Abstract
A screen for imprinted genes on mouse Chromosome 7 recently identified Inpp5f_v2, a paternally expressed retrogene lying within an intron of Inpp5f. Here, we identify a novel paternally expressed variant of the Inpp5f gene (Inpp5f_v3) that shows a number of unusual features. Inpp5f_v3 initiates from a CpG-rich repeat region adjoining two B1 elements, despite previous reports that SINEs are generally excluded from imprinted promoters. Accordingly, we find that the Inpp5f_v3 promoter acquires methylation around the time of implantation, when many repeat families undergo de novo epigenetic silencing. Methylation is then lost specifically on the paternally derived allele during the latter stages of embryonic development, resulting in imprinted transcriptional activation on the demethylated allele. Methylation analyses in embryos lacking maternal methylation imprints suggest that the primary imprinting mark resides within an intronic CpG island ∼1 kb downstream of the Inpp5f_v3 transcriptional start site. These data support the hypothesis that SINEs can influence gene expression by attracting de novo methylation during development, a property likely to explain their exclusion from other imprinted promoters.
INTRODUCTION
Genomic imprinting in mammals results in a small number of genes being silenced on one allele, depending on the gender of the parent from which the allele is inherited. Imprinted genes typically occur in clusters, in regions of the genome containing allele-specific DNA methylation marks which are established in the germ line. These germ-line methylation imprints can subsequently act after fertilization to establish allele-specific epigenetic asymmetry at the promoters of adjacent genes (1,2).
Mutations in members of the de novo methyltransferase gene family lead to disruptions in imprinted gene expression and to retrotransposon animation (3,4), suggesting that the two processes are controlled by a common mechanism (5). Dnmt3l encodes a regulatory protein that stimulates de novo methylation by Dnmt3a and Dnmt3b, but lacks the catalytic motifs necessary for methyltransferase activity. Male mice lacking functional copies of the Dnmt3l gene are sterile due to meiotic arrest, which is associated with the upregulation of endogenous retrotransposons (3). Females carrying null mutations in the Dnmt3l gene fail to establish imprinted methylation marks during oogenesis, but show no obvious effects on retrotransposon activity (6).
There is evidence to suggest that at least five known imprinted domains originated from retrotransposition events that generated novel genes during mammalian evolution. The imprinted Peg10 gene is essential for placental development (7), and is derived from a sushi-ichi-type LTR retrotransposon that originated in a common ancestor of placental mammals (8,9). We recently reported the identification of a group of four retrotransposed gene copies (retrogenes), each of which originated from X-linked source genes and generated elements that form germ-line differentially methylated regions (gDMRs) in the mouse genome (10). Parent-of-origin effects on DNA methylation have also been extensively studied at the agouti locus, where stochastic methylation at an IAP insertion generates coat colour variegation (11).
Two independent studies have reported that short interspersed nuclear element (SINE) repeats are significantly underrepresented at imprinted promoters (12,13). It has been suggested that this may reflect selection against insertions within imprinted regions due to the ability of SINEs to disrupt local methylation patterns. Imprinted genes would be particularly susceptible to this epigenetic disruption due to their dependence on allele-specific CpG methylation for normal expression levels (12). However, detailed developmental methylation analyses in the region of individual SINE repeats have not been reported.
Here, we identify a novel imprinted transcript variant of the murine Inpp5f gene (Inpp5f_v3) which initiates from a CpG-rich region adjacent to inverted B1 SINE repeats. The promoter is situated 1 kb upstream from an imprinted and retrotransposed copy of the X-linked gene, Tmem114a. Both imprinted transcripts are expressed only from paternally derived alleles. A developmental analysis of CpG methylation at the two imprinted promoters suggests that the CpG island associated with the X-derived retrogene carries the primary imprint mark at the locus. The upstream repeat region undergoes de novo methylation at around the time of implantation, before undergoing demethylation specifically on the paternal allele during the latter stages of embryonic development. Allele-specific demethylation is a common feature of genomic imprinting in plants (14), but has not, to our knowledge, previously been reported in mammals.
MATERIALS AND METHODS
Tissue sources
To obtain oocytes and morulae, 4-week-old C57BL/6J (B6) females were superovulated using procedures described in (15). For oocyte collection, ovaries were dissected and cumulus clumps were flushed into hyaluronidase solution (300 μg/ml; Sigma Cat No. H4272) and left for 10 min at room temperature. To remove cumulus cells, oocytes were passed through a series of 100 μl drops of PBS in a glass Petri dish using a fine bore Pasteur pipette, and then collected in aliquots of 50 in 10 µl TE buffer. Morulae were obtained from females at embryonic day (E)3.5, and collected in aliquots of eight. Over 90% were at the 16 cell stage. E8.5 embryos were collected in pools of five for RNA purification, and individually for the isolation of genomic DNA. Embryos and tissues collected at later stages were not pooled.
