Abstract
The number of regulatory RNAs with identified non-canonical structures is increasing, and structural transitions often play a role in their biological function. This stimulates interest in internal motions of RNA, which can underlie structural transitions. Heteronuclear NMR relaxation measurements, which are commonly used to study internal motion, only report on local motions of few sites within the molecule. Here we have studied a 27-nt segment of the human hepatitis B virus (HBV) pregenomic RNA, which is essential for viral replication. We combined heteronuclear relaxation with the new off-resonance ROESY technique, which reports on internal motions of H,H contacts. Using off-resonance ROESY, we could for the first time detect motion of through-space H,H contacts, such as in intra-residue base-ribose contacts or inter-nucleotide contacts, both essential for NMR structure determination. Motions in non-canonical structure elements were found primarily on the sub-nanosecond timescale. Different patterns of mobility were observed among several mobile nucleotides. The most mobile nucleotides are highly conserved among different HBV strains, suggesting that their mobility patterns may be necessary for the RNA’s biological function.
INTRODUCTION
The functions of regulatory RNAs are often mediated by striking structural changes (1,2). These changes can either be induced by deformation of a rigid RNA target during binding (induced fit), or can be anticipated by inherent mobility of the RNA target, such that the structural ensemble of the free RNA samples the conformation of the bound state (‘conformational capture’) (3). Defining binding mechanisms between these limiting cases requires information on both structure and internal motions, which has been achieved in only a few cases (4–6).
The hepatitis B virus (HBV) is a DNA virus with a narrow host-range (7). It replicates via reverse transcription of an RNA intermediate. After transcription of the viral DNA by the host RNA polymerase II, the pregenomic RNA is transported into the cytoplasm and encapsidated into immature core particles together with HBV reverse transcriptase (RT). Encapsidation is a key step in HBV replication and is triggered by binding of viral RT to the encapsidation signal, epsilon, a conserved 60-nt bulged RNA structure located at the 5′-end of the RNA pregenome. A highly conserved part of epsilon is the 27-nt apical stem-loop, investigated in this work. After RT binding, a 4-nt DNA primer is synthesized from a bulge within epsilon (8,9). The RT–primer complex is then transferred to the 3′-end of the pregenome, and full-length (−)-DNA is synthesized.
We have previously determined the secondary structure (10) of the apical stem-loop of epsilon by NMR, and its 3D structure (11). The structure contains two helical stems separated by the bulged U23, and is capped by a pseudo-triloop (Figure 1). In vivo, the apical loop is essential for viral replication, although its exact role has not been established yet. In agreement with its vital function for the virus, many nucleotides of the apical stem-loop (for example G13, U14, C16 and U23) are completely conserved among 1200 sequenced strains of human HBV (10). The apical stem-loop of the epsilon structure of duck HBV opens upon forming a priming-competent complex with the RT (8), but it is not known if the apical stem-loop of human HBV undergoes a similar structural change during viral replication. During the structure determination of the apical loop of human HBV, we observed that several nucleotides in the pseudo-triloop (Figure 1) of the apical stem-loop showed few NOE contacts and systematically reduced C–H residual dipolar couplings compared to the stem nucleotides. This suggested that the nucleotides in the pseudo-triloop might show internal motions in the vicinity of their respective average positions in the molecular structure. Conservation of nucleotides in the stem and the pseudo-triloop together with the suspected internal motions raised the question of whether these motions might play a role in the biological function (12) of this RNA structure.
Figure 1.

Secondary structure of the apical loop of epsilon of human HBV. The 27-nt loop contains a pseudo-triloop with the sequence C11U12G13U14G15C16 with a C11 : G15 closing base pair and with C16 being bulged out. Between the two helical stems, U23 is not base paired and forms a bulge.
Dedicated NMR experiments are the best way of deriving information on the internal motions of biomacromolecules at atomic resolution (13). The most widely applied experiments determine relaxation rates of the heteronuclei 15N in N–H or 13C in C–H groups. 15N is used more commonly for proteins, but does not yield much information on the internal motions of nucleic acids, because they contain few N–H groups. 13C could in principle report on motion at many more sites, but is technically demanding (14). Recently, relaxation experiments of the hydrogen isotope deuterium have also been proposed (15). All heteronuclear relaxation experiments have the limitation that they only give information on local motion of bond vectors. Because of these limitations, there have been only a few comprehensive relaxation studies on RNA to date (4,14–18). Frequently either only ribose or base moieties were isotope labeled, precluding comparison of the motions of both moieties (17).
We have shown that the off-resonance ROESY experiment can be used to map mobility of a large number of H,H contacts in proteins (19), by analyzing cross peaks of all sufficiently short H,H contacts. Thus, compared to heteronuclear relaxation experiments, many more contacts in a molecule can be analyzed, including long-range contacts such as tertiary structure interactions. In off-resonance ROESY, the integrals of cross peaks between rigid H,H contacts become zero at an angle θ0max, while cross peaks that are influenced by internal motion show reduced θ0 values. Thus, the difference Δθ0 = θ0 − θ0max represents a measure of motional amplitude.
Here, we have combined 13C relaxation and off-resonance ROESY to investigate the internal motion of the apical stem-loop RNA of human HBV. For the first time, we have applied off-resonance ROESY to RNA and have thus been able to analyze the motions of non-covalent contacts in nucleic acids, as we will demonstrate by data on H1′–H4′, base-ribose, and inter-nucleotide contacts. We investigated whether the apical loop shows motions that might predispose it to structural changes required for its biological function.
