Abstract
Caulobacter crescentus is widely used as a powerful model system for the study of prokaryotic cell biology and development. Analysis of this organism is complicated by a limited selection of tools for genetic manipulation and inducible gene expression. This study reports the identification and functional characterization of a vanillate-regulated promoter (Pvan) which meets all requirements for application as a multi-purpose expression system in Caulobacter, thus complementing the established xylose-inducible system (Pxyl). Furthermore, we introduce a newly constructed set of integrating and replicating shuttle vectors that considerably facilitate cell biological and physiological studies in Caulobacter. Based on different narrow and broad-host range replicons, they offer a wide choice of promoters, resistance genes, and fusion partners for the construction of fluorescently or affinity-tagged proteins. Since many of these constructs are also suitable for use in other bacteria, this work provides a comprehensive collection of tools that will enrich many areas of microbiological research.
INTRODUCTION
Caulobacter crescentus has emerged as a powerful model system to study bacterial cell cycle control, chromosome dynamics and cell differentiation. Its hallmark is an asymmetric cell division that gives rise to both a sessile stalked cell and a motile swarmer cell. A straighforward synchronization protocol and the existence of many well-defined developmental markers have allowed the identification of various molecular circuits that control and execute the underlying differentiation process, acting at the level of transcription, translation, proteolysis and dynamic protein localization (1–4).
Cell biological studies are critically dependent on the application of fluorescent protein fusions to follow the subcellular distribution of proteins in vivo (5). They also rely heavily on the availability of inducible promoters that allow overproduction and depletion of proteins in a controlled manner, thus facilitating functional analyses and the characterization of essential genes. However, despite the excellent genetic amenability of Caulobacter (6) and years of intensive research, the molecular biological toolset for this organism is still poorly developed. So far, only a small number of plasmids is dedicated to the construction of protein fusions, offering a very limited choice of cloning sites, resistance markers and fusion partners. Moreover, regulatable gene expression in Caulobacter is currently based on a single, xylose-inducible promoter system (7), which has not yet been implemented into a standard set of broadly applicable vectors.
As the number of proteins identified as mediators of Caulobacter's developmental program rises, unraveling the interactions between the different factors has become increasingly complex and frequently requires the construction of strains bearing multiple genetic modifications. To respond to this new technical challenge, it is essential to improve the tools available for genetic manipulation of Caulobacter. Here, we report the identification and characterization of a vanillate-inducible promoter, which is tightly regulated and can be used independently of the existing xylose-regulated system. Moreover, we present a comprehensive set of integrating and replicating shuttle vectors that are designed for the straightforward construction of protein fusions and the convenient expression of genes under control of the vanillate- and xylose-inducible promoters.
MATERIALS AND METHODS
Chemicals and enzymes
Vanillic acid (≥97%) was purchased from Fluka. Vanillin (99%), vanillyl alcohol (≥ 98%) and d-(+)-xylose (>99%) were obtained from Sigma-Aldrich. The following stock solutions were prepared: 50 mM vanillic acid/NaOH (pH 7.5 in H2O), 50 mM vanillin/NaOH (pH 7.0 in H2O), 50 mM vanillyl alcohol (in 70% ethanol) and 20% xylose (in H2O). For long-term storage, the vanillate and vanillin stocks were kept at −20°C. Restriction and DNA-modifying enzymes were purchased from New England Biolabs and MBI Fermentas. PCR reactions were performed using KOD polymerase (Novagen). Oligonucleotides were synthesized by the Stanford PAN Facility.
Bacterial strains and growth conditions
Caulobacter crescentus CB15N (8) and its derivatives were grown in PYE rich or M2G minimal medium (6) at 28°C. For cloning purposes, plasmids were propagated in Escherichia coli TOP10 (Invitrogen), which was cultivated in Luria-Bertani medium at 37°C. When appropriate, media were supplemented with antibiotics at the following concentrations (liquid/solid media for C. crescentus; liquid/solid media for E. coli; in μg/ml): spectinomycin (25/50; 50/100), kanamycin (5/25; 30/50), rifampicin (2.5/5; 25/50), gentamicin (0.5/5; 15/20), oxytetracycline (1/1; 12/12), chloramphenicol (2/1; 20/30), apramycin (10/60; 30/30). Plasmid transfer into C. crescentus was achieved by electroporation (6). Escherichia coli was transformed using a chemical method (9). The CB15N derivatives MT219 (▵vanR) and MT231 (▵vanA) were generated with the help of plasmids pMT422 and pMT487, respectively, following a previously described gene replacement protocol (10). Strains MT232, MT236 and MT240 were created by transforming strain CB15N with integration plasmids pMT627, pMT704 or pMT760, respectively, and selecting for homologous recombination of the constructs into the chromosomal vanA or xylX locus.
Vanillate degradation assay
Cells were grown in M2G minimal medium containing 0.5 mM vanillate. At the indicated timepoints, aliquots were withdrawn from the culture. The cells were pelleted by centrifugation for 5 min at 16 000 g, and the concentration of vanillate left in the supernatant was determined spectrophotometrically using a wavelength of 286 nm (the absorption maximum of vanillate).
Microscopy
Cells were grown to exponential phase in M2G medium, induced for 1–2 h with vanillate or xylose at the indicated concentrations, immobilized on pads of 1% agarose in M2 salts (6) and visualized using a Nikon Eclipse E800 microscope equipped with a Nikon Plan Apo 100×/1.40 differential interference contrast objective and a 5 MHz MicroMax cooled CCD camera (Princeton Instruments). Images were processed with Metamorph 4.5 (Universal Imaging Group) and Photoshop 6.0 (Adobe Systems).
