Abstract
Little is known about the regulation of the DNA damage-mediated gene expression in archaea. Here we report that the addition of actinomycin D to Sulfolobus solfataricus cultures triggers the expression of the radA paralogue sso0777. Furthermore, a specific retarded band is observed when electrophoretic mobility shift assays (EMSAs) with crude S. solfataricus cell extracts and the sso0777 promoter were carried out. The protein that binds to this promoter was isolated and identified as Sta1. Footprinting experiments have shown that the Sta1 DNA-binding site is included in the ATTTTTTATTTTCACATGTAAGATGTTTATT sequence, which is located upstream the putative TTG translation starting codon of the sso0777 gene. Additionally, gel electrophoretic mobility retardation experiments using mutant sso0777 promoter derivatives show the presence of three essential motifs (TTATT, CANGNA and TTATT) that are absolutely required for Sta1 DNA binding. Finally, in vitro transcription experiments confirm that Sta1 functions as an activator for sso0777 gene expression being the first identified archaeal regulatory protein associated with the DNA damage-mediated induction of gene expression.
INTRODUCTION
Transcription in archaea has been studied for several years, and has similarities with transcription in both eukarya and bacteria (1,2). The basal transcriptional apparatus of archaea (RNA polymerase and transcriptional factors) has an eukaryal nature, while transcriptional regulation resembles that of bacteria. For the basal transcription initiation, the archaeal RNA polymerase needs two factors: TBP and TFB. TBP binds in a sequence-specific manner to the TATA-box, a T/A-rich sequence located ∼24 bp upstream of the start codon. TFB has its counterpart in the eukaryotic TFIIB and it binds specifically to a purine-rich B-recognition element (BRE) located immediately upstream of the TATA-box (3). This element is crucial for the orientation of the transcription pre-initiation complex (4). The recruitment of the RNA polymerase takes place by interaction with the amino terminal domain of TFB once TBP and TFB are bound to the promoter region (5). Some archaea have several homologues of TFB and/or TBP which points to the possibility of gene expression regulation by differential usage of these factors (7,3,6). However, it is worth noting that this is not the only way to regulate gene expression in archaea since they have multiple putative regulators similar to bacterial ones (8). Many of them belong to the bacterial Lrp-like regulator family (9), nevertheless none of these regulators have been related to DNA damage regulation.
In Escherichia coli, the DNA damage response is well defined, and it is known that more than 40 genes are involved in the SOS response (10,11). DNA repair in archaea is still poorly understood and its regulation has not been elucidated. The increasing number of sequenced archaeal genomes showed that most of the DNA-repair proteins are more similar to eukarya rather than bacteria, but many DNA repair-related proteins are absent from their genomes (12). Recently, several microarray studies with Halobacterium NRC-1 and Pyrococcus furiosus have shed light on this area showing that not many DNA repair-related genes are upregulated after DNA damage in these organisms, suggesting that a SOS-like response does not exist for archaea (13,14). In such microarray studies some transcriptional regulators were upregulated but their function is still unknown. Here we report the first transcriptional regulator clearly implicated in DNA damage regulation.
The RecA protein family, which includes RadA in archaea and Rad51 in eukarya, is one of the most highly conserved DNA-repair proteins known, and plays a fundamental role in DNA strand exchange during homologous recombination and double-strand break repair. In Sulfolobus solfataricus and related species, there are two clear homologues of RecA, the well-characterized RadA protein (Sso0250) (15–17) and a less-conserved RadA paralogue of unknown function named Sso0777 (18). In this work, we describe the effect in the expression of S. solfataricus radA and sso0777 genes of actinomycin D, which binds to DNA duplexes interfering with transcription, replication and DNA/RNA processing also being able to induce DNA damage (19). We also show that in vitro, the Sta1 protein, which has been recently described as an activator of some promoters from the Sulfolobus rudivirus SIRV1 (20), positively regulates the expression of sso0777 in this organism.
MATERIALS AND METHODS
Microbial strains growth and actinomycin D treatment
The S. solfataricus strain P2 used in this work was a generous gift from Prof. Francesca M. Pisani (Instituto di Biochimica delle Proteine, Consiglio Nazionale delle Ricerche, Napoli, Italy). S. solfataricus cultures were grown in DSM 182 medium with glucose (0.1%), being its pH adjusted to 3.7 with H2SO4, and incubated at 70°C with vigorous shaking. For DNA damage induction experiments, 10 μg/ml of actinomycin D (Sigma) was added to 10 ml culture when OD600 reached 0.3 and incubated for 2 h at 70°C. As a negative control, the same procedure was carried out with another culture without actinomycin D. All plasmid constructions and cloning experiments were performed in E. coli DH5α (21). Protein overexpression was carried out using either E. coli BL21 (DE3) CodonPlus RIL or E. coli BL21 (DE3) Rosetta cells as described below. E. coli growth conditions were as described elsewhere (21). When needed, antibiotic concentrations were used for each bacterial strain as reported previously (22).
