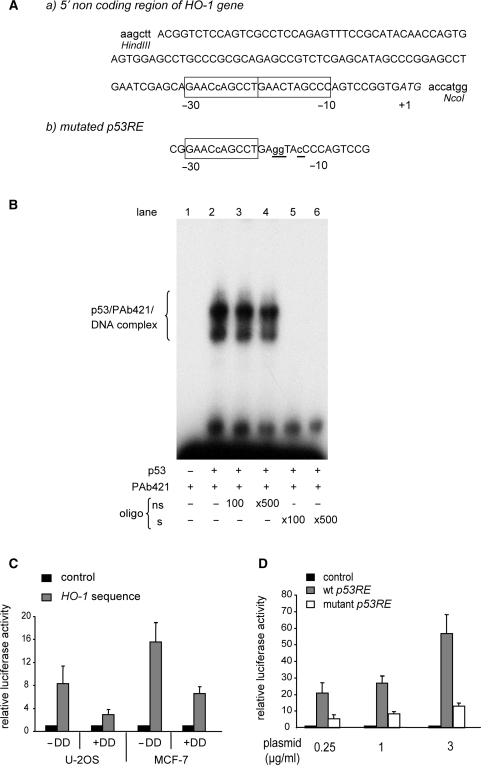
If HO-1 is a true target of p53, one or more p53RE should exist in this gene. Using an in silico approach developed by Bourdon et al. (40), we found one potential p53RE (GAACcAGCCTGAACTAGCCC) in exon 1 of the mouse HO-1 gene, located 10 nt upstream of the ATG initiation codon (Figure 2A). This sequence contains a single mismatch only, compared to the consensus sequence defined by el-Deiry and collaborators (41).
To explore the functionality of this putative p53RE, we first performed a band shift assay to test whether p53 specifically binds to this element. An extract of Sf21 cells overexpressing wt-p53 protein (see Materials and Methods section) was incubated with a radiolabelled double-strand oligonucleotide corresponding to the potential p53RE in presence of anti-p53 PAb421 antibody (Figure 2B, lane 2). PAb421 activates p53 and led to an increased binding of this protein to the probe (42). To confirm the specificity of the protein–DNA interaction, competition assays were performed. Addition of a 100- and 500-fold excess of non-radioactive oligonucleotide devoid of a p53-binding site did not affect the specific complex (lanes 3 and 4), while the formation of this complex was inhibited by 100- and 500-fold excess of unlabelled consensus probe (lanes 5 and 6). This result clearly identifies specific binding of p53 to the putative p53RE identified in the 5′non-coding region of HO-1 gene.
Figure 2.
The mouse HO-1 gene is directly transactivated by p53. (A-a) Sequence of the 5′ non-coding region of the mouse HO-1 gene with, in the open boxes, the putative p53RE that consists of two adjacent decamers that respond to the definition of a p53-binding site according to El Deiry et al. (41). Nucleotide numbering starts at the A of the translation initiation codon. The lower case letter within the first decamer indicates the variant nucleotide as compared to the consensus decamer: PuPuPuC(A/T)(A/T)GPyPyPy). The two HindIII and NcoI restriction sites were added by PCR amplification in order to clone this sequence within the pluc.min vector to obtain the pGL3-HO1 reporter gene plasmid (see Materials and Methods section). (b) Three mutations (underlined lower case letters) have been introduced within the second decamer by PCR-directed mutagenesis. (B) EMSA shows a specific binding of baculovirus-produced human wt-p53 to the two decamers identified in the mouse HO-1 gene. 32P-labelled DNA probe corresponding to the two decamers was incubated with p53 and the antibody anti-p53 PAb421 (lane2). Competitions were performed by using 100- or 500-fold molar excess of unlabelled double-stranded oligonucleotides either non-specific (ns, lanes 3 and 4); or homologous (s, lanes 5 and 6), as described in Materials and Methods section. (C) Luciferase assay showed that the transcriptional activity of the 5′ non-coding sequence of the mouse HO-1 gene is stimulated by wt-p53. U-2 OS and MCF-7, two cell lines that express wt-p53, were transfected with either pluc.min (control) or pGL3-HO1 (HO-1 sequence) and cotransfected with (+DD) or without (−DD) the plasmid pDDm-TO that encodes a dominant negative mutant of p53. Luciferase activities of cells transfected with pGL3-HO1 were expressed relative to the luciferase activity of cells transfected with pluc.min. The averaged results are of three independent experiments performed in duplicate. Standard deviations are indicated. (D) mHO-1 p53RE confers the ability to be transactivated by wt-p53. U-2OS were transfected with either pGL3-HO1 (wt p53RE) or the plasmid pGL3-HO1mut that is mutated within the p53RE as indicated above (mutant p53RE). Luciferase activities of cells transfected with these two plasmids were expressed relative to the luciferase activity of cells transfected with pluc.min (control). The averaged results are of four independent experiments performed in duplicate.
