Abstract
In all organisms, precursor tRNAs are processed into mature functional units by post-transcriptional changes. These involve 5′ and 3′ end trimming as well as the addition of a significant number of chemical modifications, including RNA editing. The only known example of non-organellar C to U editing of tRNAs occurs in trypanosomatids. In this system, editing at position 32 of the anticodon loop of tRNAThr(AGU) stimulates, but is not required for, the subsequent formation of inosine at position 34. In the present work, we expand the number of C to U edited tRNAs to include all the threonyl tRNA isoacceptors. Notably, the absence of a naturally encoded adenosine, at position 34, in two of these isoacceptors demonstrates that A to I is not required for C to U editing. We also show that C to U editing is a nuclear event while A to I is cytoplasmic, where C to U editing at position 32 occurs in the precursor tRNA prior to 5′ leader removal. Our data supports the view that C to U editing is more widespread than previously thought and is part of a stepwise process in the maturation of tRNAs in these organisms.
INTRODUCTION
Transfer RNAs (tRNAs) play a central role in connecting the genetic information found in DNA with the protein synthesizing machinery at the far end of the genetic information cascade. Inevitably in the process of information transfer, a tRNA must also play a crucial role in ensuring the overall fidelity of the system and clearly even the subtlest changes in the sequence of a tRNA can have drastic effects in downstream cellular processes. Within cells, however, tRNAs are not inert molecules and in the process of maturation may undergo a number of changes, most notably the acquisition of a large variety of chemical groups collectively known as post-transcriptional modifications (1,2). To date over 100 different post-transcriptional modifications have been described in tRNAs from organisms belonging to all three domains of life: Bacteria, Archaea and Eukarya (3,4). The majority of modifications are apparently necessary to alter the structure of a tRNA to ensure proper folding. These alterations may increase either flexibility or rigidity to portions of the tRNA structure to meet demands set forth by the translational machinery (5–9). Remarkably, a majority of modifications, when tested individually, are not by themselves essential for cell viability and can in fact be viewed as structural modulators, whose full impact on cellular metabolism is not easily appreciated. This does not call into question their general importance, but rather highlights the fact that modifications play roles in global cell function that are often delicate at best.
An essential sub-set of modifications known as tRNA editing involve the ‘programmed alteration’ of tRNA sequences, permitting decoding of many more codons than what is implicit in the tRNA gene sequence (10). Although editing events may indirectly affect tRNA function by repairing otherwise non-functional tRNAs (11–14), a number of editing events have direct effects in expanding a tRNA's decoding capacity (15–17). Decoding changes imparted by tRNA editing thus provide a mechanism to effectively accommodate genetic code degeneracy. Recently, we showed that a single threonyl tRNA (tRNAThrAGU) in trypanosomatids undergoes cytidine (C) to uridne (U) and adenosine (A) to inosine (I) editing at the same anticodon loop (18). In vitro, C to U editing stimulates A to I formation, while in vivo every inosine containing tRNAThr(AGU) also has the C to U edit at position 32 (18). In turn, the heterodimeric ADAT2/3p enzymes that specify A to I in tRNA have conserved motifs that resemble cytidine deaminases yet in nature behave like adenosine deaminases (both in vivo and in vitro), leading to the proposal that both enzymes have a common ancestor dating back to the bacterial cytidine deaminases (19,20). Recently, we showed that in Trypanosoma brucei the tRNA A to I enzyme plays a role in both C to U and A to I editing of tRNAThrAGU, providing, what is to date, the only direct biochemical and genetic evidence for a common ancestry to these enzymes (21). Together, these observations have raised the question of how these reactions are specified. One possibility, at least in the case of the double-edited tRNA of T. brucei, is that inosine at position 34 is required for C to U editing.
In the present manuscript, we have followed the fate of edited tRNAs by analyzing their editing states in RNA purified from different sub-cellular fractions. We demonstrate that C to U and A to I editing of tRNAThr isoacceptors are ordered processes where C to U editing localizes to the nucleus and thus occurs prior to tRNA export to the cytoplasm. We also show that every tRNAThr isoacceptor undergoes C to U editing in both Trypanosoma and Leishmania suggesting that C to U editing is widespread, evolutionarily conserved and may serve important roles in tRNAThr function in trypanosomatids.
MATERIALS AND METHODS
Cell culture and nuclei isolation
Trypanosoma brucei cells were grown in SDM-79 medium supplemented with 10% fetal bovine serum (Fisher, USA) and 1 μg/ml hemin (Calbiochem, USA). Exponentially growing cultures (2 × 106 cells/ml) were harvested by centrifugation at 4000g and washed with phosphate-buffered saline. For nuclei preparations, cultures were harvested at ∼10 × 106 cells/ml. Cells were washed in phosphate-buffered saline; suspended in lysis buffer (0.5 M hexylene glycol (Sigma, USA), 1 mM PIPES (pH 7.4) and 1 mM CaCl2) (22); and broken using a Stansted Fluid Power apparatus set at 50 p.s.i. (23). The cell lysate was cleared by two successive centrifugations at 2500g for 20 min to obtain the cytosolic cell fraction (supernatant). The pellet was suspended in lysis buffer and nuclei isolated in a density gradient created by centrifugation of 35% Percoll (Amersham Pharmacia Biotech, USA) at 60 000g for 35 min. The nuclei were collected by side puncture, washed with nuclei wash solution [0.5 M hexylene glycol (Sigma), 1 mM PIPES (pH 7.4), 1 mM CaCl2 and 0.75 M sucrose] and pelleted by centrifugation at 2500g for 20 min. Each step of cell fractionation was monitored by phase-contrast and fluorescence microscopy by staining with 4′,6-diamidino-2-phenylindole (DAPI) (23).
