Abstract
Type II restriction-modification (R-M) systems comprise a restriction endonuclease (REase) and a protective methyltransferase (MTase). After R-M genes enter a new cell, MTase must appear before REase or the chromosome will be cleaved. PvuII and some other R-M systems achieve this delay by cotranscribing the REase gene with the gene for an autogenous transcription activator (the controlling or ‘C’ protein C.PvuII). This study reveals, through in vivo titration, that C.PvuII is not only an activator but also a repressor for its own gene. In other systems, this type of circuit can result in oscillatory behavior. Despite the use of identical, symmetrical C protein-binding sequences (C-boxes) in the left and right operators, C.PvuII showed higher in vitro affinity for OL than for OR, implicating the spacer sequences in this difference. Mutational analysis associated the repression with OR, which overlaps the promoter −35 hexamer but is otherwise dispensable for activation. A nonrepressing mutant exhibited poor establishment in new cells. Comparing promoter-operator regions from PvuII and 29 R-M systems controlled by C proteins revealed that the most-highly conserved sequence is the tetranucleotide spacer separating OL from OR. Any changes in that spacer reduced the stability of C.PvuII-operator complexes and abolished activation.
INTRODUCTION
Many bacteria possess restriction-modification (R-M) systems (1), at least in part for defense against DNA bacteriophages. The great abundance of R-M systems in the prokaryotic world reflects their mobility via transformation, transduction or conjugation (2,3). The PvuII R-M system is carried on a plasmid (4,5). Like other type II systems (6), it includes two separate enzymes: a restriction endonuclease (REase) that cleaves DNA at a target sequence, and a methyltransferase (MTase) that modifies the same sequence to protect it from the cognate REase (7–12). There is evidence that some R-M systems behave as addiction modules, with REase as ‘toxin’ and MTase as protective ‘antitoxin’ (13) [and, for that matter, that some addiction modules play anti-bacteriophage roles (14)].
The REase and MTase must be carefully balanced, ideally in a relatively host-independent manner, to minimize killing of new host cells that initially have completely unmethylated chromosomes. Even after establishment in a new host, on the one hand too much methylation could lead to several problems. First, overmethylation would increase the ‘escape rate’ (bacteriophage becoming methylated before restriction can occur) (15). Second, overexpression of the MTase would undermine the post-segregational killing associated with the selfish behavior of R-M systems (16). Third, changes in DNA methylation can have broad effects on gene expression patterns (17–19). Finally, under some circumstances, overmethylation can even lead to mutation (20,21). On the other hand, too much REase would lead to possibly-lethal DNA damage (22,23). Despite the importance of this balance, and the roles of restriction in modulating gene exchange as well as defense against bacteriophages (24), the regulation of R-M system gene expression is still not well understood, though progress is being made [e.g. (25,26)].
In addition to the MTase and REase genes, a subset of type II R-M systems contains regulatory genes. The regulatory C (controlling) gene was first discovered in the PvuII (27) and BamHI (28) R-M systems. Subsequently, active regulatory genes have been demonstrated in the BclI (29), BglII (30), Esp1396I (31), EcoO109I (32), EcoRV (33), Eco72I (34), HgiAI (35), BstLVI (36), Kpn2I (37) and SmaI (38) R-M systems. The C proteins appear to have a remarkably broad host range; for example a C-protein from the Gram-positive bacterial genus Bacillus activates transcription in Escherichia coli (38).
C proteins, where tested, activate their own transcription (‘autogenous’ activation), and are believed to be responsible for the delay in REase activity that is crucial when an R-M system enters a new host cell. In R-M systems having a C gene, the REase gene typically does not have its own promoter (an exception is LlaI (39)). The C and REase open reading frames usually overlap (as in the PvuII system; Figure 1A), and the REase gene is completely dependent on transcription from the upstream autogenously regulated C gene (40). Disruption of pvuIIC leads to a drastic reduction in REase expression that is restored by supplying the C gene in trans (27,41). Thus, in a new cell, REase expression should be low until C protein accumulates. The role of this activation requirement in delaying REase expression is indicated by the observation that pre-expressing C protein prevents transformation by the intact cognate R-M system, presumably due to premature REase expression and cleavage of recipient cells’ chromosomal DNA (13,40).
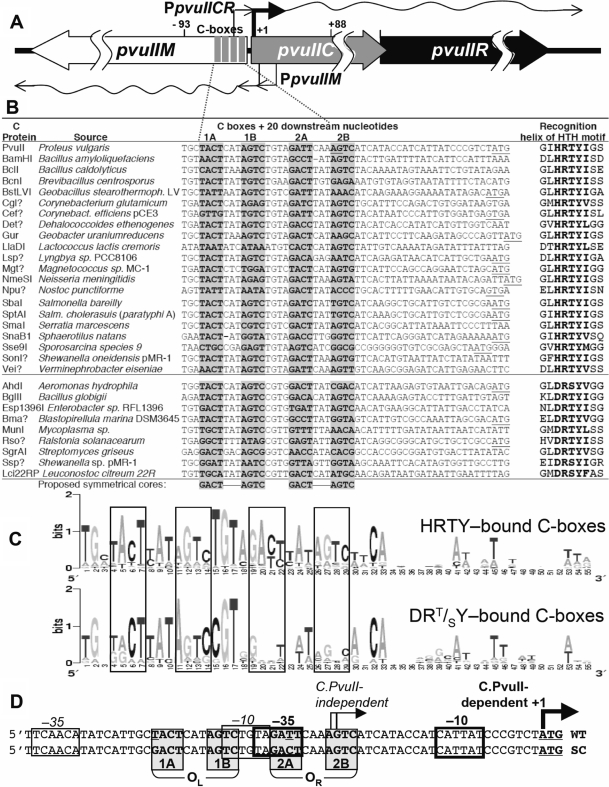
Figure 1.
PvuII R-M system control region and alignment with regions upstream of C.PvuII orthologs. (A) Genetic map of the PvuII R-M system. Numbering is relative to the initiation codon of pvuIIC. The four gray rectangles represent the C-half-boxes (shaded in part A). The two transcription starts for pvuIICR are identified by rightward bent arrows: from the C-independent weak promoter (thin) and C-dependent strong promoter (thick) (43). The two pvuIIM promoters are also shown (leftward bent arrows). (B) Comparison of the regions upstream of confirmed and putative C protein genes. Genes were identified by using the Blink function of Entrez (92), and REBASE (1), with the C.PvuII amino acid sequence as seed. This set was then screened for two features: proteins with predicted recognition helix sequences (of the helix–turn–helix motif) similar to that of C.PvuII (HRTYI) (41) or of C.AhdI (DRT/SY) (52), and being part of a putative R-M system (usually indicated by an adjacent MTase gene, as REase genes are difficult to recognize). A question mark indicates an ORF not yet formally named (93). The C protein initiation codon is underlined, unless it starts farther downstream. For C.PvuII (top), the underlined nucleotides (in half-box 2B and at the C.PvuII initiation codon) indicate the previously-identified transcript starts (40,43). The shaded nucleotides indicate actual or predicted C-boxes, and the proposed symmetrical core (40,43) is shown at the bottom. (C) Sequence Logos for the two subsets of C-box/promoter regions associated with C proteins having HRTY (upper) or DRT/SY (bottom) in the recognition helix. The Logo (46) was obtained from the two subsets of sequences in panel A, generated by the server at: http://weblogo.berkeley.edu. In Logo analysis, the full height (two bits of information content) applies to completely conserved positions. The C-box elements are boxed. (D) The pvuIIC promoter region sequence showing the C-box and promoter elements. The palindromic operators each contain a pair of C-boxes, designated as boxes 1AB or OL (operator left), and 2AB or OR (operator right). Conserved elements of the stronger, C-dependent promoter are indicated by heavy rectangles, while thinner rectangles indicate the weak C-independent promoter. Transcript starts are indicated as in (A). The upper sequence is from the wild-type (WT) PvuII R-M system, while the lower sequence has one substitution each in half-boxes 1A and 2A such that they match the symmetrical cores (SC) shown at the bottom of (B).
R-M systems-associated C proteins fall into several groups based on their sequences and (to the extent this is known) DNA sequence specificities. The archetypes of three currently recognized groups are C.PvuII, C.EcoRV and C.EcoO109I (R.M.B., unpublished data). All of them share a helix–turn–helix DNA-binding motif (predicted, in most cases). The first solved crystal structure of a C protein, C.AhdI of the C.PvuII group, strongly resembles the N-terminal DNA-binding domains of the Xre family of transcription factors, a group that includes the well-studied λcI and 434 repressors (42). The similarity has been confirmed by the structure of C.BclI (29). Comparisons of C and RpoD amino acid sequences (41,43) and structures (C.AhdI structure docked with those of DNA from the λcI–DNA complex and of RpoD) (42) suggest that C proteins may activate transcription through direct contact with the σ70 subunit of RNA polymerase.
The C proteins act on conserved operator sequences called ‘C-boxes’ (40,44). Conserved sequences upstream of C genes were first noted in a study of the Eco72I RM system (34). Functional C-boxes have been determined for C.PvuII (40), C.AhdI (45), C.EcoRV (33), C.EcoO109I (32) and C.BclI (29), though details of C protein interaction with this DNA and with RNA polymerase have yet to be reported. Recently sequenced bacterial genomes are revealing many putative C protein-like open reading frames (ORF), and upstream C-box sequences. We have updated and expanded an analysis of C-box/promoter regions for the PvuII-like group of C proteins (40,43), as shown in Figure 1B, C. The C-box position varies relative to the C ORF start codon, but the box regions all resemble two pairs of symmetrical inverted repeats linked by a 4 nt spacer (43): GACT-(N3)-AGTC-(N4)-GACT-(N3)-AGTC (Figure 1B). Logo analysis (46) reveals that the central base pairs (GT) of this 4 nt spacer are more highly conserved than the symmetry elements themselves (Figure 1C) (43). Strikingly, when the C proteins are divided based on their recognition helices, the group falls into two distinct subsets, in which the central spacer is predominantly TGTA for the HRTY helix and CGTG for the DRT/SY helix (Figure 1B, C).
The palindromic nature of C-boxes suggests that each of the two C-boxes (designated as 1AB or OL - operator left, and 2AB or OR - operator right; Figure 1D) could each be bound by the helix–turn–helix motifs of a C protein homodimer (47). The two operators have center-to-center spacing of 15 bp (Figure 1B) (40,43), meaning that the two C homodimers would occupy opposite faces of the DNA double helix (48), similar to repressor DtxR (49).