MATERIALS AND METHODS
Sample preparation
RNA samples of the apical stem-loop were synthesized as previously described (20). Sample concentration was 0.5 mM (unlabeled) and 0.15 mM (13C-labeled), in D2O containing 100 mM NaCl, 0.1 mM EDTA and DSS as chemical shift standard, with pH 7. 13C-labeled samples used for 13C relaxation measurements were either uniformly labeled in the uridines, or uniformly labeled in cytidines and labeled in the ribose for guanosines.
NMR
All NMR measurements were performed at 25°C on a DRX600 spectrometer equipped with a 1H, 15N, 13C cryoprobe with a shielded z gradient or a conventional 1H, 15N, 13C TXI probe. NMR data were processed using XWINNMR version 2 and Topspin 2.0 (Bruker, Karlsruhe, Germany), and analyzed using Sparky 3.112 (21). Peak integrals were used to fit the relaxation data as described elsewhere (14,22).
13C relaxation
For each sample, we carried out the following experiments using pulse sequences for 13C relaxation measurements with gradient sensitivity enhancement (23), and shared-time evolution (24) with a maximum t1 of ∼4 ms to minimize 13C–13C coupling evolution. Carbon T1 relaxation times were measured as described (13), using a 2.2 s relaxation delay, 44 complex t1 points and 96 scans per free induction decay (FID). Ten T1 delays up to 900 ms were sampled in a total experiment time of 2.5 days. Carbon T1ρ relaxation times were measured (25,26), using a relaxation delay of 2.4 s, 64 scans per FID and 15 complex t1 points. In separate experiments, the carrier frequency of the 13C spin lock was placed on resonance on the C6, C5 and C1′ with a field strength of 1700 Hz, and on the other ribose carbons with a field strength of 505 Hz. Twelve spectra with T1ρ delays of up to 55 ms were recorded, and the duration of the experiment was 18 h for each nucleus. Raw T1ρ values were corrected for resonance offset according to standard procedures (14). In all T1ρ experiments, the energy dissipated in the sample was kept constant by a suitable spin lock applied at the beginning of the pulse sequence. 13C-{H} NOE spectra, with and without 1H saturation, were recorded interleaved in a total time of 34 h, with the delay for relaxation or 1H saturation set to 4.5 s.
ModelFree analysis
The 13C spin relaxation parameters T1, T2 (calculated from T1ρ) and heteronuclear 13C-{H} NOE were analyzed for the two 13C-labeled samples using the program ModelFree 4.15 (27,28). Fitting errors were in the range of 2% for T1 and T1ρ. Errors for 13C-{H} NOE were ∼5%, as estimated from duplicate measurements. 13C chemical shift anisotropies (CSAs) for ribose carbons (C1′ = 45 p.p.m., C2′ = 21.6 p.p.m., C3′ = 58.3 p.p.m., C4′ = 61.8 p.p.m.) and C5 (144 p.p.m.) were taken from Stueber and Grant (29), and for C6 (208 p.p.m.) from Ying et al. (30), and were used without correction for asymmetry of the CSA tensor or its non-colinearity with the C–H bond. The 1H–13C bond length was set to 1.09 Å for ribose C–H and 1.104 Å for base C–H, respectively. It was recently shown that the influence of 13C–13C interactions on 13C relaxation parameters is negligible for RNA molecules of up to at least 30 nt (14,16,31), therefore the C–H groups in the apical loop can be treated as isolated two-spin systems. For data analysis, a rotational correlation time (τc) was estimated by HydroNMR 5a (32), based on a representative NMR structure (11). This τc served as starting value to derive an axially symmetric diffusion model in ModelFree, based on the ribose 13C relaxation data. Base carbons were excluded from the initial fit because of their biased orientation relative to the diffusion tensor. Using this diffusion model, all 13C data were then analyzed with ModelFree; selection of models of internal motion was performed using F-statistical testing and the AIC—penalty model AIC = χ2 + 2k, where k is the number of degrees of freedom of each model (33). Errors derived from the fitting of the relaxation data were used in ModelFree. The ‘wobbling-in-a-cone’ model (34) was used to illustrate motion in the structure of the apical loop.
Off-resonance ROESY
A series of off-resonance ROESY experiments was recorded using the unlabeled RNA sample. The spin-lock field strength was 6800 Hz, the mixing time 120 ms, and the relaxation delay 3.8 s. Nine spectra with spin lock angles spaced equally between 26° and 50° were recorded, using 32 scans per FID and 200 complex t1 points. The total experiment time was 90 h. The data were processed and analyzed as described previously (19). The dependence of Δθ0 on the order parameter 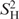 of H,H contacts and the correlation time of internal motion, τe, was obtained from simulations (Figure S3). As discussed (19),
of H,H contacts and the correlation time of internal motion, τe, was obtained from simulations (Figure S3). As discussed (19), 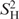 values obtained in this way represent upper limits. Because θ0 values are affected by experimental offsets (19), a reference value of θ0 of rigid H,H contacts is needed. To obtain this reference value, we assumed that for rigid H5, H6 contacts in the stem (θ0 = 35.51 ± 0.14°),
values obtained in this way represent upper limits. Because θ0 values are affected by experimental offsets (19), a reference value of θ0 of rigid H,H contacts is needed. To obtain this reference value, we assumed that for rigid H5, H6 contacts in the stem (θ0 = 35.51 ± 0.14°), 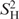 equaled
equaled 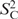 of C5–H and C6–H bonds (0.97 ± 0.07), excluding U5 (see Supplementary Data). This consensus
of C5–H and C6–H bonds (0.97 ± 0.07), excluding U5 (see Supplementary Data). This consensus 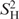 corresponds to Δθ0 = −0.04° (Figure S3), therefore θ0max (of ideally rigid contacts,
corresponds to Δθ0 = −0.04° (Figure S3), therefore θ0max (of ideally rigid contacts, 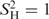 ) equaled 35.55°. Statistical significance was calculated using Student's t-test for paired data and the correlation option of the program KaleidaGraph 3.6.