cDNA synthesis and PCR amplification
RNA was isolated from cells (total RNA) and/or nuclear fractions by the guanidinium thiocyanate/phenol/chloroform extraction method (24). RNA was further treated with RQ1 (RNA qualified) RNase-free DNase I (Promega, USA). Two picomoles of the desired reverse oligonucleotide primer (ThrAGU57R: 5′-AGGCCACTGGGGGGATCGAACCC-3′), (ThrCGU830R: 5′-AGAATCGAACTCGCGACCCCCTC-3′), (ThrUGU832R: 5′-AGAATTGAACTCGGGACCCCTGG-3′), or (preThrUGU817R: 5′-CCGAAGTGTCAATAGGCGCG-3′) complementary to the 3′-end of the tRNA of interest was added to 5 μg of total or nuclear RNA with 10 μmol of all four deoxynucleotide triphosphates and heated at 65°C for 5 min and then quick-cooled at 4°C for 1 min followed by the addition of 1 μl of SuperScript II reverse transcriptase (RT) in 1× first strand buffer followed by incubation at 50°C, as described (Invitrogen, USA). Following the RT reaction, the cDNA was amplified using 2 μl of the 20 μl RT reaction as a template in a final volume of 100 μl (PCR) with 40 pmol of the appropriate forward oligonucleotide primer (ThrAGU56F: 5′-GGCCGCTTAGCTCAATGGCAGAG-3′), (ThrCGU536F: 5′-GGCCGCTTAGCACAGTGGCAGT-3′), (ThrUGU538F: 5′- GGCCTCGTAGCACAGTGGCAGT-3′), or (preThrUGU818F: 5′-ATATCTATTAGCCCTTTCCGC-3′) and 40 pmol of reverse oligonucleotide primer previously used for the RT. PCR reactions were performed using Taq DNA polymerase and incubated in a thermal cycler at a 94°C denaturation step, a 50°C annealing step for 40 s, and an elongation step of 72°C repeated for a total of 20 cycles, following manufacturer's instructions (Perkin-Elmer Life Sciences, USA). Controls included a mock reaction in which the RT was left out of the reaction and used as a negative control to test for DNA contamination in the RNA samples and a reaction in which total genomic DNA was used as a template serving as a positive control for amplification. RT-PCR products were cloned into pCR2.1-TOPO (Invitrogen, USA). Independent clones were isolated after transformation of DH5α Escherichia coli and sequenced using Sequenase™ Version 2.0 DNA polymerase (USB, USA), per manufacturer's instructions. The dideoxynucleotide terminated sequencing reactions were separated in a 7 M urea/6% acrylamide denaturing gel, and the resulting sequences were used to ascertain the state of editing for each clone.
Acid urea gels and northern blot analysis
Acid urea gels allow the separation of aminoacylated and deacylated species of tRNA by an electrophoretic mobility shift (25). Individual tRNAs can then be detected by northern blot analysis. Total RNA maintained in acidic conditions by suspension in 100 mM sodium acetate (pH 4.5) was separated in an acid gel (6.5% polyacrylamide gel containing 7 M urea and 0.1 M sodium acetate pH 5.0). A lane where the sample was treated for 1 h at 37°C under basic conditions (10 mM Tris, pH 9.0) to deacylate the tRNA before loading was routinely used as deacylated control and also served as a size marker. Following electrophoresis, samples were transferred to a Zeta-Probe membrane (Bio-Rad, USA) according to manufacturer's directions. Alternatively to assess fraction cross-contamination, RNA (1.5 μg) from each cell fraction was separated on a 7 M urea/6% acrylamide denaturing gel. The gel was stained with ethidium bromide for RNA visualization and photography, and the RNA was transferred to Zeta-Probe membranes (Bio-Rad) according to the manufacturer's directions. The membranes were hybridized at 45°C with the appropriate oligonucleotides, which were 5′-end-labeled with [γ-32P]ATP (GE Biosciences, USA) using T4 polynucleotide kinase (New England Biolabs, USA). The hybridization and wash conditions were as per the manufacturer's directions. The following oligonucleotide probes were utilized for northern analysis: U6-snRNA 856R: 5′-GATTGACATCAGCCTTGCGC-3′, SL-RNA 988R: 5′-GCTGCTACTGGGAGCTTCTCATAC-3′, ThrUGU838R: 5′-CCAGTGCACTGCCACTGTG-3′.
Coupled oxidation RT-PCR assays (oxopap assays)
To corroborate the editing state of aminoacylated species, we perform coupled oxidation-RT-PCR assays (oxopap assay) (18). Total aminoacyl-tRNAs were extracted under acidic conditions (using phenol equilibrated with 0.3 M sodium acetate, pH 4.5 and 10 mM EDTA), ethanol-precipitated, and suspended in 10 mM sodium acetate, pH 4.5 and 1 mM EDTA. The RNA was then split into two fractions. One fraction was deacylated by incubation at 37°C for 1 h in a basic buffer (10 mM Tris, pH 9.0) followed by oxidation of the 3′-ribose by treatment with 40 mM NaIPO4 in ice for 90 min. The second fraction was directly oxidized by NaIPO4 followed by deacylation as above. Both fractions were individually polyadenylated by incubation of the RNA at 37°C for 45 min in buffer containing 20 mM Tris, pH 7.0, 50 mM KCl, 0.7 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.1 mg/ml bovine serum albumin, 10% glycerol, 500 μM ATP and 1700 U of yeast poly(A) polymerase in 100 μl of reaction buffer. The reaction was then supplemented with 30 μl of 5 × E. coli poly-A buffer (200 mM Tris, pH 7.0, 1 M NaCl and 25 mM MgCl2), 15 μl of 5 mM ATP, 1 μl of 0.1 M DTT, 3.5 μl of MnCl2 and 3 U of E. coli poly(A) polymerase and incubated further for 45 min at 37°C. The reactions were phenol-extracted and ethanol-precipitated. Both samples were then used in RT-PCR reactions. First, a 3′-primer specific for the poly(A) tail was used to RT-PCR the polyadenylated RNA followed by PCR with the RT primer and a 5′-specific primer specific for the tRNA of interest in a 100 μl PCR reaction as above. One microliter of this reaction was used as a template for a second PCR reaction in which both primers were specific for the tRNA of interest. The resulting product was purified, cloned into pCR2.1-TOPO (Invitrogen), and transformed into DH5α E. coli, and individual clones were sequenced to establish editing levels.