The C-boxes are located amid two overlapping promoters for C gene transcription (33,43). For the PvuII R-M system (40,43), the two transcription start sites have been determined by primer extension and nuclease S1 mapping (arrows in Figure 1D). The transcript starting within half-box 2B (OR) is produced from a weak, C-independent promoter believed to be responsible for initial C.PvuII production when the R-M system enters a new cell. The second, leaderless mRNA (50,51) comes from a strong, C-activated promoter (40,43). The complexity of this regulatory region is reflected by the fact that a 12 bp DNA segment includes the 1B and 2A half-box elements, together with the central spacer (TGTA), the −10 hexamer for the weak promoter, and the −35 hexamer for the strong, C-dependent promoter (Figure1D).
The roles of particular C-box elements have not been clear. Two C proteins, C.AhdI and C.EcoRV, have higher in vitro affinities for OL than OR (33,45), and OL is sufficient for activation of ecoRVCR expression (33). The close proximity of C-box and promoter sites led to a proposal that C protein and RNAP compete for binding, such that C protein binding to OL activates transcription, while binding to OR leads to repression (45,52). However the C proteins are remarkably small (monomer MW of 9.4 kDa, for C.PvuII), and at OR might act like the AsiA/MotA coactivators of bacteriophage T4, allowing RpoD interactions at −10 but replacing them at −35 by ‘remodeling’ the RpoD (53,54). Other activator–RNAP complexes, or certain −35/−10 spacer sequences, can also functionally replace the −35 hexamer (55,56), which is one possible explanation of the observation that the spacing between pvuIIC C-boxes/putative −35 hexamer and the −10 hexamer could be varied by ±4 bp with little effect (43), in sharp distinction to most promoters (57). Thus, we could not rule out an alternative regulatory model in which the occupation of all four half-boxes is required for full activation; this arrangement could in theory make the control system respond to rising C.PvuII levels as a relatively irreversible switch (58). To distinguish between this model and the activation-repression model, and to study the roles of the C-boxes, we carried out a series of in vivo and in vitro experiments. Our results strongly support the autogenous activator-repressor model, consistent with what had been proposed on theoretical grounds for C.AhdI (45,52), and indicate that this regulatory circuit is more than a simple switch.
MATERIALS AND METHODS
Strains and plasmids
The E. coli K-12 strains used in this study are described below. All strains into which pvuIIM is introduced must lack the mcrBC restriction system (4,59,60). MC1061 [araD139 Δ(ara, leu)7697, ΔlacX74, galU, galK, hsdR, strA] (61) transports arabinose but is deficient in its metabolism; it was used as the host for in vivo titrations with C.PvuII. TOP10 [F− mcrA (mrr-hsdRMS-mcrBC) Φ80lacZΔM15 lacX74 recA1 ara139 (ara-leu)7697 galU galK rpsL (StrR) endA1 nupG] (Invitrogen) was used for all other purposes including cloning steps and CAT assay. BL21(DE3) was used for C.PvuII purification, as it contains the gene for phage T7 RNA polymerase under an inducible promoter (62). The plasmids used are listed in Table 1. The oligodeoxyribonucleotides used are shown in Table S1 of the Supplementary Data.
Table 1.
Plasmids used in this study
| Name | Relevant feature(s) | Reference |
|---|---|---|
| pACYC177 | Cloning vector (AmpR, KanR) | (90) |
| pBAD24 | Arabinose inducible araBAD promoter, araC, AmpR, ColE1 ori | (68) |
| pBR322 | Cloning vector (AmpR, TetR) | (91) |
| pDK200 | wt pvuIIC gene with its own promoter, ▵pvuIIM, ▵pvuIIR, TetR, p15A ori | (43) |
| pDK201 | As pDK201, but pvuIIC-Esp19 mutation | (43) |
| pDK178 (WWWW) | Transcriptional fusion of pvuIIC promoter including C-boxes with the symmetrical core - WWWW (GATCcat AGCTtgtaGACTcaaAGCT) to the cat gene in pKK232-8 | D. Knowle, unpublished |
| pDK435 | Transcriptional fusion of pvuIIC promoter including wt C-boxes (positions −93 to +88) in front of promotorless lacZ reporter gene in pKK232-8, TetR, p15A ori | D. Knowle, unpublished |
| pIM1 | pvuIIC under control of araBAD promoter in pBAD24, AmpR, ColE1 ori | This study |
| pIM2 | Kan cassette inserted to break bla gene in pIM1, KanR, ColE1 ori | This study |
| pIM4 | As pIM2 but p15A ori | This study |
| pIM6 | Nonrepression mutant, as pPvuRM3.4, but C-box (GACTcatAGTCtgtaGACTcaaGATC) | This study |
| pIM8 | As pDK435, but symmetrized C-box, as for pDK178 | This study |
| pIM9 | As pDK435, but C-box sequence as for pIM6 | This study |
| pKK232-8 | Promotorless cat reporter gene | (66) |
| pMal-c2x | Vector for purification of protein fused to malE (maltose-binding protein) | New England Biolabs |
| pMal.CPvII | Translational fusion of pvuIIC::malE in pMal-c2x | This study |
| pPvuRM3.4 | wt PvuII R-M system in pBR322 backbone | (4) |
| pPvuM1.9-ACYC | As pPvuM1.9 (M+R-C-), but p15A ori | (4) |
| pRWWW | As pDK178, C-box–RWWW (TCAGcatAGTCtgtaGACTcaaAGCT) | D. Knowle, unpublished |
| pWWWR | As pDK178, C-box–WWWR (GACTcatAGTCtgtaGACTcaaCTGA) | D. Knowle, unpublished |
| pWWRW | As pDK178, C-box–WWRW (GACTcatAGTCtgtaTCAGcaaAGTC) | D. Knowle, unpublished |
| pWRWW | As pDK178, C-box–WRWW (GACTcatCTGAtgtaGACTcaaAGCT) | D. Knowle, unpublished |
| UL3 (WWRR) | As pDK178, C-box–WWRR (GACTcatAGTCtgtaTCAGcaaCTGA) | This study |
| UL4 (RRWW) | As pDK178, C-box–RRWW (TCAGcatCTGAtgtaGACTcaaAGCT) | This study |
In vivo titration of C.PvuII
All experiments were performed in MOPS-minimal medium (Teknova) with 0.2% glycerol or 0.2% glucose as carbon source (63). The minimal media were supplemented with 2 μg/ml thiamine and 40 μg/ml each of the l-amino acids Ala, Arg, Glu, Gly, His, Ile, Leu, Lys, Met, Pro, Ser and Val. Single colonies were used to inoculate overnight cultures in MOPS media containing 0.2% glucose and appropriate antibiotics. These cultures were diluted 1:50 into the same medium but without antibiotics, and grown with shaking to an OD600nm of 0.2–0.3. The cells were then gently pelleted, resuspended and divided among flasks containing MOPS-minimal media with varied concentrations of l-arabinose.
β-Galactosidase assay
The LacZ assay was based on hydrolysis of O-nitrophenyl-β-d-thiogalactoside (64) as modified by others (65). Briefly, β-galactosidase activity and culture density were measured at 20–30 min intervals during exponential growth. The units for this assay were calculated by dividing the measured A420nm (released nitrophenol) by the time allowed for the reaction and by the volume of permeabilized cells used for the reaction. The units of β-galactosidase activity are arbitrary units: 1000 × ΔA420nm min−1 ml−1. The specific activity was obtained by determining the slope of a plot of β-galactosidase activity versus the OD600nm culture density via linear regression.
C protein purification
C.PvuII was expressed from plasmid pMal.CPvuII, in which the PCR-amplified pvuIIC gene (primers C1, C2) from pPvuRM3.4 (4) was ligated into the XmnI and SalI sites of plasmid pMal-c2x (New England Biolabs, MA). In pMal.CPvuII, pvuIIC is translationally fused to malE (maltose-binding protein, MBP). The signal sequence for malE has been deleted, so the fusion protein remains in the cytoplasm. The host was E. coli strain BL21(DE3), cultivated in 1 l LB medium supplemented with ampicillin (100 µg/ml) and 0.2% glucose at 37°C. When the cell density reached an OD600nm of 0.5, overproduction of C.PvuII was induced by adding IPTG to 0.5 mM. After 3 h incubation, cells were pelleted and stored at −70°C until used. Frozen cells were thawed in C buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA) and sonicated (20 × 10 s in a cup horn probe). The centrifugally clarified lysate was applied to a 2.5 × 10 cm amylose resin column (New England Biolabs), and bound fusion protein eluted with 60 ml of 10 mM maltose in C buffer. Protein-containing fractions were pooled and digested overnight with Factor Xa protease (1 mg per 100 mg fusion protein) at room temperature to cleave the MBP from C.PvuII. The digest was concentrated in an Amicon Ultra-15 centrifugal filter unit (5 kDa MW cutoff; Millipore). Then gel filtration chromatography was used to separate C.PvuII from MBP. A Superose-12 column (Amersham-GE) was equilibrated with S buffer (20 mM Tris, pH 8.0 and 100 mM NaCl), and the protein sample was loaded. C.PvuII fractions were pooled, concentrated again in Amicon filter units and stored in 20% glycerol at −20°C. About 2.2 mg C.PvuII was obtained with purity >99% as assessed by SDS–PAGE.
Generation and analysis of randomized libraries
Oligodeoxyribonucleotides L1, L2 and L3 (IDT, Iowa) span the region upstream of the pvuIIC initiation codon, and include four consecutive randomized positions each: L1: the spacer between half-boxes 1B and 2A, L2: half-box 2A (with 2B wt) and L3: half-box 2A (with 2B as the reverse complement), as described in Table S1 and Figure S1. Oligodeoxyribonucleotide UP was used to generate the complementary strand. The two oligodeoxyribonucleotides (L1 and UP for the spacer library, L2 and UP for the WWNW library, or L3 and UP for the WWNR library) were hybridized, and the shorter primer was extended with Klenow polymerase. The DNA was digested with BamHI and SalI, and ligated into pDK178 (which contains PpvuIICR from −91 to −1 relative to the initiation codon of pvuIIC, in the cat promoterless vector pKK232-8). This plasmid was electroporated into E. coli TOP10 cells, and plated onto LB agar containing carbenicillin (Cbn, 50 μg/ml). Transformants were pooled, and plasmid DNA was recovered with the midi plasmid kit (QIAGEN). The library DNA was electroporated into an E. coli strain containing pDK200 (a pACYC184-based plasmid that specifies pvuIIC, but not pvuIIR or pvuIIM), and colonies were selected on plates containing Cbn and chloramphenicol (Cml), with the latter at either 20 or 120 µg/ml. To isolate nonrepressing mutants, the library plasmids were introduced into E. coli MC1061 carrying pIM4, and transformants were plated onto solid MOPS-minimal medium supplemented with 0.2% arabinose and 80 µg/ml Cml. Under these conditions, a large (repressing) amount of C.PvuII protein is made, so only nonrepressing mutants will express cat, and C.PvuII-independent mutants are eliminated by screening. Plasmid DNA was isolated from resultant colonies, and the 4 nt randomized region was determined by sequence analysis.