β-Galactosidase activity assays
After growth of the cells to exponential phase, synthesis of β-galactosidase was induced by addition of sodium vanillate, vanillin or vanillyl alcohol to the indicated final concentrations. Unless indicated otherwise, cultivation was continued for 2 h (cultures in PYE medium) or 3 h (cultures in M2G medium), respectively, and β-galactosidase activities were determined according to Miller (11), with 0.01% SDS included in the reaction mixtures.
RNA extraction
Caulobacter crescentus CB15N was grown in M2G medium to an OD660 of 0.15. Subsequently, two 25 ml-aliquots were withdrawn and incubated for 3 h in the absence or presence of 0.5 mM vanillate, respectively. The cells were harvested by centrifugation, resuspended in 1 ml of Trizol (Invitrogen), and incubated for 10 min at 65°C. After addition of 0.2 ml chloroform, the samples were shaken by hand for 15 s and incubated for 5 min at 4°C. Following centrifugation, the aqueous phase was collected and transferred to a fresh tube. RNA was then precipitated by addition of 0.5 ml isopropanol and incubation at −80°C for 1 h, and collected by centrifugation for 30 min at 4°C and 16 000 g. The pellets were washed with 75% ethanol and dried for 10 min at room temperature. The isolated RNA was dissolved in 50 μl RNase-free water, incubated for 10 min at 55°C, and stored at −80°C until further use.
Primer extension analysis
Extension of the oligonucleotide 5′-CATACCAGGCGTCAAGCGGC-3′, which hybridizes to the coding region (bases 36–55) of the vanA gene, was performed as described (12), using SuperScript II Reverse Transcriptase (Invitrogen) and an annealing temperature of 45°C. The Thermo Sequenase Cycle Sequencing Kit (USB) was used for DNA sequencing, with plasmid pMT437 as the DNA template.
Plasmid construction
Reporter plasmids and primer extension templates
To construct the PvanA reporter plasmid pMT122, a chromosomal fragment comprising the vanR gene, the vanR-vanA intergenic region, and the first four codons of vanA was PCR-amplified, with a HindIII site added to its 5′ end and a PstI site added to its 3′ end. The reaction product was cleaved with HindIII and PstI and ligated into the equally treated lacZ reporter plasmid pPR9TT (13). The reporter plasmid pMT129 was created by restriction of pMT122 with HindIII and SphI, blunting the newly generated ends with T4 DNA polymerase, and self-ligation of the plasmid backbone, thereby deleting bases 136–735 of the vanR gene. For the construction of pMT437, a chromosomal fragment comprising vanR, the vanR-vanA intergenic region, and the first 355 bases of vanA was PCR-amplified. The reaction product was then ligated into pCR4Blunt-TOPO using the ZERO Blunt TOPO PCR Cloning Kit (Invitrogen).
Plasmids for generating in-frame deletions in vanR and vanA
To construct plasmid pMT422, the upstream flanking region and first twelve codons of vanR were PCR-amplified, using primers that add a HindIII site to the 5′ end and a SacI site to the 3′ end of the reaction product. In parallel, a fragment comprising the last 12 codons and the downstream flanking region of vanR was PCR-amplified, with a SacI site added to its 5′ end and an EcoRI site added to its 3′ end. The PCR products were treated with HindIII/SacI or SacI/EcoRI, respectively, and ligated into allele exchange vector pNPTS138 (M. R. Alley, unpublished), which had been cut with HindIII and EcoRI.
The knockout construct pMT487 was generated in an analogous way. First, a fragment comprising the upstream flanking region and first twelve codons of vanA was PCR-amplified, with a HindIII and an NheI site added to its 5′ and 3′ end, respectively. In parallel, the last twelve codons plus the downstream flanking region of vanA were PCR amplified using primers that add an NheI site to the 5′ end and an EcoRI site to the 3′ end of the reaction product. The two fragments were digested with HindIII/NheI or NheI/EcoRI, respectively, and then ligated into plasmid pNTPS138 which had been treated with HindIII and EcoRI.
Plasmids for the inducible expression of fluorescent protein fusions
In order to create plasmid pMT627, the mipZ gene (10) was PCR-amplified, replacing its start codon with an NdeI restriction site and the stop codon with a VspI restriction site. The reaction product was cut with the corresponding enzymes and ligated into the NdeI/VspI-treated integration vector pVYFPC-1. Plasmid pMT704 was generated as follows: the mipZ gene was PCR-amplified, using primers that add a SacI restriction site to its 5′ end and an EcoRI restriction site its 3′ end. The reaction product was cut with the corresponding enzymes and then ligated into equally treated pXGFPN-2. For the construction of pMT760, a fragment carrying the ftsZ gene was isolated from plasmid pMT217 (10) by restriction with NdeI and SmaI, and ligated into NdeI/Ecl136II-treated pVCHYC-3.
High-copy vectors for vanillate- and xylose-inducible gene expression
To generate the high-copy number expression vector pBVMCS-4, the vanR gene and the vanR-vanA intergenic region were PCR-amplified, introducing an NdeI restriction site at the vanA start codon. The product was phosphorylated and ligated into the KpnI/SphI-treated and blunted broad-host-range vector pBBR1MCS-5 (14) such that Pvan was adjacent to the plasmid-borne multiple cloning site (MCS). To generate pBVMCS-2 and pBVMCS-6, a fragment containing vanR, Pvan, and the MCS was isolated from pBVMCS-4 by restriction with SspI and ligated into SspI-treated pBBR1MCS-2 (14) or PflFI/VspI-treated and blunted pBBR1MCS (15), respectively, in an orientation analogous to that in pBVMCS-4.
