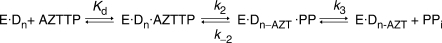Abstract
Native nucleotides show a hyperbolic concentration dependence of the pre-steady-state rate of incorporation while maintaining concentration-independent amplitude due to fast, largely irreversible pyrophosphate release. The kinetics of 3′-azido-2′,3′-dideoxythymidine (AZT) incorporation exhibit an increase in amplitude and a decrease in rate as a function of nucleotide concentration, implying that pyrophosphate release must be slow so that nucleotide binding and incorporation are thermodynamically linked. Here we develop assays to measure pyrophosphate release and show that it is fast following incorporation of thymidine 5′-triphosphate (TTP). However, pyrophosphate release is slow (0.0009 s−1) after incorporation of AZT. Modeling of the complex kinetics resolves nucleotide binding (230 µM) and chemistry forward and reverse reactions, 0.38 and 0.22 s−1, respectively. This unique mechanism increases selectivity against AZT incorporation by allowing reversal of the reaction and release of substrate, thereby reducing kcat/Km (7 × 10−6 μ M−1 s−1). Other azido-nucleotides (AZG, AZC and AZA) and 8-oxo-7,8-dihydroguanosine-5′-triphosphate (8-oxo-dGTP) show this same phenomena.
INTRODUCTION
Nucleoside reverse transcriptase inhibitors (NRTIs) continue to be an integral part of the highly active antiretroviral therapy (HAART) used to combat HIV infections (1). The primary limiting factors of the current treatments are the ability of HIV to rapidly develop resistance to the drugs and the toxic side effects of the NRTIs. Each of the eight US Food and Drug Administration (FDA) approved NRTIs are given orally as inactive prodrugs that require subsequent phosphorylation by host cell kinases. In the triphosphate form they act as substrates for HIV reverse transcriptase (RT) and terminate DNA polymerization because they lack a 3′-OH (2). Under most circumstances, the NRTIs cannot be removed because RT lacks a proofreading exonuclease activity, although mutations affording resistance have evolved by optimization of pyrophosphorolysis, the reversal of the polymerization reaction but with ATP as the pyrophosphate donor (3,4). Moreover, toxic side effects limit the dose and extent of therapy with many NRTIs.
The toxicity of nucleoside analogs varies widely across the series of eight NRTIs currently approved by the FDA. For the most toxic analogs, side effects are reminiscent of heritable mitochondrial diseases and are the consequence of duration-dependent mitochondrial dysfunction (5–11). These analogs are thought to act as alternative substrates for the mitochondrial DNA polymerase (Pol γ), thus slowing the replication of the mitochondrial genome. In order to quantify the inhibition of mitochondrial genome replication, we previously reconstituted and characterized recombinant human Pol γ holoenzyme in vitro (12,13). The overall fidelity of replication was established using transient state kinetic methods (14–16). Measurements of the kinetics of incorporation and exonuclease removal of each of the FDA-approved NRTIs were used to compute a ‘toxicity index’ which correlated well with the severity of the clinically observed toxicities over six orders of magnitude (17,18). This correlation held true for all of the analogs except 3′-azido-2′,3′-dideoxythymidine (AZT or zidovudine). That is, the low toxicity of AZT towards mitochondrial DNA replication seen in vitro was at odds with the clinical experience, thereby suggesting a different site of toxic action. Moreover, AZT primarily causes bone marrow toxicity, while ddC (zalcitabine) and ddI (didanosine) produce peripheral neuropathy, a hallmark of mitochondrial toxicity (19). There is a growing body of literature supporting the notion that the toxicity of AZT may arise due to inhibition at a site other than the mitochondrial DNA polymerase, and so both clinical and enzymatic data suggest that AZT does not significantly inhibit human mitochondrial DNA replication.
Some questions remained concerning the incorporation of AZT by Pol γ. The ‘toxicity index’ was based upon the specificity constant (kcat/Km) for incorporation of 3′-azido-2′3′-dideoxythymidine 5′-triphosphate (AZT-TP) relative to that of TTP and also the rate of exonuclease removal of AZT. For all of the natural deoxynucleoside triphosphates (dNTPs) and the NRTIs (in their triphosphate form), the specificity constant was measured under single turnover conditions ([E] > [DNA]) by measuring the concentration dependence of the rate of incorporation in a single nucleotide extension reaction. Following the standard paradigm (20), the hyperbolic concentration dependence of the observed rate of incorporation defined the maximum rate of polymerization (kpol) and the ground state dissociation constant (Kd). Normally, the kpol/Kd can be equated to kcat/Km, because the initial binding reaction is a rapid equilibrium relative to the rate of incorporation and is followed by a single, effectively irreversible, rate-limiting step. However, the kinetics of incorporation using AZT-TP were complex and precluded an accurate estimate for the specificity constant. The rate of incorporation appeared to decrease slightly with increasing concentration, whereas the amplitude followed a hyperbolic dependence (17). This suggested that the formation of enzyme-bound product (EDNAn+1 inorganic pyrophosphate, PPi) was reversibly linked to the nucleotide ground state (EDNAndNTP). Qualitatively, this would explain the fact that higher concentrations of AZT-TP appeared to drive product formation at the active site of the enzyme, but relied upon the unusual prediction that pyrophosphate release was slow.
The most striking aspect of this hypothesis is that it suggests a novel mechanism for the polymerase to correct an aberrant incorporation event by holding on tightly to the products, thereby favoring reversal of the reaction and release of the substrate. In this report, our aim was to further explore the mechanism governing the unusual kinetics of incorporation of AZT and to better define the specificity constant. Direct measurement shows that pyrophosphate release after incorporation of AZT is indeed slow, allowing binding and chemistry to be reversibly linked. The kinetic behavior observed with AZT-TP is also shown to be general for other azido-analogs: 3′-azido-2′,3′-dideoxyadenosine-5′-triphosphate (AZA-TP), 3′-azido-2′,3′-dideoxycytidine-5′-triphosphate (AZC-TP) and 3′-azido-2′,3′-dideoxyguanosine-5′-triphosphate (AZG-TP). This report also provides insights for the recent observations concerning the incorporation of (–)-β-l-2′-3′-dideoxy-5-fluoro-3′-thiacytidine ((–)FTC) (21) and the potent mutagen 8-oxo-7,8-dihydroguanosine-5′-triphosphate (8-oxo-dGTP) (22).
MATERIALS AND METHODS
NRTI triphosphates and oligonucleotides
AZT-TP was obtained from Moravek Biochemicals (Brea, CA, USA). AZA-TP, AZC-TP and AZG-TP were purchased from TriLink Biotechnologies (San Diego, CA, USA). All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA).