Chloramphenicol acetyltransferase (CAT) assay
The CAT ELISA kit (Roche) was used to colorimetrically quantitate CAT reporter levels based on a sandwich ELISA method. The recommended protocol was followed with some modifications. The culture was grown in LB or MOPS-minimal medium until the OD600nm reached 0.3. Samples (1 ml each) were removed and, after pelletting, the cells were resuspended in 0.5 ml of lysis buffer (100 mM K/PO4 pH 7.8, 1% Triton X-100, 5 mg/ml BSA, 1 mM DTT, 5 mg/ml lysozyme) and kept for 20 min at RT. The centrifugally cleared cell extracts were diluted 1 : 50 and loaded onto microplate modules with anti-CAT antibodies prebound to the surface. CAT concentration was calculated as nanogram per microgram of total protein. The total protein concentration was determined with the RC/DC kit and protocol (Bio-Rad).
Electrophoretic mobility shift assays (EMSA)
DNA substrates were 5′-biotinylated, double-stranded PCR-amplified fragments (primers: emsa1, 2), that included the entire PpvuIICR region (wt or mutant as indicated). For a nonspecific DNA substrate, having no C-boxes but of the same size as the other substrates, primers emsa3, 2 were used (Table S1). This primer pair amplified DNA from the polylinker region of the vector pKK232-8 (GenBank #U13859)(66). Reactions containing 20 nM DNA and the indicated protein concentrations were prepared in binding buffer [50 mM Tris–HCl (pH 8.0), 1 mM DTT, 10 mM MgCl2, 2.5% glycerol] in a final volume of 20 µl, and incubated for 20 min at 22°C. Samples were electrophoresed on 10% native polyacrylamide gels in 0.5 × TBE buffer for 90 min at 100 V at 22°C. The location of dsDNA in the gels was determined either via ethidium bromide staining and photography with UV transillumination, or DNA was transferred by electroblotting to positively charged nylon membranes (Ambion), and the transferred DNA fragments were immobilized onto the membrane by ultraviolet (UV) cross-linking. Detection of the biotin end-labeled DNA was performed using the North2South Chemiluminescent Hybridization and Detection Kit (Pierce) as recommended, and the CCD camera of the Omega Molecular Imaging System (UltraLum). For competition experiments, the unlabeled DNAs were used at 1-, 2.5-, 5-, 10- or 20-fold molar excess followed by addition of C protein, electrophoresis and analysis as described above.
Western blot analysis
For each sample, equal volumes of culture were centrifuged at 16 000 g for 2 min. The supernatants were removed and the cell pellets stored at −80°C until analysis. Pellets were resuspended in 1× SDS buffer (Novagen), and lysed by heating to 98°C for 10 min. Total protein concentration was determined using the RC/DC kit and protocol (Bio-Rad). Equal amounts of protein were loaded onto a 4–12% Bis–Tris NuPAGE Novex gradient gel (Invitrogen) and electrophoresed at 100 V in 1 × NuPAGE MES buffer (Invitrogen). Proteins were then electroblotted to PVDF membranes at 30 V for 1 h using an Xcell apparatus (Invitrogen). Proteins were detected by fluorescence using the ECL-plus Western Blotting Detection System (GE Health Sciences) as per the manufacturer's protocol, with 1:1000 dilution of rabbit anti-C.PvuII polyclonal serum (Strategic Biosolutions), and a 1 : 25000 dilution of horseradish peroxidase-conjugated goat ant-rabbit IgG. Protein bands were visualized on an Omega Molecular Imaging System (UltraLum). The prestained MW markers used were SeeBluePlus (Invitrogen).
Efficiency of transformation (EOT) assay
EOT is defined in this study as the relative number of transformants obtained, using the same amount of plasmid DNA, from the test strain compared with the number of transformants obtained from the reference strain. This term is equivalent to the term ‘relative transformation efficiency’. For plasmid transformation, the standard CaCl2-heat shock method was used (67) with plasmid DNA amounts that were nonsaturating.
RESULTS
Accumulation of C.PvuII leads to strong repression
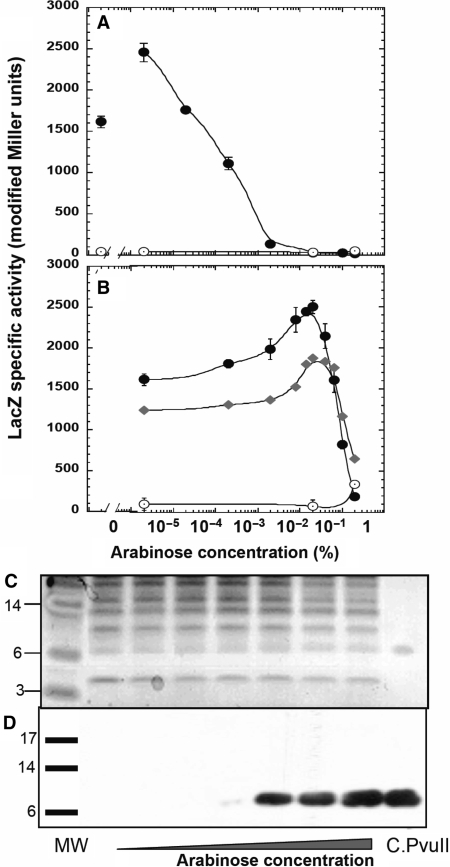
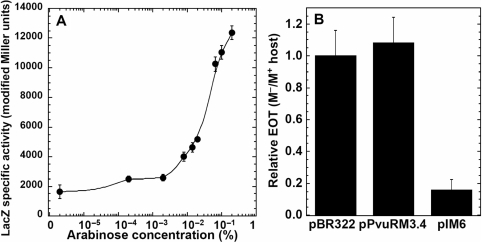
To examine the DNA-binding sites important for activation of the promoter for the C and REase genes (PpvuIICR) by C.PvuII, we first tested the effects of an in vivo titration in which intracellular C.PvuII protein levels were varied from undetectably low to overexpression. We used the E. coli araBAD promoter (PBAD), as its activity can be modulated over a wide range (68–70). We fused pvuIIC to PBAD, followed by strong transcription terminators, to generate plasmid pIM1 (Table 1). This plasmid also carries the gene for the bifunctional AraC protein, that represses PBAD in the absence of arabinose and activates it when arabinose is present. To reveal the effects of the C.PvuII titration, we cloned the PpvuIICR region (including wild-type C-boxes, positions −93 to +88, Figure 1A) upstream of the lacZ reporter gene (plasmid pDK435 or its variant pIM8 with the symmetrized C-boxes), generating transcriptional fusions. The host was E. coli MC1061, which lacks the lac and ara operons but carries araE for arabinose transport. Experiments were carried out in minimal media with concentrations of arabinose from 0 to 0.2% (saturating induction) (Figure 2A and B). The effects of induction on C.PvuII levels were confirmed by western blot (Figure 2D, and I.M. unpublished data), and show the strong nonlinear response to arabinose concentration characteristic of PBAD (68). Expression of the PpvuIIC-lacZ transcriptional fusion was measured by β-galactosidase assay.
Figure 2.
In vivo titration with C.PvuII and its effect on pvuIICR transcription. Cells were grown in minimal media with 0.2% glycerol and the indicated concentration of arabinose, and supplemented as described in Materials and Methods section. In addition, the cells were grown without (A) or with (B) 0.2% glucose. Expression from transcriptional fusion PpvuIICR:lacZ (pDK435) was measured as β-galactosidase specific activity, determined by linear regression of the slopes of the lines generated by plotting LacZ activity (modified Miller units) versus optical density of the culture (Platko et al., 1990). Unconnected points at the beginning of the curve (panel A) indicate values obtained in glucose with no arabinose. Closed symbols represent LacZ activity from cells with the native PvuII C-boxes (circles, pDK435) or its symmetrized variant (diamonds, pIM8), in the presence of PBAD–pvuIIC (plasmid pIM1); open circles represent LacZ fusion activity when the vector plasmid (lacking pvuIIC) is present (plasmid pBAD24). (C) Extracts from the same cultures shown in (A) were resolved on a 4–12% acrylamide SDS gel. Ten micrograms of total protein was loaded per lane, except for the MW protein markers in the far left lane and 150 ng of purified C.PvuII in the far right lane. Lanes 1–7 contained culture samples: 1-glucose, 0% arabinose; 2–0% arabinose; 3–7-increasing concentrations of arabinose, in 10-fold steps, from 0.00002 to 0.2%. The gel was then silver stained and photographed. (D) A parallel gel to that shown in (C) was electroblotted to a PVDF membrane, blocked, and probed with polyclonal rabbit anti-C.PvuII antiserum and then with horseradish peroxidase-coupled goat anti-rabbit IgG. The positions of the prestained MW markers are indicated by bars. The expected subunit size of native C.PvuII is 9.4 kDa.
The results were unexpected for a known transcription activator (27,40). In the absence of glucose, the lowest achieved induction level of C.PvuII gave the maximal observed expression of the fusion, with higher levels of C.PvuII (Figure 2C, D) associated with progressively lower LacZ expression (Figure 2A). Increases in the intracellular level of C.PvuII were associated with substantial reductions in LacZ expression from PpvuIICR. Specifically, increasing arabinose from ∼10−5 to ∼10−3% gave a several 100-fold decrease in LacZ expression, ultimately dropping to the range of the vector control at 0.02% arabinose. Even the absence of arabinose (where C.PvuII levels were undetectably low) was associated with significantly more LacZ activity than was seen at high levels of arabinose (Figure 2A). This may reflect the activity of the known C-independent promoter (Figure 1D), believed to provide starting amounts of C.PvuII for this positive feedback system (40,43), but the results indicate the occurrence of repression.
As C.PvuII is known to activate transcription (27,40), we tested an even lower range of C.PvuII levels to determine whether this in vivo titration system shows progressive activation. For this purpose, we used the same minimal medium and arabinose concentrations, but added 0.2% glucose. Glucose causes catabolite repression, that indirectly affects the PBAD promoter via effects on the promoter for the AraC repressor-activator gene (71). The combination of glucose and arabinose resulted in an increase in LacZ expression between 0 and 0.02% arabinose, where it reached a maximum 1.6-fold increase over the initial expression level (Figure 2B). Further induction of C.PvuII levels with arabinose led as before to a sharp drop in LacZ activity, down to background levels at 0.2% arabinose. Both the WT and symmetrized PvuII C-boxes yielded the same activity profile, with slightly lower values for the variant (Figure 2B). This result clearly indicates that C.PvuII is both an activator and a repressor. This is the first in vivo demonstration of concentration-dependent activation-repression by C proteins.