RNA purification and RT-PCR experiments
Total RNA was extracted using RNAeasy kit (Qiagen) following the manufacturer's instructions. The resultant RNA was treated with RNAse-free DNAse I (Roche) and the absence of residual DNA was determined by PCR. RNA integrity was tested by agarose gel electrophoresis and quantified spectrophotometrically. RT-PCR experiments were performed using Titan One Tube RT-PCR System (Roche) with the suitable oligonucleotides (listed in Table S1), following the supplier instructions. Real-time quantitative RT-PCR analysis of total RNA was carried out in a LightCycler apparatus (Roche), using the LCRNA master SYBR green I kit (Roche) according to the manufacturer's instructions. 23S RNA, radA, sso0777 and sta1 reverse and forward oligonucleotides (Roche) are shown in Table S1 of Supplementary Data. The 23S ribosomal RNA gene was used as a control as its expression is not affected by DNA damage (19). The concentration of total RNA of both treated and untreated cultures were adjusted to the same value, and the genes to be tested, as well as the control, were assayed simultaneously using a set of standard samples for each one.
Electrophoretic gel mobility shift assays
DNA probes were generated by PCR using one of the primers labelled at its 5′ end with digoxigenin (DIG) (Table S1). After PCR, electrophoresis in a 2–3% low-melting-point agarose gel was performed and the PCR amplicons were purified using Wizard PCR preps DNA purification Resin (Promega). Binding reactions (20 μl) containing 20 ng of the DIG-DNA-labelled probe and 1 μl of S. solfataricus crude extract (0.5 μg/μl) or 100 ng of purified Sta1 protein were incubated 10 min at 65°C using the previously described binding buffer: 10 mM N-2-Hydroxyethyl-piperazine-N′ 2-ethanesulphonic acid (HEPES), NaOH (pH 8), 10 mM Tris–HCl (pH 8), 5% glycerol, 50 mM KCl, 1 mM EDTA, 1 mM DTT, 50 μg/ml BSA and 1 mg/ml of salmon DNA that was added to the binding mixture to avoid the activity of either non-specific DNA-binding proteins or nucleases present in the crude extract (23), and loaded onto a 6% non-denaturing polyacrylamide gel buffered with Tris–glycine (25 mM Tris–HCl (pH 8.5), 250 mM glycine, 1 mM EDTA). DNA–protein complexes were separated at 100 V for 90 min, and transferred to a Biodine B nylon membrane (Pall Gelman Laboratory). DIG-labelled DNA–protein complexes were detected following the manufacturer's protocol (Roche). For the competition EMSA experiments, a 300-fold molar excess of either specific or non-specific unlabelled competitor DNA was also included in the mixture. All EMSA were repeated a minimum of three times to ensure reproducibility of results. For EMSA studies using either the radA or the sta1 probe, other described experimental conditions were also tested (5,24–27).
Preparation of crude S. solfataricus cell extracts and identification of the sso0777 promoter-binding protein
A S. solfataricus culture (120 ml) was grown to mid-exponential phase, pelleted and sonicated in 1.5 ml of sonication buffer (50 mM Tris–HCl pH 7.5, 5% glycerol, 2 mM EDTA) plus protease inhibitors (Complete Mini; Roche). Cell debris was removed by centrifugation, and the soluble fraction was quantified (28).
A DNA oligonucleotide containing two copies of the sso0777 promoter region (from −63 to + 1 position with regard to the TTG translational start codon) was constructed, and used as a template for a PCR with biotinylated primers. The crude extract binding capacity of this PCR fragment was tested in a competition EMSA assay. The PCR product was purified with QIAquick PCR purification kit (Qiagen), and 400 ng of this DNA was bound to 50 μl of streptavidin-coated magnetic beads (10 mg/ml) (Dynabeads M-280 Streptavidin; Invitrogen) following the manufacturer's description for 15 min at room temperature. After washing with the previously described binding buffer (23), the beads were incubated for 15 min at 65°C, in 150 μl of binding mixture containing 50 μl of S. solfataricus cells crude extract (0.5 μg/μl) and binding buffer. Bound proteins were eluted with binding buffer containing 2.5 M KCl. Eluted proteins were desalted with Microcon YM-3 (Amicon; Millipore), loaded in a 15% SDS–PAGE gel, and stained with either Brilliant Blue G Colloidal (Sigma) or Bio-Rad Silver Stain kit (Bio-Rad). As a control, the same process was carried out using streptavidin-coated magnetic beads with no biotinylated DNA bound to the beads. In all cases, the whole process was repeated three times and the same bands were always detected.
The identification of the eluted proteins was done in the Servei de Proteómica i Bioinformàtica (SepBio) of the Universitat Autònoma de Barcelona. The eluted proteins gel bands were in-gel digested with trypsin. Montage In-Gel Digestzp Kit manufacturer's recommendations were followed for band destaining and digestion. Peptide elution was performed by centrifugation and the eluted peptides were stored at −20°C until they were analysed by mass spectrometry. For MALDI-MS analysis, a microlitre of the sample was mixed with the same volume of a solution of a-cyano-4-hydroxy-transcinnamic acid matrix (0.3 mg/ml in ethanol: acetone 6 : 3) and spotted onto a 600 μm AnchorChip MALDI target plate (Bruker) and allowed to air dry at room temperature. MALDI-mass spectra were recorded in the positive ion mode on an Ultraflex time-of-flight instrument. Ion acceleration was set to 25 kV. All mass spectra were externally calibrated using a standard peptide mixture. For PMF analysis, the MASCOT search engine (Matrix Science. London, UK) was used.