The Dnmt3l mutant allele was generated on a 129/SvJ background. E8.5 embryos were obtained from crosses of Dnmt3l homozygous null mothers and wild-type (wt) cast fathers as previously described (6). The Dnmt1 mutant allele was generated on a 129/SvJ background, and homozygous mutant, heterozygous mutant and wt E8.5 embryos were generated as described previously (16). Neonatal brains from mice carrying uniparental duplications for Chromosome 7 [proximal to the T65H breakpoint; (17)] were generated as described (Beechey et al., 2004). All tissues were flash-frozen in liquid nitrogen and stored at –80°C.
Expression studies
RNA was prepared using the RNAeasy kit (Qiagen) according to the manufacturer's standard protocol, and then quantified using an Agilent 2100 Bioanalyser. cDNA was prepared from 1 μg of total RNA using the superscript II cDNA synthesis kit (Invitrogen). RT-PCR was conducted using Reddymix PCR mix (Abgene), under the following cycling conditions: 95°C for 5 min, followed by n cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min. For the amplification of Inpp5f, Inpp5f_v2, Bag3 and 111007A13Rik, n = 32 cycles. For Inpp5f_v3, n = 33 cycles. In every case, control reactions were performed using cDNA prepared in the absence of reverse transcriptase, to eliminate the possibility of genomic DNA contamination. RT-PCR primer sequences are listed in Table S1. For the allele-specific RT-PCR sequencing assays, PCR products were purified using the ExoSapIT reagent (USB) and sequenced using standard ABI sequencing technology (Bigdye v3.1, Applied Biosystems). RACE (rapid amplification of cDNA ends)-ready cDNA was generated using the SMART RACE cDNA kit (Clontech Cat. No. 634914) according to the manufacturer's standard protocol, using 1 μg total RNA per reaction. Touchdown PCR was performed using reagents supplied with the Advantage 2 PCR kit (Clontech Cat. No. 639207) according to the manufacturer's instructions.
Bisulphite mutagenesis
One microgram of genomic DNA in 30 µl H2O was denatured by adding 5.3 µl 2 M NaOH (0.3 M final concentration) followed by incubation at 37°C for 15 min. Sodium metabisulphite measuring 2.1 g was dissolved in 2.5 ml of H2O, 720 µl of 2 M NaOH and 350 µl of 0.1 M hydroquinone prior to heating at 37°C for 20 min. Four hundred microlitres of this solution was added to the denatured DNA and the mixture was overlaid with 100 µl light mineral oil. Tubes were incubated in the dark for 5 h at 55°C and mineral oil was removed by serial transfer through eppendorf tubes. DNA was purified using the QiaEX II kit (Qiagen Cat No. 20021) and care was taken to shield the bisulphite-treated DNA from the light. DNA was recovered in 20 µl H2O, and desulphonated by the addition of 3.5 µl NaOH (0.3 M final concentration) followed by incubation at 37°C for 15 min. DNA was recovered by ethanol precipitation and re-suspended in 50 µl H2O.
Due to the low quantity of material available, aliquots of oocytes (50) and morulae (8) were converted using the agarose bead technique (18,19). Two percent LMP agarose was melted in a 65°C water bath, and added to the oocytes/embryos at a final concentration of 1.6%. The tubes were flicked to mix the contents and spun briefly in a benchtop centrifuge. Tubes were placed on ice for 5 min to solidify the agarose, which was overlaid with 100 µl of cold mineral oil. Four hundred microlitres of this solution (10 mM Tris–HCl pH 7.6, 10 mM EDTA, 1% SDS) was added, and 4 µl proteinase K (100 mg/ml, Sigma), prior to incubation for 15 h at 50°C. Tubes were placed on ice to resolidify the agarose and then the lysis buffer and mineral oil were removed. Agarose beads were washed for 3 × 15 min in TE buffer, 2 × 15 min in 0.3 M NaOH then finally 1 × 15 min in 0.1 M NaOH. Sodium metabisulphite/hydroquinone solution was prepared as described above and added to the agarose beads, and the mixture was overlaid with mineral oil. After incubation in the dark at 55°C for 5 h, solutions were removed and the beads were washed for 5 × 15 min in TE buffer. After the last wash, DNA was desulphonated by adding 500 µl 0.2 M NaOH and incubating at 37°C for 15 min. NaOH was then neutralized by adding 100 µl 1 M HCl and leaving to stand for 5 min. Beads were finally washed for 2 × 10 min in H2O and all liquid was removed. The dry beads were heated at 70°C for 5 min and flicked gently to mix, then at 80°C for a further 5 min or until the agarose had melted completely. Agarose from each bead was split into two separate tubes, and used directly as a template for the subsequent PCR step. Regions 1 and 2 were amplified in nested PCR reactions, in which 2 μl of the first round reaction were carried forward to the second round. Cycling conditions were identical for each round of amplification: 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min. Bisulphite PCR primers were designed to be specific for the same strand from which transcription occurs, and are listed in Table S2.
