Abstract
The S-adenosylmethionine (AdoMet) analog sinefungin is a natural product antibiotic that inhibits nucleic acid methyltransferases and arrests the growth of unicellular eukarya and eukaryal viruses. The basis for the particular sensitivity of fungi and protozoa to sinefungin is not known. Here we report the isolation and characterization of spontaneous sinefungin-resistant mutants of the budding yeast Saccharomyces cerevisiae. In all cases, sinefungin resistance was attributable to a loss-of-function mutation in Sam3, the yeast high-affinity AdoMet transporter. Overexpression of wild-type Sam3 increased the sensitivity of yeast to growth inhibition by sinefungin. Thus, Sam3 is a tunable determinant of sinefungin potency. The shared ability of protozoan parasites to import AdoMet might determine sinefungin's anti-infective spectrum. Insights to the intracellular action of sinefungin stem from the finding that increased gene dosage of yeast AdoMet synthase plus cap guanine-N7 methyltransferase afforded greater resistance to sinefungin than either enzyme alone. These results are consistent with the proposal that mRNA cap methylation is a principal target of sinefungin's bioactivity.
INTRODUCTION
Sinefungin is a natural product analog of S-adenosylmethionine (AdoMet) that has antifungal, antiprotozoal and antiviral activities (1–10). Sinefungin differs from AdoMet in that the S–CH3 sulfonium moiety is replaced by a C–NH3 amine. Sinefungin inhibits a wide spectrum of AdoMet-dependent nucleic acid methyltransferases in vitro, by competing with AdoMet for occupancy of the methyl donor site on the enzyme (9,11–14). Several studies suggest that sinefungin's antiviral and antifungal properties derive from selective inhibition of pathogen-encoded mRNA cap-methylating enzymes, specifically RNA (guanine-N7) methyltransferase, which adds a methyl group from AdoMet to GpppRNA to form the m7GpppRNA cap (3,8–10). The cap promotes eukaryal translation initiation and protects RNAs from degradation by 5′-exoribonucleases.
The budding yeast Saccharomyces cerevisiae cap methyltransferase Abd1 is exquisitely sensitive to sinefungin in vitro (IC50 of 24 nM sinefungin at 25 µM AdoMet); indeed, sinefungin is 900-fold more potent than the product AdoHcy in inhibiting cap methylation by Abd1 (9). Two recent studies implicated cap methylation as an antifungal target of sinefungin in vivo. First, isogenic S. cerevisiae strains containing fungal versus mammalian mRNA capping systems (15) displayed 5-fold differences in sensitivity to growth inhibition by sinefungin that correlated with the source of the cap (guanine-N7) methyltransferase component—the strain with the human cap methyltransferase being more resistant to sinefungin (8). Second, the susceptibility of budding yeast to growth inhibition by sinefungin was diminished when Abd1 was overexpressed by increased gene dosage (9).
To better understand sinefungin's antifungal mechanism, we attempted to identify missense mutations in the yeast cap methyltransferase that confer sinefungin resistance in vivo. The approach was to select for outgrowth of sinefungin-resistant (sfr) cells from a pool of yeasts containing mutagenized (but functional) ABD1 genes on a single-copy plasmid. Although forced passage in liquid culture in the presence of 10 µM sinefungin yielded resistant strains, it quickly became apparent that the resistance determinants did not reside on the ABD1 plasmid. Genetic tests assigned sinefungin-resistance to SAM3, which encodes the yeast high-affinity AdoMet transporter (16,17). Spontaneous sfr Sam3 mutations resulted in loss of function in AdoMet uptake, as gauged by growth on medium containing exogenous AdoMet as the sulfur source. We found that overexpression of wild-type Sam3 increased the sensitivity of yeast to growth inhibition by sinefungin. Our results highlight sinefungin uptake by an AdoMet transporter as a critical determinant of its antifungal activity. The shared capacity of many unicellular parasites to import AdoMet (18–20) is likely to underlie sinefungin's broad anti-infective spectrum. The finding that overexpression of Abd1 plus AdoMet synthase confers sinefungin resistance in a wild-type SAM3 background provides further support for the proposal that cap methylation is the principal intracellular target for sinefungin.
MATERIALS AND METHODS
Yeast strains and media
All S. cerevisiae strains used in this study are haploid derivatives of W303 with the following genotype: ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,112 can1-100. The nonessential SAM3 gene was replaced with a sam3::kanMX cassette in which the SAM3 open reading frame was deleted and substituted by a bacterial gene that confers resistance to the antibiotic G418. Minimal B medium lacking exogenous sulfur was prepared as described in (21). B medium plates contained 1% agarose (from EMD Chemicals, Inc.) instead of agar. Sinefungin was purchased from Sigma.