Enzymes
Recombinant human Pol γ catalytic and accessory subunits were obtained as previously described (12,13). Holoenzyme was reconstituted at a 1:5 stoichiometric ratio of catalytic subunit to accessory subunit in order to saturate the binding with an apparent Kd of 35 nM, and no inhibition was seen due to excess accessory protein (13). All of the experiments were conducted using an exonuclease-deficient mutant (E200A). HIV-1 RT was obtained similarly to previously published methods (23). Yeast inorganic pyrophosphatase (PPase), purine nucleotide phosphorylase and nucleoside diphosphate kinase (NDP kinase) were purchased from Sigma-Aldrich Corp (St Louis, MO).
Preparation of DNA
A DNA primer/template of 25 and 45 nt, respectively, was employed for all experiments except when terminated with 3′-azido-2′,3′-dideoxythymidine 5′-monophosphate (AZT-MP) or TMP for pyrophosphorolysis reactions, in which case they were 26 and 45 nt, respectively. The primer sequence was 5′-GCCTCGCAGCCGTCCAACCAACTCA-3′, and the template sequence was 5′-GGACGGCATTGGATCGAGGXTGAGTTGGTTGGACGGCTGCGAGGC-3′, where the bold X in the template was modified to provide the desired base pair for the particular experiment being conducted. For most experiments the primer was 5′-32P-labeled using T4 polynucleotide kinase as previously described (22). Primers containing 3′-terminal AZT-MP were created to examine the pyrophosphorolysis reaction. A polymerase reaction was performed using 1μM HIV-1 RT, 3 μM duplex DNA (containing the appropriate template base opposite the 26th primer position), and 50 μM AZT-TP in the following reaction buffer: 50mM Tris–Cl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2. The reaction was allowed to ensue for 30 min at 37°C and then the product was purified by denaturing polyacrylamide gel electrophoresis (PAGE) to obtain the AZT-MP terminated primer.
Incorporation and pyrophosphorolysis reactions
Unless otherwise stated incorporation assays were performed at 37°C in a buffer consisting of 50 mM Tris–Cl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2. The methods for rapid mixing experiments were conducted as previously reported (17).
Stopped-flow measurement of PPi release
A SF 2004 series stopped-flow apparatus from KinTek Corp. (Austin, TX, USA) was used for the measurement. Exonuclease-deficient holoenzyme (100 nM) was preincubated for 5 min with 2.5 90 mM Mg2+, 90 nM 25/45-mer DNA, 1.5 μM Escherichia coli phosphate-binding protein (PBP) mutant labeled at Cys197 with N-[2-(1-maleimidyl)ethyl]-7-(diethylamino)coumarin-3-carboxamide (MDCC), 100 μM 7-methylguanosine, 0.02 U/ml purine nucleotide phosphorylase and 0.005 U/μl yeast inorganic PPase. The solution was then rapidly mixed at 20 C with 2.5 mM Mg2+, 50 μM TTP, 100 μM 7-methylguanosine and 0.02 U/ml purine nucleotide phosphorylase. The fluorophore was excited at 425 and a 450 nm long pass filter was used to observe emission. The coupled assay for phosphate release was previously described but needed to be modified here by the inclusion of yeast PPase (24). Control experiments verified that the rate of hydrolysis of pyrophosphate to phosphate was sufficiently fast so that the kinetics was solely limited by the release of PPi from Pol γ.
Enzymatic synthesis of γ-32P labeled AZT-TP
ATP γ-32P-labeled was obtained from Perkin Elmer (Wellesley, MA, USA). AZT-DP was purchased from Moravek Biochemicals (Brea, CA, USA). The phosphorylation reaction was carried out in the same buffer as described for incorporation assays at room temperature for 1 h with 600 nM γ-32P-labeled ATP (3000 Ci/mmol), 600 nM AZT-DP and 0.1 U/μl NDP kinase, at which time, conventional rat kinesin was added to a final concentration of 5 μM to rid the solution of unreacted γ-32P-labeled ATP. The sample was loaded onto a 1 ml DEAE Sepharose FF (Amersham Biosciences, Piscataway, NJ, USA) column at flow rate of 1 ml/min and a gradient of 0.1–1 M triethylammonium (pH 7.5) was run to elute γ-32P-labeled AZT-TP. The elution of product was monitored by counting 10 μl of each fraction using a scintillation counter by standard methods. Peak fractions were lyophilized to constant weight and resuspended in 10 mM Tris–Cl, pH 7.5. Labeled and unlabeled AZT-TP were combined at a ratio of 1:10 000 for a stock concentration of 3 mM.
Measurement of PPi release during AZT-TP incorporation
Reactions were performed by manual mixing under single turnover conditions (exact reaction conditions are listed in the main text). The Pol γ/DNA complex was mixed with γ-32P-labeled AZT-TP and Mg2+ to start the reaction. At each time point a 40 μl aliquot was taken and passed through a BioSpin 30 (BioRad, Hercules, CA, USA) gel filtration column by centrifuging for 30 s at 4000 r.p.m. The elution from each time point was spotted onto a PEI cellulose F 20 × 20 cm glass backed TLC plate (EM Science) along with various amounts of γ-32P-labeled AZT-TP to generate a standard curve. Imaging was performed using a phosphorimager, Molecular Dynamics Storm 860, and quantified using ImageQuaNT software (Amersham Biosciences, Uppsala, Sweden).
Data fitting and analysis
The data for the time dependence of substrate depletion or product formation for the various pre-steady-state experiments reported were analyzed by non-linear regression using the software GraFit 5 purchased from Erithacus Software (Horley Surrey, UK). Data were fitted using exponential functions with the general form of the following equation:
| 1 |
where Y is a value corresponding to the concentration or signal observed for the species of interest, C is the offset, Ai is the observed amplitude (Aobs) and ki is the observed rate constant (kobs). In the main text the specific equations used are referred to as single, double or triple exponential equations and correspond to values of i = 1, 2 or 3, respectively.
Data from the single nucleotide incorporation reactions were fitted using single or double exponential equations to extract the observed reaction amplitude(s) and rate(s) and when appropriate the data were fitted using the following hyperbolic functions describing the concentration dependence:
| 2 |
In the above equation, Amax is defined as the maximum amplitude at infinite substrate (S) concentration and Kd is the ‘apparent’ dissociation constant.
It was seen that for incorporation reactions using AZT-TP, the observed rate of reaction decreased hyperbolically as a function of increasing substrate concentration and was therefore fitted using the equation shown below. Also, a direct competition experiment was performed where the rate of incorporation was measured using TTP in the presence of increasing concentrations of AZT-TP and was analyzed similarly.
| 3 |
In the above equation ▵kobs is the predicted overall change in the observed rate over the concentration series, Kd,app is the ‘apparent’ dissociation constant and C is the Y-intercept. The true Kd for AZT-TP binding was obtained according to the following relationship:
| 4 |
The kinetic parameters for TTP incorporation were previously measured under identical conditions (15).