DNA sequence analysis
CpG islands were defined according to the criteria proposed by Gardiner-Garden and Frommer (20) :>200bp, > 50% C + G and an observed/expected ratio of CpG dinucleotides of > 0.6 when compared to the total proportion of G + C in the same DNA segment. All repeat elements were identified using the repeatmasker tool (http://www.repeatmasker.org/), which is integrated into the UCSC genome browser (http://genome.ucsc.edu/). The region of X/autosome homology was determined using the self-chain track (Kent et al., 2003) on the UCSC genome browser (http://genome.ucsc.edu/), which is currently available only for mouse genome build v34.
All sequence files generated by the ABI 3730xl machine were analysed using the Sequencher software package (GeneCodes). For allele-specific RT-PCR sequencing assays, the relative height of the sequence trace corresponding to each parental allele of a SNP gives an indication of the relative contribution of maternally and paternally derived alleles to the total transcript pool. For the analysis of bisulphite PCR data, strands with thymine nucleotides at positions occupied by cytosine in unconverted DNA were presumed to have been unmethylated prior to bisulphite conversion. Cytosines in CpG dinucleotides that were unconverted following treatment were presumed to have originally been methylated. Where F1 hybrid DNA was used, the parental origin of each strand was determined from a SNP within the PCR product.
RESULTS
The Inpp5f transcription unit
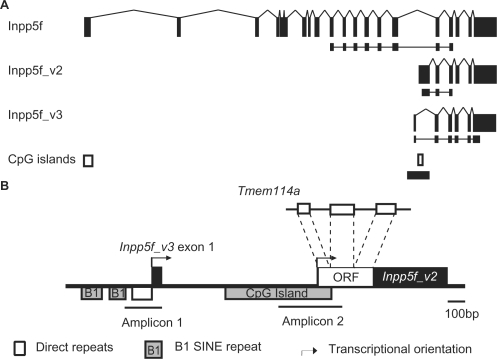
Recent evidence suggests that a number of mammalian imprinted domains were formed by the retrotransposition of X-linked genes into autosomal regions, generating sequences that undergo methylation in the maternal germ line (9,10). In order to gain insights into the regulation of imprinting at this class of imprinted domain, we chose to study the murine Inpp5f_v2 locus, searching for additional imprinted transcripts and putative regulatory elements. Inpp5f_v2 is a paternally expressed transcript variant of the murine Inpp5f gene and is transcribed at high levels in the developing nervous system (21). The unique first exon of the imprinted isoform contains a retrotransposed ORF originating from the X-linked Tmem114a gene, which is spliced onto five downstream exons of the non-imprinted Inpp5f gene (Figure 1A and B). A CpG island overlapping the retrotransposed sequence undergoes sexually dimorphic patterns of DNA methylation during gametogenesis, with maternal allele-specific methylation established during oogenesis (10). EST evidence suggests the presence of an additional promoter ∼1 kb upstream of Inpp5f_v2, generating a transcript variant of Inpp5f that has not previously been described (Inpp5f_v3). RT-PCR assays demonstrate that Inpp5f_v3 contains a unique first exon which, like the first exon of Inpp5f_v2, is spliced onto the five downstream exons of Inpp5f (Figure 1A). The unique first exon of Inpp5f_v3 lacks putative translational start codons. However, a number of in-frame start codons occur within the five exons that are shared with Inpp5f, potentially giving rise to C-terminal Inpp5f polypeptides.
Figure 1.
Genomic and transcriptional organization at the Inpp5f locus. (A) Exonic structure of Inpp5f, Inpp5f_v2 and Inpp5f_v3. Exons are black rectangles and splice patterns are indicated. The RT-PCR products for the allele-specific assays in Figure 2 are indicated below each transcript. The positions of two CpG islands are shown at the bottom, and the region enlarged in panel B is indicated by a thick horizontal line. (B) Genomic features within Inpp5f intron fifteen. The ORF within the first exon of Inpp5f_v2 is a retrotransposed duplicate of the X-linked Tmem114a gene. The two internal promoters are represented by arrows. The region upstream of Inpp5f_v3 is rich in repeats, which are examined further in Figure 3. Amplicon 1 and Amplicon 2 denote the regions within which DNA methylation is examined in Figure 4.