For the construction of pBXMCS-2, a fragment comprising the E. coli rrnB T1T2 transcriptional terminator (ter) and Pxyl was isolated from pMT27 (10) by restriction with BamHI, blunting of the overhang, and subsequent treatment with NdeI. It was then ligated into pBVMCS-2, which had been cut with AscI, blunted, and treated with NdeI. Plasmid pBXMCS-4 was created analogously, using pBVMCS-4 as the target vector. To generate pBXMCS-6, a fragment comprising ter and Pxyl was released from pBXMCS-2 by restriction with SspI and ligated into PflFI/VspI-treated and blunted pBBR1MCS (14).
Low-copy vectors for vanillate- and xylose-inducible gene expression
To construct the low-copy number expression vector pRVMCS-5, an SspI-fragment from pBVMCS-4 containing vanR and Pvan was ligated into the PstI/SphI-digested and blunted plasmid pMT288 (10) such that vanR was placed next to the plasmid-borne rrnB T1T2 terminators (16). A fragment containing ter, vanR and Pvan was released from the resulting plasmid (pMT355) by restriction with NotI. It was blunted and ligated into pJB3Tc20 (17) whose NdeI site had been eliminated by a silent mutation in the trfA gene. The resulting plasmid, which carried vanR adjacent to the plasmid-borne tetR gene, was then treated with NdeI and PstI and ligated with a synthetic DNA duplex bearing an MCS. To create plasmid pRVMCS-6, a fragment comprising ter, vanR, PvanA and the MCS was released from pRVMCS-5 by restriction with NotI, blunting of the overhang, and subsequent restriction with SfiI. It was then ligated into pJB3Cm6 (17), which had been cut with SapI, blunted, and subsequently cleaved with SfiI. Plasmid pRVMCS-2 was generated by ligating BstBI/Bpu10I-treated and blunted pRVMCS-6 with a blunted PagI-fragment from pMT27 (10) containing the neo gene.
For the construction of pRXMCS-2, -5 and -6, a fragment containing ter and Pxyl was isolated from pMT27 by restriction with NotI and NdeI and ligated into NotI/NdeI-treated pRVMCS-2, -5 and -6, respectively.
Integration vectors for the one-step generation of depletion strains
A fragment comprising ter, vanR, Pvan and the MCS was isolated from pRVMCS-5 by restriction with NotI and PvuII and ligated into NotI/BsrBI-treated pMT27 (10). The resulting plasmid (pMT384) was then cut with NotI and ligated with a PCR-fragment which carried the RK2 origin of transfer (oriT) (18) and possessed engineered NotI and EagI sites at the 5′ and 3′ end, respectively. Colony PCR was used to identify a construct (pMT434) with oriT positioned immediately upstream of the plasmid-borne neo gene. A Spc/StrR-derivative of pMT434 (named pMT510) was created by restriction of the plasmid with PagI, blunting of the ends, and ligation with a blunted BstBI/StuI-fragment from pHPV465 (19) containing the aadA gene of plasmid R100.1.
Deletion of vanR from pMT510 by restriction with HindIII and NcoI, blunting of the ends, and self-ligation of the plasmid backbone resulted in pVMCS-1. Plasmids pVMCS-3 and -6 were created by ligating PagI/AvrII-treated pVMCS-1 with PCR fragments that comprised (i) the parr-2 gene from plasmid pMW1409 (20) or (ii) the Tn9 cat gene from p34S-Cm (21), respectively, and carried an engineered PagI site followed by a ribosomal binding site at the 5′ end and an engineered AvrII site at the 3′ end. To generate pVMCS-2, a fragment comprising oriT, ter and Pvan was isolated from pVMCS-1 by restriction with NotI and PstI and ligated into equally treated pMT434. Plasmids pVMCS-4, -5 and -7 were subsequently generated by ligating PagI-treated and blunted pVMCS-2 with (i) a blunted SacI-fragment from pUCGm (22) containing the Tn1696 aacC1 gene, (ii) a blunted BssSI-fragment from pJB3Tc20 (17) carrying the tetR and tetA genes of pFL129, and (iii) a DraI/PmlI-fragment from pHP45Ωaac (23) containing the E. coli aacC4 gene, respectively.
To construct pXMCS-1, the Pxyl fragment (including the NdeI restriction site) of pMT27 was PCR amplified, adding a HindIII restriction site to the 5′ end. The reaction product was cut with HindIII and NdeI and ligated into equally treated pMT510. Ligation of a NotI/PstI-fragment from pXMCS-1 containing oriT, ter, and Pxyl into equally cut pVMCS-2, -3, -4, -5, -6 and -7 then resulted in plasmids pXMCS-2, -3, -4, -5, -6 and -7, respectively.
Vectors for generating N- and C-terminal protein fusions encoded at the vanA or xylX locus
In order to extend the Pvan fragment carried by pMT510 to a length of 1586 bp, the 3′ region of vanR (containing an AscI site) and ∼ 0.7 kb of the chromosomal vanR downstream region were PCR amplified, adding a HindIII restriction site to the 5′ end. The reaction product was cut with HindIII and AscI and ligated into equally cut pMT510, resulting in pMT517. To create an integration plasmid with a 2316 bp Pxyl promoter region, a fragment containing Pxyl and its upstream region was isolated from pXGFP4 (M. R. Alley, unpublished) by restriction with NheI, blunting of the overhang, and subsequent restriction with NdeI. It was then ligated into pMT510 which had been cut with HindIII, blunted, and subsequently treated with NdeI, giving rise to pMT519. Synthetic MCS were inserted between the NdeI and AgeI restriction sites of pMT517 and pMT519, respectively, resulting in pMT517MCS-N, pMT519MCS-N, pMT517MCS-C and pMT519MCS-C.