Initial selection for sinefungin resistance
We generated a new library of PCR-mutated ABD1 alleles (ABD1*) in a CEN TRP1 plasmid under the control of the ABD1 promoter according to methods described previously (22), with minor modifications of the PCR reaction conditions. The pABD1* library was transformed into a S. cerevisiae abd1Δ pABD1 (CEN URA3 ABD1) strain and Trp+ colonies were subjected to two rounds of replica-plating on medium containing 5-fluoroorotic acid (FOA) to select for loss of the URA3-marked pABD1 plasmid. Approximately 2500 FOA-resistant abd1Δ pABD1* colonies were harvested directly from agar plates and then divided into 15 separate pools that were stored in 17% glycerol at −80°C. Aliquots (50 µl) from each pool were inoculated into 1 ml of YPD medium containing 10 µM sinefungin. A control aliquot was inoculated into YPD medium with no drug. The cultures were incubated for 24 h at 30°C with constant shaking. Whereas the control culture became turbid after 24 h (A600 > 4.0), the sinefungin-containing cultures did not. Cells were collected by centrifugation from the sinefungin-containing cultures and resuspended in 1 ml of fresh YPD medium with 10 µM sinefungin. Incubation was continued for another 24 h, at which time the cells were again collected by centrifugation and resuspended in fresh medium with 10 µM sinefungin. After the third 24 h incubation, several of the sinefungin-containing cultures had become moderately turbid. Aliquots of those cultures were added to 1 ml of fresh medium with 10 µM sinefungin to attain an A600 of 0.1. After incubation for another 24 h, these cultures were fully turbid. Single colonies were obtained from the cultures by plating on YPD agar. Sinefungin resistance was confirmed by inoculation of single colonies into YPD medium with 10 µM sinefungin and assessment of turbidity after overnight growth. Several of the cultures that had grown out in the presence of 10 µM sinefungin were subjected to dose escalation by serially inoculating them at 1-day intervals into YPD containing 20, 40, and 80 µM sinefungin. Single colonies were recovered at each step by plating on YPD agar.
pABD1* DNA was recovered from individual sfr strains and amplified clonally by transformation in Escherichia coli. pABD1* DNAs isolated from bacteria were transformed into S. cerevisiae abd1Δ pABD1 (CEN URA3 ABD1) and exchanged for the wild-type pABD1 plasmid by selection on medium containing FOA. In no instance was the resulting pABD1* yeast strain capable of growing during a 24 h incubation in YPD medium with 10 µM sinefungin. Thus, the sinefungin-resistant phenotype was apparently not linked to the ABD1* plasmid. This conclusion was verified by transforming several of the sfr strains with wild-type pABD1 and then screening for spontaneous loss of the pABD1* (TRP1) plasmid by tryptophan auxotrophy. The reintroduction of wild-type ABD1 did not ameliorate the sinefungin-resistance, implying that the putative resistance-conferring mutation(s) reside in one or more yeast chromosomal genes.
The sfr abd1Δ pABD1 strains were backcrossed to a sinefungin-sensitive ABD1 strain and the diploids were subjected to sporulation and tetrad dissection. The haploid progeny of four-spore tetrads were screened for sensitivity to sinefungin by spotting aliquots of the drug on a freshly plated lawn of cells and assessing the size of any resulting zone of growth inhibition (9). Sensitivity and resistance segregated in a 2:2 pattern (Figure 1), suggesting that resistance was conferred by mutation of a single genetic locus. Haploid sfr Mata progeny that contained a wild-type chromosomal ABD1 locus were selected for further study. These sfr strains were provisionally designated sfr-1, sfr-2, sfr-3, etc.
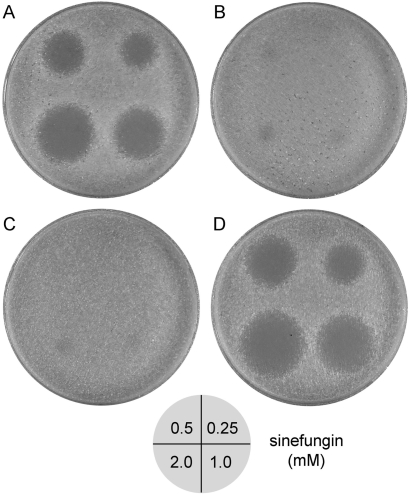
Figure 1.
Meiotic segregation of sinefungin resistance. The four haploid progeny (A, B, C and D) were grown in YPD medium to mid-log phase and aliquots (106 cells) were spread on YPD agar plates (15-cm diameter). After incubation of the plates for 1 h at 30°C to allow the cell suspension to dry, 2 µl aliquots of aqueous solutions of sinefungin (0.25, 0.5, 1.0 or 2.0 mM as diagrammed at the bottom) were spotted on the agar plates. The plates were incubated for 2 days at 30°C and then photographed.
RESULTS
Sinefungin-resistant yeast mutants
We isolated sfr strains by forced passage in liquid cultures of YPD medium containing 10 µM sinefungin. Although our selection strategy was designed to identify sfr versions of the yeast cap methyltransferase Abd1, initial genetic linkage tests showed that none of the yeast strains that grew in the presence of sinefungin did so because of resistance-conferring changes in the plasmid-borne ABD1 gene. By backcrossing the sfr strains to a sinefungin-sensitive yeast strain, and then subjecting the diploids to sporulation and tetrad analysis, we obtained evidence that drug-resistance was caused by a mutation that segregated 2:2 with drug sensitivity. Figure 1 illustrates the use of a simple spot test to gauge sinefungin's effect on the growth of the four haploid progeny of one such backcross. Aliquots (2 µl) of an aqueous solution of sinefungin (either 0.25, 0.5, 1.0, or 2.0 mM) were spotted on a freshly plated lawn of yeast cells on YPD agar medium. In the case of two of the haploid progeny (Figure 1A and D), diffusion of the drug from the site of application resulted in a circular zone of growth inhibition with a sharp demarcation at the circumference. The diameter of the zone of inhibition increased with the increasing sinefungin concentration, as expected. The two other haploid progeny (Figure 1B and C) showed hardly any zone of growth inhibition at even the highest sinefungin dose. Three other tetrads from the same diploid also yielded two sinefungin-sensitive and two sfr haploid progeny (data not shown). This pattern of meiotic segregation is consistent with sinefungin resistance being caused by a single-gene mutation.