Purification of recombinant Sso0110 and Sso048 proteins
The sta1 (sso0048) and sso0110 genes were amplified by PCR using either SSO0048Nde and SSO0048Bam primers or SSO0110Nde and SSO0110Bam oligonucleotides, respectively (Table S1), containing the suitable endonuclease restriction sites and were cloned into a pGEM-T vector (Promega) followed by the subcloning into a pET15b expression vector. For overexpression, either the pET15b-sta1 or pET15b-sso0110 constructs were transformed into the E. coli BL21(DE3) CodonPlus RIL strain (Stratagene, CA, USA) for further expression of the recombinant proteins. Transformant cells were grown in LB at 30°C and a 3 h induction was carried out adding 1 mM IPTG to the culture. His-tagged proteins were purified using the Talon™ Metal Affinity Resin Kit (Clontech) following the manufacture's instructions. To remove the His-tag, the purified proteins were cleaved with thrombin. Either Sta1 (Figure 5A) and Sso0110 (data not shown) recombinant proteins obtained were above 95% purity as determined with Coomassie Blue staining of SDS–PAGE (15%) gel.
Figure 5.
(A) Overexpression and purification of the S. solfataricus Sta1 protein. Lanes 1 and 2 contain crude extracts from E. coli BL21(DE3) pET15b/sta1 in the absence or in the presence of 10 mM IPTG, respectively. Lane 3 contains the trombin digested Sta1 purified protein. The molecular weight of protein markers are indicated on the left side. (B) EMSA experiments with 100 ng of pure Sta1 protein plus the FrgB sso0777 promoter fragment as a probe. The specificity of the binding was tested using 300-fold molar excess of either unlabelled sso0777 or T6 promoters, as a specific or non-specific competitor fragment, respectively. As a control, crude S. solfataricus cell extracts were also used revealing the same mobility shift as the purified Sta1 protein.
DNAse I footprinting
DNAse I footprinting assay was performed as described previously (23) with slight modifications. Briefly, 100 ng of Cy5 labelled DNA from the sso0777 promoter [obtained by PCR using –63 sso0777 and +61 sso0777 oligonucleotides (Table S1)] was bound to 2.5 μg of Sta1 and incubated for 10 min at 65°C. Afterwards, 0.02 U of RNAse-free DNase I (Roche) were added and allowed to act for 3 min at 30°C. The reaction was stopped and precipitated with ethanol, and analysed in an ALF DNA sequencer (Pharmacia Biotech) as previously reported (23).
Cloning and purification of TFB1 and TBP factors
tfb1 and tbp genes were amplified from S. solfataricus genomic DNA by PCR and cloned into a modified pDEST Gateway vector for expression with a Tev protease-cleavable N-terminal his-tag. Full details of this methodology will be published elsewhere, and are available from the corresponding author on request.
pDEST-TFB1 and pDEST-TBP were transformed into E. coli BL21 (DE3) Rosetta. Cells were grown in LB medium at 37°C to an OD600 of 0.6. At this point, induction of His6-tagged TFB1 and TBP was carried out by 200 μM IPTG overnight at room temperature. Cells were harvested and re-suspended in lysis buffer [20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 0.1% Triton X-100, 1 mM MgCl2 and Complete EDTA-free protease inhibitors (Roche)], lysed by sonication and clarified by centrifugation. The supernatant was heated to 70°C for 10 min and re-centrifuged. The resultant supernatant was diluted two-fold in buffer A [20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 30 mM NaH2PO4] plus 30 mM imidazole and was then applied to a column containing Ni-NTA-Agarose (HiTrap 5 ml Chelating HP; GE Healthcare) pre-equilibrated with buffer A plus 30 mM imidazole. The proteins were eluted with a linear gradient of imidazole (buffer A plus 500 mM imidazole). Fractions containing the His-TFB1 and His-TBP were identified by SDS–PAGE and pooled. His-TFB1 and His-TBP were dialysed against Tev cleavage buffer [20 mM Tris–HCl (pH 7.0), 500 mM NaCl, 1 mM DTT, 30 mM NaH2PO4 and 10% glycerol] overnight at 4°C. The following day, the proteins were cleaved with Tev protease overnight at room temperature by adding a final concentration of 200 ng/μl of Tev protease. Cleaved TFB1 and TBP were re-purified by loading onto the same column pre-equilibrated with buffer A plus 30 mM imidazole and collecting the flow through (FT). Positive fractions were pooled and dialysed extensively against freezing buffer [50 mM Tris–HCl (pH 7.5), 200 mM KCl, 1 mM DTT, 1 mM EDTA, 0.01% Triton X-100 and 50% glycerol].
Purification of S. solfataricus RNA polymerase
Ten litres of S. solfataricus P2 cells were grown up to mid-log phase. At this point, cells were harvested and re-suspended in lysis buffer [50 mM MES (pH 6.0), 100 mM NaCl, 1 mM EDTA, 1 mM DTT and complete protease inhibitors (Roche)], lysed by sonication and clarified by centrifugation. The resultant supernatant was diluted two times in buffer 1 [20 mM MES (pH 6.0), 1 mM EDTA and 0.5 mM DTT] plus 10 mM NaCl and was then applied to a pre-equilibrated Hi-Trap 5 ml Heparin column (Amersham). The proteins were eluted with a linear gradient of NaCl (buffer 1 + 1M NaCl). Fractions containing the RNAP were identified by dot-blot immunodetection with RpoB1-specific antibodies and pooled. RNAP was purified to homogeneity by using a HiLoad 26/60 Superdex 200 pg (GE Healthcare) size exclusion column (GE Healthcare) equilibrated with gel filtration buffer [20 mM MES (pH 6.0), 200 mM NaCl, 1 mM EDTA and 0.5 mM DTT]. Fractions containing the RNAP were identified by SDS–PAGE and confirmed by mass spectrometry. Positive fractions were pooled and dialysed extensively against freezing buffer.