The concentration dependence of the rate of pyrophosphorolysis of TMP from the 3′-terminus of a primer followed a hyperbolic relationship and was analyzed by fitting the data using the following equation:
| 5 |
where, kmax is the predicted rate of pyrophosphorolysis at infinite PPi concentration.
Global data fitting
Pre-steady state and steady-state data were fitted simultaneously using the program KinTek Global Kinetic Explorer (KinTek Corp, Austin, TX, USA). The best fit was obtained using the Levenberg–Marquat method of least-squares fitting based upon numerical integration of the rate equations. Data were normalized to reduce unequal weighting of errors between the two experiments. Error estimates were based upon the covariance matrix.
RESULTS
Kinetics of incorporation
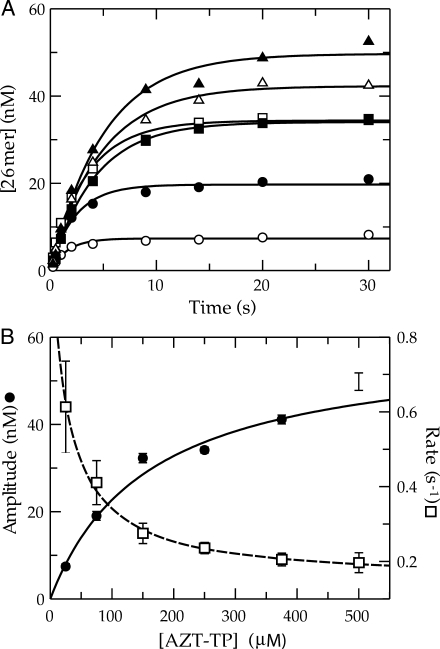
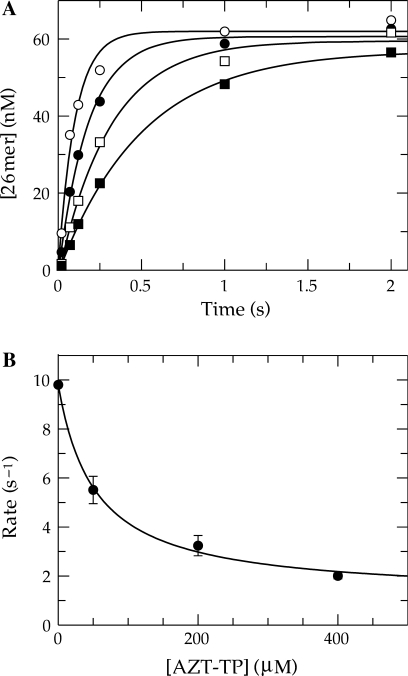
We previously reported that the single nucleotide incorporation of AZT-TP catalyzed by Pol γ under single turnover conditions was complex compared to that of the other NRTIs examined (17). The kinetics indicated that the amount of product formed in a single turnover was a function of the concentration of AZT-TP in solution. This implied that product formation was reversibly linked to substrate binding. Given the importance of AZT in treating HIV infections and the current debate over the site of the observed clinical toxicities, we thought that it was important first to exclude the possibility that the abnormal kinetics were a result of an artifact due to the incomplete or insufficient quenching of enzyme activity upon the addition of 0.5 M EDTA, as was routinely employed. Therefore, we carried out a similar series of reactions, but instead quenched with a final concentration of 0.5 M HCl. The results are shown in Figure 1A. The data appear to be similar to our previous data, in that, the amplitude of product formation increased in a hyperbolic fashion as a function of AZT-TP concentration (Figure 1B). A fit of the data using a hyperbolic function [Equation (2)] defined an apparent Kd for AZT-TP binding of 160 ± 40 μM and a maximum amplitude of 59 ± 7 nM. In addition, we observed a decrease in the observed rate of incorporation over the concentration series, although the rate measurements at the two lower concentrations were less accurate because of the lower amplitudes. The observed rates of incorporation appeared to decrease hyperbolically over the concentration series and were therefore fitted using Equation (3) to obtain an apparent Kd of 25 ± 10 μM and an overall decrease in the observed rate from ∼1.1 to 0.2 s−1 (net change of 0.95 ± 0.23 s−1). This experiment shows that there is no obvious artifact due to using EDTA to quench the reaction and confirms our previous observation that the kinetics are complex and not explained by the conventional model.
Figure 1.
Kinetics of AZT-TP incorporation by Pol γ. (A) Exonuclease-deficient holoenzyme (100 nM) was preincubated with 90 nM 25/45-mer DNA (radiolabeled primer) and then rapidly mixed with Mg2+ and various concentrations of AZT-TP [25 (open circle), 75 (filled circle), 150 (open square), 250 (filled square), 375 (open triangle) and 500 μM (filled triangle)]. Each data set was fitted using a single exponential equation to obtain the observed rate and reaction amplitude. (B) The reaction amplitudes were plotted as a function of AZT-TP concentration (filled circle). The data were fitted by non-linear regression using Equation (2) to yield an apparent Kd of 160 ± 40 μM and maximum amplitude of 59 ± 7 nM. The observed rates were also plotted as a function of AZT-TP concentration (open square) and analyzed according to Equation (3) to yield an apparent Kd of 25 ± 10 μM, an overall predicted decrease in the observed rate of 0.95 ± 0.23 s−1, and a Y-intercept of 1.1 ± 0.2 s−1.
Although the standard pathway of the minimal steps involving nucleotide binding, chemistry and product release is presumably maintained as shown in Scheme 1, the rate constants observed with AZT must be very different. The data suggest that the binding of AZT-TP is reversibly linked to product formation, a conclusion that requires either pyrophosphate is able to rebind to the enzyme and react at a rate comparable to the forward reaction (0.2 s−1), or, alternatively, that rate of pyrophosphate release must be very slow. To explore these possibilities, we first examined the kinetics of the reverse reaction.
Scheme 1.
Minimal model for AZT incorporation.
Kinetics of pyrophosphorolysis
To measure the rate constant for the reverse reaction (pyrophosphorolysis), a primer strand was synthesized with AZT-MP located at the 3′-terminus (26-mer) then purified, radiolabeled and annealed to a complementary template strand (45-mer). Pol γ (100 nM) was incubated with the DNA duplex (90 nM) and then mixed with various concentrations of inorganic pyrophosphate (PPi) and Mg2+ to initiate the reaction. To our surprise, no pyrophosphorolysis reaction was observed. The experiment was repeated using various conditions at concentrations of PPi up to 10 mM with no observable depletion of 26-mer. To determine whether this was an intrinsic property of Pol γ, the same experiment was repeated with a TMP-terminated primer strand. The results were quite different with nearly complete depletion of the 26-mer to lengths as short as 6-mer (data not shown). Unfortunately, this experimental format did not allow the accurate quantification of the reaction kinetics because lengthening of the primer strand was also fairly substantial, due to the production of dNTPs over time.