Sinefungin-resistant mutants are defective in utilization of exogenous AdoMet
sfr haploids that contained a wild-type chromosomal ABD1 locus after backcrossing were selected for further study. These sfr strains were provisionally designated sfr-1, sfr-2, sfr-3, sfr-4, etc. We considered several possible mechanisms for drug resistance including: (i) defective uptake of sinefungin from the medium; (ii) enhanced export of sinefungin out of the cell; (iii) accelerated metabolism of sinefungin to an inactive derivative; and (iv) resistance-conferring changes in an AdoMet-dependent methyltransferase other than Abd1. Our first suspicions focused on drug export in light of the well-studied phenomenon of yeast pleiotrophic drug resistance (PDR), which is caused by gain-of-function mutations in transcription factors that increase the level of expression of the ATP-dependent plasma membrane pumps that expel various xenobiotics from the cell (23). However, two preliminary results obtained by spot-testing for drug inhibition hinted that sinefungin resistance might not be mediated via PDR. First, the yeast sfr strains were no less sensitive than the parental wild-type to cycloheximide, a substrate for export by the PDR system. Second, a yeast pdr1Δ strain, which lacks a master transcriptional regulator of expression of the export pumps, was not more sensitive than a PDR1 strain to sinefungin.
The prospect that sinefungin-sensitivity might rely on the same pathway used to assimilate exogenous AdoMet was suggested by an initial finding that the zone of inhibition of the wild-type yeast strain in the sinefungin spot test was diminished progressively by including increasing concentrations of AdoMet in the spotted solution along with a fixed concentration of sinefungin (data not shown). One explanation for this effect (though by no means the only one) is that sinefungin and AdoMet compete for a common yeast transport system. Saccharomyces cerevisiae can take up AdoMet from the medium; this capacity is not essential for growth under normal conditions, but becomes critical when the paralogous yeast SAM1 and SAM2 genes encoding AdoMet synthase are inactivated simultaneously, resulting in AdoMet auxotrophy (24). The uptake of AdoMet by S. cerevisiae requires Sam3, a 587-aa integral membrane protein that belongs to the amino acid permease superfamily (16). Although a yeast sam3Δ mutant is viable, the strain is defective for growth on B medium (a minimal medium lacking sulfur) when exogenous AdoMet is included as the lone sulfur source (16).
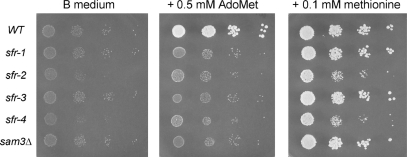
To test whether the four sfr strains were defective in utilizing exogenous AdoMet, we spotted serial dilutions of wild-type, sfr, and sam3Δ cells on unsupplemented B medium and on B medium containing either AdoMet or methionine as the sulfur source (Figure 2). All of the strains were impaired for growth on B medium. The sam3Δ mutant was unable to grow using AdoMet as the sulfur source, yet it was able to grow in the presence of methionine, as reported previously (16). The wild-type strain was the only one for which growth was restored by AdoMet. Each of the sfr strains phenocopied sam3Δ with respect to their ability to utilize methionine but not AdoMet (Figure 2). These results underscore a correlation between impaired AdoMet uptake and sinefungin-resistance in yeast. Yet, they do not reveal whether the sfr mutations elicit a sam3-like phenotype directly, via mutations in Sam3, or indirectly, through mutations in genes that regulate Sam3 expression or function.
Figure 2.
The sfr strains are defective in utilization of exogenous AdoMet. The indicated yeast strains were grown in YPD medium at 30°C until A600 reached ∼0.7. The cells were harvested by centrifugation, suspended in water, recentrifuged and resuspended in water. Serial 10-fold dilutions (in water) of the washed cells were prepared and aliquots (2 µl) were spotted on B medium agarose plates or B medium supplemented with 0.5 mM AdoMet or 0.1 mM methionine. The plates were photographed after incubation for 2 days at 30°C.
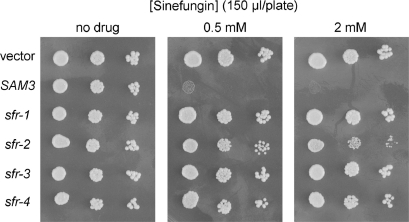
sfr mutants are allelic to SAM3
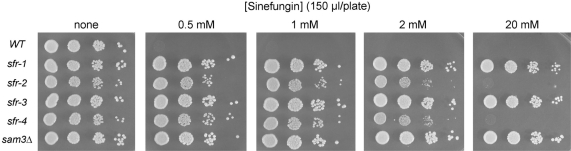
To better quantify sinefungin resistance and simultaneously compare the sensitivities of various mutants strains, we applied 150 µl aliquots of 0.5, 1, 2 and 20 mM sinefungin stock solutions to the surfaces of YPD agar plates (containing 25 ml of medium) and allowed the drug to adsorb into the agar before spotting serial dilutions of wild-type, sfr, and sam3Δ cells. (If one assumes that the applied drug is evenly distributed through the z-plane of the agar plate, then these doses correspond to net sinefungin concentrations of 3, 6, 12 and 120 µM, respectively.) Wild-type yeast cells failed to grow on plates that had received ≥0.5 mM sinefungin (Figure 3). Back titration of the applied dose revealed that 150 µl of a 0.2 mM sinefungin solution sufficed to prevent growth of the wild-type yeast strain on a YPD agar plate (data not shown). The sam3Δ strain was impervious to the 20 mM dose of sinefungin, which translates into at least 100-fold resistance compared to the wild-type strain (Figure 3). This result signifies that sinefungin is transported into yeast cells by the same permease that transports AdoMet. The sfr-1 and sfr-3 strains phenocopied sam3Δ with respect to resistance to the highest level of sinefungin (Figure 3). However, growth of the sfr-2 and sfr-4 strains, though resistant to the 0.5 mM sinefungin dose, was slowed at the 2 mM dose and inhibited fully by the 20 mM dose (Figure 3). These results highlight heterogeneity of the resistance phenotypes of different sfr strains.