For the construction of C-terminal fusion vectors, the egfp, eyfp, ecfp (Clontech), venus (24), cerulean (25) and mCherry (26) genes were PCR-amplified, adding an AgeI restriction site followed by a short linker to the 5′ end and an NheI restriction site to the 3′ end, respectively. The AgeI/NheI-treated fragments were then ligated into pMT517MCS-N, resulting in pVGFPC-1, pVYFPC-1, pVCFPC-1, pVVENC-1, pVCERC-1 and pVCHYC-1. Ligation of the same fragments into pMT519MCS-N, on the other hand, gave rise to the plasmids pXGFPC-1, pXYFPC-1, pXCFPC-1, pXVENC-1, pXCERC-1 and pXCHYC-1. Plasmid pXTCTC-1 was obtained by restriction of pXGFPC-1 with AgeI and NheI and ligation of the plasmid backbone with a synthetic double strand encoding the tetracysteine tag (27).
To create N-terminal fusion vectors, egfp, eyfp, ecfp (Clontech), venus (24), cerulean (25) and mCherry (26) were PCR-amplified, with an NdeI restriction site engineered at the respective start codon. After digestion with NdeI and BsrGI, the reaction products were ligated into equally treated pMT517MCS-C, yielding plasmids pVGFPN-1, pVYFPN-1, pVCFPN-1, pVVENN-1, pVCERN-1 and pVCHYN-1. Ligation of the same restriction fragments into pMT519MCS-C, by contrast, gave rise to plasmids pXGFPN-1, pXYFPN-1, pXCFPN-1, pXVENN-1, pXCERN-1 and pXCHYN-1.
In order to extend the range of resistance markers, the plasmids described above were cut with NheI and NotI, releasing a fragment that contained the aadA Spec/StrR resistance gene. Subsequently, they were ligated with NheI/NotI-fragments from plasmids pVMCS-2, -3, -4, -5 or -6 carrying the neo, parr-2, aacC1, tetRA and cat genes, respectively.
Plasmid pXFLGC-2 was obtained by restriction of pXGFPC-2 with AgeI and NheI, and ligation of the plasmid backbone with a synthetic double strand encoding the FLAG-tag (28).
Vectors for generating C-terminal protein fusions encoded at a site of interest
To generate the integration vectors pGFPC-1, pYFPC-1, pCFPC-1, pVENC-1, pCERC-1 and pCHYC-1, the Pxyl promoter fragment was released from pXGFPC-1, pXYFPC-1, pXCFPC-1, pXVENC-1, pXCERC-1 and pXCHYC-1 by restriction with HindIII and NdeI, and replaced by a synthetic double-stranded linker. Plasmids pFLGC-1, pSTRC-1 and pTCYC-1 were obtained by restriction of pGFPC-1 with AgeI and NheI, and ligation of the plasmid backbone with synthetic double strands encoding the FLAG-tag (28), the Strep-tag II (29) or the tetracysteine tag (27) peptides.
Derivatives of these plasmids bearing alternative resistance markers were constructed by replacement of the original NheI/NotI-fragment carrying the aadA gene with NheI/NotI-fragments from pVMCS-2, -3 and -4 carrying the neo, parr-2, and aacC1 genes, respectively.
Vectors for integration at a site of interest
For the construction of pMCS-1, the Pvan promoter fragment of plasmid pMT517MCS-C was released by restriction with HindIII and NdeI, and replaced with a synthetic linker. To create derivatives carrying alternative resistance markers, the aadA gene was excised from pMCS-1 by restriction with NheI and NotI. The plasmid backbone was then ligated with NheI/NotI-fragments from pVMCS-2, -3, -4, -5, -6 and -7 carrying the neo, parr-2, aacC1, tetRA, cat or aacC4 genes, respectively.
Low-copy vectors for generating N- and C-terminal protein fusions under the control of PvanA
Fragments carrying the MCS and the fluorescent protein gene were released from pVGFPN-1, pVYFPN-1, pVCFPN-1, pVGFPC-1, pVYFPC-1, pVCFPC-1, pVVENC-1 and pVCHYC-1 by restriction with NdeI and NheI. They were then ligated with NdeI/NheI-treated pRVMCS-2, -4 and -5, respectively, giving rise to the pRV-series of gene fusion vectors.
RESULTS
Identification of a vanillate-inducible promoter in Caulobacter
Caulobacter crescentus thrives in oligotrophic freshwater habitats (30), where structural polymers derived from decaying vascular plant material represent the predominant source of organic carbon (31). Its genome indeed encodes a number of hydrolases with the potential to degrade cellulose and hemicellulose (32), and it can grow on xylose, an abundant constituent of hemicellulose, as the sole carbon source (30). The presence of xylose serves as a general signal for the availability of plant polysaccharides and induces synthesis of a suite of proteins that mediate polymer degradation and funneling of the hydrolysis products into the general catabolic pathways (33). One of the most highly activated members of the xylose regulon is the xylX gene, encoding a protein of unknown function, whose promoter (Pxyl) has been widely used for inducible gene expression in Caulobacter (7).