C.PvuII exhibits higher affinity for OL than for OR
As there are two C-box symmetry pairs (Figure 1), a straightforward model is that occupancy of one operator leads to activation, while binding to the other (or to both) leads to repression. This has been directly proposed for the AhdI C-dependent R-M system (and indirectly for the EcoRV system) (33,45). However this model predicts that there is a significant difference in C.PvuII affinity for the two operators. Further, C.AhdI had been found to occupy its two operators with such high cooperativity that no intermediate (with C.AhdI bound to just one operator) could be found [see Figure 1 in (45)], that would give a very narrow concentration range in which activation could occur. To test the relative affinity of C.PvuII for OL and OR, we performed C.PvuII–C-box binding experiments via EMSA.
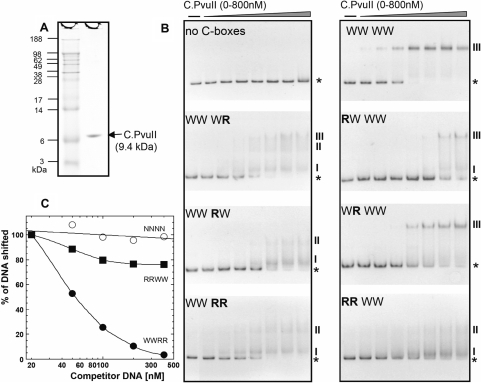
C.PvuII was purified to >95% homogeneity (Figure 3A). For this, pvuIIC was fused at its N-teminus to malE, which specifies the E. coli maltose-binding protein (MBP). The fusion protein was purified by affinity chromatography and cleaved to separate C.PvuII from MBP (see Materials and Methods section). Subsequent gel filtration yielded 35 ng of C.PvuII protein per milliliter of starting culture. The binding reactions varied C.PvuII concentrations from 0 to 800 nM.
Figure 3.
C.PvuII protein and its in vitro interaction with wild-type and altered C-boxes. (A) C.PvuII was purified as described in Materials and Methods section. MW markers and 1.5 µg of C.PvuII were resolved on a 4–12% acrylamide SDS gel and silver stained. (B) A series of 126 bp dsDNA binding targets were prepared by PCR amplification, and included in each binding reaction at 20 nM. The DNAs were a control sequence (‘no C-boxes’; amplified from the polylinker region of the vector plasmid), or contained WT or variant C-boxes flanked on either side by 50 bp of native PvuII sequence. The increasing concentrations of C.PvuII were 0, 100, 150, 200, 300, 400, 600 and 800 nM. Reactions were processed as outlined in Materials and Methods section. ‘WWWW’ indicates that all four C-boxes are WT, where this refers to the proposed symmetrical C-box sequences (underlined): GACTCATAGTCTGTAGACTCAAAGTC. ‘R’ refers to a reversed variant C-half-box (GACT→TCAG or AGTC→CTGA), and the position of the R or W refers to C-half-boxes 1A, 1B, 2A and 2B in that order (see Figure 1D). The PvuII background sequence remains intact. Reactions were resolved on 10% native polyacrylamide gels, and DNA was visualized by staining with ethidium bromide. The stars and Roman numbers denote positions of unbound DNA and the C.PvuII–DNA complexes, respectively. Bands with the same numbers correspond to one another when run side by side on the same gel (data not shown). (C) EMSA competition assays were performed using 300 nM of C.PvuII and 20 nM of biotin-labeled WWWW C-box 126-mer (as described in the Figure 2 legend). Competitions contained increasing amounts of unlabeled 126-mer DNA fragments (from 1- to 20-fold molar excess); competitors were RRWW (squares), WWRR (closed circles), or no C-boxes (open circles, negative control). Following EMSA and electroblotting, the shifted bands for each reaction were visualized and quantified via chemiluminescent detection of the biotinylated DNA as described in Methods section.
The target DNAs were based on the 26 nt sequence containing symmetrical C-boxes, GACT-(N3)-AGTC-(N4)-GACT-(N3)-AGTC, derived from a comparison of 29 R-M systems that specify known or putative C.PvuII orthologs (Figure 1B, C). The spacers are listed here as ‘N’ to distinguish them from the symmetry elements, but are themselves conserved (see below), and were unchanged from the wild-type PvuII sequence (Figure 1D). The WT PvuII C-boxes differ from this pattern at just two of 16 positions: in half-boxes 1A (GACT→TACT) and 2A (GACT→GATT). This symmetrized sequence is used as the reference ‘wild-type’ in subsequent experiments. Reporter fusion assays indicated that this symmetrized promoter/operator region was transcribed at similar levels to the native sequence (43), and with the same activation/repression profile (Figure 2B); our rationale is that identical C-box elements in the two operators would facilitate characterizing any position-specific differences in their roles.
In the first set of mutants we prepared, one or more 4 nt half-boxes were substituted by the reversed sequence (GACT→TCAG or AGTC→CTGA). These mutants were named by indicating W (‘Wild’ symmetrized) or R (Reversed) for each of the four half-box elements, 5′→3′ (e.g. RWWW where half-box 1A is reversed). For gel mobility shift experiments, we used 126 bp PCR products amplified from the various mutagenized plasmids at a fixed concentration of 20 nM. The C-boxes were located at the center of the amplification product, such that the 26 bp C-box segment was flanked on each side by 50 bp of DNA from the PvuII R-M system.
The top two panels of Figure 3B show positive (WWWW) and negative controls (a 126 bp fragment of an irrelevant sequence). The C.PvuII protein bound WWWW DNA, yielding a single complex at concentrations ≥400 nM. The binding of C.PvuII to the symmetrized C-box region (WWWW, complex marked ‘III’) was characterized by a Hill coefficient of 4.1 (Figure S2). This result indicates strong positive cooperativity for binding, and suggests [but does not prove (72)]) that one C.PvuII monomer binds to each C-half-box, consistent with results for another C-protein, C.AhdI (45).
We next tested binding to the DNA derivatives having one or more half-sites reversed. First, an intact OL was necessary and sufficient for enough binding to eliminate free DNA, at least at the highest concentrations of C.PvuII (Figure 3B, bottom three panels on left, including WWRW, WWWR and WWRR). However, mutation of OR had three obvious effects. First, the amount of C.PvuII required for mobility shift was higher for the mutants; for example 200 nM C.PvuII shifted about half of the WWWW and virtually none of the WWRW DNA. Second, complexes with WWRW DNA yielded multiple bands, suggesting loss of cooperativity; and third, the bands are smeared, suggesting that the WWRW complexes are less stable than WWWW complexes.
Any mutation within OL (RWWW, WRWW or RRWW, with OR left intact) significantly reduced the association of C.PvuII as revealed by the amount of unbound DNA remaining (Figure 3B, lower three panels on right). Nevertheless, the OL single half-box variants RWWW and WRWW still yield some of the distinct, major complex (III) corresponding to that observed with WWWW DNA.
These results imply that C.PvuII has greater binding affinity for OL than for OR. To confirm this, EMSA competition studies were performed. Biotin-labeled WWWW DNA was used with increasing concentrations of unlabeled competitor WWRR (OL intact), RRWW (OR intact) or no C-box (as a negative control) DNA with 1-, 2.5-, 5-, 10- or 20-fold molar excess. The results (Figure 3C) directly demonstrate the higher binding affinity of OL as WWRR DNA was the stronger competitor, where a 20-fold excess of competitor resulted in a 95% loss of the shifted WWWW band. In contrast, the same amount of RRWW competitor DNA led to only a 25% loss of the shifted WWWW complex.
C.PvuII-dependent activation of transcription requires the intact 4 nt spacer between operators OL and OR
The binding studies described above suggest that OL (half-boxes 1AB) is the primary binding site for C.PvuII. Since activation precedes repression as C.PvuII levels rise, both logically and according to the data in Figure 2B, OL is the likely activation site for pvuIICR transcription. The simplest model is that OL binding alone mediates activation, while OR binding is responsible for repression. However, this simple model provides no role for the 4 nt spacer between OL and OR, that includes the most highly conserved positions among C-box regions (Figure 1C). Over 75% (16/21) of the identified C-box spacer sequences, from C proteins with an HTRY recognition helix, have the sequence TGTA, and 28/30 of all sequences have GT in the central two positions (Figure 1B). Accordingly, we tested the role of the operator spacer (TGTA in the PvuII R-M system). This spacer overlaps the predicted −35 hexamer of the C-dependent promoter (Figure 1D), raising the question of whether its conservation is coincidental with this overlap or if the spacer plays some role in C.PvuII binding.
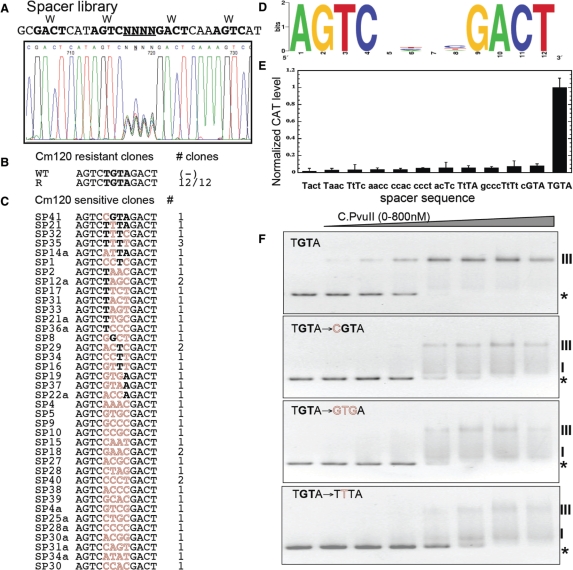
To test the role of the operator spacer, we generated a plasmid library in which the 4 nt spacer was replaced with a randomized sequence, and the promoter region including this modification was cloned in front of a promotorless cat (chloramphenicol acetyltransferase) reporter gene (details in Materials and Methods section, and in Table S1 and Figure S1). Electroporation into E. coli produced about 104 transformants, giving a theoretical ∼40-fold coverage of the 256-possibility library. The pooled library was purified and sequence confirmed, consistent with randomization where expected and with no other changes (Figure 4A). This confirmed library was then electroporated into E. coli cells containing pDK200, a compatible plasmid that supplies intact pvuIIC in trans. The objective was to determine which spacer sequences conferred high-level resistance to Cml, due to increased cat expression, in the presence, but not the absence of C.PvuII.