Allele-specific expression of Inpp5f isoforms
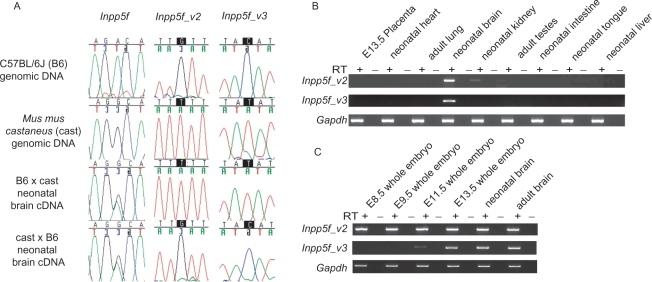
To assess the allele-specific expression of Inpp5f_v3 transcripts, RT-PCR sequencing assays were performed in neonatal brain cDNA from F1 hybrids of the C57BL/6J and Mus musculus castaneus sub-species. Like Inpp5f_v2 (21), Inpp5f_v3 is expressed exclusively from the paternally derived allele (Figure 2A). These findings were confirmed by the lack of Inpp5f_v3 expression in neonatal brains from mice carrying maternally inherited uniparental duplications for the proximal portion of Chromosome 7 (Figure S1). The 20 exon Inpp5f transcript is expressed from both parental alleles, as previously reported (21).
Figure 2.
(A) Allele-specific expression of Inpp5f transcript variants, assessed by RT-PCR sequencing assays. PCR primers were designed to specifically amplify each transcript (Figure 1A) over regions containing SNPs between two inbred strains of mice: C57BL/6J (B6) and Mus mus castaneus (cast). Allele-specific expression can be determined in F1 hybrids by the relative height of the sequence trace corresponding to each parental allele at the polymorphic site. The strain of the mother is given first in each cross. (B). Tissue-specific expression of Inpp5f_v2 transcript variants, assessed by qualitative RT-PCR. All primers were designed to span introns (Figure 1A), and each reaction was conducted using cDNA prepared in the presence (RT+) or absence (RT–) of reverse transcriptase, to eliminate the possibility of template contamination. cDNA was prepared from 1 μg of RNA extracted from B6 × cast F1 hybrid tissues. Adult material was prepared from animals sacrificed at 6 weeks. In every case, Inpp5f_v2 and Inpp5f_v3 underwent 33 cycles of amplification, whereas Gapdh was amplified for 30 cycles. (C) Developmental expression screen for Inpp5f_v2 and Inpp5f_v3 transcript variants under the same experimental conditions as panel B.
The two genes flanking Inpp5f in the 5′ (Bag3) and 3′ (1110007A13Rik) direction were examined for evidence of imprinting. In each case, both parental alleles were expressed at approximately equal levels in cDNA from neonatal brain, E13.5 whole embryo and E13.5 placenta (Figure S2). These data suggest that imprinting at this locus is limited to within the Inpp5f transcriptional unit, but cannot exclude the imprinting of adjacent genes in a highly tissue-specific manner.
RT-PCR expression screens were conducted to determine the tissue-specific expression profile of Inpp5f_v3. It has previously been shown that Inpp5f_v2 is expressed primarily in neural tissues (21). RT-PCR assays confirmed this finding, with expression also detected in adult testes and kidney (Figure 2B). Inpp5f_v3 transcripts were detected only in brain (Figure 2B). Further RT-PCR analyses were conducted in cDNA from whole embryo and brain tissues at various developmental stages. Inpp5f_v2 transcripts were detected in every cDNA sample assessed, indicating that expression initiates prior to E8.5 (Figure 2C). No Inpp5f_v3 transcripts were detected at E8.5, but expression was first detected at E11.5 and continued into adulthood (Figure 2C).
The Inpp5f_v3 promoter
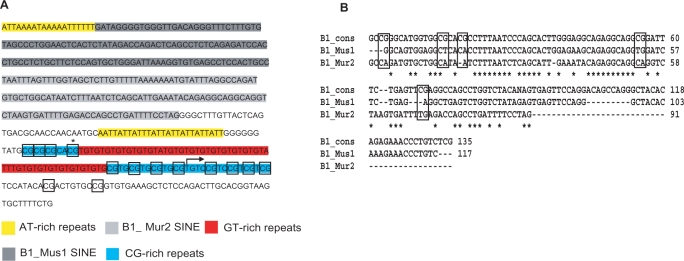
EST evidence suggests that Inpp5f_v3 initiates from within a CpG-rich simple repeat ∼150 bp downstream of two B1 SINEs (Figure 3A). Attempts to identify the transcriptional start site by 5' RACE were unsuccessful, likely due to the CpG- and repeat-rich nature of the 5′ region (Figure 3A). RT-PCR using a forward primer upstream of the GT simple repeats but downstream of the B1 SINEs fails to generate a product, whereas products are readily amplified when the forward primer overlaps the 3′ terminus of the CG repeats (data not shown). This suggests that Inpp5f_v3 initiates from within the simple repeat region.
Figure 3.