Apart from polysaccharides, lignin represents the second most abundant constituent of the vascular plant cell wall. It is a heterogeneous, highly crosslinked polymer derived from the aromatic monolignols coniferyl, coumaryl and sinapyl alcohol (34). Due to its complex chemical structure, its initial, oxidative cleavage can only be performed by a small group of organisms, most importantly fungi (35). The soluble phenolic intermediates, however, released during this process are subsequently metabolized by a variety of different bacterial species. One of the compounds commonly produced from lignin is vanillate (Figure 1A). To allow its utilization as a carbon source, it first has to be converted into protocatechuate by removal of its O-methyl group. Cleavage of the aromatic ring system and further degradation via the β-ketoadipate pathway then yield succinyl-CoA and acetyl-CoA, which can directly enter the citric acid cycle (36). In many bacteria, O-demethylation of vanillate is carried out by a two-component monooxygenase, composed of a terminal oxygenase (VanA) and a ferredoxin-like reductase (VanB) subunit. Usually, these two proteins are encoded in an operon, with their synthesis being regulated by a transcriptional repressor, VanR, whose gene is transcribed together with or divergently from the vanAB trancriptional unit (37–40) (Figure 1B).
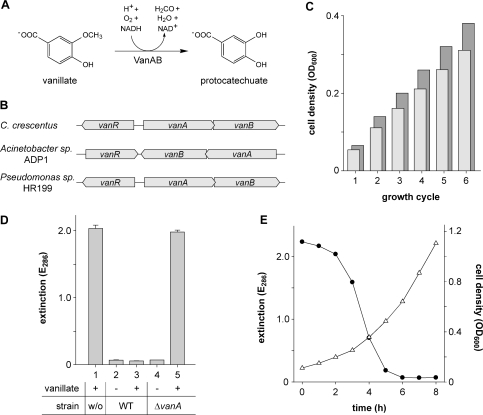
Figure 1.
Vanillate degradation by Caulobacter. (A) Conversion of vanillate into protocatechuate, as catalyzed by the vanillate demethylase (VanAB) complex. (B) Arrangement of the vanR, vanA and vanB genes in C. crescentus, Acinetobacter sp. ADP1 and Pseudomonas sp. HR199. (C) Cell densities achieved with vanillate or glucose as the sole carbon source. Minimal media composed of 0.5 mM vanillate (light bars) or glucose (dark bars), respectively, in M2 salts (6) were inoculated with washed cells of wild-type strain CB15N. The cultures were grown until the carbon source was depleted, and their optical densities were determined (growth cycle 1). Subsequently, the cells were subjected to another five growth cycles, each of which started with replenishment of the carbon source (0.5 mM final concentration), followed by growth to stationary phase and determination of the respective cell densities. Note: Stepwise addition of the carbon source was necessary due to a negative effect of vanillate on growth at concentrations higher than 0.5 mM. (D) Involvement of CC2393 (VanA) in vanillate degradation. M2G minimal medium containing no (−) or 0.5 mM (+) vanillate, respectively, was inoculated with wild-type strain CB15N (WT) or its ▵vanA-derivative MT231 and incubated for 24 h. Subsequently, the cells were pelleted, and the supernatant was analyzed spectrophotometrically at 286 nm (the absorption maximum of vanillate) to determine the amount of vanillate left in the medium. Sterile medium (w/o) was used as a control. Data represent the mean of three independent experiments (±SD). (E) Induction kinetics of vanillate degradation. Cells of wild-type strain CB15N were grown to exponential phase in M2G minimal medium, washed, and resuspended in M2G medium containing 0.5 mM vanillate to an OD600 of 0.11. Samples were taken at one-hour intervals and analyzed for cell density (OD600, open triangle) and vanillate content (E286, filled circle).
Motivated by the ability of Caulobacter to utilize plant material for growth and to degrade a variety of different aromatic compounds (41), we analyzed the Caulobacter genome (32) for homologs of the vanAB genes. This search identified two overlapping open reading frames, CC2393 and CC2394, whose deduced products show 67% and 48% identity with the well-characterized VanA and VanB proteins, respectively, from Acinetobacter sp. ADP1 (38). The polypeptide encoded by CC2393 contains sequence motifs (42) that are characteristic of the mononuclear and the Rieske-type iron-binding sites of class IA terminal oxygenases. CC2394, on the other hand, gives rise to a protein with the NAD-ribose-, flavin mononucleotide-, and chloroplast-type ferredoxin [2Fe-2S]-binding domains typically found in class IA reductases (42). This domain composition indicates that the Caulobacter VanA and VanB homologs could indeed constitute a two-component monooxygenase with vanillate demethylase activity. Encoding all components of the β-ketoadipate pathway (CC2405-CC2411) (32), Caulobacter might thus possess the full complement of enzymes necessary for the degradation of vanillate. Growth experiments indeed showed that it can utilize this aromatic compound as the sole carbon source, reaching cell densities comparable to those obtained in glucose minimal medium (Figure 1C). Metabolism of vanillate still occurred in the presence of excess glucose (Figure 1D, bars 2 and 3), which indicates that the degradative pathways involved are not subject to catabolite repression. By contrast, vanillate metabolism was completely abolished in a mutant strain carrying an in-frame deletion in the vanA homologue (MT231), thus strongly supporting a function of Caulobacter VanAB in vanillate demethylation (Figure 1D, bars 4 and 5). In order to investigate the kinetics of its degradation, vanillate was added to a culture of wild-type strain CB15N growing exponentially in glucose minimal medium and quantified in regular intervals (Figure 1E). Maximal degradation rates were reached after a short lag phase, leading to complete removal of vanillate from the growth medium within a few hours. The lag phase observed in these measurements suggested that the enzymatic pathway mediating vanillate utilization is not constitutively active but specifically induced in the presence of the substrate. Vanillate concentrations higher than 0.5 mM noticeably decreased the growth rate of Caulobacter (reduction at 1 mM : 25% in M2G medium and 2% in PYE medium). This effect might be attributable to a weak uncoupling activity, as observed for many other aromatic monocarboxylic acids (43).