Western blots and immunofluorescence analysis
For western blots, protein fractions isolated as above were separated by 10% SDS–PAGE and transferred onto nitrocellulose filters as described (21). Membranes were incubated with antibodies specific for enolase (cytoplasm-specific marker) and/or antibodies specific to the TY tag (below) or antibodies specific to TbADAT2p (21).
For immunofluorescence detection, the protocol was carried out as described previously with minor modifications (Munoz-Jordan and Cross, 2001). Briefly, 1 × 106 procyclic forms were washed with phosphate buffer saline (PBS) before fixing in 2% formaldehyde in PBS for 10 min at 4°C. The fixed cells were attached onto glass cover slips and permeabilized with 0.2% NP-40 in PBS for 10 min at room temperature. Monoclonal TY BB2 antibody was used to detect ADAT2-TY, DAPI and Mitotracker were used to stain DNA and mitochondria, respectively. Monoclonal antibody was visualized with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody. Cells were mounted in anti-fade mounting solution and analyzed with fluorescence microscope (Zeiss, Axioplan 2). Images were captured using MetaVue acquisition software (Universal Imaging, USA).
RESULTS
All three threonyl-tRNA isoacceptors undergo C to U editing in the anticodon loop
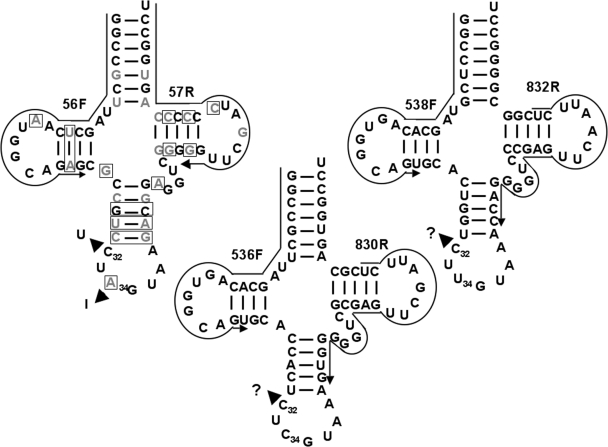
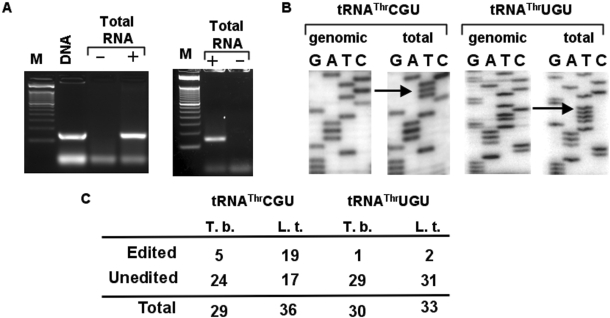
In all eukaryotes and most bacteria (but not archaea), tRNAs that are encoded with an adenosine at position 34 (the wobble nucleotide) are the subject of adenosine (A) to inosine (I) editing. Kinetoplastids are not an exception and in both T. brucei and Leishmania tarentolae all A34-containing tRNAs get edited, where I34 then permits decoding of the C-ending codons for the amino acids Ala, Arg, Ile, Leu, Pro, Ser, Thr and Val (Gaston and Alfonzo, unpublished data). Previously, we showed that tRNAThrAGU undergoes two distinct editing events in the anticodon loop, whereby every I34 containing tRNAThr also contains a C to U editing at position 32 in vivo (18). This observation raised the question of a possible interrelation between the two editing events, where either C to U is necessary for A to I formation or the reverse is true (i.e. A to I is necessary for C to U formation). Establishment of an efficient A to I editing assay led us to conclude that in vitro the presence of U32 (and no other nucleotide substitution at position 32) had a stimulatory effect, but it was not required, in the further formation of I34 (18). Still a standing question is whether the reverse is true, that A to I maybe required for C to U formation. The lack of an efficient in vitro C to U editing assay for tRNAs in any system (be it plants, marsupials or trypanosomatids) has thus far precluded answering some questions about editing and/or modification and their possible interrelation. To address the issue of editing site interrelation, we have used sequence comparative analysis coupled with an in vivo approach. We compared the sequence of the double-edited tRNA (tRNAThrAGU) to that of the two remaining tRNAThr isoacceptors (anticodon UGU and CGU) (Figure 1). All three tRNAs differ at a number of nucleotide positions in their backbone sequences (sequences not including the anticodon arm). Importantly, all three isoacceptors contain nearly identical anticodon loop sequences, including a C at position 32, the edited position in tRNAThrAGU, raising the possibility that the other two isoacceptors also undergo C to U editing in vivo. In addition, the L. tarentolae homologous isoacceptors have identical anticodon arm sequences to that of the T. brucei tRNAThr. We designed oligonucleotide primers specific for each of the two additional tRNAThr isoacceptors (Figure 1), whereas a 3′-specific oligomer was used to reverse transcribe tRNAThr from total T. brucei RNA. The resulting cDNA was then used as a template for PCR amplification with the same 3′ primer and a 5′-specific primer. Specific amplification products were obtained with these sets of primers when the reaction was performed in the presence of RT (Figure 2A), but was absent in ‘mock’ controls where the enzyme was omitted from the reactions. Indicating that the observed products are derived from reverse transcription of the RNA template and not from genomic DNA contamination. A product of identical size was obtained when both primers were used to amplify tRNAThr from total genomic DNA used as a positive control for amplification (Figure 2A). Similar results were obtained when L. tarentolae RNA and/or DNA was used in the RT-PCR and PCR reactions (data not shown). Both the cDNA-derived and the genomic DNA-derived products were then cloned into a plasmid vector, transformed into E. coli and a number of independent clones sequenced (Figure 2). We found that both tRNAThrCGU and -UGU undergo C to U editing at position 32 of the anticodon loop despite lacking an encoded A34 (Figure 2B). The observed editing (5 out of 29 clones, 17%) for the tRNAThrCGU and UGU (1 out of 30 clones, 3%) isoacceptors is lower than that observed for the double-edited tRNAThrAGU (18) (Figure 2C). Again similar numbers were obtained when the analogous products were sequenced from L. tarentolae sub-cellular fractions (Figure 2C). These results show that C to U editing of cytoplasmic tRNAs is more widespread than previously thought and is conserved among different trypanosomatid species. We also tested the possibility that other C32 containing tRNAs may undergo C to U editing at position 32, no editing was found in either tRNAArg or tRNAIle (data not shown). Notably these two tRNAs undergo A to I at position 34. Still our findings suggest that C to U editing may occur in other tRNAs in these organisms, perhaps at different positions, but this will remain an open question. The occurrence of C to U editing in tRNAs which lack an encoded A34 also rules out the possibility of inosine as a pre-requisite for C to U formation in these organisms.