(A) Sequence of the region upstream of and including Inpp5f_v3 exon one in the B6 genome. Five distinct classes of repeat, as defined by RepeatMasker (www.repeatmasker.org), are shown. DNA methylation at individual CpG dinucleotides (boxed) is assessed in Figure 4. The asterisk denotes a CpG that is polymorphic between B6 and cast, and was thus excluded from methylation analyses. The furthest 5′ point to which ESTs corresponding to Inpp5f_v3 extend is indicated by an arrow. (B) Clustalw alignment showing sequence homology between the inverted B1 SINE repeats upstream of Inpp5f_v3 exon 1 (B1_Mus1 and B1_Mur2) and the consensus sequence for the B1 family [B1_cons; (22)]. CpG dinucleotides in the B1 consensus sequence are boxed. Note that all five consensus CpGs have been lost in the Mus1 and Mur2 sequences shown.
The SINEs belong to the Mus1 and Mur2 clades of the B1 family, and consist of 117 and 91 bp, respectively. The full-length B1 consensus sequence defined in (22) consists of 135 bp (Figure 3B). The Mus1 repeat shows 10.3% divergence from the consensus sequence annotated by repeatmasker (www.repeatmasker.org), whereas the Mur2 element shows 27.5% divergence. Full-length B1 elements consist of ∼135 bp, making it unlikely that either of the repeats upstream of Inpp5f_v3 are replication competent. Within the aligning region, the Mus1 and Mur2 elements exhibit 68% nucleotide homology with one another. The elements are arranged in an inverted 3′–5′:5′–3′ orientation with respect to Inpp5f_v3. Although several CpG dinucleotides are present in the B1 consensus sequence (Figure 3B), all have been lost in the Mus1 and Mur2 sequences under discussion, presumably due to the spontaneous deamination of methylated cytosines to thymine nucleotides.
DNA methylation at the Inpp5f_v2 and Inpp5f_v3 promoters
Both imprinted promoters are associated with clusters of CpG dinucleotides, for which the methylation profile was assessed by the sequencing of bisulphite-modified genomic DNA from C57BL/6J germ cells or F1 inter-subspecific hybrid embryos. Maternal germ-line methylation at the Inpp5f_v2 promoter has been reported previously (10), and the Inpp5f_v3 promoter is also methylated in oocytes but unmethylated in sperm (Figure 4A). Allele-specific methylation is maintained at both regions at the 16 cell morula stage (Figure 4B). By E8.5, the Inpp5f_v3 promoter has undergone de novo methylation on the paternally derived allele, and so allele-specific methylation differences are lost (Figure 4C). In contrast, the CpG island overlapping the retrotransposed ORF maintains maternal allele-specific methylation at this stage and throughout somatic development [Figure 4C and D, and (21)], suggesting that the primary imprinting mark lies within this region. In genomic DNA extracted from E13.5 head, the paternally derived Inpp5f_v3 promoter has undergone loss of methylation relative to E8.5 (Fisher's Exact P < 0.01; Figure 4D). No significant differences in methylation levels are observed on maternally derived alleles between E8.5 and E13.5 (Fisher's Exact P = 0.76), which are hypermethylated at both stages (Figure 4C and D). The allele-specific demethylation at CpG's surrounding the Inpp5f_v3 promoter shows a temporal correlation with the onset of expression (Figure 2C).
Figure 4.
Allele-specific CpG methylation at the two imprinted promoter regions shown in Figure 1B. At embryonic stages, the parental origin of each strand was determined on the basis of SNPs between the B6 and cast alleles. Germ cell material was from purebred B6 adults. Horizontal lines represent individual strands of DNA and circles depict CpG dinucleotides. Closed circles are methylated CpGs, open circles are unmethylated. Strands derived from the same PCR amplification are connected by brace symbols to the right of the figure. The germ cell data for Inpp5f_v2 have been published previously (10). (A) Gametes, (B) 16 cell morulae, (C) E8.5 whole embryo and (D) E13.5 head.
Tissue-specific methylation differences at the Inpp5f_v3 promoter
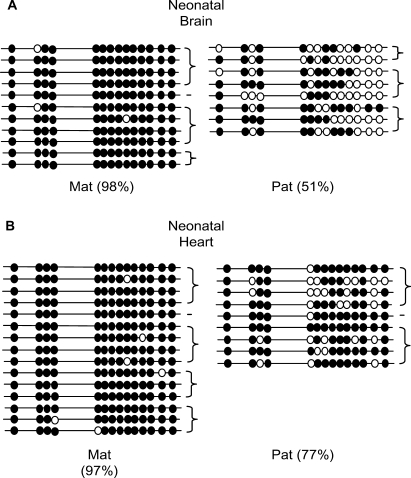
Inpp5f_v3 is expressed in the brain, but silent in all other tissues tested (Figure 2B). To determine whether tissue-specific differences in DNA methylation could contribute to the tissue-specific expression profile of this transcript, bisulphite-modified DNA from neonatal brain and heart was sequenced over the Inpp5f_v3 promoter region (Figure 5). Allele-specific differences were observed in both the expressing and non-expressing tissue, but the difference was significantly less pronounced in heart than in brain (77% versus 51%; Fisher's Exact P < 10−5). Thus, the Inpp5f_v3 promoter shows differences in methylation both between alleles and between tissues. Tissue-specific variation in CpG methylation levels has been reported previously at the imprinted Igf2 locus (23).