To determine whether p53 can transactivate gene expression via this element, a 131-bp DNA fragment containing the response element (Figure 2A) was cloned into a minimal promoter luciferase reporter gene plasmid as described in the Materials and Methods section. Expression of the luciferase gene was analysed under physiological concentration of p53 by transfecting the reporter plasmid in either U-2OS or MCF-7, two cell lines expressing an endogenous wt-p53 that could be activated by the stress of transfection. Results are presented in Figure 2C. The luciferase activity was significantly higher when the reporter gene was driven by HO-1 sequence as compared to the empty vector. The inhibition of luciferase activity by cotransfecting the reporter plasmid with the p53 dominant negative form p53DD, strongly suggest that expression from HO-1 sequence depends on the wt-p53 expressed in MCF-7 and U-2OS cells. This was further supported by introducing inactivating mutations within the p53RE as indicated in Figure 2A. These mutations abolished the specific binding of wt-p53 to DNA (data not shown) and strongly reduced the transcriptional activity of the HO-1 sequence independently of the concentration of the reporter plasmid transfected in U-2OS cells (Figure 2D).
Taken together these results indicate that p53 could directly transactivate the mouse HO-1 gene both in vitro and in vivo.
HO-1 p53RE is conserved between mice, human and rat
An alignment of the exon 1 of HO-1 gene of three species (mouse, human and rat) revealed marked similarities between rat, human and mouse sequences in particular at the level of the p53RE, strongly suggesting that the p53-dependent transactivation of HO-1 may be conserved between species (Figure 3A).
To further support this assumption, we investigated whether if the expression of HO-1 is stimulated in human cells in response to wt-p53 activation. To this aim, endogenous HO-1 gene expression was analysed in two pairs of cells expressing either a functional or an inactive form of p53. The first series corresponds to the two related cell lines MCF-7 and MCF7/R-A1 that express wt-p53 and the transcriptional inactive mutant p53R280K, respectively (32). The second pair corresponds to two SK-N-SH-derived cell lines, SKNSH-CMVneo that expresses wt-p53 and SKNSH-DDp53 that overexpresses the p53DD truncated protein that inhibits the endogenous p53 transcriptional activity (31). Our unpublished results revealed that a high irradiation dose of 10 Gy is required to obtain a significant stimulation of Bax, a well-characterized proapoptotic p53 target gene, in SKNSH-CMVneo cells. Therefore, the subconfluent cell cultures were exposed to a γ-irradiation dose of 10 Gy, total RNA was extracted 3 h after irradiation and HO-1 mRNA levels were estimated by quantitative real-time PCR as described in Materials and Methods section. Results are presented in Figure 3B. As compared to non-irradiated cells (NI), irradiation (IR) led to a significant increase of HO-1 mRNA levels in both MCF-7 and SKNSH-CMVneo cells that express wt-p53, but not in their related cell lines, MCF7/R-A1 and SKNSH-DDp53 defective in functional p53. These results show that the expression of the human endogenous HO-1 gene is stimulated in response to an activation of physiologic levels of wt-p53. It is of interest to note that a relative high irradiation dose is needed.