) equaled 35.55°. Statistical significance was calculated using Student's t-test for paired data and the correlation option of the program KaleidaGraph 3.6.
RESULTS AND DISCUSSION
In structure calculations on the apical loop of HBV epsilon (11), structural variability was observed for several nucleotides, which raised the question of whether these nucleotides are mobile or whether structural information was missing. We have addressed this question with off-resonance ROESY and complementing 13C relaxation measurements. Because it is difficult to quantify timescales of internal motion precisely using 13C relaxation, we applied dedicated experiments to test for microsecond (35) and millisecond motion. The results of these experiments for both 13C-labeled samples are presented in Supplementary Data (Figures S1 and S2). Microsecond motion was only observed for C1′ of C11 (Figure S2), and only C5 of C16 and U23 showed slight motion on the millisecond timescale (Figure S1). Although these experiments do not exhaustively probe all timescales longer than nanoseconds, their results make it likely that most internal motions of the apical loop lie on the picosecond to nanosecond timescale, and are therefore well suited for analysis by 13C relaxation and off-resonance ROESY.
Analysis of 13C relaxation data
13C relaxation parameters T1, T1ρ and 13C-{H} NOE were measured for the C5–H and C6–H groups of the bases and all ribose carbons of the 13C-labeled C, G and U nucleotides (Table S1). Representative T1 relaxation data are shown in Figure 2. The relaxation data were analyzed by plotting and according to the Lipari-Szabo (36) approach using the program ‘ModelFree’ (28). This analysis resulted in a rotational diffusion anisotropy of 1.73 and an averaged rotational correlation time of 5.9 ns, in reasonable agreement with the anisotropy of 2.3 and averaged correlation time of 5.9 ns, estimated from hydrodynamic calculations using HydroNMR (32). The ModelFree results are also in good qualitative agreement with an analysis of the relaxation data by inspection, and mobility can in the present case be unequivocally detected by ModelFree analysis or by direct inspection of relaxation data. This indicates that the ModelFree analysis of the relaxation data neglecting the asymmetry of the base carbon CSA tensors and their non-colinearity with the C–H bond is permissible in our case, although the molecule is at the upper end of the molecular weight range for this approximation. In higher molecular weight systems, it has been demonstrated that mobility can still be detected by direct comparison of relaxation data (37).
Figure 2.
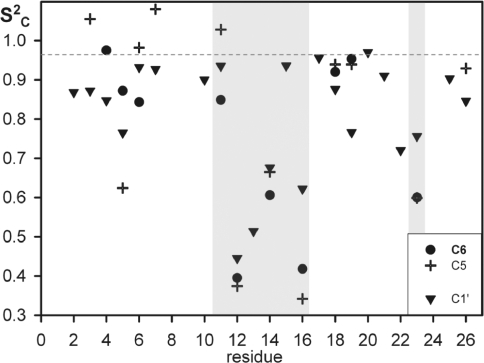
Representative 13C T1 decay curves for C1′ of C16, C7 and G13 (for data, see Table S1).
Complete ‘ModelFree’ results are given in Table S1, and order parameters 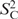 , describing mobility of C–H bonds, are plotted as function of nucleotide sequence in Figure 3. According to this analysis, the stem nucleotides are rigid in the base and in the ribose moieties, with
, describing mobility of C–H bonds, are plotted as function of nucleotide sequence in Figure 3. According to this analysis, the stem nucleotides are rigid in the base and in the ribose moieties, with 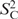 values of 0.97 ± 0.07 and 0.87 ± 0.07, respectively. The loop nucleotides U12, U14, C16 and the bulged U23 show strong motion, which is less pronounced in C1′
values of 0.97 ± 0.07 and 0.87 ± 0.07, respectively. The loop nucleotides U12, U14, C16 and the bulged U23 show strong motion, which is less pronounced in C1′ 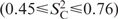 , compared to the bases
, compared to the bases 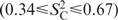 . G13 is highly mobile at C1′
. G13 is highly mobile at C1′  . The closing base pair C11:G15 of the pseudo-triloop is as rigid as the stem nucleotides, with
. The closing base pair C11:G15 of the pseudo-triloop is as rigid as the stem nucleotides, with 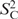 of 0.85 or higher. For the majority of mobile C–H bonds, the internal motion was described by a model with a correlation time (τe) of the order of 400 ps (Table S1).
of 0.85 or higher. For the majority of mobile C–H bonds, the internal motion was described by a model with a correlation time (τe) of the order of 400 ps (Table S1).
Figure 3.
13C order parameter (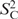 ) as function of nucleotide sequence. Circles represent C6, crosses C5 and triangles C1′ atoms, respectively. The dashed line represents the rigid average
) as function of nucleotide sequence. Circles represent C6, crosses C5 and triangles C1′ atoms, respectively. The dashed line represents the rigid average 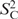 of base C–H groups of stem nucleotides. Gray shading marks the loop region C11–C16 and the bulged U23.
of base C–H groups of stem nucleotides. Gray shading marks the loop region C11–C16 and the bulged U23.