Figure 5.
Tissue-specific methylation differences at the Inpp5f_v3 promoter in an expressing and non-expressing tissue. The overall percentage of methylated CpGs on maternally (Mat) and paternally (Pat) derived alleles is indicated in each case. Strands derived from the same PCR amplification are connected by brace symbols to the right of the figure. (A) Neonatal brain and (B) Neonatal heart.
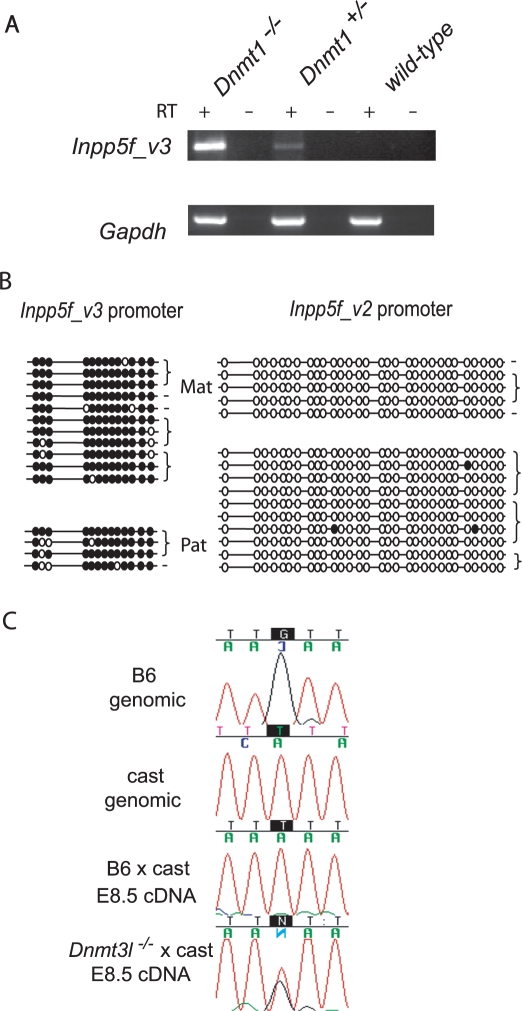
Activation of Inpp5f_v3 in Dnmt1 mutants
Dnmt1 is responsible for the faithful propagation (or ‘maintenance’) of CpG methylation patterns during DNA replication (24). To establish whether DNA methylation is required for the silencing of Inpp5f_v3 at E8.5, expression was assessed in embryos that were homozygous or heterozygous for null mutations in Dnmt1 (16), and stage-matched wt controls. Qualitative RT-PCR showed expression in Dnmt1−/– embryos, with a weak product also generated from Dnmt1+/– cDNA (Figure 6A). No expression was detected in wt E8.5 embryos. These data indicate that all of the trans-acting regulatory factors required for the expression of Inpp5f_v3 are present at E8.5, and that maintenance methylation is sufficient to silence the repeat-rich promoter. Demethylation of the paternal allele later in development (Figures 4D and 5) may therefore be sufficient for imprinted transcriptional activation in neural tissues. However, the lack of Inpp5f_v3 expression in heart tissue (Figure 2B), which also undergoes some degree of paternal allele demethylation (Figure 5B), indicates that neural-specific transcription factors also play a role.
Figure 6.
Disrupted expression of Inpp5f_v2 and Inpp5f_v3 in Dnmt1 and Dnmt3l mutants. (A) Qualitative RT-PCR expression assay for Inpp5f_v3 in E8 embryos carrying null mutations in Dnmt1. Inpp5f_v3 was amplified for 34 cycles, Gapdh was amplified for 30 cycles. Parallel amplifications were performed using cDNA generated in the presence (+RT) or absence (–RT) of reverse transcriptase. (B) Allele-specific CpG methylation at the two promoter regions in E8.5 embryos derived from Dnmt3l−/– mothers (on a Mus musculus domesticus background) and wild-type cast fathers. (C) Allele-specific RT-PCR sequencing assay for Inpp5f_v2 in E8.5 embryos derived from mothers carrying homozygous null mutations in the Dnmt3l gene, on a Mus domesticus background. The maternal strain or genotype is given first in the hybrid crosses.