Next, we carried out a ChIP assay to ensure that the human HO-1 gene is directly targeted by endogenous wt-p53 in MCF-7 cells. Using specific primers that amplify a 98-bp DNA fragment that includes the putative p53RE of the human HO-1 gene, we were able to detect DNA that was immunoprecipitated by an anti-p53 antibody, but not by a non-specific antibody. As a positive control, a 105-bp DNA fragment that included p53RE identified in the p21/Waf1 gene was amplified from chromatin immunoprecipitated by the anti-p53 antibody but not by the control antibody (Figure 3C).
Taken together, these results identify HO1 as a bona fide p53 target gene both in mice and human cells.
Dose-response and kinetics of HO-1 stimulation in lymphoid organs of whole body irradiated mice—comparison to the stimulation of p21/Waf1
We have shown previously that the expression of Puma, an essential mediator of p53-induced apoptosis, is readily stimulated in the lymphoid organs of mice exposed to a dose of irradiation as low as 0.1 Gy. Moreover, induction of the major proapoptotic p53 target genes was observed as early as 1 h post-irradiation increasing to a maximum at 3 h (39). We were then interested to determine the sensitivity of HO-1 stimulation in vivo in comparison to p21/Waf1, a downstream target of p53 and an universal inhibitor of cyclin-dependent kinase.
HO-1 is stimulated at higher doses of irradiation than p21/Waf1
Relative mRNA levels were evaluated by quantitative real-time RT-PCR, in the thymus and spleen of p53+/+ mice taken 3 h after whole body irradiation at 0.1, 0.2, 0.5, 1 or 2 Gy. From the results presented in Table 2, it appears that a clear up-regulation of p21/Waf1 transcription can already be detected at 0.1 Gy in spleen and at 0.2 Gy in thymus, showing that the in vivo sensitivity of p21/Waf1 to irradiation is comparable to that determined previously for the main proapoptotic genes (39). Nevertheless, the stimulation of HO-1 gene expression required higher irradiation doses of 0.5 Gy in thymus and of 1 Gy in spleen. At higher doses of irradiation, the stimulation of HO-1 and p21/Waf1 gene expression reached comparable levels in the thymus but not in the spleen, where HO-1 was noticeably less induced than p21/Waf1.
Table 2.
Dose effect of γ-irradiation on the p53-dependent induction of the HO-1 and Waf1/p21 in thymus and spleen of mice after whole body irradiation
| Organs | Gy | H0-1 | Waf1/p21 | ||
|---|---|---|---|---|---|
| Stimulation* | P | Stimulation* | P | ||
| Thymus | 0.1 | nd | – | 1.4 ± 0.15 | 0.03 |
| 0.2 | 1.8 ± 0.3 | <0.05 | 2.8 ± 1.3 | 0.01 | |
| 0.5 | 3.1 ± 1.1 | <0.05 | 5.4 ± 1.7 | <0.001 | |
| 1 | 9.7 ± 2.1 | <0.005 | 8.9 ± 3.8 | 0.001 | |
| 2 | 31.3 ± 6.6 | <0.005 | 20.4 ± 6.6 | <0.001 | |
| Spleen | 0.1 | nd | – | 2.5 ± 0.04 | <0.0001 |
| 0.2 | 1.2 ± 0.3 | ns | 3.4 ± 1.1 | <0.01 | |
| 0.5 | 1.4 ± 0.4 | ns | 6.0 ± 1.9 | <0.005 | |
| 1 | 2.3 ± 0.8 | <0.05 | 10.7 ± 5.8 | <0.02 | |
| 2 | 2.9 ± 0.8 | <0.005 | 16.2 ± 2.9 | <0.0001 | |
Three hours following whole body irradiation, mice were sacrificed and total mRNA was extracted from thymus and spleen taken from p53 wild-type (p53+/+) mice exposed to irradiation doses of 0.1, 0.2, 0.5, 1 and 2. Non-irradiated mice were treated in parallel. Transcriptional induction was evaluated by real-time RT-PCR as described in Materials and Methods section and expressed relative to mRNA level measured from non-irradiated mice. One-way ANOVA analysis was used to determine significance between stimulation factors of irradiated versus non-irradiated mice. A stimulation factor of 2 with a P value <0.05 was considered as showing a significant up-regulation of the gene expression. Asterisk: mean of 4 (thymus) or 5 (spleen) mice ± mean deviation; nd = not determined; ns = not significant.