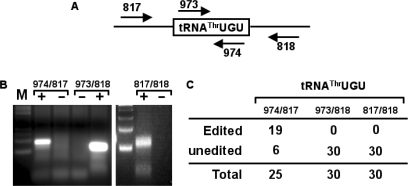
Figure 1.
The threonyl tRNA isoacceptors from T. brucei. The tRNAThrAGU was previously shown to undergo two editing events in the same anticodon loop (denoted by arrowheads in the figure). The other two isoacceptors (anticodon CGU and UGU, respectively) have a number of nucleotide differences in their backbone sequence but bear nearly identical anticodon loops, position 34 being the exception. Gray letters and boxed nucleotides mark positions that differ between the double-edited tRNA and the UGU and CGU isoacceptors, respectively. Arrows mark the position of the isoacceptor-specific primers used for RT-PCR and PCR reactions presented in this work.
Figure 2.
The T. brucei tRNAThrCGU and tRNAThrUGU undergo C to U editing in the anticodon loop. (A) RT-PCR with tRNAThrCGU and tRNAThrUGU-specific primers. The reactions were separated in a 3% agarose gel and stained with ethidium bromide. M, refers to a size marker lane. ‘DNA’ denotes a positive control lane in which a PCR reaction was performed with genomic DNA from T. brucei. ‘+’ and ‘−’ refer to RT-PCR reactions with the same primer but performed in the presence and/or absence of reverse transcriptase, where the ‘−’ reaction is a mock control to check for DNA contamination in our RNA preparation. (B) A representative sequence of either a genomic DNA PCR product or one derived from the RT-PCR reaction. The arrow indicates the presence of the C to U editing event at position 32 of the anticodon loop, which is only present in the cDNA but not in the genomic DNA sequences. (C) A number of independent clones were sequenced from the RT-PCR products above, where 5 out 29 and 1 out of 29 clones were found edited for tRNAThrCGU in T. brucei (T.b.). Similar reactions as above yielded 19 out of 36 and 2 out of 33 clones edited in L. tarentolae (L.t.). No editing was detected in 30 clones derived from the genomic DNA PCR reaction.
Both edited and unedited isoacceptors are functional in T. brucei
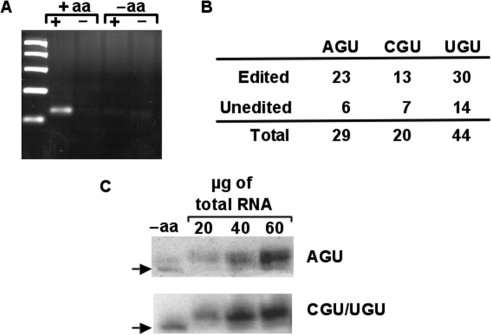
Previously we showed that both the double-edited and unedited versions of tRNAThrAGU were actively aminoacylated. We also showed that although in vivo neither editing event played a major role as an aminoacylation determinant, the fact that both species are substrates for aminoacylation indicated that both were functional in translation. To examine the fate of the two edited isoacceptors, we performed similar experiments using our oxopap assay as previously described (18). Briefly, total RNA was isolated under acidic conditions and treated with sodium periodate which can oxidize free vicinal hydroxyls at the 3′ ends of RNA to form a dialdehyde, whereas tRNAs that bear an amino acid remain intact. Following oxidation, total tRNA was deacylated and incubated with ATP and poly(A) polymerase and used in a reverse transcription reaction with a tagged oligo-T primer specific for the poly(A) tail. Followed by PCR with a forward primer specific for the tRNA of interest and a reverse oligo complementary to the tag added during reverse transcription. The resulting products were cloned, transformed into E. coli and a number of independent clones analyzed by sequencing to assess the editing states of tRNAs that are aminoacylated in vivo. In this assay, only aminoacylated tRNAs are protected from periodate oxidation and can be amplified. A negative control of deacylated tRNA was used to show that only aminoacylated tRNAs are substrates for polyadenylation. (Figure 3A). We found that a majority of the clones sequenced were edited at C32 (65% and 68% for tRNAThrCGU and –UGU, respectively) (Figure 3B). Surprisingly these numbers are higher than those observed with total RNA. It is possible that during isolation the unedited tRNAs are preferentially deacylated, over the edited ones, which will skew the numbers from the oxidation-RT-PCR assay. To rule out this possibility, we also separated total RNA in an acid polyacrylamide gel for analysis of their aminoacylation extent by northern blots with radioactive probes specific for each tRNA. This experiment showed that a majority of the tRNA is aminoacylated in vivo and remained aminoacylated throughout the purification process (Figure 3C), as indicated by the shifted band observed during acid-gel electrophoresis as compared to a control RNA sample that was deacylated prior to electrophoresis. Alternatively, it is possible that the higher numbers may indicate a preference for the synthetase to charge the edited tRNAs, however we deem this possibility unlikely, given that aminoacylation experiments with the double-edited tRNA showed no such preference. Furthermore, should a preference exist, the acid gel northern analysis would have shown charging efficiencies commensurate with the 17–21% editing levels. Likely, the higher editing levels may be due to the presence of either oxidation-labile modifications (or some other modifications) that under normal RT-PCR conditions prevent amplification of all the tRNA species in a given sample and as such leads to a misrepresentation of the actual editing levels. Regardless, qualitatively these results show that, like in the case of the double-edited isoacceptor, these tRNAs are also functional in vivo in that they are efficiently utilized by the synthetase as a substrate.