Loss of methylation at Inpp5f_v2, but not Inpp5f_v3, in maternal imprint-free embryos
Dnmt3l−/– mothers fail to establish germ-line methylation imprints, and their heterozygous offspring die before midgestation showing disrupted expression of maternally imprinted genes (6). Maternal imprint-free embryos were generated by crossing Dnmt3l−/– females with Mus mus castaneus males, and were collected at E8.5. Bisulphite-modified genomic DNA was sequenced over the CpG-rich promoter regions of Inpp5f_v2 and Inpp5f_v3. The Inpp5f_v2 promoter region shows a lack of maternal allele-specific methylation in the maternal imprint-free embryos (Figure 6B), which is associated with reactivation of the normally silent maternal allele (Figure 6C). Surprisingly, the repeat-rich Inpp5f_v3 promoter is methylated on both alleles in maternal imprint-free embryos (Figure 6B). This region is methylated on both alleles in wt embryos at E8.5 (Figure 4C) and is hence unaffected by maternal Dnmt3l deficiency. Taken together, these data suggest that the epigenetic mark conferring memory of parental origin is located at the Inpp5f_v2 promoter, and that the onset of imprinted Inpp5f_v3 expression is likely to be a secondary event occurring after E8.5.
DISCUSSION
Imprinted genes commonly occur in clusters, and genes within these clusters are coordinately regulated by one or a few DNA elements termed imprinting control regions [ICRs; (25)]. One property shared by all eight ICRs that have been defined by gene targeting, (Igf2, Kcnq1, Igf2r, Snrpn, Gnas1a, Nespas, Rasgrf1, Dlk1) is the ability to undergo differential CpG methylation during gametogenesis, and subsequently maintain these allele-specific differences after fertilization during somatic development.
The CpG island at the promoter of Inpp5f_v2 overlaps a retrotransposed ORF with homology to the X-linked Tmem114a gene (10). A detailed methylation analysis of this region revealed that the allele-specific markings, which are established during gametogenesis, are maintained throughout embryonic development (Figure 4). Furthermore, embryos derived from Dnmt3l−/– mothers showed activation of the normally silent maternally derived allele (Figure 6C). These observations suggest that the CpG island with homology to the X chromosome is integral to the imprinting mechanism at this locus.
Despite also undergoing differential methylation during gametogenesis, the Inpp5f_v3 promoter undergoes de novo methylation on the paternally derived allele between E3.5 and E8.5 (Figure 4B and C). The genome undergoes extensive epigenetic reprogramming during this period (26), including the acquisition of methylation at Alu elements in primates (27,28). Murine B1 repeats are homologous to primate Alu elements; both families originated from 7SL, the RNA component of the signal recognition particle (29). Thus, the inverted B1 elements situated <100 bp upstream of the CpG-rich repeats (Figure 3A) may attract methylation to the surrounding region at the implantation stage. A similar B1 repeat upstream of the murine Aprt gene is capable of attracting de novo methylation to surrounding sequences in a cell culture system (30).
The activation of Inpp5f_v3 in Dnmt1−/– embryos shows that maintenance DNA methylation is required for Inpp5f_v3 silencing in vivo (Figure 6A). The proximity of the promoter region to tandem B1 elements suggests that the SINEs may contribute to the developmental expression profile of this gene. This provides an attractive explanation for the paucity of murine B1 and primate Alu elements found in the vicinity of imprinted promoters (12,13). Appropriate DNA methylation at imprinted loci is essential for embryonic viability (6), but may be disrupted by SINE insertions with potentially deleterious consequences. In this regard, Inpp5f_v3 can be viewed as exceptional. Here, the epigenetic regulation of the repeat-rich sequences may be utilized by the host organism to achieve the appropriate developmental expression pattern for Inpp5f_v3. Alternatively, the imprinting and tissue-specific expression of Inpp5f_v3 may serve no intrinsic function but may occur as a downstream consequence of paternal-allele-specific Inpp5f_v2 expression, which is highest in neural tissues (21).
Despite biallelic hypermethylation at E8.5, the paternal Inpp5f_v3 promoter subsequently loses methylation and is expressed while the maternal allele remains hypermethylated and silent (Figures 2C and 4D). Demethylation is more pronounced in the brain than in non-expressing heart tissue, suggesting a model in which active chromatin ‘spreads’ from the Inpp5f_v2 promoter region which is active primarily in neural tissues (Figure 2B). This could occur due to the binding of transcription factors, which is known to cause demethylation of local sequences in dividing cells (31,32). To our knowledge, Inpp5f_v3 is the first example in mammals of an imprinted gene that undergoes allele-specific demethylation during somatic development.