Figure 3.
Sinefungin resistance of sfr and sam3Δ strains. The indicated yeast strains were grown in YPD medium at 30°C until A600 reached ∼0.7. The cells were harvested by centrifugation and suspended in water. Serial 10-fold dilutions were prepared and aliquots (2 µl) were spotted on an unsupplemented agar plate (‘none’) and on YPD agar plates onto which 150 µl aliquots of a sinefungin solution (0.5, 1, 2 or 20 mM) had been applied and spread. If one assumes that the applied drug is evenly distributed through the depth of the agar plate, then these doses correspond to net sinefungin concentrations of 3, 6, 12 and 120 µM, respectively. The plates were photographed after incubation for 2 days at 30°C.
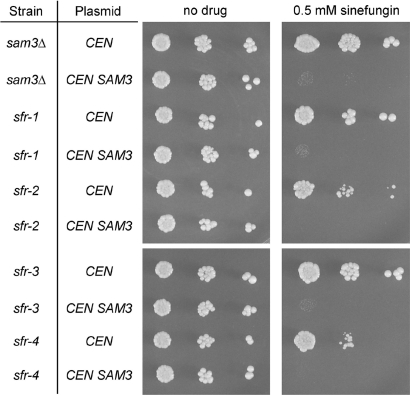
To test whether the sfr mutations were allelic to SAM3, the sfr and sam3Δ strains were transformed with a CEN TRP1 plasmid bearing the wild-type SAM3 gene under the control of its native promoter. The strains were transformed in parallel with the empty CEN TRP1 plasmid vector. The SAM3 plasmid restored sinefungin sensitivity to the sam3Δ strain (Figure 4). The sfr-1, sfr-2, sfr-3, and sfr-4 strains were also rendered sinefungin-sensitive by the plasmid-borne SAM3 gene (Figure 4), implying that the sfr mutations are recessive and allelic to SAM3. The results are not consistent with the sfr mutations affecting a regulator of SAM3 expression or function, insofar as a single copy of the SAM3 gene on a plasmid would not be expected to restore Sam3 activity in such a scenario.
Figure 4.
SAM3 restores sinefungin sensitivity to yeast sfr strains. The indicated yeast strains were transformed with either the control CEN TRP1 plasmid or a derivative bearing wild-type SAM3. Cells were grown in Trp− medium at 30°C until A600 reached ∼0.7, then harvested by centrifugation and suspended in water. Serial 10-fold dilutions were prepared and aliquots (2 µl) were spotted on an unsupplemented Trp− agar plate (‘no drug’) and on a Trp− plate onto which 150 µl of a 0.5 mM sinefungin solution had been applied and spread. The plates were photographed after incubation for 3 days at 30°C.
Sinefungin resistance is caused by a variety of mutations in the SAM3 gene
Convincing evidence that mutations in the SAM3 gene were the cause of spontaneous sinefungin-resistance was obtained by PCR-amplifying, cloning, and sequencing the SAM3 locus from the wild-type, sfr-1, sfr-2, sfr-3, and sfr-4 strains. Four plasmid clones were sequenced from each strain to avoid specious attribution of phenotypes to PCR-generated changes. The sequence of the wild-type SAM3 open reading frame encoded a 587-aa polypeptide identical to the wild-type Sam3 reported previously (16). The sfr-1 strain had a single nucleotide coding change in the SAM3 gene that resulted in a stop codon in lieu of the codon for Ser444 (Table 1). The sfr-3 strain had a 2-nucleotide deletion in the codons for Val371 and Ser372; the −2 frameshift created a mutant peptide sequence from Leu373 to the truncated C-terminus at Gly404 (Table 1). Note that the protein truncations caused by sfr-1 and sfr-3 mutations mimicked the null phenotype of the sam3Δ deletion with respect to extreme sinefungin-resistance (Figure 3). By contrast, the sfr-2 and sfr-4 strains, which were less resistant to sinefungin than sam3Δ (Figure 3), had single-nucleotide missense mutations in the SAM3 gene that resulted in E213K and S476F changes in the Sam3 protein, respectively. We surmise that the defective Sam3 E213K and S476F proteins retained some low level of transport function in vivo that allowed uptake of sinefungin when the drug was present at very high concentration in the medium.
Table 1.
Spontaneous SAM3 mutations that confer sinefungin resistance
| sfr allele | Sam3 protein mutations | SAM3 DNA mutations |
|---|---|---|
| sfr-1 | Ser444 → ter | TCA → TAA |
| sfr-2 | Glu213 → Lys | GAG → AAG |
| sfr-3 | 371VSVCNSCVYASSRLIQALGASGQLPSVCSYMDRK … → 371VSLQFLRLCFFKTNSSFRCIWPTSFGMFLHGQKG404(ter) | −2 frameshift GTGTCAGTT → GTCAGTTTG |
| sfr-4 | Ser476 → Phe | TCC → TTC |
| sfr-5 | Ser180 → ter | TCA → TAA |
| sfr-6 | Gln126 → Pro | CAG → CCG |
| sfr-7 | Ser171 → Arg | AGC → AGA |
| sfr-8 | 262PVFKNLCNTFVSAAFSFGGSEL … → 262LSSRICVTHSFLLFPLVVS282(ter) | −1 frameshift CCTGTC → CTGTCT |
| sfr-9 | Ser171 → Arg | AGC → AGG |
| sfr-10 | Tyr400 → ter | TAC → TAA |
| sfr-11 | Glu60 → ter | GAG → TAG |
| sfr-12 | 90TLGTGLFIGL … → 90PWGRDCSLV98(ter) | −1 frameshift and T → C ACTTTG → CCTTGG |
The sfr strains were selected for growth by passage in YPD medium with 10 µM sinefungin, except for sfr-1 and sfr-5, which were subjected to dose escalation and selection for growth in 80 µM sinefungin. The nucleotide changes in the SAM3 genes of each strain are shown in the right column. Single missense or nonsense nucleotide changes in the sfr strains are underlined. The nucleotides of the wild-type SAM3 gene that were deleted in the sfr-3, sfr-8, and sfr-12 strains are shown in italics. The resulting mutations in the Sam3 protein are indicated in the middle column.