Transcription assays and primer extension
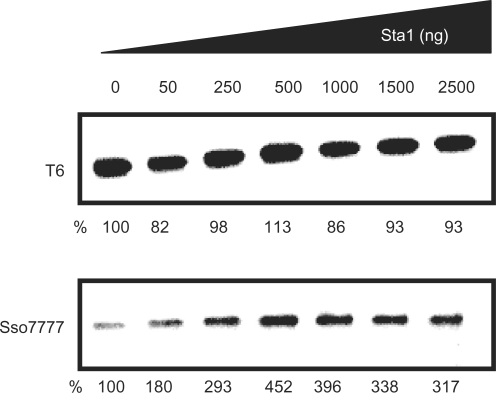
Plasmid carrying T6 promoter was generated as described previously (29) and plasmid carrying the sso0777 promoter was generated from genomic S. solfataricus P2 DNA using standard conditions and oligonucleotides psso0777for and psso0777rev (Table S1). The PCR products were cloned directly into pCR2.1-TOPO (Invitrogen). In vitro transcription reactions were performed in a 50 μl mixture containing 50 ng of the corresponding linear plasmid containing the T6 or the sso0777 promoters (XhoI digestion of pT6 and psso0777), 200 μM of each rNTP, 40 nM of RNAP, 300 nM of TFB-1, 300 nM of TBP and Sta1 in amounts indicated in Figure 8. Sta1 was incubated with the DNA for 15 min at 65°C prior to the transcription assay. The reactions were carried out for 10 min at 70°C in transcription buffer [20 mM Tris–HCl (pH 8.0), 220 mM KCl, 10 mM MgCl2 and 2 mM DTT] and then rNTPs were added to the reaction, which was allowed to continue for 20 min. The reactions were stopped by chilling on ice. The in vitro synthesized RNA (1.5 μl for T6 and 12.6 μl for sso0777) was then used as a template for primer extension reactions. Transcription products were detected by primer extension using 300 fmol of [γ-32P]ATP-labelled primers, T6R primer or sso0777RV primer (Table S1) for T6 and sso0777 transcripts, respectively.
Figure 8.
Effect of Sta1 on T6 and sso0777 promoters. The in vitro transcription assay was performed in the presence of increasing concentrations of Sta1 (0, 50, 250, 500, 1000, 1500 and 2500 ng). The products of the in vitro transcription of promoters indicated were detected by primer extension analysis and electrophored on a 12% denaturing polyacrylamide gel. RNAP : TFB-1 : TBP concentrations used were 40 : 300 : 300 (nM). Percentage of transcription referred to the first lane (no Sta1) is shown under each panel.
RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas) was used according to the manufacturer's instructions. After incubation for 1 h at 42°C, 10 μl of loading buffer (95% formamide and 0.025% each of bromophenol blue and xylen cyanol) was added, the sample boiled for 2 min at 95°C, chilled on ice and 10 μl were loaded on a 12% polyacrylamide sequencing gel containing 7 M urea.
RESULTS
Analysis of radA and sso0777 gene expression following DNA damage by actinomycin D
As a first approach to analyse the regulation of putative damage-induced genes in S. solfataricus, we assayed the induction of two genes: radA (sso0250) and a radA paralogue (sso0777). These genes are homologues of bacterial recA and eukaryal rad51/DMC1 recombinases, which respond to DNA damage in some mesophilic archaea (Methanococcus voltae, Methanococcus maripaludis, Halobacterium NRC-1) and thermophilic archaea (Methanococcus jannaschii, P. furiosus) (30,13,14). We used actinomycin D as a DNA damaging agent, since it has been previously shown that this compound is an effective DNA damage generator agent in S. solfataricus (19). As shown in Figure 1, addition of actinomycin D to S. solfataricus gives rise to a significant increase in the levels of mRNA of both radA and sso0777 genes detected by real-time RT-PCR, demonstrating that these two genes are involved in the S. solfataricus DNA damage-inducible response.
Figure 1.
Actinomycin D-mediated induction of S. solfataricus radA and sso0777 genes measured by quantitative RT-PCR. For each gene, the induction factor in the presence (+) or in the absence (−) of actinomycin D is shown. The induction factor is the ratio between the relative mRNA concentration for either radA or sso0777 genes measured by quantitative RT-PCR in the presence or absence of actinomycin D cultures and that obtained in the S. solfataricus untreated culture. The relative mRNA concentration for each gene is normalized with the 23S ribosomal RNA. In each case, the mean value from three independent experiments (each in triplicate) is shown.