Internal motion detected by off-resonance ROESY
The θ0 values observed in the apical stem-loop are given in Supplementary Data, Table S2. In total, 58 H,H contacts could be analyzed using one unlabeled RNA sample, while only the same number of C–H bonds could be analyzed using the two specially made 13C-labeled samples. A maximum value of θ0 = 35.51 ± 0.14° was observed for base H5,H6 contacts in base pairs. We identify this value with rigid parts  , see Materials and Methods section) of the molecule. The precision of θ0 measurements was ca. 0.2°, which was the mean difference in θ0 between symmetry-related H5,H6 cross peaks. Internal motion reduces θ0 values as a function of τe and
, see Materials and Methods section) of the molecule. The precision of θ0 measurements was ca. 0.2°, which was the mean difference in θ0 between symmetry-related H5,H6 cross peaks. Internal motion reduces θ0 values as a function of τe and 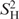 , where
, where 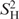 is the order parameter for H,H contacts (Figure S3). We therefore used Δθ0 values, as described in Materials and Methods section, as a measure of motion of H,H contacts. Simulations of Δθ0 as a function of τe and
is the order parameter for H,H contacts (Figure S3). We therefore used Δθ0 values, as described in Materials and Methods section, as a measure of motion of H,H contacts. Simulations of Δθ0 as a function of τe and 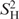 (Figure S3) show that off-resonance ROESY is most sensitive for detecting motion on a timescale of 150 ps, which is similar to the timescale of motions detected by 13C relaxation in the apical loop. Inspection of Figure S3 reveals that a reduction of θ0 by Δθ0 = 0.2° (the precision of θ0) corresponds to a minimal change of 0.1 in
(Figure S3) show that off-resonance ROESY is most sensitive for detecting motion on a timescale of 150 ps, which is similar to the timescale of motions detected by 13C relaxation in the apical loop. Inspection of Figure S3 reveals that a reduction of θ0 by Δθ0 = 0.2° (the precision of θ0) corresponds to a minimal change of 0.1 in 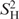 , and we therefore estimate that a precision of 0.1 in
, and we therefore estimate that a precision of 0.1 in 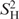 can be achieved.
can be achieved.
In the RNA stems, H,H contacts in ribose moieties and intra-nucleotide base-ribose contacts showed Δθ0 values of −0.08 ± 0.27° and −0.18 ± 0.39°, respectively. These values are not significantly different from the value Δθ0 = −0.04° observed for the rigid H5,H6 contacts, indicating that these groups of H,H contacts are rigid in RNA helices. In contrast, inter-nucleotide base-ribose contacts between H1′ or H2′ and the base of the consecutive nucleotide in the stems showed significantly (P = 0.04, single-factor ANOVA test) more negative Δθ0 values of −0.22 ± 0.19°, corresponding to an 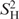 of 0.83. Strongly negative Δθ0 values were observed for inter-nucleotide contacts between G15H2′ and C17H6 (Δθ0 = −0.6°) and G22H2′ and U23H6 (Δθ0 = −0.86°), corresponding to
of 0.83. Strongly negative Δθ0 values were observed for inter-nucleotide contacts between G15H2′ and C17H6 (Δθ0 = −0.6°) and G22H2′ and U23H6 (Δθ0 = −0.86°), corresponding to 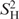 of 0.67 and 0.6, respectively. Inter-nucleotide H,H contacts are used in NMR structure determination, and their motion must underlie structural transitions. Therefore the observation of motion of H,H contacts in secondary structure elements in RNA is important for understanding biological function and to improve structure calculations.
of 0.67 and 0.6, respectively. Inter-nucleotide H,H contacts are used in NMR structure determination, and their motion must underlie structural transitions. Therefore the observation of motion of H,H contacts in secondary structure elements in RNA is important for understanding biological function and to improve structure calculations.
Results from 13C relaxation and off-resonance ROESY are compared in Figure 4. The correlation between observed 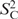 and Δθ0 for all atom groups is highly significant (P < 0.001), and the same holds for separate correlations for base, ribose or base-ribose contacts. The correlation is also in agreement with a theoretical correlation (dashed line), but there are also strong differences in motion as detected by both techniques. These differences are not surprising, given that 13C relaxation and off-resonance ROESY are sensitive to motion on different timescales and around different axes. Furthermore, a larger amplitude of motion of H,H contacts compared to C–H bonds is to be expected, because more degrees of freedom can contribute to H,H motion.
and Δθ0 for all atom groups is highly significant (P < 0.001), and the same holds for separate correlations for base, ribose or base-ribose contacts. The correlation is also in agreement with a theoretical correlation (dashed line), but there are also strong differences in motion as detected by both techniques. These differences are not surprising, given that 13C relaxation and off-resonance ROESY are sensitive to motion on different timescales and around different axes. Furthermore, a larger amplitude of motion of H,H contacts compared to C–H bonds is to be expected, because more degrees of freedom can contribute to H,H motion.
Figure 4.
Correlation of 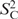 and Δθ0 for C–H groups and H,H contacts. Circles: C5–H or C6–H bonds and H5,H6 contacts; diamonds: C–H bonds and H,H contacts in riboses; triangles: base–ribose H,H contacts and C–H bonds involved. Filled symbols indicate H,H contacts for which
and Δθ0 for C–H groups and H,H contacts. Circles: C5–H or C6–H bonds and H5,H6 contacts; diamonds: C–H bonds and H,H contacts in riboses; triangles: base–ribose H,H contacts and C–H bonds involved. Filled symbols indicate H,H contacts for which 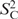 is available for both C–H bonds. A theoretical dependence of Δθ0 on
is available for both C–H bonds. A theoretical dependence of Δθ0 on 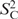 is shown as a dashed line. This correlation is based on the assumption that motion of a H,H contact is caused exclusively by motion of one C–H bond, and that
is shown as a dashed line. This correlation is based on the assumption that motion of a H,H contact is caused exclusively by motion of one C–H bond, and that 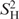 equals
equals 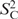 . Assuming that the motion has a τe at the sensitivity maximum of off-resonance ROESY, Δθ0 values corresponding to S2 values were derived from Figure S3. Data points where
. Assuming that the motion has a τe at the sensitivity maximum of off-resonance ROESY, Δθ0 values corresponding to S2 values were derived from Figure S3. Data points where 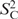 and
and 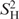 are available and which are discussed in text were connected by horizontal lines. These data points illustrate how correlation of H,H and C–H motion can cause deviations from the theoretical dependence.
are available and which are discussed in text were connected by horizontal lines. These data points illustrate how correlation of H,H and C–H motion can cause deviations from the theoretical dependence.