Figure 3.
All tRNAThr isoacceptors are substrate for aminoacylation in vivo. (A) Total RNA from T. brucei was extracted under acidic conditions and aminoacylation levels were correlated to editing levels by the oxopap assay as described. (B) Independent clones derived from the ‘+aa’ reaction (in A) were used in the OXOPAP assay (Materials and Methods section) and analyzed by sequencing. In all cases both edited and unedited species are substrates for aminoacylation in vivo. (C) Total RNA from the same fraction as above was also separated by acid denaturing polyacrylamide electrophoresis and probed with radioactive oligonucleotides specific for either tRNAThrAGU (top panel) or tRNAThrCGU/UGU. The probe used does not discriminate between the CGU and UGU isoacceptors. ‘−aa’ refers to a control reaction in which the RNA was deacylated by incubating under basic conditions prior to analyses. ‘+aa’ refers to the aminoacylated fractions purified and kept under acid pH.
Differential intracellular localization of C to U and A to I editing
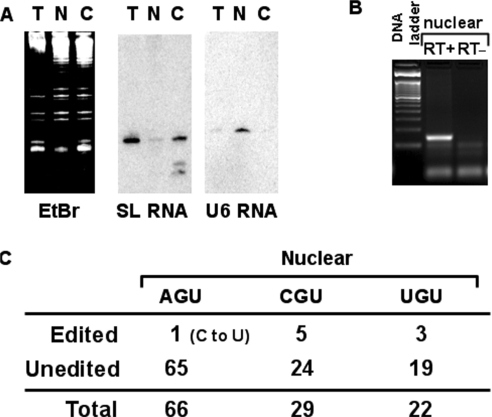
Although A to I editing of tRNA has been known for many years, little is known about the intracellular distribution of these editing events. In the case of A to I mRNA editing, it is well established that the editing enzyme localizes to the nucleus/nucleolus of mammalian cells (26). We decided to probe the intracellular distribution of the two editing events in trypanosomatids. We have previously developed purification methods that generate sub-cellular fractions with negligible cross-contamination (23). We have used similar methods to isolate total nuclear and cytoplasmic RNA fractions from T. brucei. These fractions were used in northern analysis to assess the relative purity of the fractions. RNAs were transferred to nitrocellulose membranes and hybridized to compartment-specific radioactive probes as described in the ‘Materials and Methods’ section. These probes were either specific for U6 snRNA (a nuclear marker) or spliced leader RNA (a nucleo-cytoplasmic marker which is predominant in the cytoplasm). As expected hybridization signals were observed which established that these fractions have little cross-contamination from other cellular compartments (Figure 4A). RNAs from these preparations were used for RT-PCR analysis as described earlier, where once again a number of independent clones were sequenced to assess editing levels (Figure 4B and C). All three isoacceptors showed C to U editing in the nuclear fractions with 17%, 14% and 1.5% edited for tRNAThrCGU, tRNAThrUGU and tRNAThrAGU, respectively. However, since every sub-cellular marker used in northern analysis to assess fraction purity corresponds to an RNA that is inevitably transcribed in the nucleus and transits to the cytoplasm, it could be argued that the observed values could be due to cytoplasmic contamination of our nuclear fractions. Notably, however, in the case of the tRNAThrAGU, no single clone out of a total of 66 analyzed contained the A to I editing event. Similar RT-PCR reactions were performed with tRNAValAAC, which also contains an adenosine at the first position of the anticodon and undergoes A to I editing. Again, no inosine-containing clone was observed with this particular tRNA, which in the cytoplasm is 97% edited from A to I (29 out of 30 clones) (data not shown). In addition, negligible A to I editing activity was detected when labeled tRNAThrAGU was incubated with nuclear protein fractions (data not shown). Taken together, this observation suggests that C to U editing of tRNAs occurs in the nucleus prior to export to the cytoplasm, while A to I editing is a cytoplasmic event.
Figure 4.
C to U but not A to I editing is a nuclear event in tRNAThrAGU. (A) Total (T), nuclear (N) and cytoplasmic (C) RNA was purified as previously described (Materials and Methods section) and analyzed by northern blots. The same membrane was probed with either a spliced-leader RNA-specific probe, which is a mostly cytoplasmic RNA or with a U6 RNA-specific probe, a nucleus-specific marker, to assess the level of purity of each fraction. (B) The nuclear fraction from (A) was used for RT-PCR with tRNAThrAGU-specific primer. ‘RT+’ and ‘RT−’ refer to reactions performed either in the presence or absence of reverse transcriptase, where the RT− reaction serves as a control for DNA contamination. (C) The RT-PCR product from (B) was cloned into a plasmid vector and 65 independent clones sequenced. Only 1 out 66 clones analyzed contained the C to U editing at position 32 but none had the A to I editing at 34, these numbers were compared to our previous results with total RNA were 18 out of 30 clones were double edited. Similarly, a number of clones for the other two isoacceptors were analyzed and both were edited to significantly higher levels.