The ORF of Inpp5f_v2 is one of a group of at least four imprinted retrogenes that originated from X-linked source genes (10). The data presented here show that the promoter region exhibits the epigenetic characteristics of an ICR, which could control the imprinting of both the Inpp5f_v2 and Inpp5f_v3 transcripts. U2a1-rs1 and Mcts2, two other X-to-autosome retrogenes, also physically overlap gDMRs that are likely to control the imprinting of adjacent genes (10,33). We have recently identified additional imprinted transcripts at the fourth locus: Nap1l5 (Wood et al., unpublished data). Thus, all of the four known gDMRs that overlap X-to-autosome retrogenes are associated with multiple imprinted transcripts. These observations, combined with recent findings at the Peg10 locus (9), suggest that the (retro)transposition of elements from the X chromosome is one mechanism by which mammalian imprinted domains were generated during evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Colin Beechey for providing tissues from mice carrying uniparental partial disomies proximal to the T65H translocation (UpDp7), Ben Woodman and Michelle Lupton for the collection of embryos and Reiner Schulz and Ruth McCole for critical reading of the manuscript. Funding sources are the Generation Trust (A.J.W.), the Wellcome Trust (R.J.O.) and The National Institutes of Health (THB). Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lerchner W., Barlow D.P. Paternal repression of the imprinted mouse Igf2r locus occurs during implantation and is stable in all tissues of the post-implantation mouse embryo. Mech. Dev. 1997;61:141–149. doi: 10.1016/s0925-4773(96)00630-2. [DOI] [PubMed] [Google Scholar]
- 2.Bhogal B., Arnaudo A., Dymkowski A., Best A., Davis T.L. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics. 2004;86:961–970. doi: 10.1016/j.ygeno.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Bourc’his D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 4.Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 5.Yoder J.A., Walsh C., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 6.Bourc’his D., Xu G.-L., Lin C.-S., Bollman B., Bestor T.H. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 7.Ono R., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- 8.Ono R., Shiura H., Aburatani H., Kohda T., Kaneko-Ishino T., Ishino F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003;13:1696–1705. doi: 10.1101/gr.906803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki S., Ono R., Narita T., Pask A.J., Shaw G., Wang C., Kohda T., Alsop A.E., Marshall Graves J.A., et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood A.J., Roberts R.G., Monk D., Moore G.E., Schulz R., Oakey R.J. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:192–203. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blewitt M.E., Vickaryous N.K., Paldi A., Koseki H., Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2:e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greally J.M. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl Acad. Sci. USA. 2002;99:327–332. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luedi P.P., Hartemink A.J., Jirtle R.L. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehring M., Huh J.H., Hsieh T.F., Penterman J., Choi Y., Harada J.J., Goldberg R.B., Fischer R.L. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan B.L.M., Constantini F., Lacy E. Manipulating the Mouse Embryo, a Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 16.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 17.Beechey CV, Ball ST, Townsend KM, Jones J. The mouse chromosome 7 distal imprinting domain maps to G-bands F4/F5. Mammalian Genome. 1997;8:236–240. doi: 10.1007/s003359900400. [DOI] [PubMed] [Google Scholar]
- 18.Olek A., Oswald J., Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenherr C.J., Levorse J.M., Tilghman S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 20.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 21.Choi J.D., Underkoffler L.A., Collins J.N., Williams P.T., Golden J.A., Loomes K.M., Schuster E.F., Jr, Wood A.J., Oakey R.J. A novel variant of Inpp5f is imprinted in brain and its expression is correlated with differential methylation of an internal exonic CpG island. Mol. Cell. Biol. 2005; 25:5514–5522. doi: 10.1128/MCB.25.13.5514-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quentin Y. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res. 1994;22:2222–2227. doi: 10.1093/nar/22.12.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber M., Milligan L., Delalbre A., Antoine E., Brunel C., Cathala G., Forne T. Extensive tissue-specific variation of allelic methylation in the Igf2 gene during mouse fetal development: relation to expression and imprinting. Mech. Dev. 2001;101:133–141. doi: 10.1016/s0925-4773(00)00573-6. [DOI] [PubMed] [Google Scholar]
- 24.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 25.Lewis A., Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 26.Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 27.Hellmann-Blumberg U., Hintz M.F., Gatewood J.M., Schmid C.W. Developmental differences in methylation of human Alu repeats. Mol. Cell. Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin C.M., VandeVoort C.A., Teplitz R.L., Schmid C.W. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 1994;22:5121–5127. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriegs J.O., Churakov G., Jurka J., Brosius J., Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Yates P.A., Burman R.W., Mummaneni P., Krussel S., Turker M.S. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 1999;274:36357–36361. doi: 10.1074/jbc.274.51.36357. [DOI] [PubMed] [Google Scholar]
- 31.Brandeis M., Frank D., Keshet I., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo K., Silke J., Georgiev O., Marti P., Giovannini N., Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Joh K., Masuko S., Yatsuki H., Soejima H., Nabetani A., Beechey C.V., Okinami S., Mukai T. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 2004;24:270–279. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.