While the correlation line in Figure 4 assumes that motion of an H,H contact depends on motion of one C–H bond only, more likely the motions of both C–H bonds and their correlation will affect H,H motion. Hence the nature of the motion determines its effect on motional parameters of H,H contacts and C–H bonds. If both C–H bonds are mobile, the H,H contact can be even more mobile than either C–H bond, as indicated by a data point below the correlation line (C16 H1′,H4′ and H1′,H2′ contacts in Figure 4). Mobile H,H contacts with no detectable C–H bond motion can occur for motions which reorient the H,H vector, but not the C–H bonds, such as a rotation around an axis parallel to both C–H bonds. The ribose of G15 may be such a case, because the C–H vectors appear rigid according to the ModelFree analysis and by inspection of the relaxation data (long T1 relaxation times and—besides C2′—only weakly increased NOE, Table S1). The mobile contact between H2′ and H8 of G13 deviates most strongly from the correlation line, which most likely reflects mobility of the unpaired base, while the C2′–H bond is rigid. Data points located above the correlation line (U12 H5,H6; U14 H5,H6; C16 H5,H6) may represent a rotating motion of the base round the χ angle, which reorients the C–H bonds in pyrimidines, but not the connecting H,H vector. Molecular dynamics simulations can be used to derive motional models that can explain the relaxation data.
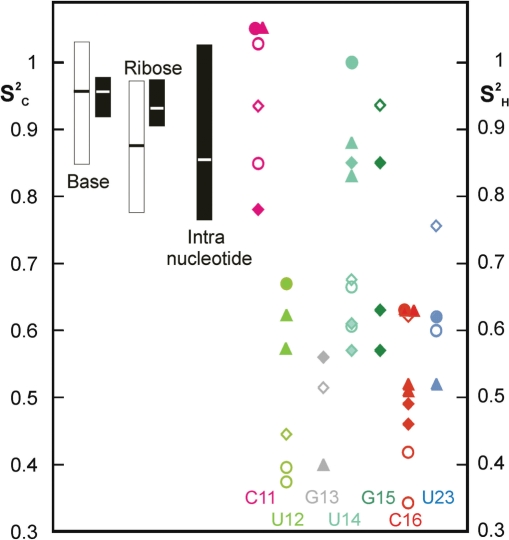
General patterns of motion can be derived from the combination of 13C relaxation and off-resonance ROESY measurements (Figure 5). For the stem nucleotides, 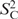 and Δθ0 agree not only for the H5–H6 vectors that were used for calibration, but also for the ribose moieties (see Materials and Methods section and Figure 5). The nucleotides found to be mobile according to 13C relaxation, U12, G13, U14, C16 and U23, showed an overall strong decrease in
and Δθ0 agree not only for the H5–H6 vectors that were used for calibration, but also for the ribose moieties (see Materials and Methods section and Figure 5). The nucleotides found to be mobile according to 13C relaxation, U12, G13, U14, C16 and U23, showed an overall strong decrease in 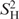 (Figure 5), and the ranges of
(Figure 5), and the ranges of 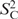
 and
and 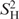
 were similar. Note that base–ribose contacts can show the full range of behavior, from rigid (C11) to very mobile (U23 and G13). This underlines the need for motional information at many sites, which was here provided by off-resonance ROESY.
were similar. Note that base–ribose contacts can show the full range of behavior, from rigid (C11) to very mobile (U23 and G13). This underlines the need for motional information at many sites, which was here provided by off-resonance ROESY.
Figure 5.
Summary and comparison of internal motions in the apical loop, derived from 13C relaxation and off-resonance ROESY. 13C-derived order parameters (from Table S1) are displayed with open symbols. Order parameters from off-resonance ROESY (derived as in Figure 4 from θ0 values listed in Table S2) are displayed as filled symbols. Bars to the left represent the average and spread of order parameters of nucleotides in helical stems, open and filled bars denote 13C and off-resonance ROESY data, respectively. The bar for intra-nucleotide contacts represents 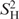 of contacts between base and H1′ or H2′ atoms. On the right side, order parameters of individual nucleotides are presented, the nucleotides are color-coded as in Figure 1. Circles, diamonds and triangles represent contacts in the bases, riboses and ribose–base contacts, respectively.
of contacts between base and H1′ or H2′ atoms. On the right side, order parameters of individual nucleotides are presented, the nucleotides are color-coded as in Figure 1. Circles, diamonds and triangles represent contacts in the bases, riboses and ribose–base contacts, respectively.