Delimitation of the protein-binding sequence in the sso0777 promoter
To characterize the regulation of radA and sso0777 genes their promoters were used as a probe in electrophoretic mobility shift assays (EMSA) with crude S. solfataricus cell extracts. Upstream and in very close proximity to the S. solfataricus radA gene (sso0250), there are two open-reading frames (sso0251 and sso0252). Each of these genes overlaps the following one. Several RT-PCR experiments were carried out to determine the transcription pattern of this region using a subset of primers (see Table S1 in Supplementary Data). mRNA was reverse transcribed and amplified when using primers annealing to consecutive genes (data not shown). These data demonstrated that radA, sso0251 and sso0252 are part of the same transcriptional unit as they were transcribed as a single polycistronic message (data not shown). For this reason, a fragment containing the upstream region of the sso0252 ORF was used in the EMSA experiments. Nevertheless, no change in the electrophoretic mobility of this fragment was detected in the presence of S. solfataricus crude extracts (data not shown). However, and to further discard the existence of internal promoters within this transcriptional unit, the upstream region of either sso0251 or radA ORFs were also analysed as probes in EMSA experiments. None of these fragments changed their electrophoretic mobility pattern under our experimental conditions (data not shown), which are those usually employed for S. solfataricus in these kind of experiments (5,24–27).
In contrast, a change in the electrophoretic mobility of the sso0777 upstream region was detected when incubated in the presence of crude S. solfataricus cell extracts (Figure 2). The presence of the unlabelled competitor sso0777 DNA fragment in the reaction mixture did prevent the band shift (Figure 2), while the addition of 300-fold of unlabelled competitor DNA from the T6 promoter region (26) did not affect the retardation band formation, confirming the binding specificity. Regarding all of these data, we attempted to identify the protein(s) responsible for the change in the mobility pattern of the sso0777 promoter region as well as its (their) DNA-binding sequence. For that purpose, several fragments of different lengths of the promoter region were generated and designated as FrgA (−107 to +61), FrgB (−63 to +61) and FrgC (−23 to +61) (Figure 3A). A retarded band was observed when FrgA and FrgB fragments were incubated in the presence of crude S. solfataricus cell extracts, in contrast the mobility of the fragment covering positions from −23 to +61, FrgC, was unaffected under the same conditions (Figure 3B). Taken all these data together, the region spanning from −63 to −23 is the minimal region required for the band shift of sso0777 promoter.
Figure 2.
EMSA of the S. solfataricus sso0777 promoter in the absence or in the presence of crude S. solfataricus cell extracts. To determine the specificity of binding, a 300-fold molar excess of either unlabelled sso0777 or T6 promoters was used as a specific or non-specific competitor fragment, respectively.
Figure 3.
(A) Schematic diagram of the sso0777 promoter. The position of the primers used to obtain several promoter fragments (FrgA, FrgB and FrgC) is indicated. (B) EMSA experiments using FrgA, FrgB and FrgC fragments from sso0777 promoter shown above in the presence (+) or absence (−) of crude S. solfataricus cell extracts.
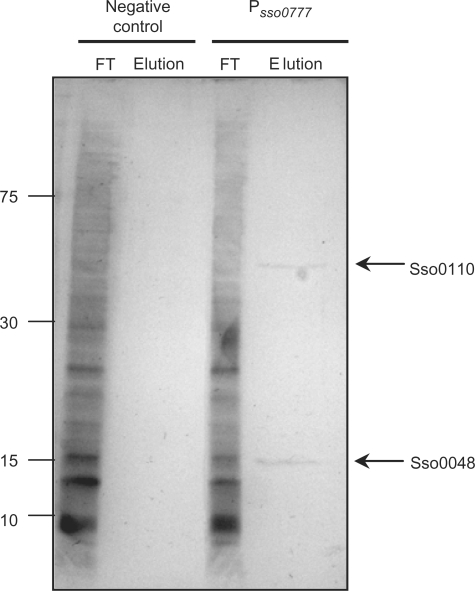
Figure 4.
SDS–PAGE silver-stained gel showing the pull-down experiment using streptavidin-coated magnetic beads. Magnetic beads were bound to biotinylated DNA from the promoter region of sso0777 (lanes 3 and 4), and incubated with S. solfataricus cell extracts. The FT contains all S. solfataricus proteins which were not bound to the sso0777 promoter; and the bound proteins where eluted (Elution) as described in Materials and Methods section. As a control, the same procedure was carried out using non-biotinylated DNA (lanes 1 and 2). Arrows indicate the two identified proteins, Sso0110 and Sso0048.
Purification of the DNA-binding protein
Once the binding region was delimited, and in order to pull-down the protein(s) responsible for the change in the electrophoretic mobility of the promoter probe, a DNA fragment containing two copies of the sso0777 promoter region was synthesized (from positions −63 to +1 upstream the putative translational starting codon) to increase the binding efficiency of this DNA probe. This fragment was tested in a competitive EMSA assay against FrgB and, as expected, it abolished the retardation of FrgB (data not shown). Afterwards, this fragment was amplified by PCR using biotinylated primers, and bound to streptavidin-coated magnetic beads (Dynabeads; Dynal Biotech), following manufacturer's instructions with slight modifications as indicated above (see Materials and Methods section). These beads were incubated with both whole-cell extracts of S. solfataricus and 20 μg of sonicated herring sperm DNA as a non-specific competitor. After washing, bound proteins were eluted with 2.5 M KCl and loaded on two independent SDS–PAGE gels. One of them was stained using Bio-Rad Silver Stain kit and is shown in Figure 4. Two bands showing an approximate molecular weight of 14 and 35 kDa were detected. The second gel was stained with Brilliant Blue G Colloidal (Sigma) to avoid any stain interference with the subsequent procedures. In this gel, the same bands were observed (data not shown). Both protein gel bands were in-gel digested with trypsin. Eight and seven peptides were identified using MALDI-TOF mass spectrometry for the 14 and 35 kDa gel bands, respectively. These peptides corresponded to sso0048 and sso0110 ORF products that have been annotated as a hypothetical transcriptional regulator and a hypothetical conserved protein, with a score of 90.8 and 90.2 (using the MASCOT search engine, Version 2.1), respectively. The experiment was repeated three times, with the same gene products identified on each occasion. The predicted products of those two genes are proteins of 14.3 and 33.5 kDa molecular weight, respectively, in agreement with the apparent molecular mass seen on SDS–PAGE (Figure 4). Both proteins were overexpressed and purified and their binding ability to sso0777 promoter region was tested through EMSA assays. Data obtained showed that only the Sso0048 protein was able to reproduce specifically the shift band observed when the crude extract from S. solfataricus was used (Figure 5). Moreover, the fact that the purified Sso0048 protein was able to bind to the sso0777 labelled promoter fragment generating the same mobility shift as crude cell extracts indicates that it does not need prior binding of the TBP protein to bind to that promoter region. However, to obtain the shift mobility it was necessary to add a great amount of recombinant Sta1 protein, probably because, as it has been reported, the native Sta1 protein is modified by N-terminal acetylation in S. solfataricus (31), a modification that does not occur in E. coli.