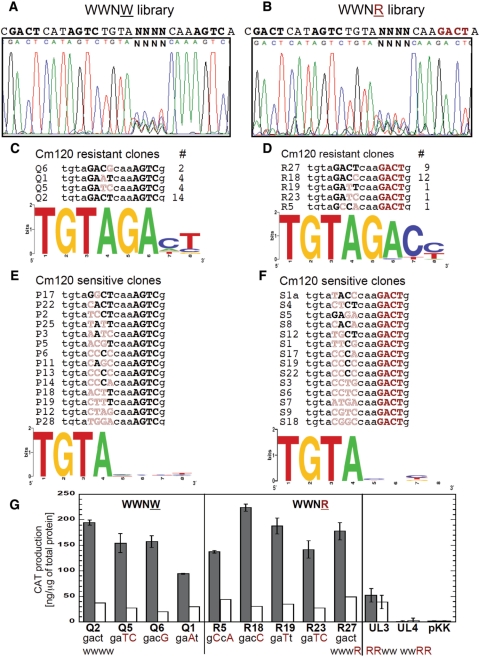
Figure 4.
Randomized operator spacer library and selected variants. (A) Sequencing trace of the pooled randomized spacer plasmid library before selection. The randomized spacer lies between C-half-boxes 1B and 2A. The promoter region library is upstream of a promoterless cat gene (chloramphenicol acetyltransferase). (B) The plasmid library was used to transform an E. coli strain that already carried a compatible plasmid producing C.PvuII at physiological levels, and transformants were plated onto agar with a high Cml concentration (120 µg/ml) to select functional C.PvuII-activated variants, followed by a screen for reduced resistance in the absence of C.PvuII. The sequencing result is shown, along with the WT sequence for comparison. (C) Non-activated variants, that grew at a low Cml concentration (20 µg/ml) but not at 120 µg/ml. Sequences are shown, with matches to the WT sequence in bold, and alterations in outline. (D) Logo analysis of the Cml-sensitive variants shown in (C). (E) Promoter activity for selected variants in the presence of physiological steady-state levels of C.PvuII (pvuIIC under native control from plasmid pDK200). CAT levels were determined in triplicate via immunoassay as described in Material and Methods section. Error bars indicate the SD. CAT levels are expressed as nanograms of protein per microgram of total protein and then normalized to the WT PpvuIICR-cat level (TGTA). (F) EMSA performed for WT (TGTA) and three representative variants. The experiments were carried out as in Figure 3B. The stars and Roman numerals denote positions of unbound DNA and C.PvuII–DNA complexes, respectively.
Pilot experiments revealed that the plasmid carrying the wild-type (TGTA) spacer confers resistance to 120 μg of Cml/ml in the presence of C.PvuII, but to only 20 μg/ml in the absence of C protein or in the presence of inactive C protein (mutant pvuIIC Esp19, (41)). The latter low-level resistance is presumably due to the weak C-independent promoter (see Figure 1D). Similar results were obtained for both solid and liquid media. We thus plated the plasmid library, independently in pvuIIC± hosts, onto 20 and 120 μg of Cml/ml in parallel. We obtained about 80-fold more colonies on the lower than on the higher Cml concentration (data not shown).
Next, individual resistant colonies were picked and their plasmid DNAs were isolated and digested with a restriction enzyme to selectively inactivate the pvuIIC-carrying plasmid. The resultant DNA was used to transform cells carrying plasmid pDK201, that produces inactive C.PvuII, followed by screening for loss of the high-level Cml resistance. Such plasmids were considered to be subject to C-dependent activation, and were sequenced. Surprisingly, in 12/12 sequenced variants giving C.PvuII-dependent activation, the randomized 4 nt spacer combination was strictly limited to TGTA (Figure 4B). The random probability of obtaining the single 4 nt sequence from this library is 1/256 (1/44). The probability of getting 12/12 variants with the identical sequence by chance is ∼1.2 × 10−29, and strongly indicates that all four positions are required, at least in the PvuII R-M system.
We also sequenced >40 clones that were sensitive to 120 µg Cml/ml, even in the presence of C.PvuII, but resistant to 20 µg/ml. The resulting Logo shows that none of the four positions is strongly biased (Figure 4D). Two variants among the sequenced non-activated clones maintained 3 out of the 4 WT nt, TtTA and cGTA, even though these changes did not overlap the −35 promoter hexamer.
To quantitate the effects of these changes on transcription activation, we measured CAT protein levels in the cells by an ELISA assay. The results (Figure 4E) show that even changing a single base in the operator spacer reduced promoter activity by 90–95%. To determine whether this effect is due to impaired C.PvuII binding, EMSA was performed. The results clearly indicate that altering even one position in the spacer (cGTA or TtTA; Figure 4F) impairs C.PvuII binding about as much as reversing one of the C-box half-sites (Figure 3B). A nucleoprotein complex formed and migrated at the position characteristic of the WT DNA sequence. However, as with the half-site reversals, disappearance of unshifted DNA occurs at higher C.PvuII concentrations, an intermediate complex is more pronounced, and band smearing suggests that the complexes are unstable. Other tested variants (from among those shown in Figure 4E) gave similar binding patterns (data not shown). These results indicate that the TGTA spacer plays a role independent of the overlapping −35 hexamer.
Half-box 2B in OR is dispensable for C.PvuII-dependent activation
If C.PvuII binding to OR is associated exclusively with repression, then altering that site should not reduce activation by C.PvuII. To test this prediction, we again prepared a randomized plasmid library for selection on plates and CAT reporter assays. We generated two separate libraries, each of them containing a randomized half-box 2A in OR. In one library, designated WWNW (where ‘N’ indicates the randomized 4 nt sequence), the background sequence of the other three half-boxes and the TGTA spacer remain intact (Figure 5A). In contrast, the second library, labeled WWNR, includes a reversed complement of half-box 2B (Figure 5B).
Figure 5.
Randomized C-half-box 2A libraries and selected variants. (A) Sequencing trace of the pooled randomized C-half-box 2A plasmid library before selection. All other C-half-boxes were WT (WWNW, where ‘N’ indicates the randomization), and the promoter region library is upstream of a promoterless cat gene. (B) Sequencing trace as in (A), except that in this library C-half-box 2B is replaced by the reversed complement (AGTC→GACT; WWNR). (C) The selection for C.PvuII-activated variants was as described in Figure 4B. The resulting sequences from the WWNW library, number of variants showing each recovered sequence, and Logo analysis are shown. (D) Sequences that could be activated detectably as in (C), but for the WWNR library. (E) The isolation of variants that are not detectably activated by C.PvuII was as described in Figure 4C. The resulting sequences from the WWNW library and Logo analysis are shown. (F) Sequences that are not detectably activated as in (E), but for the WWNR library. (G) Quantitative evaluation of C.PvuII-activated variants from WWNW (panel C) or WWNR (panel D) libraries. CAT levels were measured either in the presence of physiological steady-state levels of C.PvuII (pvuIIC under native control on plasmid pDK200; black bars), or without pvuIIC (white bars). CAT levels were determined via triplicate immunoassays as described in Material and Methods section. For comparison, mutants with inactive OR (WWRR) or OL (RRWW) were also analyzed. pKK represents vector (pKK232-8) control. The error bars indicate SDs.
The rationale for producing two libraries (WWNW and WWNR) derives from the overlap between half-box 2A and the −35 hexamer of the C.PvuII-activated promoter (Figure 1D). If OL binding leads to activation and OR binding leads exclusively to repression, then C.PvuII-activated variants from the WWNW library should include sequences that preserve −35 functionality but do not resemble the GACT C-binding consensus. That is, the sequences should retain RNAP binding but not necessarily repression-associated C.PvuII binding. The −35 hexamer consensus sequence is 5′-TTGACA-3′, with a 5′→3′ gradient of decreased conservation (57,73,74). However, if cooperative binding to both OL and OR (or OL and half-box 2A) is required for full activation, the randomized half-box 2A should retain similarity to GACT (as well as to the -35 consensus). The second library (WWNR) has disrupted C.PvuII binding to OR in all cases, and should yield C.PvuII-activated variants only if OR (or at least half-box 2B) plays no role in activation.
The WWNW and WWNR library analysis followed the same protocols as for the TGTA spacer library (described above). Electroporation into E. coli produced about 8000 transformants per library, again assuring full coverage given the complexity of 256 sequences. Each pooled library was purified and sequence confirmed, indicating randomization at each of the four positions with no other changes (Figure 5A, B). For each library, 24 clones showing C.PvuII-dependent resistance to 120 µg Cml/ml were isolated (Figure 5CD).
For the WWNW library, nearly 60% of variants had the wild-type symmetrical consensus GACT at half-box 2A. The other activated sequences were GATC, GAAT and GACG. It thus appears that the C.PvuII binding sequence GACT is favored (and certainly not disfavored). If OR is associated exclusively with repression, then sequences associated with better C.PvuII binding should not have emerged from a selection for C.PvuII-dependent activation. C.PvuII-activated variants were also obtained from the WWNR library, demonstrating that half-box 2B is not required for activation. From this library five different half-box 2A sequences were recovered, but again the GACT sequence was favored (38%). A second preferred sequence, unique to that library, was GACC (50%) with small amounts of GATT (the PvuII wild type) and GCCA.
We next carried out quantitative analysis of the isolated WWNW and WWNR library variants, using ELISA to measure CAT production. The assay was carried out in cells with or without plasmid pDK200, which supplies WT C.PvuII. The highest cat expression, in the presence of C.PvuII, was obtained when half-box 2A had the sequence GACT (WT) in the WWNW context, or GACT (WT), GATT, GACG and in the WWNR context (Figure 5G). Again, it is noteworthy that there is no expression penalty for a strong C.PvuII-binding sequence at box 2 (or OR overall).
Surprisingly, while WWWR mutants had substantially-impaired C.PvuII binding, as judged by EMSA (Figure 3B), some variants in the WWNR context produced CAT protein at WT levels (Figure 5G). All tested variants and WT produced similar basal CAT levels in the absence of C.PvuII, presumably driven by the weak C-independent promoter (Figure 1D). RRWW (UL3) and WWRR (UL4) C-box-cat combinations were also generated and analyzed as controls (Figure 5G). The RRWW mutant (inactive OL) cannot drive transcription in a C-dependent manner, as CAT expression remains at the basal level whether or not C.PvuII is provided in trans (Figure 5G). The WWRR mutant (inactive OR) has no promoter activity at all, presumably due to disruption of the overlapping -35 hexamer of the C-dependent promoter and changes immediately adjacent to the −10 hexamer of the C-independent promoter (Figure 1D).