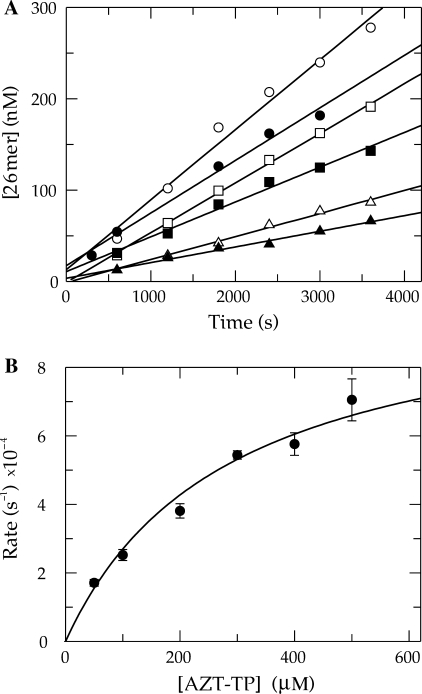
It was clear that the radiolabel must be present in the terminal base rather than the 5′-terminus to make an accurate rate measurement. Therefore, a double mixing experiment was performed where, in the first mixing event, Pol γ/DNA complex (coding for T incorporation) was mixed with α-32P-TTP and allowed to react for 5 s to obtain full incorporation. In the second mixing event this solution was combined with varying concentrations of PPi and 2.5 mM Mg2+, plus 20 μM TTP. The cold TTP was necessary to trap radiolabeled TTP in solution. The results are shown in Figure 2. The time dependence of the disappearance of 26-mer was monophasic and therefore fitted using a single exponential equation to extract the observed rate for each concentration of PPi used. The observed rates of reaction followed a hyperbolic concentration dependence to yield a maximum rate of pyrophosphorolysis of 1.4 ± 0.3 s−1 and a Kd of 240 ± 80 μM. Pol γ was able to catalyze the reverse reaction efficiently, but only using a naturally terminated primer strand. These results were counter to our hypothesis that a substantial reverse rate constant for phosphoryl transfer was the cause of the abnormal kinetics of incorporation using AZT-TP.
Figure 2.
Pyrophosphorolysis of TMP-terminated primer. (A) Exonuclease-deficient holoenzyme (100 nM) was preincubated with 90 nM 25/45-mer DNA (radiolabeled primer) and mixed with 400 nM α-32P-TTP in the presence of 2.5 mM Mg2+. The reaction was aged for 5 s to permit radiolabeled TTP incorporation. A second mixing event then combined this with varying concentrations of PPi [15 (filled circle), 30 (open circle), 75 (filled square), 200 (open square), 400 (filled triangle) and 1000 (open triangle) μM] in solution with 2.5 mM Mg2+ and 20 μM unlabeled TTP. The unlabeled TTP was included to prevent the undesired incorporation of radiolabeled TTP following the pyrophosphorolysis reaction. Shown is the concentration of the 26-mer plotted as a function of time and the data were analyzed by non-linear regression using a single exponential equation to obtain the observed rate constants at the various PPi concentrations. (B) The observed reaction rates were plotted versus the concentration of PPi used and fitted using Equation (5) to yield a maximum rate of pyrophosphorolysis of 1.4 ± 0.3 s−1 and a Kd of 240 ± 80 μM.
Role of free PPi
Some other approaches were taken in an attempt to probe any possible effect of free PPi on the kinetics of the incorporation reaction. First, the incorporation reaction was performed in the presence of yeast inorganic PPase included in the reaction mixture in order to hydrolyze PPi to Pi rapidly. The kinetics of the incorporation reaction using 200 μM AZT-TP were nearly identical regardless of whether or not PPase was included (data not shown). This suggests that any role PPi may have on the abnormal kinetics of the incorporation must occur prior to dissociation from the enzyme. To further test this result, the same incorporation reaction was performed except that PPase was omitted and varying concentrations of PPi were included. There was no obvious difference in the reaction kinetics obtained at concentrations of PPi up to 1 μM. At concentrations greater than 1 μM PPi, the DNA substrate degraded at a rate comparable to that of the incorporation reaction and rendered the data difficult to interpret rigorously. In any event, we would expect a maximum concentration of only 60 nM PPi to build up in solution during the incorporation reaction. Therefore, these experiments clearly showed that free PPi has no effect on the apparently reversible nature of the incorporation reaction.
Hydrolysis of AZT-TP
Unable to observe a role for free PPi on the reaction kinetics it seemed possible that perhaps the concentration-dependent amplitude when using AZT-TP could be explained by hydrolysis rather than pyrophosphorolysis. However, the experiments were performed with exonuclease-deficient Pol γ so hydrolysis by the usual mechanism would not be likely, but may be occurring at the polymerase-active site. If this was taking place, one might expect to see an accumulation of AZT-MP in solution. In order to investigate this, a single-turnover experiment ([E]>[DNA]) was performed using a relatively low concentration of AZT-TP (75 μM). As judged by anion-exchange chromatography of the reaction mixture, no conversion of AZT-TP to either AZT-DP or AZT-MP was observed up to 110 min of incubation time (data not shown).
Direct measurement of PPi release following incorporation using TTP
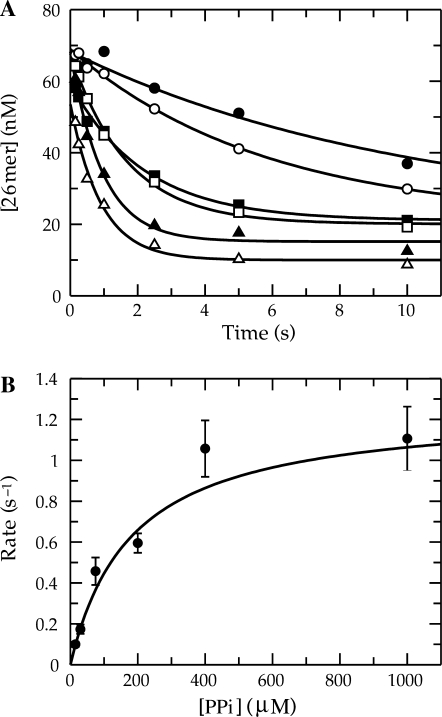
The next obvious question was regarding the kinetics of PPi release following the chemical reaction. It appeared from the previous experiments that free PPi had no influence on the reaction amplitude. The data seemed to be suggesting that the dissociation of PPi was not occurring rapidly following the formation of product. Therefore, it was of interest to measure the rate of PPi release during the forward reaction. An assay was developed using TTP in order to be sure that it was capable of quantifying PPi dissociation accurately. The fundamental component in obtaining a signal was the mutant MDCC-labeled phosphate-binding protein (MDCC-PBP) that was developed for monitoring the release of Pi (24). This protein exhibits a rapid and large fluorescence increase upon binding Pi. However, PPi does not bind and induce a fluorescence change, so in order for this assay to function, yeast inorganic PPase was included. Shown in Figure 3A (smooth line) is the fluorescent signal collected when an incorporation reaction was performed under single turnover conditions with a saturating concentration of TTP. To compare this to the kinetics of the incorporation reaction, an experiment was performed in the chemical quench flow with a radiolabeled primer strand. The results are plotted along with the stopped-flow data in Figure 3A. The quench-flow data overlay the fluorescence trace even though one is measuring PPi release and the other is measuring the formation of DNAn+1. In addition, there is no observable lag phase in the stopped-flow fluorescence data. These results indicate that the rate constant for PPi dissociation must be at least five times greater than the maximum rate of incorporation using TTP (∼25 s−1 at 20 C). It is quite possible that a conformational change of the protein actually limits the rate of PPi dissociation, but this data alone does not provide such evidence, but sets a lower limit on the rate of PPi release of approximately five times the observed rate, 125 s−1.