To check that the SAM3 alleles obtained from these four sfr strains were indeed functionally compromised, we cloned them into CEN TRP1 plasmids and then used these vectors, and a wild-type SAM3 control, to transform the sam3Δ strain. The transformants were tested for growth on Trp− plates that had been overlaid with 0.5 or 2 mM sinefungin. The wild-type SAM3 gene restored sinefungin-sensitivity, but the empty vector control did not, nor did any of the four mutated sam3 genes from the sfr strains (Figure 5).
Figure 5.
Sinefungin phenotype of sfr sam3Δ strains. sam3Δ cells transformed with a CEN TRP1 vector containing the genes specified at left were grown in Trp− medium at 30°C until A600 reached ∼0.7. The cells were harvested by centrifugation and suspended in water. Serial 10-fold dilutions were prepared and aliquots (2 µl) were spotted on an unsupplemented Trp− agar plate (‘no drug’) and on Trp− plates onto which 150 µl of a 0.5 mM or 2 mM sinefungin solution had been applied and spread. The plates were photographed after incubation for 3 days at 30°C.
To better survey the spectrum of spontaneous Sam3 lesions that cause sinefungin-resistance, we sequenced the SAM3 locus from eight additional sfr strains isolated in the screen. The sfr-5, sfr-10 and sfr-11 strains had single nucleotide changes that resulted in translation stops at the codons for amino acids 180, 400 and 60, respectively (Table 1). The sfr-6, sfr-7 and sfr-9 strains had single nucleotide mutations that resulted in Q126P, S171R and S171R missense changes in Sam3, respectively. Note that the identical S171R protein mutations in the sfr-7 and sfr-9 strains were the result of independent spontaneous mutations of the Ser171 AGC codon to AGA and AGG arginine codons, respectively (Table 1). The sfr-8 and sfr-12 strains had single nucleotide deletions in the codons for Pro262 and Thr90, respectively; the −1 frameshifts created mutant C-terminal Sam3 peptides that stopped after Ser282 and Val98, respectively (Table 1). These results underscore that complete inactivation of Sam3 by a protein-truncating stop codon is the predominant route to sinefungin resistance.
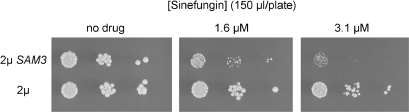
Overexpression of Sam3 sensitizes yeast to sinefungin
We reasoned that if loss of Sam3 transporter function results in sinefungin resistance, then overexpression of Sam3 might enhance the sinefungin sensitivity of S. cerevisiae. Previous studies had established that introducing SAM3 on a high copy plasmid into a wild-type SAM3 strain elicited a 3.5-fold increase in AdoMet uptake (16). We expected that SAM3 overexpression would also increase sinefungin bioavailability. Thus, we cloned the wild-type SAM3 gene into a high-copy 2 µ HIS3 plasmid and then introduced the 2 µ SAM3 plasmid into wild-type yeast, in parallel with the empty 2 µ vector. The transformants were tested for growth on His− plates with no added drug and His− plates that had been overlaid with sinefungin. Preliminary experiments established that growth of wild-type yeast cells was suppressed by applying 150 µl of a 25–50 µM solution of sinefungin to the minimal agar medium. Thus, we tested for the effects of Sam3 overexpression at lower doses of sinefungin (150 µl of a 1.6 µM or 3.1 µM sinefungin solution) and observed that increasing the copy number of the SAM3 gene sensitized the cells to inhibition of growth by a sinefungin dose that had little impact on cells that had only a single copy of SAM3 (Figure 6). Thus, Sam3 is a tunable determinant of sinefungin sensitivity and resistance.
Figure 6.
Sam3 overexpression sensitizes yeast to sinefungin inhibition. Wild-type SAM3 cells were transformed with 2 µ SAM3 or 2 µ plasmids and grown in His− medium at 30°C until A600 reached ∼0.7. The cells were harvested by centrifugation and suspended in water. Serial 10-fold dilutions were prepared and aliquots (2 µl) were spotted on an unsupplemented His− agar plate (‘no drug’) and on His− plates onto which 150 µl of a 1.6 µM or 3.1 µM sinefungin solution had been applied and spread. If the applied drug is evenly distributed through the depth of the agar plate, these doses correspond to net sinefungin concentrations of about 10 and 19 nM, respectively. The plates were photographed after incubation for 3 days at 30°C.