Several clones from each library that were sensitive to 120 µg/ml Cml (Figure 5E and F) were also sequenced and analyzed by CAT assay. None of them is able to drive cat expression above the basal level in the presence of C.PvuII (data not shown; variants tested were CGTA, TTTA, TTTC and TTTT). We assessed C.PvuII binding for selected sensitive variants having one or 2 nt changed in half-box 2A (Figure S3). In all three variants the shifted complex was smeared, suggesting reduced stability.
Repression is associated with OR
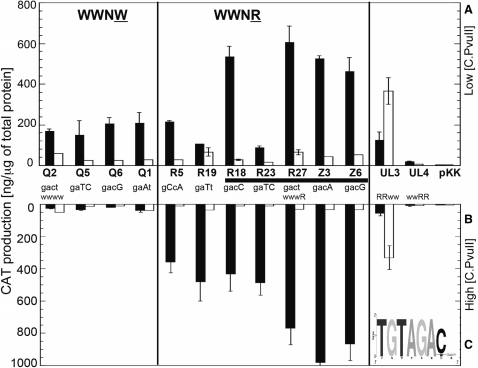
The library selection results shown in Figure 5 did not address the role of OR in the repression we observed (Figure 2A and B). To study this, we again used our randomized plasmid libraries WWNW and WWNR. These libraries were introduced into cells that contained plasmid pIM4, that has pvuIIC under the arabinose-inducible PBAD promoter (Figure 2, Table 1).
Our strategy was to select for Cml resistance in the presence of high-level C.PvuII induction, so as to isolate nonrepressing variants (if possible) from the libraries. Transformants were plated onto MOPS-minimal media supplemented with 0.2% glycerol, 0.2% arabinose and 80 µg Cml/ml. This concentration of arabinose led to very low cat expression in our titration experiments (Figure 2A).
We obtained no Cml-resistant variants from the WWNW library, but got many from the WWNR library (not shown). This result strongly suggests a key role for half-box 2B in repression. The majority of Cml-resistant WWNR variants contain half-box 2A sequences that were already obtained from screening C-dependent variants as shown in Figure 5D. The two new sequences were GACA and GACG.
CAT production was determined quantitatively for selected WWNR variants and, for comparison, some previously obtained C-dependent variants from the WWNW library (selected under lower C.PvuII expression levels; Figure 5G). The measurements were carried out under two conditions, chosen based on the in vivo titrations (Figure 2A). The first condition favors activation and maximum cat expression (0% arabinose; Figure 6A). The second condition favors repression (0.2% arabinose; Figure 6B). The results for all tested WWNW variants reveal similar levels of cat expression in low levels of C.PvuII (Figure 6A and B; white bars). When the C.PvuII level was increased by induction of its PBAD promoter, cat expression was significantly reduced in all cases.
Figure 6.
The effects of altered OR on activation and repression by C.PvuII. Quantitative evaluation of selected C.PvuII-activatable variants from the WWNW and WWNR libraries, upstream of a promoterless cat gene, was determined with C.PvuII produced from PBAD-pvuIIC (pIM4; black bars) or without pvuIIC (white bars). Furthermore, experiments were carried out at two levels of C.PvuII that, for the WT C-boxes, are respectively associated with activation (A, upper panel) and repression (B, lower panel). In panel A, cultures were grown in the absence of arabinose, leading to lower C.PvuII levels, while for panel B, cultures were grown in 0.2% arabinose, associated with high levels of C.PvuII (see Figure 2A). CAT levels were determined in triplicate immunoassays as described in Material and Methods section. For comparison, variants having inactive OR (WWRR) or OL (RRWW) were also analyzed. pKK represents vector (pKK232-8) control. The error bars indicate SDs. For each variant, the sequence of C-half-box 2A is shown (WT = GACT); the horizontal black bar indicates those variants that were selected from the library on the basis of nonrepressibility. (C) Logo analysis of the nonrepressing variants from the WWNR library, showing the interoperator spacer and C-half-box 2A.
In contrast, all Cml-resistant variants from the WWNR library, which lack an active half-box 2B, show no effect of the different C.PvuII levels on cat expression (Figure 6). For the half-box 2A variants GACT, GACG and GACA, the reporter gene expression is even increased (∼3-fold) by high levels of C.PvuII. This confirms that we have selected non-repressing variants, and again strongly implicates half-box 2B in repression. The half-box 2A logo for these non-repressing WWNR variants is consistent with half-box 2A providing part of the −35 hexamer in the absence of repression (Figure 6C).
Impaired establishment of the PvuII R-M system in nonrepressing mutants
The nonrepressing mutant WWWR gave about triple the expression of symmetrized pvuIICR promoter, even at a low concentration of C.PvuII, as revealed by the cat reporter gene fusions (Figure 6A). The same mutation (AGTC→GATC in half-box 2B of OR) was placed upstream of a lacZ reporter (pIM9), and the in vivo C.PvuII titrations were performed as had been done with the WT (pDK435) or symmetrized (pIM8) C-boxes (Figure 2B). The results (Figure 7A) confirmed that disruption of half-box 2B prevents repression, and leads to even greater than wt autogenous activation at higher C.PvuII concentrations.
Figure 7.
The effect of mutation in OR on pvuIICR transcription and on PvuII R-M system establishment. The nonrepressing C-box mutation (WWWR: 5′-GACT-CAT-AGTC-TGTA-GACT-CAA-GATC-3′) was tested in two ways. (A) In vivo titration with C.PvuII on arabinose inducible plasmid pIM1, where mutated C-box was fused to lacZ gene (pIM9). Cells were grown in minimal media with 0.2% glycerol, 0.2% glucose and the indicated concentration of arabinose as described in Figure 2B. The transcriptional activity was measured as β-galactosidase specific activity as described in the Figure 2 legend. (B) Equal amounts of three plasmid DNAs were used to determine the efficiency of transformation (EOT) in each of two host strains. The three plasmids were pPvuRM3.4 (WT PvuII R-M system), pIM6 (symmetrized nonrepressing variant), and pBR322 (vector control). These were introduced into competent E. coli TOP10 cells that already carried either the gene for the PvuII MTase (pvuIIM; plasmid pPvuM1.9-ACYC) or a vector control (pACYC177). Relative EOT was determined as the fraction of M.PvuII− transformants obtained relative to the number of transformants for the M.PvuIIM+ strain, and then normalized to the pBR322 EOT ratio. Error bars indicate the SD.
As C.PvuII-C-box interactions also control the downstream pvuIIR gene, we investigated the physiological effects of the repression. We introduced the same WWWR mutation into the complete PvuII R-M system via site-directed mutagenesis. The resulting plasmid (pIM6) retains the WT PvuII R-M system sequence intact, aside from this change and the symmetrization of the other C-boxes (see Figure 7 legend). To study the effect of eliminating repression on establishment in new cells, the intact or nonrepressing versions of the R-M system were introduced and we determined the EOT. Our hypothesis was that lack of repression increases amounts of REase relative to MTase, and thus increases the risk of lethal damage to the new host's chromosome. Accordingly, two sets of competent cells were prepared: one strain carried the pvuIIM gene (M+; plasmid pPvuM1.9-ACYC), so the cells were fully protected before the R-M system was introduced, and the other strain lacked pvuIIM (M-; pACYC184). Equal concentrations of three types of plasmid DNA were introduced: pPvuRM3.4 (wt R-M systems), pIM6 (nonrepressing R-M system) or pBR322 (vector control). The relative EOTs were expressed as the fraction of obtained colony-forming units (CFUs) for M− versus M+, and then normalized to the pBR322 values. As predicted by the hypothesis, pIM6 plasmid appears to have a preferentially lethal effect on M− cells, as the EOT was ∼5-fold lower than that in M+ cells. (Figure 7B). In contrast, neither the wild-type PvuII R-M system nor the vector control plasmid showed any significant M.PvuII-dependent difference in EOT (Figure 7B).
This result helps to explain our observation that the nonrepressing clone (pIM6) could be obtained only when we used premethylated (M+) competent cells for transformation with our ligation reactions (I.M., unpublished data). In M.PvuII− competent cells, the clones we obtained had large rearrangements near the introduced mutation. The transformation results clearly indicate that the observed repression plays an important physiological role, and is required for efficient establishment in new host cells. This is the first demonstration that the repression part of the regulatory circuit has in vivo significance.
DISCUSSION
We present direct in vivo as well as in vitro evidence for a model of PvuII R-M system regulation that could also apply to the other RM systems controlled by orthologous C genes (Figure 1B). In this model, C protein acts as both transcription activator and repressor for its own gene (pvuIIC, in this case) and that of the downstream REase gene (pvuIIR). While bifunctional regulators such as AraC switch between activation and repression based on small-molecule coregulators, C.PvuII apparently makes this switch based on its concentration alone, like the cI activator-repressor of bacteriophage λ (75). Our results are consistent with in vitro studies on C.AhdI and C.EcoRV (33,45) and suggest that the regulatory switch depends on C protein occupancy within the C-box region that, in turn, is highly sensitive to the intracellular C protein concentration.
Role of repression in control of R-M systems such as PvuII
R-M systems appear to undergo horizontal gene transfer at a substantial rate (2). C proteins are believed to facilitate this process by delaying REase expression, so that the cognate MTase has time to protectively modify the new host's DNA during the establishment in a new cell (40,43). The delay is thought to be a direct consequence of the requirement of the pvuIICR promoter for autogenous activation by C.PvuII. Evidence supporting this view includes the inability to transform cells with the PvuII R-M system if the cells already contain a copy of pvuIIC (13,40). However this delayed activation is apparently necessary but not sufficient to ensure efficient establishment. Our data demonstrate that impairing the repression portion of the C.PvuII regulatory circuit significantly reduces the efficiency with which the entire R-M system is transferred into new cells (Figure 7B).