Figure 3.
Comparison of PPi release following incorporation using TTP or AZT-TP. (A) Exonuclease-deficient holoenzyme (100 nM) was preincubated for 5 min with 2.5 mM Mg2+, 90 nM 25/45-mer DNA, 1.5 μM E. coli PBP mutant labeled at Cys197 with the fluorescent compound MDCC, 100 μM 7-methylguanosine, 0.02 U/ml purine nucleotide phosphorylase and 0.005 U/μl yeast inorganic PPase and then rapidly mixed in the stopped-flow apparatus at 20 ° C with 2.5 mM Mg2+, 50 μM TTP, 100 μM 7-methylguanosine, and 0.02 U/ml purine nucleotide phosphorylase. PPi dissociation was monitored by observing the change in fluorescence that results from Pi binding to the MDCC-labeled PBP (smooth line). Shown also are quench-flow data from a parallel experiment monitoring the DNAn+1 formation using a radiolabeled primer strand (filled circle). (B) Exonuclease-deficient holoenzyme (1 μM) was preincubated with 900 nM 25/45-mer DNA and mixed with 300 μM γ-32P-labeled AZT-TP and 2.5 mM Mg2+ to start the reaction. The amount of PPi (or PPi in equilibrium with AZT-TP) that remained associated with Pol γ as a function of time is shown on the plot. The data were fitted to a double exponential equation to obtain an observed fast phase rate and amplitude of 1.2 ± 0.3 min−1 and 700 ± 70 nM, respectively. The observed rate and amplitude of the slow phase were 0.007 ± 0.002 min−1 and 700 ± 50 nM, respectively.
The overall goal of developing this assay was to measure the rate of PPi dissociation using AZT-TP as the substrate. Therefore, the same experiment was performed with AZT-TP but it failed to produce any clear signal. This provided the first direct evidence that PPi dissociation was not rapid following incorporation. It is important to note that PPi release occurring at a rate comparable to that of the rate of incorporation using AZT-TP (∼0.2 s−1) would have been easily measured under these conditions, but a much slower reaction could not have been measured because of the presence of the phosphate mop included to scavenge traces of phosphate and improve sensitivity (24).
PPi release using AZT-TP as a substrate
To examine the apparently much slower rate of release of PPi following AZT-TP incorporation, a different experimental approach was needed. The experiment required the synthesis of γ-32P-labeled AZT-TP to perform this measurement. We then used rapid size exclusion chromatography to monitor the release of 32P-PPi from the enzyme–DNA complex after incorporation. Pol γ (1 μM) was then incubated with DNA (900 nM) and mixed with γ-32P-AZT-TP (300 μM) to start the reaction. Then as a function of time, samples of the reaction mixture were applied to a small centrifugal size exclusion column in order to remove any free or loosely associated radiolabeled small molecules. As a result, the quantity of radioactivity in the eluant was due to only tightly associated enzyme-bound PPi (E▪DNAn▪AZT-TP plus E▪DNAn+1▪PPi). The concentration of bound label was quantified according to a standard curve to give the results shown in Figure 3B.
There was a transient rise and fall in the concentration of enzyme-bound radioactivity. The rate and amplitude of the fast phase were 1.2 ± 0.3 min−1 and 700 ± 70 nM, respectively, and the rate and amplitude of the slow phase were 0.007 ± 0.002 min−1 and 700 ± 50 nM, respectively. This experiment clearly demonstrates that PPi remains tightly associated with the enzyme on a time scale far greater than that of the incorporation reaction. The absolute amplitude of the radioactivity also shows that less than one radiolabeled molecule associated with Pol γ. From this and previous experiments, it can be concluded that PPi is not released rapidly from the active site following the incorporation reaction in contrast to the reaction using TTP. The outcome of very slow PPi release is that it must allow the chemical reaction to occur in the reverse direction and therefore provides an avenue for enzyme-bound product to come to equilibrium with free substrate.
The observed rate of the rise (∼0.02 s−1) was slower than the observed rate of incorporation (0.2 s−1), but the time resolution of the method was insufficient to resolve the rate of incorporation directly. The rate of the slow decline provides a semi-quantitative estimate of the rate of PPi release of ∼0.0001 s−1.
Estimates of Kd for AZT-TP in competition with TTP
Because the concentration dependence of the kinetics of incorporation of AZT are complex (Figure 1), estimates for the Kd for ground state nucleotide binding of AZT-TP were dependent upon the model used to fit the data. Therefore, we measured the Kd for AZT-TP by competition against TTP in a single nucleotide incorporation. This assay was possible because of the dramatic difference in the rate of incorporation of TTP (kpol ∼ 25 s−1) compared to that for AZT-TP so that AZT-TP binds but is not incorporated on the time scale of a single turnover of incorporation of TTP. The observed rate of TTP incorporation at a concentration of 1 μM (close to the Kd for TTP) was measured in the presence of various concentrations of AZT-TP under single turnover conditions (Figure 4). The observed rate of incorporation decreased hyperbolically as a function of increasing AZT-TP concentration with an apparent Kd of 53 ± 13 μM [Equation (3)]. The Kd for TTP (0.63 μM) was previously measured under these conditions and was used according to the relationship shown in Equation (4) to determine a true Kd of 20 ± 7 μM for AZT-TP. This estimate of the Kd obtained by this method is 8-fold tighter than that predicted previously by analyzing the obvious amplitude change of the single nucleotide incorporation reactions (Figure 1B). However, this estimate appears to mirror the apparent Kd obtained from the hyperbolic fit of the decrease in the observed rate (Figure 1B).
Figure 4.
Determination of the Kd for AZT-TP in competition with TTP. (A) Exonuclease-deficient holoenzyme (100 nM) was preincubated with 90 nM 25/45-mer DNA (radiolabeled primer) and then rapidly mixed with Mg2+, 1 μM TTP and various concentrations of AZT-TP [0 (open circle), 50 (filled circle), 200 (open square) and 400 (filled square) μM]. Each data set was fitted using a single exponential equation to obtain the observed rate of incorporation. (B) The observed rates were plotted as a function of AZT-TP concentration and fitted using Equation (3) to obtain an apparent Kd of 53 ± 13 μM for AZT-TP binding, a Y-intercept of 9.7 ± 0.3 s−1, and an overall decrease in the observed rate of 8.6 ± 0.6 s−1. According to Equation (4), a true Kd of 20 ± 7 μM for AZT-TP in competition with TTP can be defined.