HO-1 gene induction is delayed in relation to p21/Waf1
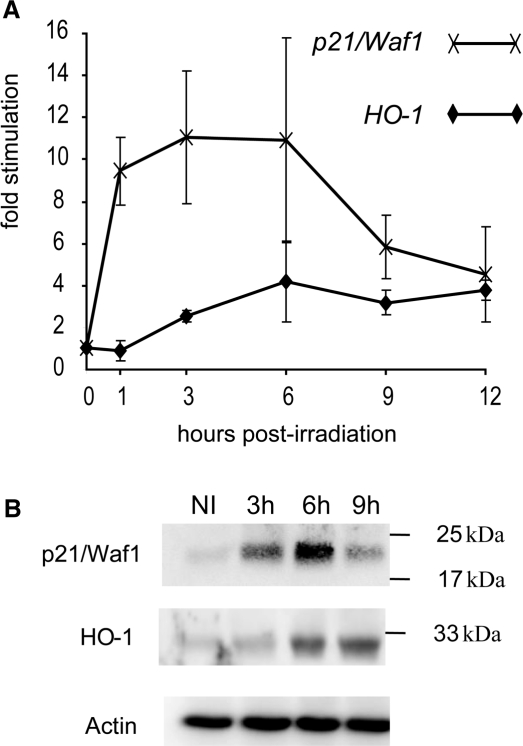
Kinetics of HO-1 and p21/Waf1 mRNA accumulation was measured in the spleens of p53+/+ mice following whole body irradiation at 1 Gy. Results are presented in Figure 4A. The stimulation of p21/Waf1 reached a maximum as soon as 1 h after irradiation, while a significant stimulation (2.5 ± 0.3) of HO-1 was detectable starting at 3 h post-irradiation. Similar results were obtained while assaying the RNA extracted from thymus of irradiated mice. As in the spleen, stimulation of p21/Waf1 is readily detectable at 1 h post-irradiation, while a significant stimulation of HO-1 was noticeable at 3 h only (data not shown).
Figure 4.
Kinetics of HO-1 and p21/Waf1 stimulation in the spleen of whole body-irradiated mice. Mice irradiated at a dose of 1 Gy were sacrificed at the indicated time post-irradiation. (A) HO-1 and p21/Waf1 mRNA levels determinate by real-time quantitative RT-PCR (see Material and Methods section) and normalized relative to GAPDH mRNA. The graphs represent the mean of four independent experiments with their respective standard deviation. (B) HO-1 and p21/Waf1 protein levels analysed by western blot using anti-p21/Waf1, anti-p53 and anti-actin (to control loading).
Finally, we addressed whether the stimulation of HO-1 transcripts is also reflected in a stimulation of HO-1 protein levels. Western blot analysis was performed with protein extracts prepared from the spleens of mice subjected to 1 Gy of whole body irradiation. As shown in Figure 4B, the level of the p21/Waf1 protein increased 3 h after irradiation, to reach a peak at 6 h, and then declined at 9 h. The HO-1 protein was found to increase 6 h after irradiation and the increased level was sustained up to 9 h. These results further support the results of mRNA clearly showing that the stimulation HO-1 gene expression in response to γ-irradiation is delayed compared to p21/Waf1.
DISCUSSION
As more has been learned about p53, it has become apparent that there is a reaction to nearly every p53 action leading to an array of autoregulation feedback loops. The first described was the p53-Mdm2 negative feedback loop that represents the best-studied relationship between a tumour suppressor gene, which functions as a transcription factor and an oncogene. p53, activates transcription of the Mdm2 gene. Mdm2, once produced, binds to p53 preventing it from acting as a transcription factor, and subsequently ubiquitinates it, which enhances its proteolytic breakdown [review in Oren et al., (43)]. Another example is the role of p53 in the ROS regulation. As reported in the Introduction section, p53 can activate numerous genes involved in increased generation of ROS, which contributes to apoptosis and also functions in a feedback loop in which ROS can signal to the further activation of p53. More recent evidence suggests however, that p53 may also induce the expression of proteins that function to lower ROS levels thereby protecting from oxidative-stress-induced DNA damage and apoptosis [review by Vousden (44)]. In this vein, we identified HO-1 as a direct p53 target gene.