C to U editing of tRNAThr precedes 5′ end maturation
Marchfelder and co-workers showed that in plant mitochondria, C to U editing of tRNAs follow a specific sequence of events, where editing is required for 3′ processing of a 5′ matured tRNA. Therefore, in plant mitochondria C to U editing restores base pairing in the anticodon necessary for 3′ trailer removal. Our nuclear localization results have thus led us to explore the possibility that like in plant organelles nuclear tRNA editing also follows an orderly cascade. We generated oligonucleotide primers specific for either the 5′ or 3′ precursor tRNAs and used them in RT-PCR reactions (Figure 5). These reactions generated products of a size consistent with that of a pre-tRNA containing, either the 5′ leader, the 3′ trailer or both. These products were cloned into a plasmid vector individually analyzed, by cloning and sequencing a number of independent clones. We found that 76% (19 out of 25) of the 5′-precursor containing clones had undergone C to U editing at position 32, whereas none of the clones containing either a 3′ trailer or both the 5′ and 3′ extensions were edited (i.e. all have a C at position 32). Our observations reinforce the view that C to U editing is a nuclear, orderly process that occurs prior to 5′ end maturation and likely following 3′ end removal. This observation also suggests that in vivo the pre-tRNA is in fact the natural target of C to U editing.
Figure 5.
C to U editing precedes 5′ leader removal but occurs after 3′ maturation. (A) The nuclear RNA from above was the subject of RT-PCR analysis with the primers indicated, where primers 817 and 818 are specific for the 5′ leader and 3′ trailer, respectively. (B) The resulting RT-PCR reactions with all possible primer combinations were separated in a 3% agarose gel and stained with ethidium bromide. ‘+’ and ‘−’ refer to RT-PCR reactions with the same primer but performed in the presence and/or absence of reverse transcriptase, where the ‘−’ reaction is a mock control to check for DNA contamination in our RNA preparation. (C) The PCR products from (B) were cloned and sequenced. Out of 25 clones, 19 were derived from the leader containing RT-PCR reaction contained the C to U edit. No edited clones were observed in either the trailer-specific reaction (oligos 973/818) or the reaction specific for both precursors (oligos 817/818).
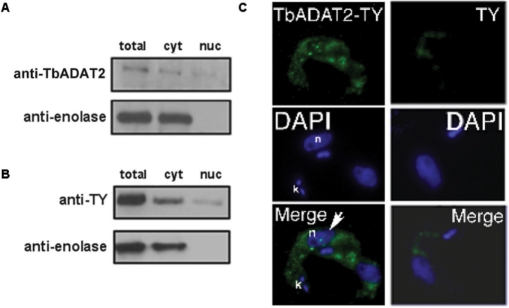
Sub-cellular localization of the catalytic sub-unit of the A to I editing enzyme (TbADAT2p)
We have recently reported that the catalytic component of the A to I editing enzyme (ADAT2p) may play a role in both A to I and C to U editing in trypanosomatids, since down-regulation of TbADAT2p led to a concomitant decrease in both A to I and C to U editing levels in tRNAThrAGU (21). In light of our current results, we decided to also explore the sub-cellular localization of the ADAT2 protein. Protein fractions were isolated by similar methods as described in Materials and Methods section and then analyzed by western blot with antibodies specific for ADAT2. We observed a low but significant signal for this protein in both the cytoplasmic and nuclear fractions (Figure 6). However, due to the relative low titer of this antibody, we also performed similar analyses but with protein extracts from cells transformed with an epitope-tagged copy of ADAT2. Again we found a measurable amount of TbADAT2p localized to the nucleus (Figure 6). This finding was further supported by immunofluorescence assay that showed the co-localization of the ADAT2p signal with DAPI. Taken together, these observations support the view that ADAT2p's role in both editing events and its intracellular localization correlates with the distribution of C to U and A to I editing in these cells.
Figure 6.
TbADAT2p localizes to both the nucleus and the cytoplasm. (A) Western blot with polyclonal antibodies specific for TbADAT2 (anti-TbADAT2). ‘Anti-enolase’ refers to polyclonal antibodies specific for enolase (a known cytoplasmic marker). (B) TbADAT2p was tagged with a TY epitope and expressed in procyclic T. brucei. ‘anti-TY’ refers to a western blot with antibodies specific for the epitope tag. ‘Anti-enolase’ refers to the enolase antibody used as a control as described above. ‘Total’, ‘cyto’ and ‘nuc’ refer to whole-cell, cytoplasmic and nuclear protein fractions, respectively. (C) Immunofluorescence experiments where cells transformed with the recombinant epitope-tagged TbADAT2 (TbADAT2-TY) (left panels) or cells transformed with TY alone (right panels) were stained with DAPI to determine the position of the nuclei (blue) or with fluorescent anti TY-antibodies (green). Two trypanosome cells are shown; in the first, TbADAT2-TY is detected within the nucleus (arrow); in the second, TbADAT2 is excluded from the nucleus. The location of the nucleus is marked with ‘n’; ‘k’ denotes the location of the kinetoplast (mitochondria).
DISCUSSION
Due to the inherent intracellular organization of eukaryotic cells, RNA processing may take place in different cellular compartments, which in turn may set an order to RNA maturation events. The intracellular distribution of a particular enzyme, or cellular component, may lead to effects in the regulation of gene expression in a more temporal manner (27–30). In the case of mRNA editing for example, different editing enzymes have different intracellular compartmentalization like the case of the C to U editing enzyme apobec (31–37) or the A to I enzymes, ADARs (38). In particular cases even the sub-nuclear localization of an editing enzyme may have tremendous effects in the regulation of editing activity (26). For instance, nucleolar sequestration of ADAR1 has been correlated with regulation of editing activity (26). Although previous studies have focused on the mRNA editing enzymes and a number of key observations have been made in the last several years, to date little is known about the intracellular distribution of the analogous enzymes that edit tRNA.