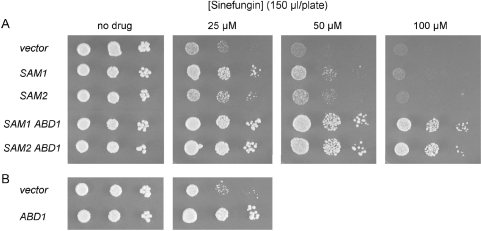
Overexpression of AdoMet synthase plus cap methyltransferase confers sinefungin resistance
The genetic studies presented above highlight Sam3 as the major determinant of sinefungin susceptibility of budding yeast, but they do not illuminate the intracellular actions of the drug once it is taken up from the medium. If sinefungin is blocking growth by acting as a competitive inhibitor of AdoMet binding to an essential yeast methyltransferase, then we reasoned that increasing the intracellular level of AdoMet might ameliorate the effects of sinefungin on its methyltransferase target. S. cerevisiae encodes two AdoMet synthases: Sam1 and Sam2 (24). Previous findings that increased SAM1 or SAM2 gene dosage can suppress temperature-sensitive abd1 mutations (22) implied that Abd1 function is responsive to intracellular AdoMet levels. Here we introduced into yeast a multicopy 2 µ URA3 plasmid containing SAM1 or SAM2 under the control of the constitutive yeast TPI1 promoter (22). Compared to an empty 2 µ URA3 vector control, overexpression of either isozyme of AdoMet synthase enhanced cell growth on minimal medium overlaid with 150 µl of 25 µM or 50 µM sinefungin, but did not allow for growth at a higher drug dosage (Figure 7A). Transformation of yeast with a 2 µ TRP1 plasmid containing wild-type ABD1 driven by the TPI1 promoter also enabled growth on minimal medium dosed with 150 µl of 25 µM sinefungin (Figure 7B), but not with ≥50 µM sinefungin (not shown). Cotransformation with 2 µ ABD1 plus either 2 µ SAM1 or 2 µ SAM2 plasmids had an additive effect on sinefungin-resistance, whereby the strains overexpressing AdoMet synthase and cap methyltransferase grew on minimal medium overlaid with 150 µl of 100 µM sinefungin, a condition in which neither wild-type yeast nor yeast overexpressing Sam1 or Sam2 alone were able to form colonies (Figure 7A).
Figure 7.
Increased gene dosage of SAM1 or SAM2 plus ABD1 confers sinefungin-resistance. (A) Yeast abd1Δ cells containing either a CEN TRP1 ABD1 plasmid (with ABD1 expression controlled by its native promoter) or a 2 µ TRP1 ABD1 plasmid (with ABD1 driven by the strong TPI1 promoter) were transformed with a 2 µ URA3 plasmid bearing either SAM1 or SAM2 or no insert. Transformants were grown in Ura− medium at 30°C until A600 reached ∼0.7. The cells were harvested by centrifugation and suspended in water. Serial 10-fold dilutions were prepared and aliquots (2 µl) were spotted on an unsupplemented Ura− agar plate (‘no drug’) and on Ura− plates onto which 150 µl of a 25, 50 or 100 µM sinefungin solution had been applied. If the applied drug is evenly distributed through the depth of the agar plate, these doses correspond to net sinefungin concentrations of 0.15, 0.3 and 0.6 µM, respectively. The plates were photographed after incubation for 3 days at 30°C. (B) Aliquots (2 µl) of serial 10-fold dilutions of wild-type yeast cells carrying either an empty CEN TRP1 vector or a 2 µ TRP1 ABD1 plasmid were spotted on an unsupplemented Trp− agar plate (‘no drug’) and on Trp− plates onto which 150 µl of a 25 µM sinefungin solution had been applied. The no drug and 25 µM sinefungin plates were photographed after incubation at 30°C for 3 days and 5 days, respectively.
DISCUSSION
A classical genetic approach to identifying the target of a drug is to screen for a drug-resistant mutant and then identify the gene responsible for the resistance phenotype. There have been several earlier reports of the isolation of sfr mutants or resistant strain variants of protozoan organisms that are normally sensitive to the drug (25–30). An sfr isolate of Leishmania displayed impaired uptake of sinefungin compared with the wild-type strain (30). The present study shows that impaired sinefungin uptake is the predominant route to spontaneous sinefungin-resistance of S. cerevisiae, which is caused by a variety of loss-of-function mutations in the yeast high-affinity AdoMet transporter Sam3. To our knowledge, sinefungin resistance-conferring genetic changes in a free-living organism had not been assigned previously to specific genes.
SAM3 was identified initially by Rouillon et al. (16) as a gene required for growth of budding yeast on sulfur-free medium containing AdoMet as the sole sulfur source. The Sam3 protein consists of 12 putative membrane-spanning segments and is classified as a member of the amino acid permease superfamily. Sam3 is capable of discriminating structurally related sulfonium derivatives of methionine, insofar as Sam3 suffices to transport AdoMet, but not S-methylmethionine (16). The genetic evidence presented here indicates that Sam3 is the principle (if not the only) yeast transporter of sinefungin. Although sinefungin has a primary amine instead of the sulfonium center found in AdoMet, the two compounds are virtually isosteric and both have a positive charge on the amine/sulfonium groups. Thus, it is sensible that Sam3 would be able to transport sinefungin. A recent study suggests that Sam3 is also able to transport other positively charged compounds such as putrescine and spermidine (17).
The Sam3 mutations that confer sinefungin-resistance also result in loss-of-function in AdoMet uptake, as gauged by impaired growth on AdoMet-containing B medium. Most of the sfr mutants isolated in the screen are functionally null for Sam3 because of premature translation stops that remove three or more of the 12 predicted membrane-spanning segments. The Sam3 missense mutations that confer sinefungin-resistance are Q126P, S171R, E213K, and S476F. Gln126, Ser171 and Ser476 are located in predicted membrane spanning segments 2, 3 and 11, respectively. Glu213 is situated between the third and fourth membrane-spanning segments. All four of these Sam3 side chains are conserved in the yeast Mmp1 permease that transports S-methylmethionine (16).