The enzyme encoded by HO-1 belongs to the haeme-oxygenase family that consists of two constitutive (HO-2 and HO-3) and one inducible (HO-1) isoforms that is also known as heat shock protein-32 (HSP-32). The haeme-oxygenase is remarkably conserved from algae to human. Its substrate, haeme, is the prosthetic group in many enzymes and proteins that transport, use, store and detoxify oxygen, such as hemoglobin, catalase and cytochromes (45). This enzyme catalyses the rate-limiting degradation of haeme into carbon monoxide (CO), iron and biliverbin, which is metabolized to bilirubin, and thus constitutes a major intracellular source of iron and CO. Although ubiquitously distributed, HO-1 is highly expressed in the spleen and other tissues that degrade senescent red blood cells, including specialized reticuloendothelial cells of the liver and bone marrow. In most other tissues, HO-1 typically occurs at low to undetectable levels under basal conditions, but its expression is rapidly activated in response to a number of stimuli that include haeme, hyperoxia, hypoxia, heavy metals, UV radiation, H2O2 and nitric oxide. The common characteristic of these numerous inducers is their ability to cause oxidative stress (46–48).
HO-1 expression is primary regulated at the transcriptional level. A multiplicity of regulatory elements has been identified in the promoter region, in particular specific binding sites for the most important regulators of the cellular stress response such as members of the heat-shock factor, nuclear factor-kappaB, nuclear factor-erythroid 2 and activator protein-1 (48,49). The presence of a p53RE within the HO-1 gene was described here for the first time.
Early induction of HO-1 expression in response to whole body irradiation has already been reported in rat liver (50) and mouse lung (51). But the possible implication of p53 was not described. Here we reported an early and dose-dependent stimulation of the HO-1 mRNA level in thymus and spleen of p53-wt but not of p53-null mice, in response to whole body irradiation, showing that the increase of HO-1 mRNA in lymphoid organs of irradiated mice required the presence of a functional p53.
Vogelstein's group has already reported a significant induction of HO-1 in colorectal cancer cells forced to express of wt-p53 (52). However, they have demonstrated that in cells expressing physiological levels of p53, stimulation of HO-1 in response to drugs known to activate p53 such as doxorubicin or 5-fluorouracil, is comparable in cells expressing either wt- or mutant-p53. The authors concluded that the induction of HO-1 was not strictly p53 dependent. The discrepancy between our results and those of Vogelstein's group could account for a different cellular context (transformed cell lines in culture versus in vivo lymphoid cells) and/or the nature of the induction signal (drug versus γ-irradiation). Indeed we have proven that HO-1 is a direct p53 target gene by showing that the p53RE identified within the 5′non-coding region of HO-1 gene binds p53 specifically, both in vitro and in vivo.
According to Sablina et al. (53), the p53-inducible genes can be broadly placed into two groups: those induced rapidly by low levels of stress (generally cell-cycle arrest such as Waf1/p21 or antioxidant targets such as SESN1, SESN2) and those that are induced by higher levels of stress (generally apoptotic targets such as Puma and Killer/DR5). These authors propose that the stimulation of SESN1, SESN2 could protect cells against oxidative DNA damage and genomic instability under physiologic condition of stress.