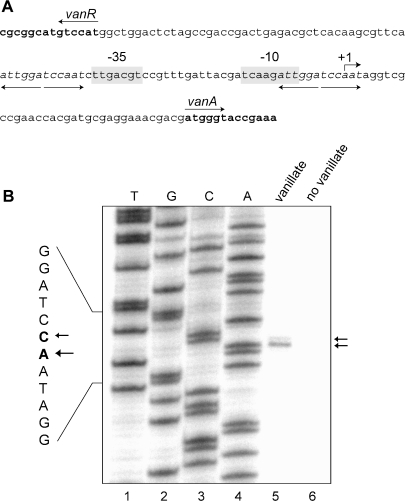
Having identified a putative vanillate degradation gene cluster, we determined if the promoter driving transcription of the vanAB operon (Pvan) could be exploited for inducible gene expression in Caulobacter. For this purpose, a detailed functional analysis of the 126 bp intergenic region shared by the vanR gene and vanAB genes (Figure 2A) was performed. Primer extension analysis of RNA isolated from wild-type strain CB15N grown in the presence of vanillate showed that transcription of vanA mostly initiates at an adenosine residue 35 bp upstream of the vanAB translational start site (Figure 2B, lane 5). In addition, a minor transcript species carrying an additional 5′ cytosine residue could be detected. The primer extension reaction consistently failed to yield a product when performed on RNA from cells that had been grown in medium lacking vanillate (Figure 2B, lane 6), indicating that expression of vanAB is induced by vanillate or one of its degradation products. Upstream of the experimentally determined +1 site, sequences corresponding to the characteristic -35 and -10 boxes of a housekeeping, σ73-dependent promoter can be identified (44,45) (Figure 2A). Their relative spacing and distance to the transcriptional start site suggests that they represent the core promoter elements responsible for vanAB transcription. The vanAB upstream region further contains two copies of a perfect inverted repeat (5′-ATTGGATCCAAT-3′), which cannot be found anywhere else in the Caulobacter genome (32). One of them immediately precedes the –35 box, whereas the other one overlaps the –10 box and the +1 site (Figure 2A). Palindromic sequences arranged in such a manner frequently reflect GntR-type transcription factor binding sites (46). The two repeats are, therefore, likely to interact with the Caulobacter VanR homolog, mediating repression of vanAB transcription in the absence of vanillate.
Figure 2.
Characteristics of the vanAB promoter. (A) Overview of the vanAB promoter region. The vanR-vanAB intergenic region is shown in normal print, whereas the 5′ ends of the flanking vanR and vanA genes are given in boldface, with their orientations indicated by arrows. The transcriptional start site of vanA (see Figure 2B) and the putative −10 and −35 motifs of the vanA promoter are labeled. Diverging arrows and italic letters mark two copies of a perfectly palindromic sequence, which is likely to represent the VanR target site. (B) Determination of the vanAB transcriptional start site. Primer extension analysis was conducted on RNA extracted from cells of wild-type strain CB15N which had been grown in M2G medium in the presence (lane 5) or absence (lane 6) of 0.5 mM vanillate. In parallel, sequencing reactions were performed (lanes 1–4). The two reaction products and the corresponding +1 sites are indicated by arrows.
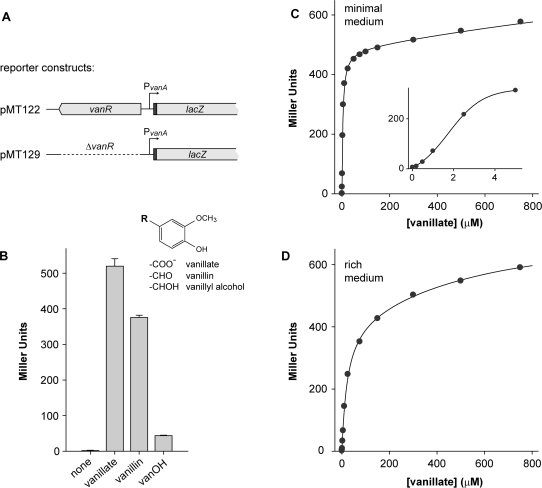
In order to investigate the regulation of vanAB expression in more detail, a fragment comprising the vanR gene, the vanR-vanAB intergenic region, and the 5′ part of the vanA gene was cloned into the low-copy promoter probe vector pPR9TT (13), generating a translational fusion between vanA and the plasmid-borne lacZ reporter gene (Figure 3A). When wild-type strain CB15N was transformed with the resulting plasmid (pMT122) and grown in glucose minimal medium lacking any inducer, no transcriptional activity was detectable. Addition of vanillate, by contrast, strongy activated the vanAB promoter, thus confirming the regulatory pattern suggested by primer extension analysis (Figure 3B). A similar response was elicited by vanillin, another common product of lignin biodegradation (35). As a structural relative of vanillate, vanillin might directly interact with the effector-binding site of VanR. Alternatively, since Caulobacter contains a gene (CC2402) whose product shows 58% identity with vanillin dehydrogenase from Pseudomonas sp. HR199 (47), the stimulating effect of vanillin could be indirect, resulting from its oxidation to vanillate, which then serves as the actual inducer. In contrast to the carboxy and aldehyde forms, vanillyl alcohol only slightly activated the vanAB promoter, possibly due to weak interaction with VanR or slow metabolic conversion into vanillate as observed in Pseudomonas sp. PN-1 (48).
Figure 3.