The sso0048 gene product has been recently described as the Sta1 protein (Sulfolobus Transcription Activator 1) a transcriptional regulator with an overall structure similar to that of both Mj233 and multiple antibiotic resistance proteins as MarR (PF01047). Sta1 has been shown to be able to activate transcription from some promoters from the S. solfataricus SIRV1 virus (20).
Identification of the Sta1 protein DNA-binding region in the sso0777 promoter
To determine the precise DNA-binding region of the Sta1 protein, a footprinting assay using a fragment from −63 to +61, with respect to the TTG start codon, was carried out. Data obtained indicated that the ATTTTTTATTTTCACATGTAAGATGTTTATT sequence was protected by the Sta1 protein (Figure 6). The above-mentioned sequence contains the previously described (20) consensus-binding site for Sta1 to the SIVR1 viral promoters (ATNTN10AT). However, the precise role of each nucleotide of the protected sequence had not been previously determined. For this reason, and to further characterize the Sta1-binding sequence and to know the most important nucleotides that are recognized by this protein, EMSA experiments using probes with specific point mutations in the protected region of the sso0777 promoter were performed. As it is shown in Figure 7, several motifs seem to be essential for Sta1 binding. First, two spaced TTATT motifs, one located between –42 and –38 and the other placed between –21 and –17 nucleotides relative to the TTG start codon. Additionally, a CANGNA sequence between the two TTATT motifs was also necessary for the binding of Sta1 (Figure 7).
Figure 6.
DNase I footprinting assays with coding and non-coding Cy5-labelled strands of the DNA fragment containing the S. solfataricus sso0777 promoter in the absence or presence of Sta1 protein. The position of the protected region, relative to the TTG translational start codon of sso0777, is shown.
Figure 7.
Single-nucleotide substitutions in the TTTATTTTCACATGTAAGATGTTTATTTA footprinting-protected sequence and their effect on the electrophoretic mobility of the S. solfataricus sso0777 promoter in the presence of Sta1 purified protein. The mobility of the wild-type sso0777 promoter in the absence (−) or presence (+) of Sta1 protein is shown as a control. Substitutions decreasing the Sta1-binding ability are indicated (*, asterisk).
Sta1 stimulates transcription from the sso0777 promoter in vitro
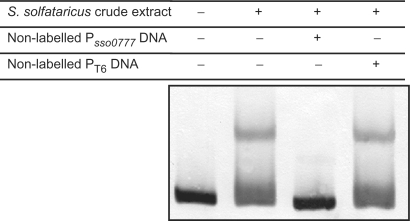
The gel-shift data presented here, together with the known property of Sta1 to act as a transcriptional activator for viral promoters, suggested that the Sta1 protein might function as a transcriptional regulator of the sso0777 gene. To determine this, in vitro transcription assays, using highly purified S. solfataricus RNAP and purified recombinant S. solfataricus TBP and TFB, were performed on the sso0777 and T6 promoters, with increasing concentrations of Sta1 (Figure 8). The T6 promoter was used as a negative control as it has been described that transcription from this promoter is not affected by the Sta1 protein (20). As seen in Figure 8, the yield of transcripts from the sso0777 promoter was stimulated 3- to 4-fold by the addition of increasing concentrations of Sta1 whilst the T6 promoter, as expected, was insensitive to the addition of Sta1. Moreover, it is worth noting that despite expression of Sta1 increases in the presence of actinomycin D showing an induction factor of 5.75 (Figure 9A), it is unlikely to be autoregulated since Sta1 is unable to bind to its own promoter (Figure 9B).
Figure 9.
(A) Actinomycin D-mediated induction of S. solfataricus sta1 measured by quantitative RT-PCR. The induction factor in the presence (+) or in the absence (−) of actinomycin D is shown. The induction factor is the ratio between the relative mRNA concentration for sta1 gene measured by quantitative RT-PCR in the presence or absence of actinomycin D cultures and that obtained in the S. solfataricus untreated culture. The relative mRNA concentration is normalized with the 23S ribosomal RNA. The mean value from three independent experiments (each in triplicate) is shown. (B) EMSA experiments of the DIG-labelled S. solfataricus sta1 promoter in the presence of increasing amounts of Sta1 protein.