Contrarily to SESN1 and SESN2, stimulation of HO-1 requires a relatively high level of stress, the threshold dose leading to a significant stimulation being even higher for HO-1 than for p21/Waf1 both in spleen and in thymus (Table 2). Moreover, induction of HO-1, measured both at the mRNA and protein levels, is delayed as compared to p21/Waf1 (Figure 4). Our previous study has revealed a high in vivo sensitivity of p53-dependent transcriptional activation of genes involved in the two main apoptotic pathways, the extrinsic (Fas, Killer/DR5) and the intrinsic (Bax, Noxa, Puma) ones. As reported here for p21/Waf1, the expression of these proapoptotic genes was induced as soon as 1 h post-irradiation and reached a maximum at 3 h. Moreover, sensitivity of Puma, Bax and Killer/DR5 to γ-irradiation is comparable to that of p21/Waf1. Indeed, a dose of 0.1 Gy of γ-irradiation gives a significant stimulation of Puma, and 0.2 Gy of γ-irradiation induces a significant increase in the expression of Bax and Killer/DR5 both in thymus and spleen of mice whole body irradiated. These results show that, in the lymphoid organs of irradiated mice, genes involved in both cell cycle arrest and apoptosis respond to p53 activation more rapidly and at lower dose of irradiation than HO-1. These results strongly suggest that the physiological function of the p53-dependent induction of HO-1 differs from that proposed by Sablina et al. for the other antioxidant p53 target genes (53).
In addition to its well-characterized role as a metabolic enzyme specialized in the breakdown of haeme, HO-1 is now generally accepted as a mediator of cyto- and tissue protection against wide variety of stress (48). The cytoprotective action of HO-1 seems to be mainly a function of the antiapoptotic effects of the enzyme. The antiapoptotic action of HO-1 derived mostly from its haeme degradation products, including bilirubin, a potent antioxidant, and CO that has been shown to possess signalling properties affecting numerous critical cellular functions including apoptotic cell death (47,48). Interestingly, Liu et al. report an antiapoptotic role of CO in vascular smooth muscle treated with a cytokine mixture that is associated with an inhibition of p53 stimulation and the suppression of cytochrome c release from mitochondria (54). Interestingly, our results show a decrease of p21/Waf1 stimulation both at the mRNA and protein levels, 9 h post-irradiation (this study) and of the proapoptotic p53 target genes, 6 h post-irradiation (our previous published data, 39) in the lymphoid organs of irradiated mice. On the contrary, no decrease in HO-1 stimulation was noticeable neither at the protein nor at the mRNA level (Figure 4). This suggests the existence of a negative feedback loop between the activation of p53 by ROS and the p53-dependent stimulation of HO-1. According to this hypothesis, increased expression of HO-1 would reduce intracellular pro-oxidant levels leading to diminution of p53 activation and consequently a decrease in p53 target genes stimulation.
ACKNOWLEDGEMENTS
We thank Dr Jean-Claude Lelong for helpful discussions, Dr Serge Boiteux for its support and Dr Pierre May for critical review of the manuscript. Centre de la Recherche Scientifique sur le Cancer; Commissariat à l’Energie Atomique; Electricité de France; Association pour la Recherche sur le Cancer. Funding to pay the Open Access publication charges for this article was provided by Commissariat à l'Energie Atomique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Immunol. Today. 1994;15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 2.Carmody RJ, Cotter TG. Signalling apoptosis: a radical approach. Redox. Rep. 2001;6:77–90. doi: 10.1179/135100001101536085. [DOI] [PubMed] [Google Scholar]
- 3.Meek DW. Multisite phosphorylation and the integration of stress signals at p53. Cell Signal. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 4.Yang X. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10:400–403. doi: 10.1038/sj.cdd.4401182. [DOI] [PubMed] [Google Scholar]
- 5.Leblanc V, May P. Activation and post-translational modifications of p53 following DNA damage. Med. Sci. 2002;18:577–584. [Google Scholar]