Steady-state kinetics of AZT-TP incorporation
Under normal circumstances, the specificity constant (kcat/Km) for the incorporation of nucleotides is more accurately obtained using transient state kinetic methods, and because DNA release is slower than incorporation conventional, steady-state methods fail to provide kinetic parameters that accurately reflect nucleotide selectivity during processive synthesis. In the present case, however, our data suggest that pyrophosphate release is much slower than DNA release, and therefore, steady-state methods should provide a valid estimate of kcat/Km. In this unusual case, the relevant kinetic parameters could be determined by measuring the steady-state kinetics of single nucleotide incorporation. The time dependence of product (DNAn+1) formation is shown under steady-state conditions with DNA in excess of Pol γ (Figure 5). The data were analyzed by linear regression, and the observed rate was plotted as a function of concentration defining a kcat of 0.001 ± 0.0001 s−1 and a Km of 280 ± 60 μM. It is important to note that the kcat obtained for this incorporation reaction is 20-fold slower than the rate of DNA release (13). The overall rate of catalysis in this case may be partially limited by a conformational change which limits the release of both PPi and DNAn+1. The specificity constant defined by this experiment is almost 300-fold less ((3.6 ± 0.9) × 10−6 μM−1 s−1) than previously reported (17) or computed from the rate of the single turnover divided by the apparent Kd defined by the amplitude dependence (Figure 1).
Figure 5.
Steady-state kinetic parameters of AZT-TP incorporation. (A) Exonuclease-deficient holoenzyme (100 nM) was preincubated with 1.5 μM 25/45-mer DNA (radiolabeled primer) and mixed with Mg2+ and various concentrations of AZT-TP [50 (filled triangle), 100 (open triangle), 200 (filled square), 300 (open square), 400 (filled circle) and 500 (open circle) μM]. The data were fitted by linear regression and the slope divided by the enzyme concentration to obtain the rate of turnover. (B) The rate of turnover was plotted against AZT-TP concentration and the data were analyzed according to the Michaelis–Menton equation to obtain a kcat of 0.001 ± 0.0001 s−1 and a Km of 280 ± 60 μM.
Previous measurement of AZT incorporation by steady-state methods provided an estimate of 1 × 10−4 μM−1 s−1 and a discrimination of 12 700 against AZT-TP relative to TTP (25). Although kcat/Km values reported by Lim et al. are reasonably close to our measurements, they grossly underestimated discrimination by reference to the artificially slow rate of TTP incorporation, limited by DNA release in the steady-state measurements. DNA release does not limit the steady-state rate of AZT-TP incorporation, but it does limit the rate of TTP incorporation.
Kinetics of incorporation of the other 3′-N3-substituted NRTIs
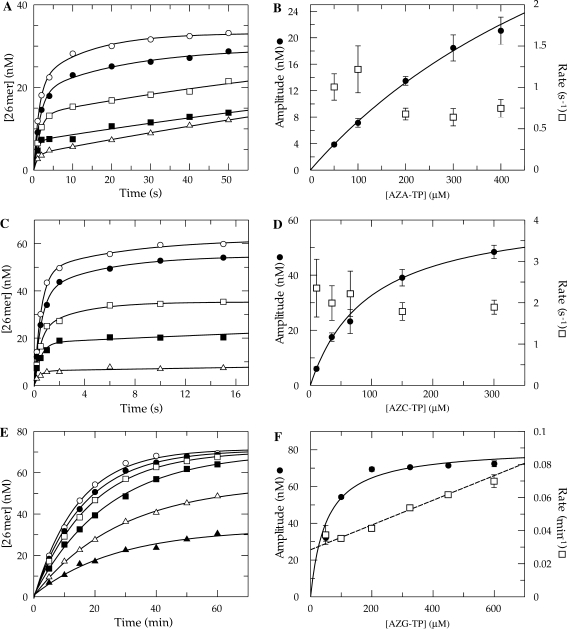
The only structural difference between TTP and AZT-TP is that the normal 3′-hydroxyl has been substituted with an azide group. Therefore, the complex kinetics of incorporation is a direct consequence of the azide substitution. In order to determine whether the effect of the azide is general, the other three nucleoside analogs were examined. The single nucleotide incorporation reactions for AZA-TP (dATP analog), AZC-TP (dCTP analog) and AZG-TP (dGTP analog) were carried out in the same fashion as that detailed for AZT-TP and the data obtained are shown in Figure 6. The kinetics of incorporation strongly resembled that of AZT-TP, even though the data collected with AZA-TP and AZC-TP were biphasic. The amplitude of the fast phase of the reaction for both analogs was clearly dependent on the concentration of substrate in solution, while the rate of the fast phase was not (Figure 6A–D). The incorporation assay using AZG-TP, on the other hand, was much slower and monophasic, although the reaction amplitude still appeared to vary with concentration (Figure 6E and F). One significant difference between AZG-TP and the other three analogs was that the rate clearly increased linearly over the concentration range examined.
Figure 6.
Kinetics of incorporation using azide-substituted analogs. (A) Exonuclease-deficient holoenzyme (100 nM) was preincubated with 90 nM 25/45-mer DNA (radiolabeled primer) and then rapidly mixed with Mg2+ and various concentrations of AZA-TP [50 (open triangle), 100 (filled square), 200 (open square), 300 (filled circle) and 400 μM (open triangle)]. Each data set was fitted using a double exponential equation. (B) The amplitude for fast phase of each reaction was plotted as a function of AZA-TP concentration (filled circle). A fit of the data to Equation (2). yields an apparent Kd of 800 ± 120 μM and a maximum amplitude of 65 ± 8 nM. The observed rate of the fast phase of each reaction was also plotted as a function AZA-TP concentration (open square). (C) Single turnover reaction performed using AZC-TP [10 (open triangle), 35 (filled square), 65 (open square), 150 (filled circle) and 300 μM (open circle)]. (D) The amplitude was plotted as a function of AZC-TP concentration (filled circle). A fit of the data predicts an apparent Kd of 96 ± 6 μM and a maximum amplitude of 64 ± 2 nM. The observed rate of the fast phase of each reaction was also plotted (open square). (E) Single turnover reaction using AZG-TP [50 (filled triangle), 100 (open triangle), 200 (filled square), 325 (open square), 450 (filled circle) and 600 μM (open square)]. Each data set was fitted using a single exponential equation. (F) The amplitude was plotted as a function of AZG-TP concentration (filled circle) to define an apparent Kd of 50 ± 10 μM and a maximum amplitude of 81 ± 3 nM. The observed rate of each reaction was also plotted and fitted by linear regression to obtain a slope of (7.5 ± 0.5) × 10−5 μM−1 min−1and a Y-intercept of 0.028 ± 0.002 min−1 (open square).