Response of the vanAB promoter to different inducers and inducer concentrations. (A) Reporter constructs used to determine Pvan promoter activity. The 5′ region of vanA, which was fused in frame to the lacZ gene, is shown in black. (B) Activity of Pvan in the presence of different inducers. Wild-type strain CB15N was transformed with reporter plasmid pMT122 and grown in M2G minimal medium. In mid-exponential phase, vanillate, vanillin or vanillyl alcohol (vanOH) were added to a final concentration of 0.5 mM, respectively, and the activity of Pvan was determined. The inset shows the structural formulas of the inducers used. (C and D) Response of Pvan to different concentrations of vanillate. Cells of strain MT231 (▵vanA) transformed with reporter plasmid pMT122 were grown in M2G minimal medium (C) or PYE rich medium (D), exposed for 3 h to different concentrations of vanillate, and used to determine the activity of Pvan. The inset in (C) shows Pvan promoter activity at vanillate concentrations of 0–5 µM. Data depicted in panels (B), (C) and (D) represent the average of two independent experiments, each performed in triplicate. Standard deviations were smaller than 11% throughout.
After having established vanillate as the most potent inducer, we analyzed vanAB promoter activity in response to different concentrations of this compound. For that purpose, cells of the vanA-deficient strain MT231 bearing reporter plasmid pMT122 were grown in either glucose minimal or rich medium and exposed to varying inducer concentrations (Figures 3C and D). The titration curves obtained show that vanillate concentrations in the nanomolar range are sufficient to activate Pvan. While promoter responsiveness was higher in minimal medium, growth conditions had no effect on the maximal promoter activity reached at saturating vanillate concentrations. These results demonstrate that the vanAB promoter is tightly regulated and can assume a wide range of activities depending on the amount of inducer added to the cultures.
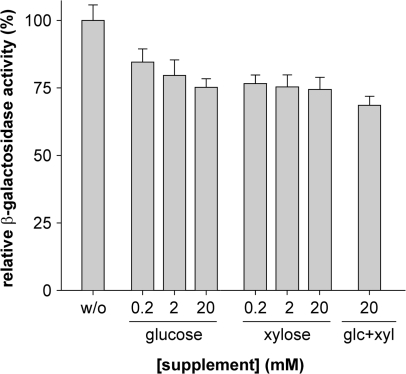
An important prerequisite for the general application of vanillate-inducible gene expression in Caulobacter is its compatibility with standard growth conditions. Many experimental setups require the use of rich media that are supplemented with glucose or xylose to repress or activate genes cloned under the control of the xylose-inducible promoter Pxyl, repectively (7). To assess the influence of sugars on the activity of the vanAB promoter at high nutrient levels, wild-type strain CB15N was transformed with reporter plasmid pMT122 (Figure 3A) and exposed to vanillate in the presence of varying concentrations of glucose, xylose or a combination thereof. In all cases, the availability of an additional carbon source reduced the promoter activity by ∼25% (Figure 4). This effect was essentially independent of the nature or concentration of the supplement and was only slightly aggravated by the combined addition of both sugars. The same percentual reduction in transcription levels was observed over a wide range of vanillate concentrations (data not shown), indicating that the overall characteristics of the vanAB promoter are resistant to changes in the composition of the growth medium. On the other hand, we could not observe any effect of vanillate on the activity of Pxyl (data not shown). These results demonstrate that the Pvan- and Pxyl-based expression systems are fully compatible and can be used in parallel and independently each other.
Figure 4.
Influence of sugars on the activity of the vanAB promoter. Cells of wild-type strain CB15N transformed with reporter plasmid pMT122 (Figure 3A) were grown in PYE medium and exposed to 0.5 mM vanillate (w/o), a mixture of 0.5 mM vanillate and glucose or xylose at the indicated concentrations, or a mixture of 0.5 mM vanillate, 20 mM glucose and 20 mM xylose. Subsequently, the cell were subjected to β-galactosidase activity assays to determine the response of Pvan under the different inducing conditions. Data represent the mean of two independent experiments (± SD), each performed in triplicate.
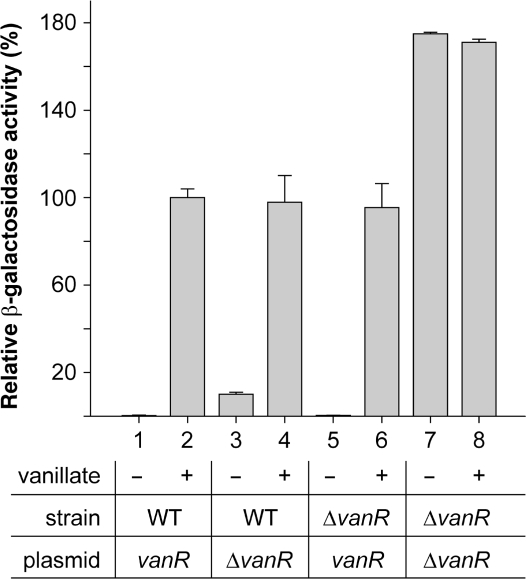
In order to improve the usability of the vanAB promoter, it was necessary to determine the identity of its cognate regulator. A gene (CC2392) encoding a GntR-type transcriptional repressor is found immediately upstream of the Caulobacter vanAB operon (Figure 1B), with its transcription oriented in the opposite direction. The corresponding protein shows 43% identity with VanR from Acinetobacter sp. ADP1 (49), suggesting a role in modulating vanAB expression in response to vanillate or structurally similar compounds. We, therefore, created a strain carrying an in-frame deletion in the chromosomal vanR gene (MT219) and analyzed the effect of this mutation on vanAB transcription, using either reporter plasmid pMT122 or its vanR-deficient derivative pMT129 (Figure 3A). When assayed in the presence of a plasmid-borne copy of vanR, promoter activities were indistinguishable from those observed in a wild-type background (Figure 5, bars 1, 2, 5 and 6). Without complementation, in contrast, transcription was fully activated irrespective of the inducer concentration, demonstrating that VanR indeed regulates expression of the vanAB operon in Caulobacter (Figure 5, bars 7 and 8). The reporter levels measured under these conditions were about 75% higher than in strains containing at least one functional copy of the vanR gene. This observation could indicate that the concentration of vanillate used in these experiments still allows for residual binding of VanR to the vanAB promoter, thus resulting in sub-maximal transcription rates. Alternatively, the induction time used might not have been long enough for vanR-containing strains to reach the plateau established by constitutive expression of the reporter construct. When transcription was measured in wild-type strain CB15N in the absence of a plasmid-borne vanR allele, significant promoter activity was detectable in the uninduced state (Figure 5, bars 3 and 4). Thus, the number of repressor molecules synthesized from the chromosomal vanR gene alone is not sufficient to ensure tight regulation of multiple promoter copies provided in trans on a replicating plasmid.