We have analyzed the distribution of edited and pre-edited tRNAThr species from different sub-cellular fractions. We show that C to U editing occurs in the nucleus prior to 5′ tRNA trimming, but following 3′ end processing. This observation suggests that C to U editing may be required for 5′ cleavage, analogous to what has been observed in plant mitochondria where tRNA editing at the acceptor stem is an ordered and required process for 3′ end cleavage (11,39,40). However, in trypanosomatids this may not be true in that even in cytoplasmic fractions matured and fully aminoacylatable pre-edited tRNAs co-exist with the edited ones (18), implying that unedited tRNAs are good substrates for end-trimming. Our findings also suggest that for all tRNAThr isoacceptors, 3′ cleavage may precede 5′ leader removal. This is unusual in that is widely accepted that 5′ maturation is a pre-requisite for 3′ cleavage. One notable exception is the case of tRNATrp in S. cerevisiae, the only tRNA in this organism for which the order is reversed (i.e. similar to tRNAThr in T. brucei) (41). However, a recent report in trypanosomatids showed that at least for the initiator tRNAMet, 3′ cleavage was required for 5′ leader removal, leading to the suggestion that unlike most organisms 3′ cleavage followed by 5′ leader removal was the prevailing pathway for tRNA end-trimming. Our data further supports this proposal and implicitly means that trypanosomatids either have a different 3′ trimming enzyme or that the enzyme is similar to that of other eukaryotes but with an alternative mode of substrate recognition (i.e. not requiring a processed 5′ end).
The observed differences in the number of edited clones between nuclear and total RNA may be partly explained by differential rates of export from the nucleus or alternatively by different rates of editing and processing. We suggest that the observed order of events described here reflects more on the requirement for 3′ end cleavage prior to editing than editing as a determinant for 5′ maturation. How then may 3′ cleavage affect C to U editing? Although currently we do not have a precise answer to this question, it is possible that the effect may be either direct, where a recessed 3′ end may be a required point of contact by the C to U editing enzyme or indirect, where the mature end is required for a modification which is in turn required for editing. Still, however, in the absence of knowledge of the complete modification set for this tRNA and a robust C to U editing assay, this will remain an open question.
We have recently shown that one of the components (TbADAT2p) of the enzyme that catalyzes A to I editing in trypanosomatids plays a role in both C to U and A to I editing of a single tRNAThrAGU in T. brucei (21). We had also previously showed that C to U editing at position 32 of the anticodon loop played a stimulatory role on the further A to I editing at the first position of the anticodon in the same tRNA. However, in vitro only A to I but not C to U editing activity could be reconstituted with recombinant proteins. This led to a scenario in which A to I could be required for C to U editing. In the absence of an efficient C to U editing assay, we explored the possibility that other tRNAThr isoacceptors (which do not contain an encoded A34) could also undergo C to U editing. We found that, in fact, in vivo under steady-state growth every tRNAThr undergoes C to U editing, ruling out the possibility that A to I editing is required for C to U. We thus suggest that the inability to recreate C to U editing in vitro (in this and other systems) may reflect the absence of other factors required for substrate recognition by the editing enzyme. Some of these factors could likely be post-transcriptional modifications and will thus require further analysis. Indeed, modifications are intermediates for further editing and/or modifications both in vivo and in vitro in archaea (42–45). Likewise even editing can be a pre-requisite for further modification like in the example of marsupial mitochondria. In this rather unique case, C to U editing of the neighboring nucleotide in tRNAAsp is required for queuosine (Q) formation at position 34 of the anticodon (46).
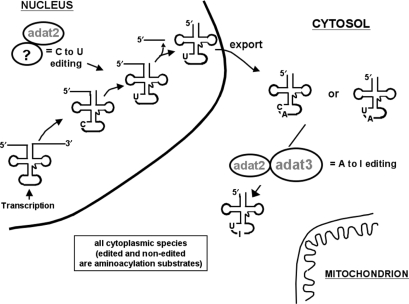
In light of our previous reports suggesting that TbADAT2p plays a role in both events, predictably this enzyme should also localize to both the nucleus and cytoplasm. In line with this proposal, we have shown that although the bulk of TbADAT2p signal is found in the cytoplasm, a measurable amount is also observed to shuttle to the nucleus. We propose a model (Figure 7) by which sub-cellular localization of the eukaryotic editing deaminases may alter their specificity. In the trypanosomatid example, this model suggests that TbADAT2p may have different specificity depending on two variables: its sub-cellular localization (i.e. nuclear, cytoplasmic and maybe even mitochondrial) or its association with different protein subunits. The first part of the model is supported by the nuclear localization experiments. The second part of the model is more difficult to test, as it requires prior knowledge of who associates with whom within a cell. However, in a way nature has already performed the experiment for us. ADAT2p and ADAT3p, cyitidine deaminases in terms of primary sequence, pair up in the form of a heterodimer and indeed act as adenosine deaminases in tRNA (19,21). Finally, beyond what sub-unit distribution and altered specificity may contribute to editing regulation, the role that either C to U editing and/or its nuclear localization plays in these cells is not yet clear. However, taken together, the data presented here show that these types of editing events may affect many more tRNA substrates than previously imagined and that sub-cellular localization as well as the order of tRNA processing may play crucial roles in the regulation of tRNA function in these cells.
Figure 7.
A schematic model of how in vivo distribution of both editing events and the localization of the editing enzyme may play a role in regulating editing levels.