By itself, the autogenous activation (positive feedback) loop of C proteins boosting transcription from their own promoters should yield an irreversible transition between two states (76). A purely positive feedback loop is not necessarily problematic for the cell, as maximal gene expression levels can be passively limited by the strength of the promoter and/or translation initiator. One could even make the argument that once an RM system has protectively methylated its new host's DNA, irreversibly switching on REase production would provide the most effective defense against bacteriophage infection. Presumably, there is a reason for the use of an active rather than a passive control mechanism to limit REase levels. Furthermore, the apparent lack of intermediate species in C.PvuII binding of WWWW DNA (Figure 3B), and the high cooperativity of this binding (Figure S2), suggest that the window between activating and repressing levels of C.PvuII is extremely narrow. However, the narrowness of this window is not necessarily problematic. The addition of a negative feedback element to the positive feedback circuit would, under some circumstances, result in an oscillator (76,77). The resultant REase cycling, that presumably would not affect levels of the MTase, may allow repeated pulses of highly protective (restriction-biased) conditions that would lead to chromosome damage if maintained continuously. On a population level, having a fraction of the cells hyperrestrictive could protect the entire population and thus be selectable. The periodic hyperrestrictive state would, in theory, also strengthen the genetic addiction (13) to this toxin–antitoxin system. This hypothesis is consistent with the reduced transformability by the non-repressing version of the PvuII RM system (Figure 7B), but requires further testing. An oscillation model would also predict that the culture-average C.PvuII level would be between the amounts giving peak activation and full repression (to the right of the peak in Figure 2B), in cultures of E. coli carrying the WT PvuII R-M system. Our preliminary results are consistent with this prediction (data not shown).
Process of repression
When the DNA for an RM system such as PvuII first enters a new host cell, there is no C protein present. C protein is expected to accumulate slowly due to activity of the weak C-independent promoter (Figure 1D) (43). Based on our mutational and EMSA data (Figure 3B, C), it appears that as C.PvuII starts to accumulate, OL (half-boxes 1A and 1B) is occupied first. Both its higher affinity, and its location immediately upstream of the −35 hexamer for the C-activated promoter (Figure 1D), implicate OL as the activation site.
As the concentration of C protein continues to rise, the relative effects of mutation on EMSA behavior (Figure 3B) suggest that half-box 2A is occupied next, and finally half-box 2B. This would be consistent with results from two other RM systems (33,45,48). At this point, C protein expression appears to reach its maximum, and the strong C-dependent promoter dominates pvuIIC/R transcription. Our experiments indicate that OR is not only dispensable for activation, but negatively affects gene expression about 3-fold (compare Q2-WWWW to R27-WWWR in Figure 6A). Specifically, reversal of half-box 2B resulted in a system that was hyperactivated but not repressed (Figure 6).
Our results do not rule out a role for C.PvuII binding to half-box 2A in activation, though it overlaps the −35 promoter hexamer. The C proteins are remarkably small regulatory proteins, with C.PvuII having a subunit MW of 9.4 kDa (41). They have a limited subunit interface (29,42), and in the case of C.AhdI homodimerization has a low affinity constant and is substantially stimulated by binding to adjacent sites on the DNA (45). Thus occupancy of half-boxes 1A, 1B and 2A (and not 2B) is possible in theory. The results shown in Figures 3B and 5C and D strongly implicate occupancy of OR (half-boxes 2A and 2B together) in the robust repression shown in Figure 2A and B. The high-frequency isolation of GACT variants of half-box 2A, from C.PvuII-activated members of the WWNR and WWNW libraries, seems inconsistent with the hypothesis that C.PvuII binding of half-box 2A plays a role exclusively in repression. In this regard it is interesting that the binding site for the activator MuC partially overlaps the −35 hexamer of its activated promoter (78).
C.PvuII binds to its C-boxes in a cooperative manner (Figures 3B and S2). This contrasts with the independent binding of two C.EcoRV dimers to its two operators (33), but is consistent with the cooperative binding found for C.AhdI (45). C.EcoRV belongs to a different C-protein family than C.PvuII, and its C-box sequence is substantially different from those shown in Figure 1B. However it is unclear why C.EcoRV binding is noncooperative.
DNA conformation and role of the spacer sequences
The spacing between the two C-binding operators (15 bp center-to-center) suggests that two homodimers interact with the opposite faces of B-form DNA (40). Such opposite-face interaction has been demonstrated for the repressor DtxR bound to its operators (79). In the case of C.AhdI this binding causes local DNA distortion (48).
While the C binding sites are very well conserved upstream of genes for C protein orthologs (Figure 1B and C), it is striking that the spacers between C-boxes are even more strongly conserved, both in sequence and orientation. This is consistent with studies of the phage 434 repressor, that demonstrated the importance of a spacer region for operator affinity despite its lack of specific contacts to the repressor (80). The spacers are also presumably responsible for the different affinities of OL and OR for C.PvuII (Figure 3B), as the OL and OR C-boxes are identical in most of our fusions. Specifically, we used the symmetrized consensus shown at the bottom of Figure 1B, such that half-box 1A = 2A and half-box 1B = 2B; this involved changing 2/16 nt and did not affect the pattern of response to in vivo titration with C.PvuII (Figure 2).
The most highly conserved sequence among the all C-box regions is the central base pairs (GT) of this 4 nt spacer, and specifically TGTA for the C proteins having recognition helices with HRTY (Figure 1B and C). We found that any changes to this tetranucleotide abolished activation by C.PvuII (Figure 4). However, in each set of spacer sequences, TGTA with HRTY C proteins or CGTG with DRT/SY C proteins, the flanking (underlined) nucleotides are complementary. This might indicate, that these nucleotide belong to the C protein recognition site, as proposed for C.AhdI: GTACT-N3-AGTCC-GT-GGACT-N3-CGACA (45). However, the PvuII spacer mutant TGTA→TtTA, which retained the proposed fifth nucleotide of the binding sites, was bound by C.PvuII more poorly than the TGTA→cGTA variant, as judged by the disappearance of unshifted DNA in (Figure 4F).
Tested variants in the TGTA spacer shared reduced stability of the shifted complex, as revealed by smearing. In considering possible explanations for TGTA conservation, we noticed that TG:CA and TA:AT dinucleotides are overrepresented in the spacers and the C-box flanking regions. This overrepresentation is obvious in the logo derived from 29 C-box regions (Figure 1C), 5′-TG-N-1A-TAT-1B-TGTA-2A-TAT-2B-N2-CA-3′). These dinucleotide sequences are the most stereochemically flexible, and confer a propensity for bending during interaction with dimeric helix–turn–helix proteins (81–84). In addition, these dinucleotides are frequently located in E. coli promoters, where they appear to facilitate the isomerization step (85). A recent report on C.AhdI–C-box binding also proposes a structural role for the asymmetrical operator spacers (CAT and TAT for C.AhdI; CAT and CAA in the case of C.PvuII) (48). In addition, C.AhdI-dependent activation involves significant twisting of the specific binding sites (48). Furthermore, the TGTA tetranucleotide itself is the single most flexible sequence among 136 tested with respect to sliding (86), consistent with its playing a structural role.
Efficient establishment of the PvuII R-M system in a new host cell requires both the activation (40,43) and repression arms (this work) of the C.PvuII control circuit. A full understanding of this regulatory system will require not only studies of C.PvuII interactions with RNA polymerase, but also the potential roles of transcriptional interference (87) between the opposing pvuIICR and pvuIIM promoters (Figure 1A), and of hybridization between their transcripts. In addition, little is currently known about how the C-controlled R-M systems respond to environmental perturbations, or even to which perturbations they have evolved to respond. The C protein-dependent regulatory systems clearly illustrate the complexities of even those extant gene regulatory circuits that only involve a single regulator (88,89). This complexity is perhaps not surprising in a system that must control both the timing and amount of expression for a potentially lethal gene.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dieter Knowle for unpublished constructs, Robert Lintner for helpful comments, and Isabel Novella for critically reviewing the manuscript. This material is based upon work supported by the National Science Foundation under Grant No. 0516692. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 3.Naderer M, Brust JR, Knowle D, Blumenthal RM. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J. Bacteriol. 2002;184:2411–2419. doi: 10.1128/JB.184.9.2411-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal RM, Gregory SA, Cooperider JS. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J. Bacteriol. 1985;164:501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvin Koons MD, Blumenthal RM. Characterization of pPvu1, the autonomous plasmid from Proteus vulgaris that carries the genes of the PvuII restriction-modification system. Gene. 1995;157:73–79. doi: 10.1016/0378-1119(94)00618-3. [DOI] [PubMed] [Google Scholar]
- 6.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams GM, Blumenthal RM. The PvuII DNA (cytosine-N4)-methyltransferase comprises two trypsin-defined domains, each of which binds a molecule of S-adenosyl-L-methionine. Biochemistry. 1997;36:8284–8292. doi: 10.1021/bi961885n. [DOI] [PubMed] [Google Scholar]
- 8.Gingeras TR, Greenough L, Schildkraut I, Roberts RJ. Two new restriction endonucleases from Proteus vulgaris. Nucleic Acids Res. 1981;9:4525–4536. doi: 10.1093/nar/9.18.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong W, O’Gara M, Blumenthal RM, Cheng X. Structure of pvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice MR, Blumenthal RM. Recognition of native DNA methylation by the PvuII restriction endonuclease. Nucleic Acids Res. 2000;28:3143–3150. doi: 10.1093/nar/28.16.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice MR, Koons MD, Blumenthal RM. Substrate recognition by the Pvu II endonuclease: binding and cleavage of CAG5mCTG sites. Nucleic Acids Res. 1999;27:1032–1038. doi: 10.1093/nar/27.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butkus V, Klimasauskas S, Petrauskiene L, Maneliene Z, Lebionka A, Janulaitis A. Interaction of AluI, Cfr6I and PvuII restriction-modification enzymes with substrates containing either N4-methylcytosine or 5-methylcytosine. Biochim. Biophys. Acta. 1987;909:201–207. doi: 10.1016/0167-4781(87)90078-9. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl Acad. Sci. USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics. 2004;272:227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 15.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi N, Naito Y, Handa N, Kobayashi I. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J. Bacteriol. 2002;184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins-Manke JL, Zraveski ZZ, Marinus M, Essigmann JM. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J. Bacteriol. 2005;187:7027–7037. doi: 10.1128/JB.187.20.7027-7037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Løbner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl Acad. Sci. USA. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, Masuda Y, Haraga S, Mori H. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 2002;45:673–695. doi: 10.1046/j.1365-2958.2002.03037.x. [DOI] [PubMed] [Google Scholar]