- 6.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 7.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 8.El Deiry WS. Regulation of p53 downstream genes. Semin. Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 9.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 10.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Johnson TM, Yu ZX, Ferrans VJ, Lowenstein RA, Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc. Natl Acad. Sci. USA. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotem J, Peled-Kamar M, Groner Y, Sachs L. Cellular oxidative stress and the control of apoptosis by wild-type p53, cytotoxic compounds, and cytokines. Proc. Natl Acad. Sci. USA. 1996;93:9166–9171. doi: 10.1073/pnas.93.17.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 14.Zhao RB, Gish K, Murphy M, Yin YX, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 15.Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810–1815. [PubMed] [Google Scholar]
- 16.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, et al. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat. Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu G, Chen XB. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21:7195–7204. doi: 10.1038/sj.onc.1205862. [DOI] [PubMed] [Google Scholar]
- 18.Bravard A, Ageron-Blanc A, Alvarez S, Drané P, Le Rhun Y, Paris F, Luccioni C, May E. Correlation between antioxidant status, tumorigenicity and radiosensitivity in sister rat cell lines. Carcinogenesis. 2002;23:705–711. doi: 10.1093/carcin/23.5.705. [DOI] [PubMed] [Google Scholar]
- 19.Tan MJ, Li SJ, Swaroop MJ, Guan KL, Oberley LW, Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J. Biol. Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 20.Pani G, Bedogni B, Anzevino R, Colavitti R, Palazzotti B, Borrello S, Galeotti T. Deregulated manganese superoxide dismutase expression and resistance to oxidative injury in p53-deficient cells [In Process Citation] Cancer Res. 2000;60:4654–4660. [PubMed] [Google Scholar]
- 21.Drané P, Bravard A, Bouvard V, May E. Reciprocal down-regulation of p53 and SOD2 gene expression – implication in p53 mediated apoptosis. Oncogene. 2001;20:430–439. doi: 10.1038/sj.onc.1204101. [DOI] [PubMed] [Google Scholar]
- 22.Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, et al. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 23.Yoon D, Wang Y, Stapleford K, Wiesmuller L, Chen J. P53 inhibits strand exchange and replication fork regression promoted by human Rad51. J. Mol. Biol. 2004;336:639–654. doi: 10.1016/j.jmb.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 24.Budanov AV, Shoshani T, Faerman A, Zelin E, Kamer I, Kalinski H, Gorodin S, Fishman A, Chajut A, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- 25.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 26.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 27.Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Willie AH. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 28.Midgley CA, Owens B, Briscoe CV, Thomas DB, Lane DP, Hall PA. Coupling between gamma irradiation, p53 induction and the apoptotic response depends upon cell type in vivo. J. Cell Sci. 1995;108(Pt 5):1843–1848. doi: 10.1242/jcs.108.5.1843. [DOI] [PubMed] [Google Scholar]
- 29.MacCallum DE, Hupp TR, Midgley CA, Stuart D, Campbell SJ, Harper A, Walsh FS, Wright EG, Balmain A, et al. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- 30.Smart P, Lane EB, Lane DP, Midgley C, Vojtesek B, Lain S. Effects on normal fibroblasts and neuroblastoma cells of the activation of the p53 response by the nuclear export inhibitor leptomycin B. Oncogene. 1999;18:7378–7386. doi: 10.1038/sj.onc.1203260. [DOI] [PubMed] [Google Scholar]
- 31.Shaulian E, Haviv I, Shaul Y, Oren M. Transcriptional repression by the C-terminal domain of p53. Oncogene. 1995;10:671–680. [PubMed] [Google Scholar]
- 32.Cai ZZ, Capoulade C, Moyretlalle C, AmorGueret M, Feunteun J, Larsen AK, Bressacdepaillerets B, Chouaib S. Resistance of MCF7 human breast carcinoma cells to TNF-induced cell death is associated with loss of p53 function. Oncogene. 1997;15:2817–2826. doi: 10.1038/sj.onc.1201445. [DOI] [PubMed] [Google Scholar]