DISCUSSION
The DNA damage response pathways of archaea are not well understood, and recent studies suggest that the induction of transcription of DNA-repair genes after DNA damage from ionizing or UV irradiation does not seem to be as extensive as the well-established bacterial SOS response (12). However, the DNA damaging agent actinomycin D has been recently shown to induce transcription of a variety of DNA repair associated proteins in S. solfataricus (19). In concordance with this, in this work, we have observed an increased expression of the S. solfataricus radA and sso0777 genes in the presence of actinomycin D.
Furthermore, we have identified here the positive regulation of the sso0777 gene, a radA paralogue, by the Sta1 transcriptional regulator that had been previously described as an activator of some genes of the Sulfolobus rudivirus SIRV1 (20). EMSA experiments performed using sso0777 derivative mutant promoters showed that, in fact, three essential motifs are required for the Sta1 binding to that promoter: TTATT, CANGNA and TTATT. These motifs match with some of the nucleotide sequences included in the generic Sta1-binding region for SIRV1 promoter regions (20). In silico searches using the PredictRegulon server (32) of the defined Sta1-binding motif throughout the S. solfataricus genome have not revealed the presence of that binding sequence in any other S. solfataricus gene promoter region (data not shown), suggesting that Sta1 controls a small regulon in comparison to that described for bacterial DNA damage response regulators (11,33).
Thus, Sta1 is the first archaeal transcriptional regulator related to the DNA damage response described to date. It is worth noting that in archaea, as in eubacteria, DNA damaging agents (like UV or Mitomycin C) also induce virus production in lysogenic strains (34). Furthermore, the fact that S. solfataricus Sta1 protein controls both viral and cellular genes is reminiscent, in some aspects, of the eubacterial SOS response where both chromosomal (controlled by LexA repressor) and viral (controlled by cI lytic cycle repressor) genes are induced after the detection of the DNA lesions by RecA which then adopts an active conformation that produces the autohydrolysis of these two molecules (35,36). On the other hand, neither sta1 nor sso0777 gene expression are stimulated following UV irradiation of S. solfataricus cells (37), suggests that perhaps several types of DNA damage elicit different transcriptional responses in this organism.
In archaea, the pathway detection of DNA lesions and the signal that activates the putative repair processes have not been clearly identified. Thus, most of the archaea lack MutS protein for mismatch detection. Besides, and although archaea have several homologues to the eukaryotic nucleotide excision repair proteins are present, they lack the damage detection proteins XPA and XPC that activate the NER pathway in those organisms (38). Likewise, recent work points to the possibility that SSB may play a role in the detection of DNA lesions (39). In this respect, pull-down experiments have demonstrated that SSB has the ability to bind to repair-related proteins like the eukaryotic XPB1 helicase (39). However, the precise mechanism for detecting DNA lesions in archaea is still unclear.
Although the radA and sta1 genes are upregulated after actinomycin D treatment, neither promoter binds Sta1 in vitro, and no gel retardation was observed using their promoter regions as a probe in EMSA experiments with crude S. solfataricus cell extracts. Thus, and although Sta1 is here reported as the first transcriptional regulator clearly involved in DNA damage response, other mechanisms must exist in S. solfataricus to control the expression of those genes whose transcription is increased after actinomycin D and other DNA damage agents (19). Further work is needed to delineate the genetic networks controlled by different regulators as well as the interactions existing among them to understand the global DNA damage response in archaea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was funded by Grants BFM2004-02768/BMC from the Ministerio de Educación y Ciencia (MEC) de España, 2005SGR533 from the Generalitat de Catalunya, and by the BBSRC in the White laboratory. S.C. is a recipient of a post-doctoral contract from INIA-IRTA. M.A. is a recipient of a pre-doctoral fellowship from the Ministerio de Educación y Ciencia (MEC). S.P. is a post-doctoral fellow of the Ministerio de Educación y Ciencia (MEC). We are deeply indebted to Prof. Francesca M. Pisani for her generous gift of the S. solfataricus strain P2. Funding to pay the Open Access publication charges for this article was provided by the BFM2004-02768/BMC of MEC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bell SD, Jackson SP. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- 2.Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol. Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 3.Reeve JN. Archaeal chromatin and transcription. Mol. Microbiol. 2003;48:587–598. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- 4.Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in archaea. Proc. Natl Acad. Sci. USA. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SD, Jackson SP. Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem. 2000;275:31624–31629. doi: 10.1074/jbc.M005422200. [DOI] [PubMed] [Google Scholar]
- 6.Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol. Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- 7.Thompson DK, Palmer JR, Daniels CJ. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol. Microbiol. 1999;33:1081–1092. doi: 10.1046/j.1365-2958.1999.01551.x. [DOI] [PubMed] [Google Scholar]