Intra-nucleotide dynamics in RNA
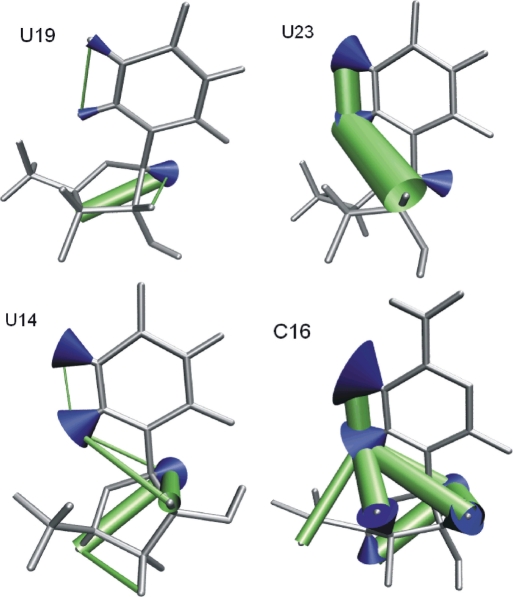
To examine the motions of the mobile nucleotides, their relaxation data are displayed in the nucleotide structures (Figure 6). Using the ‘wobbling-in-a-cone’ model (34), we translated 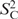 into opening angles, α, of the motions of C–H bonds. The cones are displayed in Figure 6 and the angles α are given in Table S1. For each H,H contact, the motional amplitude is displayed as a cylinder, where the diameter is proportional to the Δθ0 value. The rigid stem nucleotide U19 is shown for reference. In U14, C16 and U12, the H5,H6 contact is more rigid than the base C–H bonds, and the C1′–H bond is also more rigid than the base C–H bonds (see also Figure 5). This pattern of mobility suggests that a rotation around χ is a strong component of the motion affecting the base C–H bonds, because this rotation changes the orientation of the base C–H bonds, but not of the H5,H6 vectors. However, within this pattern there is variation. In U14, the H5,H6 contact is rigid in spite of strong mobility of C5–H and C6–H. As intermediate case, C16 and U12 show strong mobility in their H5,H6 contacts, but not as strong as their base C–H vectors. In C16, the mobility of the H5,H6 and H6,H1′ contacts instead closely matches the mobility of the C1′–H bond (Figure 5), suggesting that their motions might be correlated. Finally, in U23, base C–H bonds, the H5,H6 and an intranucleotide base-ribose contact all show similar, high mobility (Figure 6).
into opening angles, α, of the motions of C–H bonds. The cones are displayed in Figure 6 and the angles α are given in Table S1. For each H,H contact, the motional amplitude is displayed as a cylinder, where the diameter is proportional to the Δθ0 value. The rigid stem nucleotide U19 is shown for reference. In U14, C16 and U12, the H5,H6 contact is more rigid than the base C–H bonds, and the C1′–H bond is also more rigid than the base C–H bonds (see also Figure 5). This pattern of mobility suggests that a rotation around χ is a strong component of the motion affecting the base C–H bonds, because this rotation changes the orientation of the base C–H bonds, but not of the H5,H6 vectors. However, within this pattern there is variation. In U14, the H5,H6 contact is rigid in spite of strong mobility of C5–H and C6–H. As intermediate case, C16 and U12 show strong mobility in their H5,H6 contacts, but not as strong as their base C–H vectors. In C16, the mobility of the H5,H6 and H6,H1′ contacts instead closely matches the mobility of the C1′–H bond (Figure 5), suggesting that their motions might be correlated. Finally, in U23, base C–H bonds, the H5,H6 and an intranucleotide base-ribose contact all show similar, high mobility (Figure 6).
Figure 6.
Intra-nucleotide mobility of four nucleotides in the apical stem-loop. 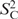 values are indicated as blue cones, with the opening angle derived from a ‘wobbling-in-a-cone’ model (data in Table S1). Δθ0 values are indicated as diameter of the green cylinders representing H,H contacts. U19 is included as a rigid stem nucleotide. U14, C16 and U23 show different distributions of mobility within the nucleotide. The graphic was created using VMD (46).
values are indicated as blue cones, with the opening angle derived from a ‘wobbling-in-a-cone’ model (data in Table S1). Δθ0 values are indicated as diameter of the green cylinders representing H,H contacts. U19 is included as a rigid stem nucleotide. U14, C16 and U23 show different distributions of mobility within the nucleotide. The graphic was created using VMD (46).
In summary, mobile nucleotides in the pseudo-triloop show strikingly variable patterns of motion. Among the base-paired nucleotides, G15 shows indications of motion in the ribose (Figure 5). Differences in the motions of ribose and base moieties might be due to the conformation of the phosphodiester backbone, which is determined by the overall fold of the molecule. It is therefore to be expected that motion of the ribose moieties reflects more global dynamics, in the case of the apical loop induced by the turn of the backbone, whereas motion of base moieties depends on their individual interactions, such as base pairing and stacking. In contrast, protein side chain motion can occur without backbone motion—but not vice versa (38). Molecular dynamics simulations can assess if the proposed components of motion exit. This evaluation is aided by the wealth of motional information accessible by the combination of off-resonance ROESY and 13C relaxation, and by their differing sensitivities for motional timescales.