DISCUSSION
Several significant observations were taken during the course of this work, but none were more significant than the discovery that the release of PPi is extremely slow following incorporation using AZT-TP. The ramifications of this can be seen in both the single turnover incorporation reactions and in the steady-state kinetic parameters. In the single turnover reactions, the data clearly demonstrate that the equilibrium amount of DNAn+1 achieved during the steady state at the active site is a function of the free AZT-TP concentration. This is in contrast to the correct incorporation reaction using TTP where, apparently, any conformational changes and PPi dissociation are extremely rapid. Clearly, we were not able to directly measure the conformational changes of Pol γ, but the fact that we were not able to force the pyrophosphorolysis reaction to take place by adding a relatively high concentration of PPi to the Pol γ/DNA complex (primer terminated with AZT-MP) suggests that when AZT-MP is bound, Pol γ is unable to productively associate with PPi from solution. In contrast, the reverse reaction using a naturally terminated DNA substrate was readily catalyzed upon addition of PPi to solution.
Here we show that the rate of PPi release after incorporation of TTP must be quite fast because the two reactions are coincident (Figure 3A). Similarly, we presume that translocation is also fast and must follow rapidly after the release of PPi, but this conclusion is based solely on our inability to show a delay in the ability of the enzyme to bind the next nucleotide after a single incorporation event (20,26,27). Our assays reported here provide the first direct evidence that PPi release is fast after incorporation of a normal nucleotide. They do not bear on the question of translocation. However, because translocation is operationally defined by the ability to bind the next nucleotide, it must follow PPi release. In addition, we presume that Pol γ, like better characterized Pol A family polymerases (28), undergoes a change in structure to a ‘closed’ state after nucleotide binding. It is therefore likely, that a conformational change to return to the ‘open’ state after incorporation limits the rates of PPi release and translocation.
The most remarkable observation reported here is the exceedingly slow release of PPi after incorporation of AZT. Because this rate is even slower than the rate of DNA release (0.02 s−1), it is reasonable to suppose that the enzyme remains in the ‘closed’ state with the products of reaction tightly bound. Maintenance of the closed state would then limit the release of PPi and DNA. Homology modeling has suggested that an active site lysine residue may sterically clash with the azido group to limit binding (29). However, we must await solution of the crystal structure of Pol γ with AZT bound to assess whether this suggestion is true, and how we might then understand the slower PPi release rates.
The effect of slow PPi release on the efficiency of AZT-TP incorporation by Pol γ is quite large. The commitment to forward catalysis is not made until PPi dissociates from the enzyme, predicting that the specificity constant for incorporation should be correspondingly low. An unambiguous measurement of kcat/Km found this to be the case and dictates that our previous calculation of the discrimination ((kcat/Km)TTP/(kcat/Km)AZT-TP) against AZT-TP be revised to (1 ± 0.5) × 107, almost 300-fold greater than previously estimated. Interestingly, the discrimination against AZT-TP is over an order of magnitude greater than that of (–)-cis-2-amino-1,9-dihydro-9-(4-hydroxymethyl)-2-cyclopenten-1-yl)-6H-purine-6-one triphosphate (CBV-TP) which was previously suggested to be the least toxic FDA approved NRTI towards mitochondrial genome replication (17,18). Therefore, we must conclude that AZT in the triphosphate form is predicted to have little to no effect on the overall rate of mitochondrial genome replication. In agreement with these studies, recent observations suggest that AZT may be a potent inhibitor of thymidine kinase and that depletion of mitochondrial DNA is actually due to much lower than normal levels of TTP (30). Of course, extremely low levels of TTP may tend to provide an unusually high probability of incorporation compared to the other NRTIs simply due to the lack of a direct competition between TTP and AZT-TP.
To get the best estimates for the kinetic parameters governing incorporation of AZT-TP, we globally fit the data from the pre-steady state and steady-state experiments (Figures 1A and 5A, respectively) by non-linear regression based upon numerical integration of the rate equations. The results of this global analysis in fitting the data to Scheme 1 are shown in Table 1. This simple model and set of rate constants adequately accounts for most, but not all of the data. Namely, it fails to account for the 12-fold tighter binding in competition with TTP (Kd = 20 μM, Figure 2) and the apparent decrease in the rate of incorporation as a function of increasing nucleotide concentration (Figure 1B). These data could be accounted for if we proposed a more complex model, such as by including isomerization of the enzyme-bound DNA between productive and non-productive states, or by proposing the binding of a second AZT-TP molecule. The simple model adequately accounts for the observed dependence of the amplitude of the burst on AZT-TP concentration but not the apparent decrease in rate (Figure 1B). However, there are large errors in the data suggesting a decrease in rate as a function of AZT-TP concentration, thereby weakening arguments in support of a more complex model. In the absence of additional data to test either of our more complex models, we settled on the minimal model (Scheme 1) as sufficient to account for the data without introducing additional undefined steps in order to achieve an incremental improvement in the quality of the fit. Further experimentation is required to look for additional AZT-binding sites or multiple states of bound DNA.
Table 1.
Globally fit kinetic parameters for AZT incorporation
| Kd (μM) | k2 (s−1) | k-2 (s−1) | k3 (s−1) |
|---|---|---|---|
| 230°±°25 | 0.38°±°0.04 | 0.22 °±°0.02 | 0.0009°±°0.00004 |
The fact that there is an equilibrium formed between free AZT-TP and the product, DNAn+1, affords the polymerase with a novel type of proofreading mechanism that, simply stated, is catalysis of the reverse reaction resulting from the slow rate of pyrophosphate release. It is reasonable to suppose that a conformational change following chemistry in the forward direction may limit PPi dissociation, thus allowing time for the reverse reaction to occur.
Previous data on the incorporation of 8-oxo-dGTP, a naturally occurring, oxidatively damaged and highly mutagenic nucleotide, displayed similar kinetic behavior (22). We proposed that Pol γ may have evolved a novel pathway to reduce incorporation of 8-oxo-dG to supplement to the effect of exonuclease proofreading activity. This mechanism would attenuate the toxic or mutagenic properties of alternative substrates that may not be as efficiently removed by the exonuclease domain compared to a typical mismatch. For reasons we do not yet understand, azido-nucleotides follow the same pathway that may have originally evolved to minimize the incorporation of 8-oxo-dG or other common damaged and mutagenic nucleotides present in the mitochondria.
ACKNOWLEDGEMENTS
Supported by National Institutes of Health (GM 044613) and the Welch Foundation (F-1604). Funding to pay the Open Access publication charges for this article was provided by The Welch Foundation.
Conflict of interest statement: K.A. Johnson is President of KinTek Corporation, the manufacturer of the stopped-flow and rapid quench-flow instruments used in this study.