- 8.Bell SD. Archaeal transcriptional regulation – variation on a bacterial theme? Trends Microbiol. 2005;13:262–265. doi: 10.1016/j.tim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker GC, Marsh L, Dodson LA. Genetic analyses of DNA repair: inference and extrapolation. Annu. Rev. Genet. 1985;19:103–126. doi: 10.1146/annurev.ge.19.120185.000535. [DOI] [PubMed] [Google Scholar]
- 11.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelman Z, White MF. Archaeal DNA replication and repair. Curr. Opin. Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 2004;14:1025–1035. doi: 10.1101/gr.1993504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams E, Lowe TM, Savas J, DiRuggiero J. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles. 2007;11:19–29. doi: 10.1007/s00792-006-0002-9. [DOI] [PubMed] [Google Scholar]
- 15.Ariza A, Richard DJ, White MF, Bond CS. Conformational flexibility revealed by the crystal structure of a crenarchaeal RadA. Nucleic Acids Res. 2005;33:1465–1473. doi: 10.1093/nar/gki288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandler SJ, Satin LH, Samra HS, Clark AJ. recA-like genes from three archaean species with putative protein products similar to Rad51 and Dmc1 proteins of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2125–2132. doi: 10.1093/nar/24.11.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naoe C, Tsunoda M, Nakamura KT. Structural study of the archaeal homologous recombinant protein complexed with DNA fragments. Nucleic Acids Symp. Ser. (Oxf.) 2004;48:115–116. doi: 10.1093/nass/48.1.115. [DOI] [PubMed] [Google Scholar]
- 18.Guy CP, Haldenby S, Brindley A, Walsh DA, Briggs GS, Warren MJ, Allers T, Bolt EL. Interactions of RadB, a DNA repair protein in archaea, with DNA and ATP. J. Mol. Biol. 2006;358:46–56. doi: 10.1016/j.jmb.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Salerno V, Napoli A, White MF, Rossi M, Ciaramella M. Transcriptional response to DNA damage in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2003;31:6127–6138. doi: 10.1093/nar/gkg831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler A, Sezonov G, Guijarro JI, Desnoues N, Rose T, Delepierre M, Bell SD, Prangishvili D. A novel archaeal regulatory protein, Sta1, activates transcription from viral promoters. Nucleic Acids Res. 2006;34:4837–4845. doi: 10.1093/nar/gkl502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 22.Miller J. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1991. [Google Scholar]
- 23.Campoy S, Fontes M, Padmanabhan S, Cortes P, Llagostera M, Barbe J. LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus. Mol. Microbiol. 2003;49:769–781. doi: 10.1046/j.1365-2958.2003.03592.x. [DOI] [PubMed] [Google Scholar]
- 24.Napoli A, van der Oost J, Sensen CW, Charlebois RL, Rossi M, Ciaramella M. An Lrp-like protein of the hyperthermophilic archaeon Sulfolobus solfataricus which binds to its own promoter. J. Bacteriol. 1999;181:1474–1480. doi: 10.1128/jb.181.5.1474-1480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enoru-Eta J, Gigot D, Glansdorff N, Charlier D. High resolution contact probing of the Lrp-like DNA-binding protein Ss-Lrp from the hyperthermoacidophilic crenarchaeote Sulfolobus solfataricus P2. Mol. Microbiol. 2002;45:1541–1555. doi: 10.1046/j.1365-2958.2002.03136.x. [DOI] [PubMed] [Google Scholar]
- 26.Brinkman AB, Bell SD, Lebbink RJ, de Vos WM, van der Oost J. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J. Biol. Chem. 2002;277:29537–29549. doi: 10.1074/jbc.M203528200. [DOI] [PubMed] [Google Scholar]
- 27.Peeters E, Thia-Toong TL, Gigot D, Maes D, Charlier D. Ss-LrpB, a novel Lrp-like regulator of Sulfolobus solfataricus P2, binds cooperatively to three conserved targets in its own control region. Mol. Microbiol. 2004;54:321–336. doi: 10.1111/j.1365-2958.2004.04274.x. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Qureshi SA, Bell SD, Jackson SP. Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich CI, McNeil LK, Brace JL, Brucker JK, Olsen GJ. Archaeal RecA homologues: different response to DNA-damaging agents in mesophilic and thermophilic archaea. Extremophiles. 2001;5:265–275. doi: 10.1007/s007920100197. [DOI] [PubMed] [Google Scholar]
- 31.Mackay DT, Botting CH, Taylor GL, White MF. An acetylase with relaxed specificity catalyses protein N-terminal acetylation in Sulfolobus solfataricus. Mol. Microbiol. 2007;64:1540–1548. doi: 10.1111/j.1365-2958.2007.05752.x. [DOI] [PubMed] [Google Scholar]
- 32.Yellaboina S, Seshadri J, Kumar MS, Ranjan A. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 2004;32:W318–W320. doi: 10.1093/nar/gkh364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erill I, Campoy S, Barbe J. [Epub ahead of print]: FEMS Microbiol. Rev.; 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. [DOI] [PubMed] [Google Scholar]
- 34.Prangishvili D, Stedman K, Zillig W. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 2001;9:39–43. doi: 10.1016/s0966-842x(00)01910-7. [DOI] [PubMed] [Google Scholar]
- 35.Little JW. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 36.Sauer RT, Ross MJ, Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J. Biol. Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- 37.Gotz D, Paytubi S, Munro S, Lundgren M, Bernander R, White M. Genome Biol. 2007. Responses of hyperthermophilic crenarchaea to UV irradiation. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White MF. Archaeal DNA repair: paradigms and puzzles. Biochem. Soc. Trans. 2003;31:690–693. doi: 10.1042/bst0310690.. [DOI] [PubMed] [Google Scholar]
- 39.Cubeddu L, White MF. DNA damage detection by an archaeal single-stranded DNA-binding protein. J. Mol. Biol. 2005;353:507–516. doi: 10.1016/j.jmb.2005.08.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.