In the ensemble of structures of the apical stem-loop, based on classical NMR restraints, U12 and G13 show all combinations of minor and major groove conformations, while U14 is restricted to the major groove side (11). C16 also shows structural variation. Thus, the structural variation U12, G13 and C16 agrees with the order parameters derived from relaxation data. The ensemble of structures shows a similar magnitude of structural variation for U14, but its base is restricted to the major groove side. This motional restriction of the base is reflected in the strong difference in motion experienced by its H5,H6 contact and base C–H bonds. This behavior is probably due to the strain in the pseudo-triloop around the sharp turn of the backbone between U14 and G15. The loop residue prior to the sharp turn, here U14, is generally found to be restricted in its motional freedom, keeping the base moiety on the major groove side. Note that the backbone must change direction over just three loop nucleotides and is stretched to bridge the gap of ca. 17 Å between the two strands, forcing the ribose of U14 into an 2′ endo conformation (39,40). In the structure calculation, available residual dipolar couplings of the loop residues were not used, because of their suspected motion, which was confirmed here. At the same time, the distinct patterns of motional amplitudes observed in the loop nucleotides and U23 (Figures 5 and 6) suggest that the underlying motions differ between these nucleotides. To describe the motions, structural data and mobility data will be combined to derive a dynamic structural ensemble, a description of the molecular structure and its motions (41).
Implications for the apical loop binding to the RT
Nucleotides G13, U14, C16 and U23 are completely conserved among different HBV strains (10). Although there is a controversy as to which step the apical loop is essential for, it is important for virus replication by forming a priming-competent complex (42). It has been proposed that the apical stem-loop binds to the viral RT during formation of the priming-competent complex (43), or to a yet unidentified host factor (9). While nucleotides in well-defined structural elements may be conserved to maintain the structure, the mobile nucleotides cannot be conserved for this reason. Thus, conservation of mobile nucleotides more directly indicates a role for biological function. We hypothesize that mobile nucleotides are completely conserved because they may adopt well-defined conformations upon binding. If this explanation holds, mobile non-paired nucleotides would contribute to the energetics of binding in two ways. First, they would make the free apical loop less stable than would be possible for RNAs without bulged nucleotides. For example, the stable GNRA tetraloop in the bacteriophage λ BoxB RNA (44) maintains its structural properties after binding to the anti-termination protein. Secondly, these nucleotides would stabilize the bound state via intermolecular contacts that cause them to be conserved. Thus, motion of conserved nucleotides would prepare the apical loop for binding via a ‘conformational capture’ mechanism (3). Testing of this hypothesis has to await availability of structural information on the functional complex of the apical stem-loop.
CONCLUSION AND OUTLOOK
We have demonstrated that combining 13C relaxation and off-resonance ROESY gives a uniquely detailed description of the internal motions of RNA (Figures 4–6). Off-resonance ROESY does not require isotope labeling, and can therefore be used at an early stage of a structure determination project to detect motion. Results from off-resonance ROESY can then guide synthesis of 13C-labeled samples and further structure determination steps.
The observation of distinct patterns of mobility among conserved nucleotides will be investigated further using molecular dynamics. The combination of 13C relaxation and off-resonance ROESY is particularly suited for this purpose, because both techniques are sensitive for motion around different axes and so their combination may allow identification of correlated motions.
The apical loop of HBV epsilon is a new example of regulatory RNAs (4,5,45) that have been found to contain mobile nucleotides that are highly conserved. Future studies of the apical loop in complex with its interaction partner could show if motion of the apical loop is necessary for the biological function, that is, if the interaction occurs via ‘conformational capture’. Disruption of the interaction between the epsilon HBV RNA and the viral RT is a promising strategy against HBV infection. Drug development along this strategy must take internal motion into account.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Swedish Research Council (621-2003-3481 to J.S.), the Wenner-Gren Foundation (G.L.), the Kempe Foundation (K.P., J.S.), Crafoord foundation (K.K.L.), the faculty of medicine Lund University (K.K.L.) and ‘Functional and Structural Genomics of Viral RNA’ (EU-FP6 to S.W.; K.K.L., K.P.). We thank Frank Nelissen for expert help with sample synthesis, the Center for Biological Magnetic Resonance (Frankfurt, Germany) for spectrometer time and two anonymous reviewers for valuable comments. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Varani G. RNA-protein intermolecular recognition. Acc. Chem. Res. 1997;30:189–195. [Google Scholar]
- 2.Williamson JR. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 3.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 5.Hoogstraten CG, Wank JR, Pardi A. Active site dynamics in the lead-dependent ribozyme. Biochemistry. 2000;39:9951–9958. doi: 10.1021/bi0007627. [DOI] [PubMed] [Google Scholar]
- 6.Dayie KT, Brodsky AS, Williamson JR. Base flexibility in HIV-2 TAR RNA mapped by solution (15)N, (13)C NMR relaxation. J. Mol. Biol. 2002;317:263–278. doi: 10.1006/jmbi.2001.5424. [DOI] [PubMed] [Google Scholar]