- 33.Munsch D, Watanabe-Fukunaga R, Bourdon JC, Nagata S, May E, Yonish-Rouach E, Reisdorf P. Human and mouse Fas (APO-1/CD95) death receptor genes each contain a p53-responsive element that is activated by p53 mutants unable to induce apoptosis. J. Biol. Chem. 2000;275:3867–3872. doi: 10.1074/jbc.275.6.3867. [DOI] [PubMed] [Google Scholar]
- 34.Bouvard V, Zaitchouk T, Vacher M, Duthu A, Canivet M, Choisy-Rossi C, Nieruchalski M, May E. Tissue and cell-specific expression of the p53-target genes: bax, fas, mdm2 and waf1/p21, before and following ionising irradiation in mice. Oncogene. 2000;19:649–660. doi: 10.1038/sj.onc.1203366. [DOI] [PubMed] [Google Scholar]
- 35.Deguin-Chambon V, Vacher M, Jullien M, May E, Bourdon JC. Direct transactivation of c-Ha-Ras gene by p53 – evidence for its involvement in p53 transactivation activity and p53-mediated apoptosis. Oncogene. 2000;19:5831–5841. doi: 10.1038/sj.onc.1203960. [DOI] [PubMed] [Google Scholar]
- 36.Delphin C, Cahen P, Lawrence JJ, Baudier J. Characterization of baculovirus recombinant wild-type p53 – dimerization of p53 is required for high-affinity DNA binding and cysteine oxidation inhibits p53 DNA binding. Eur. J. Biochem. 1994;223:683–692. doi: 10.1111/j.1432-1033.1994.tb19041.x. [DOI] [PubMed] [Google Scholar]
- 37.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 38.Eldeiry WS, Tokino T, Velculescu VE, Levy DL, Parsons R, Trent JM, Lin D, Mercer WE, Kinzer KW, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez S, Drane P, Meiller A, Bras M, Deguin-Chambon V, Bouvard V, May E. A comprehensive study of p53 transcriptional activity in thymus and spleen of gamma irradiated mouse: high sensitivity of genes involved in the two main apoptotic pathways. Int. J. Radiat. Biol. 2006;82:761–770. doi: 10.1080/09553000600949624. [DOI] [PubMed] [Google Scholar]
- 40.Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B, May E. Further characterisation of the p53 responsive element – identification of new candidate genes for trans-activation by p53. Oncogene. 1997;14:85–94. doi: 10.1038/sj.onc.1200804. [DOI] [PubMed] [Google Scholar]
- 41.El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 42.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 43.Oren M, Damalas A, Gottlieb T, Michael D, Taplick J, Leal J, Maya R, Moas M, Seger R, et al. Regulation of p53: intricate loops and delicate balances. Biochem. Pharmacol. 2002;64:865. doi: 10.1016/s0006-2952(02)01149-8. [DOI] [PubMed] [Google Scholar]
- 44.Vousden KH. Outcomes of p53 activation – spoilt for choice. J. Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 45.Padmanaban G, Venkateswar V, Rangarajan PN. Haem as a multifunctional regulator. Trends Biochem. Sci. 1989;14:492–496. doi: 10.1016/0968-0004(89)90182-5. [DOI] [PubMed] [Google Scholar]
- 46.Morse D, Choi AM. Heme oxygenase-1: the “emerging molecule” has arrived. Am. J. Respir. Cell Mol. Biol. 2002;27:8–16. doi: 10.1165/ajrcmb.27.1.4862. [DOI] [PubMed] [Google Scholar]
- 47.Fang J, Akaike T, Maeda H. Antiapoptotic role of heme oxygenase (HO) and the potential of HO as a target in anticancer treatment. Apoptosis. 2004;9:27–35. doi: 10.1023/B:APPT.0000012119.83734.4e. [DOI] [PubMed] [Google Scholar]
- 48.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol. Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 49.Alam J, Cook JL. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am. J. Respir.Cell Mol. Biol. 2007;36:166–174. doi: 10.1165/rcmb.2006-0340TR. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki K, Mori M, Kugawa F, Ishihara H. Whole-body X-irradiation induces acute and transient expression of heme oxygenase-1 in rat liver. J. Radiat. Res. 2002;43:205–210. doi: 10.1269/jrr.43.205. [DOI] [PubMed] [Google Scholar]
- 51.Risom L, Moller P, Vogel U, Kristjansen PE, Loft S. X-ray-induced oxidative stress: DNA damage and gene expression of HO-1, ERCC1 and OGG1 in mouse lung. Free Radic. Res. 2003;37:957–966. doi: 10.1080/1071576031000150788. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Zhang L, Hwang PM, Rago C, Kinzler K, Vogelstein B. Identification and classification of p53-regulated genes. Proc. Natl Acad. Sci. USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu XM, Chapman GB, Peyton KJ, Schafer AI, Durante W. Carbon monoxide inhibits apoptosis in vascular smooth muscle cells. Cardiovasc. Res. 2002;55:396–405. doi: 10.1016/s0008-6363(02)00410-8. [DOI] [PubMed] [Google Scholar]