- 20.Bandaru B, Gopal J, Bhagwat AS. Overproduction of DNA cytosine methyltransferases causes methylation and C –> T mutations at non-canonical sites. J. Biol. Chem. 1996;271:7851–7859. doi: 10.1074/jbc.271.13.7851. [DOI] [PubMed] [Google Scholar]
- 21.Shen JC, Rideout W.M., III, Jones PA. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 22.De Backer O, Colson C. Transfer of the genes for the StyLTI restriction-modification system of Salmonella typhimurium to strains lacking modification ability results in death of the recipient cells and degradation of their DNA. J. Bacteriol. 1991;173:1328–1330. doi: 10.1128/jb.173.3.1328-1330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heitman J, Zinder ND, Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc. Natl Acad. Sci. USA. 1989;86:2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milkman R. Recombination and population structure in Escherichia coli. Genetics. 1997;146:745–750. doi: 10.1093/genetics/146.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Ichige A, Kobayashi I. Regulation of the EcoRI restriction-modification system: Identification of ecoRIM gene promoters and their upstream negative regulators in the ecoRIR gene. Gene. 2007;400:140–149. doi: 10.1016/j.gene.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Kobayashi I. Negative regulation of EcoRI restriction enzyme gene associated with intragenic reverse promoters. J. Bacteriol. 2007;189:6928–6935. doi: 10.1128/JB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao T, Blumenthal RM. Sequence and characterization of pvuIIR, the PvuII endonuclease gene, and of pvuIIC, its regulatory gene. J. Bacteriol. 1992;174:3395–3398. doi: 10.1128/jb.174.10.3395-3398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohail A, Ives CL, Brooks JE. Purification and characterization of C.BamHI, a regulator of the BamHI restriction-modification system. Gene. 1995;157:227–228. doi: 10.1016/0378-1119(94)00698-r. [DOI] [PubMed] [Google Scholar]
- 29.Sawaya MR, Zhu Z, Mersha F, Chan SH, Dabur R, Xu SY, Balendiran GK. Crystal structure of the restriction-modification system control element C.BclI and mapping of its binding site. Structure. 2005;13:1837–1847. doi: 10.1016/j.str.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Anton BP, Heiter DF, Benner JS, Hess EJ, Greenough L, Moran LS, Slatko BE, Brooks JE. Cloning and characterization of the BglII restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. doi: 10.1016/s0378-1119(96)00638-5. [DOI] [PubMed] [Google Scholar]
- 31.Cesnaviciene E, Mitkaite G, Stankevicius K, Janulaitis A, Lubys A. Esp1396I restriction-modification system: structural organization and mode of regulation. Nucleic Acids Res. 2003;31:743–749. doi: 10.1093/nar/gkg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kita K, Tsuda J, Nakai SY. C.EcoO109I, a regulatory protein for production of EcoO109I restriction endonuclease, specifically binds to and bends DNA upstream of its translational start site. Nucleic Acids Res. 2002;30:3558–3565. doi: 10.1093/nar/gkf477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenova E, Minakhin L, Bogdanova E, Nagornykh M, Vasilov A, Heyduk T, Solonin A, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction-modification system. Nucleic Acids Res. 2005;33:6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157:217–219. doi: 10.1016/0378-1119(94)00794-s. [DOI] [PubMed] [Google Scholar]
- 35.Kroger M, Blum E, Deppe E, Dusterhoft A, Erdmann D, Kilz S, Meyer-Rogge S, Mostl D. Organization and gene expression within restriction-modification systems of Herpetosiphon giganteus. Gene. 1995;157:43–47. doi: 10.1016/0378-1119(94)00779-r. [DOI] [PubMed] [Google Scholar]
- 36.Vasquez CC, Saavedra CP, Pichuantes SE. Nucleotide sequence of the gene encoding the BstLVI DNA methyltransferase: comparison with other amino-DNA methyltransferases. Curr. Microbiol. 2000;40:114–118. doi: 10.1007/s002849910022. [DOI] [PubMed] [Google Scholar]
- 37.Lubys A, Jurenaite S, Janulaitis A. Structural organization and regulation of the plasmid-borne type II restriction-modification system Kpn2I from Klebsiella pneumoniae RFL2. Nucleic Acids Res. 1999;27:4228–4234. doi: 10.1093/nar/27.21.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ives CL, Sohail A, Brooks JE. The regulatory C proteins from different restriction-modification systems can cross-complement. J. Bacteriol. 1995;177:6313–6315. doi: 10.1128/jb.177.21.6313-6315.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Sullivan DJ, Klaenhammer ’ TR. Control of expression of LlaI restriction in Lactococcus lactis. Mol. Microbiol. 1998;27:1009–1020. doi: 10.1046/j.1365-2958.1998.00748.x. [DOI] [PubMed] [Google Scholar]
- 40.Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2000;182:477–487. doi: 10.1128/jb.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGeehan JE, Streeter SD, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J. Mol. Biol. 2005;346:689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 43.Knowle D, Lintner RE, Touma YM, Blumenthal RM. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2005;187:488–497. doi: 10.1128/JB.187.2.488-497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bart A, Dankert J, van der Ende A. Operator sequences for the regulatory proteins of restriction modification systems. Mol. Microbiol. 1999;31:1277–1278. doi: 10.1046/j.1365-2958.1999.01253.x. [DOI] [PubMed] [Google Scholar]
- 45.McGeehan JE, Papapanagiotou I, Streeter SD, Kneale GG. Cooperative binding of the C.AhdI controller protein to the C/R promoter and its role in endonuclease gene expression. J. Mol. Biol. 2006;358:523–531. doi: 10.1016/j.jmb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Papapanagiotou I, Streeter SD, Cary PD, Kneale GG. DNA structural deformations in the interaction of the controller protein C.AhdI with its operator sequence. Nucleic Acids Res. 2007;35:2643–2650. doi: 10.1093/nar/gkm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu X, Verlinde CL, Zhang S, Schmitt MP, Holmes RK, Hol WG. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure. 1995;3:87–100. doi: 10.1016/s0969-2126(01)00137-x. [DOI] [PubMed] [Google Scholar]
- 50.Moll I, Grill S, Gualerzi CO, Blasi U. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 51.O’Donnell SM, Janssen GR. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J. Bacteriol. 2002;184:6730–6733. doi: 10.1128/JB.184.23.6730-6733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Streeter SD, Papapanagiotou I, McGeehan JE, Kneale GG. DNA footprinting and biophysical characterization of the controller protein C.AhdI suggests the basis of a genetic switch. Nucleic Acids Res. 2004;32:6445–6453. doi: 10.1093/nar/gkh975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology. 2005;151:1729–1740. doi: 10.1099/mic.0.27972-0. [DOI] [PubMed] [Google Scholar]
- 54.Nechaev S, Geiduschek EP. The role of an upstream promoter interaction in initiation of bacterial transcription. EMBO J. 2006;25:1700–1709. doi: 10.1038/sj.emboj.7601069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar A, Grimes B, Fujita N, Makino K, Malloch RA, Hayward RS, Ishihama A. Role of the sigma 70 subunit of Escherichia coli RNA polymerase in transcription activation. J. Mol. Biol. 1994;235:405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- 56.Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S. A mutant spacer sequence between -35 and -10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc. Natl Acad. Sci. USA. 2004;101:6911–6916. doi: 10.1073/pnas.0401929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeda YT, Sano M. Regulatory dynamics of synthetic gene networks with positive feedback. J. Mol. Biol. 2006;359:1107–1124. doi: 10.1016/j.jmb.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 59.Raleigh EA. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 60.Raleigh EA, Murray NE, Revel H, Blumenthal RM, Westaway D, Reith AD, Rigby PW, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 62.Dubendorff JW, Studier FW. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 63.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 65.Platko JV, Willins DA, Calvo JM. The ilvIH operon of Escherichia coli is positively regulated. J. Bacteriol. 1990;172:4563–4570. doi: 10.1128/jb.172.8.4563-4570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 67.Sambrook J, Fritsch EF, Maniatis T. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 68.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khlebnikov A, Risa O, Skaug T, Carrier TA, Keasling JD. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 2000;182:7029–7034. doi: 10.1128/jb.182.24.7029-7034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyada CG, Stoltzfus L, Wilcox G. Regulation of the araC gene of Escherichia coli: catabolite repression, autoregulation, and effect on araBAD expression. Proc. Natl Acad. Sci. USA. 1984;81:4120–4124. doi: 10.1073/pnas.81.13.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss JN. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 73.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mitchell JE, Zheng D, Busby SJ, Minchin SD. Identification and analysis of 'extended -10′ promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kolkhof P, Muller-Hill B. Lambda cI repressor mutants altered in transcriptional activation. J. Mol. Biol. 1994;242:23–36. doi: 10.1006/jmbi.1994.1554. [DOI] [PubMed] [Google Scholar]
- 76.Ninfa AJ, Mayo AE. Hysteresis vs. graded responses: the connections make all the difference. Sci. STKE. 2004;2004:pe20. doi: 10.1126/stke.2322004pe20. [DOI] [PubMed] [Google Scholar]
- 77.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell. Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 78.Chakraborty A, Nagaraja V. Dual role for transactivator protein C in activation of mom promoter of bacteriophage Mu. J. Biol. Chem. 2006;281:8511–8517. doi: 10.1074/jbc.M512906200. [DOI] [PubMed] [Google Scholar]
- 79.Chen CS, White A, Love J, Murphy JR, Ringe D. Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry. 2000;39:10397–10407. doi: 10.1021/bi0009284. [DOI] [PubMed] [Google Scholar]
- 80.Koudelka GB, Harrison SC, Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. Nature. 1987;326:886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki M, Yagi N, Gerstein M. DNA recognition and superstructure formation by helix-turn-helix proteins. Protein Eng. 1995;8:329–338. doi: 10.1093/protein/8.4.329. [DOI] [PubMed] [Google Scholar]
- 82.Suzuki M, Yagi N, Finch JT. Role of base-backbone and base-base interactions in alternating DNA conformations. FEBS Lett. 1996;379:148–152. doi: 10.1016/0014-5793(95)01506-x. [DOI] [PubMed] [Google Scholar]
- 83.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tzou WS, Hwang MJ. Modeling helix–turn–helix protein-induced DNA bending with knowledge-based distance restraints. Biophys. J. 1999;77:1191–1205. doi: 10.1016/S0006-3495(99)76971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ozoline ON, Deev AA, Trifonov EN. DNA bendability–a novel feature in E. coli promoter recognition. J. Biomol. Struct. Dyn. 1999;16:825–831. doi: 10.1080/07391102.1999.10508295. [DOI] [PubMed] [Google Scholar]
- 86.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: tetranucleotide conformational maps. J. Mol. Biol. 2000;295:85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 87.Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hlavacek WS, Savageau MA. Rules for coupled expression of regulator and effector genes in inducible circuits. J. Mol. Biol. 1996;255:121–139. doi: 10.1006/jmbi.1996.0011. [DOI] [PubMed] [Google Scholar]
- 89.Savageau MA. Design principles for elementary gene circuits: Elements, methods, and examples. Chaos. 2001;11:142–159. doi: 10.1063/1.1349892. [DOI] [PubMed] [Google Scholar]
- 90.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 92.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dahlquist F. The meaning of Scatchard and Hill plots. Methods Enzymol. 1978;48:270–299. doi: 10.1016/s0076-6879(78)48015-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.