- 7.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 8.Beck J, Nassal M. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 1998;18:6265–6272. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 2002;30:4803–4811. doi: 10.1093/nar/gkf603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flodell S, Petersen M, Girard F, Zdunek J, Kidd-Ljunggren K, Schleucher J, Wijmenga S. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 2006;34:4449–4457. doi: 10.1093/nar/gkl582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu K, Beck J, Nassal M. SELEX-derived aptamers of the duck hepatitis B virus RNA encapsidation signal distinguish critical and non-critical residues for productive initiation of reverse transcription. Nucleic Acids Res. 2004;32:4377–4389. doi: 10.1093/nar/gkh772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korzhnev DM, Billeter M, Arseniev AS, Orekhov VY. NMR studies of Brownian tumbling and internal motions in proteins. Progr. Nuclear Magn. Reson. Spectrosc. 2001;38:197–266. [Google Scholar]
- 14.Duchardt E, Schwalbe H. Residue specific ribose and nucleobase dynamics of the cUUCGg RNA tetraloop motif by NMR 13C relaxation. J. Biomol. NMR. 2005;32:295–308. doi: 10.1007/s10858-005-0659-x. [DOI] [PubMed] [Google Scholar]
- 15.Vallurupalli P, Kay LE. A suite of 2H NMR spin relaxation experiments for the measurement of RNA dynamics. J. Am. Chem. Soc. 2005;127:6893–6901. doi: 10.1021/ja0427799. [DOI] [PubMed] [Google Scholar]
- 16.Shajani Z, Varani G. 13C NMR relaxation studies of RNA base and ribose nuclei reveal a complex pattern of motions in the RNA binding site for human U1A protein. J. Mol. Biol. 2005;349:699–715. doi: 10.1016/j.jmb.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Shajani Z, Varani G. NMR studies of dynamics in RNA and DNA by 13C relaxation. Biopolymers. 2007;86:348–359. doi: 10.1002/bip.20650. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 19.Schleucher J, Wijmenga SS. How to detect internal motion by homonuclear NMR. J. Am. Chem. Soc. 2002;124:5881–5889. doi: 10.1021/ja012040o. [DOI] [PubMed] [Google Scholar]
- 20.Flodell S, Cromsigt J, Schleucher J, Kidd-Ljunggren K, Wijmenga S. Structure elucidation of the hepatitis B virus encapsidation signal by NMR on selectively labeled RNAs. J. Biomol. Struct. Dyn. 2002;19:627–636. doi: 10.1080/07391102.2002.10506769. [DOI] [PubMed] [Google Scholar]
- 21.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; 2006. [Google Scholar]
- 22.Stone MJ, Fairbrother WJ, Palmer A.G., III, Reizer J, Saier MH, Wright PE. Backbone dynamics of the Bacillus-subtilis glucose permease-IIA domain determined from 15N NMR relaxation measurements. Biochemistry. 1992;31:4394–4406. doi: 10.1021/bi00133a003. [DOI] [PubMed] [Google Scholar]
- 23.Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, Sorensen OW, Griesinger C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 24.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki RM, Kay LE. NMR experiments for the measurement of carbon relaxation properties in highly enriched, uniformly C-13,N-15-labeled proteins - application to C-13(alpha) carbons. J. Am. Chem. Soc. 1994;116:8266–8278. [Google Scholar]
- 26.Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE. An NMR experiment for the accurate measurement of heteronuclear spin-lock relaxation rates. J. Am. Chem. Soc. 2002;124:10743–10753. doi: 10.1021/ja0204776. [DOI] [PubMed] [Google Scholar]
- 27.Palmer A.G., III, Rance M, Wright PE. Intramolecular motions of a zinc finger DNA-binding domain from XFIN characterized by proton-detected natural abundance C-13 heteronuclear NMR-spectroscopy. J. Am. Chem. Soc. 1991;113:4371–4380. [Google Scholar]
- 28.Mandel AM, Akke M, Palmer A.G., III Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 29.Stueber D, Grant DM. 13C and 15N chemical shift tensors in adenosine, guanosine dihydrate, 2'-deoxythymidine, and cytidine. J. Am. Chem. Soc. 2002;124:10539–10551. doi: 10.1021/ja012485c. [DOI] [PubMed] [Google Scholar]
- 30.Ying J, Grishaev A, Bryce DL, Bax A. Chemical shift tensors of protonated base carbons in helical RNA and DNA from NMR relaxation and liquid crystal measurements. J. Am. Chem. Soc. 2006;128:11443–11454. doi: 10.1021/ja061984g. [DOI] [PubMed] [Google Scholar]
- 31.Boisbouvier J, Wu ZR, Ono A, Kanaisho M, Bax A. Rotational diffusion tensor of nucleic acids from C-13 NMR relaxation. J. Biomol. NMR. 2003;27:133–142. doi: 10.1023/a:1024931619957. [DOI] [PubMed] [Google Scholar]
- 32.de la Torre JG, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 33.d'Auvergne EJ, Gooley PR. The use of model selection in the model-free analysis of protein dynamics. J. Biomol. NMR. 2003;25:25–39. doi: 10.1023/a:1021902006114. [DOI] [PubMed] [Google Scholar]
- 34.Kinosita K., Jr, Kawato S, Ikegami A. A theory of fluorescence polarization decay in membranes. Biophys. J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akke M, Palmer A.G., III Monitoring macromolecular motions on microsecond to millisecond time scales by R(1)rho-R(1) constant relaxation time NMR spectroscopy. J. Am. Chem. Soc. 1996;118:911–922. [Google Scholar]
- 36.Lipari G, Szabo A. ModelFree approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 37.Shajani Z, Drobny G, Varani G. Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry. 2007;46:5875–5883. doi: 10.1021/bi602658x. [DOI] [PubMed] [Google Scholar]
- 38.Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat. Struct. Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 39.van Dongen MJP, Wijmenga SS, van der Marel GA, van Boom JH, Hilbers CW. The transition from a neutral-pH double helix to a low-pH triple helix induces a conformational switch in the CCCG tetraloop closing a Watson-Crick stem. J. Mol. Biol. 1996;263:715–729. doi: 10.1006/jmbi.1996.0611. [DOI] [PubMed] [Google Scholar]
- 40.Hilbers CW, Heus HA, van Dongen MJP, Wijmenga SS. The hairpin elements of nucleic acid structure: DNA and RNA folding. In: Eckstein F, Lilley DMJ, editors. Nucleic Acids and Molecular Biology, Vol. 8. Berlin: Springer; 1994. pp. 56–104. [Google Scholar]
- 41.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 42.Wang GH, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck J, Nassal M. Hepatitis B virus replication. World J. Gastroenterol. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 45.Addess KJ, Basilion JP, Klausner RD, Rouault TA, Pardi A. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.