Our findings that sinefungin's antifungal activity is contingent on its import by the yeast AdoMet permease resonate strongly with previous studies of AdoMet and sinefungin in protozoan parasites. Sinefungin-sensitive kinetoplastid parasites such as Leishmania and Trypanosoma can import exogenous AdoMet. Inhibition of AdoMet uptake by sinefungin and direct assays of intracellular sinefungin accumulation suggest that kinetoplastids import sinefungin via the AdoMet uptake pathway (19,20,30,31). Resistance of Leishmania to sinefungin is correlated with decreased uptake of both sinefungin and AdoMet (30,31). To our knowledge, there has been no published report of the identification of the gene encoding the putative kinetoplastid AdoMet/sinefungin transporter.
To a first approximation, it appears that the anti-infective spectrum of sinefungin correlates with the presence in the susceptible organisms of an AdoMet transport system that accepts sinefungin as cargo. The acquisition of spontaneous sinefungin-resistance by inactivating mutations of the fungal AdoMet permease is a potential obstacle to sustained clinical efficacy of a sinefungin-based drug. However, this problem is less daunting if the parasite relies on AdoMet uptake for growth or persistence in the animal host, in which case mutations in the AdoMet transporter that affect sinefungin susceptibility would simultaneously diminish the virulence of the parasite. This is a plausible scenario for the fungal pathogen Pneumocystis carinii, which has no detectable AdoMet synthase activity and is naturally auxotrophic for exogenous AdoMet taken up by a high-affinity transporter (18). Sinefungin inhibits Pneumocystis growth in culture (32).
The fact that the sfr phenotype in yeast is dominated by sam3 mutations confounded our initial efforts to identify an intracellular methyltransferase target of sinefungin by screening haploid strains for drug-resistance. Because the sinefungin-sensitive trait of wild-type SAM3 is dominant in the presence of a sfr sam3 allele, it might be feasible to screen in diploid strains for non-Sam3 resistance-conferring changes. However, we observed the rapid emergence of sfr colonies in the middle of the zone of inhibition when drug was spotted on a lawn of the SAM3 sam3(sfr-1) diploid strain (data not shown). We surmise that gene conversion between homologous chromosomes resulted in transfer of the resistance mutation to the previously wild-type SAM3 locus. A potentially better way to seek resistance mutations in intracellular targets is to exploit the observation that a 2 µ SAM3 plasmid sensitizes yeast to AdoMet. The high copy number of the 2 µ plasmid makes it unlikely that an inactivating mutation in the chromosomal SAM3 gene, or any one of the plasmid-borne genes, will result in resistance in the presence of an excess of wild-type SAM3 alleles. In this background, non-Sam3 resistance-conferring mutations might be identified.
The observation that wild-type SAM3 cells become sfr upon overexpression of AdoMet synthase plus Abd1 reinforces the earlier suggestion that RNA cap methylation is a principal intracellular event targeted by the drug (8,9). It is sensible that simultaneously increasing the concentration of the enzyme responsible for AdoMet production and the level of the relevant methyltransferase enzyme target should generate greater resistance than either maneuver alone.
ACKNOWLEDGEMENTS
Supported by grant GM52470 from the U.S. National Institutes of Health. S.S. is an American Cancer Society Research Professor. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM52470.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hamill RL, Hoehn MM. A9145, a new adenine containing antifungal antibiotic: discovery and isolation. J. Antibiot. 1973;26:463–465. doi: 10.7164/antibiotics.26.463. [DOI] [PubMed] [Google Scholar]
- 2.Gordee RS, Butler TF. A9145, a new adenine containing antifungal antibiotic: biological activity. J. Antibiot. 1973;26:466–470. doi: 10.7164/antibiotics.26.466. [DOI] [PubMed] [Google Scholar]
- 3.Pugh CSG, Borchardt RT, Stone HO. Sinefungin, a potent inhibitor of virion mRNA(guanine-7-)-methyltransferase, mRNA(nucleoside-2′-)-methyltransferase, and viral multiplication. J. Biol. Chem. 1978;253:4075–4077. [PubMed] [Google Scholar]
- 4.Dube DK, Mpimbaza G, Allison AC, Lederer E, Rovis L. Antitrypanosomal activity of sinefungin. Am. J. Trop. Med. Hyg. 1983;32:31–33. doi: 10.4269/ajtmh.1983.32.31. [DOI] [PubMed] [Google Scholar]
- 5.Ferrante A, Ljungstron I, Huldt G, Lederer E. Amoebicidal activity of the antifungal antibiotic sinefungin against Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 1984;78:837–838. doi: 10.1016/0035-9203(84)90039-7. [DOI] [PubMed] [Google Scholar]
- 6.Messika E, Golenser J, Abu-Elheiga L, Robert-Gero M, Lederer E, Bachrach U. Effect of sinefungin on macromolecular biosynthesis and cell cycle of Plasmodium falciparum. Trop. Med. Parasitol. 1990;41:273–278. [PubMed] [Google Scholar]
- 7.Brassuer P, Lemetheil D, Ballet JJ. Curative and preventive anticryptosporidium activities of sinefungin in an immunosuppressed adult rat model. Antimicrob. Agents Chemother. 1993;37:889–892. doi: 10.1128/aac.37.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chrebet GL, Wisniewski D, Perkins AL, Deng Q, Kurtz MB, Marcy A, Parent SA. Cell-based assays to detect inhibitors of fungal mRNA capping enzymes and characterization of sinefungin as a cap methyltransferase inhibitor. J. Biomol. Screen. 2005;10:355–364. doi: 10.1177/1087057104273333. [DOI] [PubMed] [Google Scholar]