REFERENCES
- 1.Vandamme AM, Van Vaerenbergh K, De Clercq E. Anti-human immunodeficiency virus drug combination strategies. Antivir. Chem. Chemother. 1998;9:187–203. doi: 10.1177/095632029800900301. [DOI] [PubMed] [Google Scholar]
- 2.Squires KE. An introduction to nucleoside and nucleotide analogues. Antivir. Ther. 2001;6(Suppl. 3):1–14. [PubMed] [Google Scholar]
- 3.Meyer PR, Matsuura SE, Tolun AA, Pfeifer I, So AG, Mellors JW, Scott WA. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 2002;46:1540–1545. doi: 10.1128/AAC.46.5.1540-1545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray AS, Murakami E, Basavapathruni A, Vaccaro JA, Ulrich D, Chu CK, Schinazi RF, Anderson KS. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry. 2003;42:8831–8841. doi: 10.1021/bi034435l. [DOI] [PubMed] [Google Scholar]
- 5.Cammack N, Rouse P, Marr CL, Reid PJ, Boehme RE, Coates JA, Penn CR, Cameron JM. Cellular metabolism of (-) enantiomeric 2′-deoxy-3′-thiacytidine. Biochem. Pharmacol. 1992;43:2059–2064. doi: 10.1016/0006-2952(92)90162-c. [DOI] [PubMed] [Google Scholar]
- 6.Chang CN, Doong SL, Zhou JH, Beach JW, Jeong LS, Chu CK, Tsai CH, Cheng YC, Liotta D, et al. Deoxycytidine deaminase-resistant stereoisomer is the active form of (+/−)-2′,3′-dideoxy-3′-thiacytidine in the inhibition of hepatitis B virus replication. J. Biol. Chem. 1992;267:13938–13942. [PubMed] [Google Scholar]
- 7.Chang CN, Skalski V, Zhou JH, Cheng YC. Biochemical pharmacology of (+)- and (−)-2′,3′-dideoxy-3′-thiacytidine as anti-hepatitis B virus agents. J. Biol. Chem. 1992;267:22414–22420. [PubMed] [Google Scholar]
- 8.Gray NM, Marr CL, Penn CR, Cameron JM, Bethell RC. The intracellular phosphorylation of (–)-2′-deoxy-3′-thiacytidine (3TC) and the incorporation of 3TC 5′-monophosphate into DNA by HIV-1 reverse transcriptase and human DNA polymerase gamma. Biochem. Pharmacol. 1995;50:1043–1051. doi: 10.1016/0006-2952(95)96620-a. [DOI] [PubMed] [Google Scholar]
- 9.Hart GJ, Orr DC, Penn CR, Figueiredo HT, Gray NM, Boehme RE, Cameron JM. Effects of (–)-2′-deoxy-3′-thiacytidine (3TC) 5′-triphosphate on human immunodeficiency virus reverse transcriptase and mammalian DNA polymerases alpha, beta, and gamma. Antimicrob. Agents Chemother. 1992;36:1688–1694. doi: 10.1128/aac.36.8.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schinazi RF, Chu CK, Peck A, McMillan A, Mathis R, Cannon D, Jeong LS, Beach JW, Choi WB, et al. Activities of the four optical isomers of 2′,3′-dideoxy-3′-thiacytidine (BCH-189) against human immunodeficiency virus type 1 in human lymphocytes. Antimicrob. Agents Chemother. 1992;36:672–676. doi: 10.1128/aac.36.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalski V, Chang CN, Dutschman G, Cheng YC. The biochemical basis for the differential anti-human immunodeficiency virus activity of two cis enantiomers of 2′,3′-dideoxy-3′-thiacytidine. J. Biol. Chem. 1993;268:23234–23238. [PubMed] [Google Scholar]
- 12.Graves SW, Johnson AA, Johnson KA. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry. 1998;37:6050–6058. doi: 10.1021/bi972685u. [DOI] [PubMed] [Google Scholar]
- 13.Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39:1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- 14.Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- 15.Johnson AA, Johnson KA. Fidelity of nucleotide incorporation by human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:38090–38096. doi: 10.1074/jbc.M106045200. [DOI] [PubMed] [Google Scholar]
- 16.Lee HR, Johnson KA. Fidelity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36236–36240. doi: 10.1074/jbc.M607964200. [DOI] [PubMed] [Google Scholar]
- 17.Johnson AA, Ray AS, Hanes J, Suo Z, Colacino JM, Anderson KS, Johnson KA. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:40847–40857. doi: 10.1074/jbc.M106743200. [DOI] [PubMed] [Google Scholar]
- 18.Lee H, Hanes J, Johnson KA. Toxicity of nucleoside analogues used to treat AIDS and the selectivity of the mitochondrial DNA polymerase. Biochemistry. 2003;42:14711–14719. doi: 10.1021/bi035596s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis W, Copeland WC, Day BJ. Mitochondrial DNA depletion, oxidative stress, and mutation: mechanisms of dysfunction from nucleoside reverse transcriptase inhibitors. Lab. Invest. 2001;81:777–790. doi: 10.1038/labinvest.3780288. [DOI] [PubMed] [Google Scholar]
- 20.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 21.Feng JY, Murakami E, Zorca SM, Johnson AA, Johnson KA, Schinazi RF, Furman PA, Anderson KS. Relationship between antiviral activity and host toxicity: comparison of the incorporation efficiencies of 2′,3′-dideoxy-5-fluoro-3′-thiacytidine-triphosphate analogs by human immunodeficiency virus type 1 reverse transcriptase and human mitochondrial DNA polymerase. Antimicrob. Agents Chemother. 2004;48:1300–1306. doi: 10.1128/AAC.48.4.1300-1306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 23.Wang LZ, Kenyon GL, Johnson KA. Novel mechanism of inhibition of HIV-1 reverse transcriptase by a new non-nucleoside analog, KM-1. J. Biol. Chem. 2004;279:38424–38432. doi: 10.1074/jbc.M406241200. [DOI] [PubMed] [Google Scholar]
- 24.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 25.Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase gamma. J. Biol. Chem. 2001;276:23616–23623. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- 26.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 27.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 28.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis W, Kohler JJ, Hosseini SH, Haase CP, Copeland WC, Bienstock RJ, Ludaway T, McNaught J, Russ R, et al. Antiretroviral nucleosides, deoxynucleotide carrier and mitochondrial DNA: evidence supporting the DNA pol gamma hypothesis. AIDS. 2006;20:675–684. doi: 10.1097/01.aids.0000216367.23325.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susan-Resiga D, Bentley AT, Lynx MD, LaClair DD, McKee EE. Zidovudine inhibits thymidine phosphorylation in the isolated perfused rat heart. Antimicrob. Agents Chemother. 2007;51:1142–1149. doi: 10.1128/AAC.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]