ACKNOWLEDGEMENTS
We thank all members of the Papavasiliou and Alfonzo laboratories for helpful comments and suggestions. We also thank Paul Michels for the anti-enolase antibodies. This work was supported by grants from the American Heart Association and National Science Foundation to J.D.A. and a grant from the National Institutes of Health to F.N.P. Funding to pay the Open Access publication charges for this article was provided by National Science Foundation grant MCB0620707 to J.D.A.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grosjean H, Benne R. (eds) Modification and Editing of RNA. Washington, D.C: ASM press; 1998. [Google Scholar]
- 2.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33(Database Issue):D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 6.Sundaram M, Durant PC, Davis DR. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry. 2000;39:12575–12584. doi: 10.1021/bi0014655. [DOI] [PubMed] [Google Scholar]
- 7.Bajji AC, Sundaram M, Myszka DG, Davis DR. An RNA complex of the HIV-1 A-loop and tRNA(Lys,3) is stabilized by nucleoside modifications. J. Am. Chem. Soc. 2002;124:14302–14303. doi: 10.1021/ja028015f. [DOI] [PubMed] [Google Scholar]
- 8.Murphy FVT, Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 9.Weixlbaumer A, Murphy FVT, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covello PS, Gray MW. Editing of tRNA. In: Benne R, editor. Modification and Editing of tRNA. Washington, D.C.: ASM press; 1998. p. 596. [Google Scholar]
- 11.Kunzmann A, Brennicke A, Marchfelder A. 5′ end maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proc. Natl Acad. Sci. USA. 1998;95:108–113. doi: 10.1073/pnas.95.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fey J, Weil JH, Tomita K, Cosset A, Dietrich A, Small I, Marechal-Drouard L. Role of editing in plant mitochondrial transfer RNAs. Gene. 2002;286:21–24. doi: 10.1016/s0378-1119(01)00817-4. [DOI] [PubMed] [Google Scholar]
- 13.Reichert AS, Morl M. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res. 2000;28:2043–2048. doi: 10.1093/nar/28.10.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bullerwell CE, Gray MW. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J. Biol. Chem. 2005;280:2463–2470. doi: 10.1074/jbc.M411273200. [DOI] [PubMed] [Google Scholar]
- 15.Janke A, Paabo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993;21:1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borner GV, Morl M, Janke A, Paabo S. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 1996;15:5949–5957. [PMC free article] [PubMed] [Google Scholar]
- 17.Alfonzo JD, Blanc V, Estevez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio MA, Ragone FL, Gaston KW, Ibba M, Alfonzo JD. C to U editing stimulates A to I editing in the anticodon loop of a cytoplasmic threonyl tRNA in Trypanosoma brucei. J. Biol. Chem. 2006;281:115–120. doi: 10.1074/jbc.M510136200. [DOI] [PubMed] [Google Scholar]
- 19.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 20.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc. Natl Acad. Sci. USA. 2007;104:7821–7826. doi: 10.1073/pnas.0702394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro SZ, Doxsey SJ. Purification of nuclei from a flagellate protozoan, Trypanosoma brucei. Anal. Biochem. 1982;127:112–115. doi: 10.1016/0003-2697(82)90152-x. [DOI] [PubMed] [Google Scholar]
- 23.Kapushoc ST, Alfonzo JD, Rubio MA, Simpson L. End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae. J. Biol. Chem. 2000;275:37907–37914. doi: 10.1074/jbc.M007838200. [DOI] [PubMed] [Google Scholar]
- 24.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 26.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl Acad. Sci. USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J. Cell Sci. 2005;118:269–282. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- 28.Saxton WM. Microtubules, motors, and mRNA localization mechanisms: watching fluorescent messages move. Cell. 2001;107:707–710. doi: 10.1016/s0092-8674(01)00602-x. [DOI] [PubMed] [Google Scholar]
- 29.Lipshitz HD, Smibert CA. Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 2000;10:476–488. doi: 10.1016/s0959-437x(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 30.Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc. Natl Acad. Sci. USA. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl Acad. Sci. USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc. Natl Acad. Sci. USA. 1997;94:13075–13080. doi: 10.1073/pnas.94.24.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Sowden MP, Smith HC. Induction of cytidine to uridine editing on cytoplasmic apolipoprotein B mRNA by overexpressing APOBEC-1. J. Biol. Chem. 2000;275:22663–22669. doi: 10.1074/jbc.M910406199. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Sowden MP, Smith HC. Intracellular trafficking determinants in APOBEC-1, the catalytic subunit for cytidine to uridine editing of apolipoprotein B mRNA. Exp. Cell Res. 2001;267:153–164. doi: 10.1006/excr.2001.5255. [DOI] [PubMed] [Google Scholar]
- 35.Bennett RP, Diner E, Sowden MP, Lees JA, Wedekind JE, Smith HC. APOBEC-1 and AID are nucleo-cytoplasmic trafficking proteins but APOBEC3G cannot traffic. Biochem. Biophys. Res. Commun. 2006;350:214–219. doi: 10.1016/j.bbrc.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann DM, Galloway CA, Sowden MP, Smith HC. Metabolic regulation of apoB mRNA editing is associated with phosphorylation of APOBEC-1 complementation factor. Nucleic Acids Res. 2006;34:3299–3308. doi: 10.1093/nar/gkl417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanc V, Kennedy S, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J. Biol. Chem. 2003;278:41198–41204. doi: 10.1074/jbc.M302951200. [DOI] [PubMed] [Google Scholar]
- 38.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morl M, Marchfelder A. The final cut. The importance of tRNA 3′-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellegrini O, Nezzar J, Marchfelder A, Putzer H, Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kufel J, Tollervey D. 3′-processing of yeast tRNATrp precedes 5′-processing. RNA. 2003;9:202–208. doi: 10.1261/rna.2145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosjean H, Auxilien S, Constantinesco F, Simon C, Corda Y, Becker HF, Foiret D, Morin A, Jin YX, et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- 43.Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shigi N, Suzuki T, Terada T, Shirouzu M, Yokoyama S, Watanabe K. Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 2006;281:2104–2113. doi: 10.1074/jbc.M510771200. [DOI] [PubMed] [Google Scholar]
- 46.Morl M, Dorner M, Paabo S. C to U editing and modifications during the maturation of the mitochondrial tRNA(Asp) in marsupials. Nucleic Acids Res. 1995;23:3380–3384. doi: 10.1093/nar/23.17.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]