- 9.Zheng S, Hausmann S, Liu Q, Ghosh A, Schwer B, Lima CD, Shuman S. Mutational analysis of Encephalitozoon cuniculi mRNA cap (guanine-N7) methyltransferase, structure of the enzyme bound to sinefungin, and evidence that cap methyltransferase is the target of sinefungin. J. Biol. Chem. 2006;281:35904–35913. doi: 10.1074/jbc.M607292200. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Chorba JS, Whelan SPJ. Vesicular stomatitis viruses resistant to the methylase inhibitor sinefungin upregulate RNA synthesis and reveal mutations that affect mRNA cap methylation. J. Virol. 2007;81:4104–4115. doi: 10.1128/JVI.02681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential bind of S-adenosylmethionine, S-adenosylhomocysteine and sinefungin to the adenine-specific DNA methyltransferase M.Taq1. J. Mol. Biol. 1997;265:56–67. doi: 10.1006/jmbi.1996.0711. [DOI] [PubMed] [Google Scholar]
- 12.Thomas CB, Scavettta RD, Gumport RI, Churchill MEA. Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase. J. Biol. Chem. 2003;278:26094–26101. doi: 10.1074/jbc.M303751200. [DOI] [PubMed] [Google Scholar]
- 13.Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. The 2.2 Å structure of the rRNA methyltransferase ErmC’ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol. 1999;289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 14.Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of Dam methyltranserase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha N, Schwer B, Shuman S. Characterization of human, Schizosaccharomyces pombe and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]
- 16.Rouillon A, Surdin-Kerjan Y, Thomas D. Transport of sulfonium compounds: characterization of the S-adenolysmethionine and S-methylmethionine permeases from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:28096–28105. doi: 10.1074/jbc.274.40.28096. [DOI] [PubMed] [Google Scholar]
- 17.Uemura T, Kashiwagi K, Igarashi K. Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:7733–7741. doi: 10.1074/jbc.M611105200. [DOI] [PubMed] [Google Scholar]
- 18.Merali S, Vargas D, Franklin M, Clarkson AB. S-adenosylmethionine and Pneumocystis carinii. J. Biol. Chem. 2000;275:14958–14963. doi: 10.1074/jbc.275.20.14958. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg B, Yarlett N, Sufrin J, Lloyd D, Bacchi CJ. A unique transporter of S-adenosylmethionine in African trypanosomes. FASEB J. 1997;11:256–260. doi: 10.1096/fasebj.11.4.9068614. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg B, Yarlett N, Sufrin J, Lloyd D, Bacchi CJ. Effects of intermediates of methionine metabolism and nucleoside analogs on S-adenosylmethionine transport by Trypanosoma brucei brucei and a drug-resistant Trypanosoma brucei rhodesiense. FASEB J. 1998;56:95–103. doi: 10.1016/s0006-2952(98)00118-x. [DOI] [PubMed] [Google Scholar]
- 21.Cherest H, Surdiin-Kurjan Y. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating the sulfur metabolism pathway. Genetics. 1992;130:51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwer B, Saha N, Mao X, Chen HW, Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155:1561–1576. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer BE, Wolfger H, Kuchler K. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta. 1999;1461:217–236. doi: 10.1016/s0005-2736(99)00160-1. [DOI] [PubMed] [Google Scholar]
- 24.Thomas D, Rothstein R, Rosenberg N, Surdin-Kerjan Y. SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol. Cell. Biol. 1988;8:5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inselburg J. Induction and selection of drug resistant mutants of Plasmodium falciparum. Mol. Biochem. Parasitol. 1984;10:89–98. doi: 10.1016/0166-6851(84)90021-5. [DOI] [PubMed] [Google Scholar]
- 26.Kaminsky R, Zweygarth E. Feeder layer-free in vitro assay for screening antitrypanosomal compounds against Trypanosoma brucei brucei and T.b. evansii. Antimicrob. Agents Chemother. 1989;33:881–885. doi: 10.1128/aac.33.6.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sibley LD, LeBlanc AJ, Pfefferkorn ER, Boothroyd JC. Generation of a restriction fragment length polymorphism linkage map for Toxoplasma gondii. Genetics. 1992;132:1003–1015. doi: 10.1093/genetics/132.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phelouzat MA, Lawrence F, Robert-Gero M. Characterization of sinefungin-resistant Leishmania donovani promastigotes. Parasitol. Res. 1993;79:683–689. doi: 10.1007/BF00932511. [DOI] [PubMed] [Google Scholar]
- 29.Haughan PA, Chance ML, Goad LJ. Effects of sinefungin on growth and sterol composition of Leishmania promastigates. Exp. Parasitol. 1993;77:147–154. doi: 10.1006/expr.1993.1071. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence F, Derbecourt T, Robert-Gero M. Proton-ATPase activities involved in the uptake of an S-adenosylmethionine analogue. Mol. Biochem. Parasitol. 1998;92:99–109. doi: 10.1016/s0166-6851(97)00235-1. [DOI] [PubMed] [Google Scholar]
- 31.Phelouzat MA, Basselin M, Lawrence F, Robert-Gero M. Sinefungin shares AdoMet-uptake system to enter Leishmania donovani promastigotes. Biochem. J. 1995;305:133–137. doi: 10.1042/bj3050133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merali S, Clarkson AB. S-adenosylmethionine and pneumocystis. FEMS Microbiol. Lett. 2004;237:179–186. doi: 10.1016/j.femsle.2004.06.039. [DOI] [PubMed] [Google Scholar]