Figure 5.
Involvement of VanR in the regulation of vanAB expression. Cells of wild-type strain CB15N (WT) and its mutant derivative MT219 (ΔvanR) were transformed with reporter plasmids pMT122 (vanR) or pMT129 (ΔvanR) and grown to exponential phase in PYE medium. Subsequently, they were exposed to 0 mM (−) or 0.5 mM (+) vanillate, and used to determine the activity of Pvan. Data represent the mean of two independent experiments (± SD), each performed at least in triplicate.
Construction of plasmids for inducible gene expression in Caulobacter
The results described above demonstrate that the vanAB promoter fulfills all criteria necessary for use as a multipurpose gene expression system. We, therefore, integrated this promoter into a set of vectors that facilitates its application to a variety of problems commonly encountered in cell biological research. In parallel, we created a comparable series of Pxyl-based plasmids as well as a number of promoterless constructs for general use, thereby establishing a comprehensive and standardized toolkit for genetic manipulation in Caulobacter.
Integration vectors
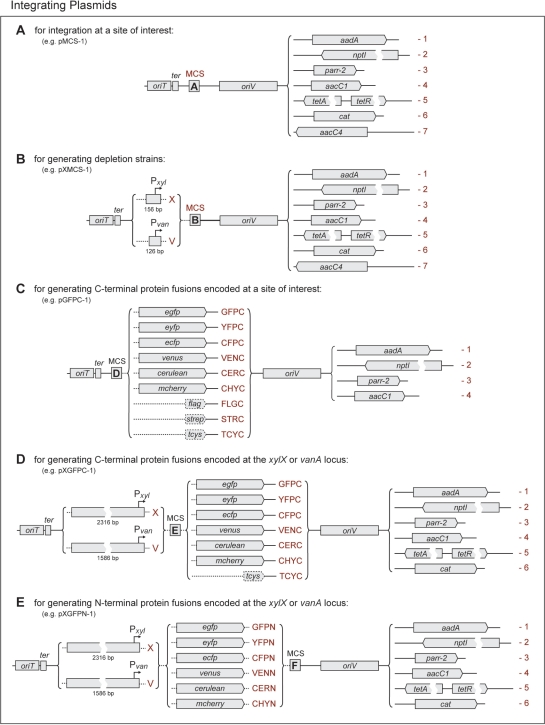
The most basic set of constructs comprises a number of general use integration vectors, which have been optimized for small size to maximize transformation yields (Figure 6A). They all have in common the RK2 origin of transfer, allowing conjugative transfer with the help of suitable host strain (18,50), the E. coli rrnB T1T2 transcriptional terminators, preventing leaky expression of cloned genes due to read-through from upstream regions (16), and a newly designed polylinker containing up to sixteen unique restriction sites (Figure 7, MCS A). Their replication is driven by the narrow host range pMB1 origin, which allows high copy numbers in E. coli but is inactive in Caulobacter. Together, these plasmids further offer a choice of seven different selective markers, thereby facilitating the generation of multiply modified strains. Five of these markers were derived from preexisting expression cassettes known to confer high-level resistance against spectinomycin, kanamycin, gentamicin, tetracycline or apramycin to Caulobacter (see Materials and Methods). In addition, novel rifampicin and chloramphenicol resistance determinants were created by combining the rickettsial parr-2 (20) and Tn9 cat genes (21), respectively, with a more efficient ribosome binding site and the R100.1 aadA promoter (51).
Figure 6.
Integration Plasmids. This schematic shows different sets of vectors designed for (A) integrating constructs at a chromosomal site of interest, (B) inserting a minimal Pxyl or Pvan fragment upstream of a defined chromosomal locus, (C) creating C-terminal protein fusions encoded at the native locus of the gene of interest, and generating inducible (D) C-terminal or (E) N–terminal protein fusions encoded at the xylX or vanA locus. Plasmid names are determined as follows: each construct carries the prefix ‘p’, followed by a string of terms that describes its characteristic features and resistance gene. The individual terms are shown in red print and are to be lined up from left to right to yield the correct designation. For each category of plasmids, the name of one construct is given as an example. The different MCS (MCS, A-E) are detailed in Figure 7. The following gene names were used: RK2 origin of transfer (oriT), E. coli rrnB T1T2 transcriptional terminator (ter), pMB1 origin of replication (oriV), aminoglycoside adenylyltransferase (Spc/StrR, aadA), neomycin phosphotransferase I (KanR, nptI), rifampicin ADP-ribosyltransferase (RifR, parr-2), AAC(3)-I aminoglycoside acetyltransferase (GentR, aacC1), tetracycline efflux permease (TetR, tetA) and repressor (tetR), chloramphenicol acteyltransferase (CamR, cat), AAC(3)-IV aminoglycoside acetyltransferase (Apr/GentR, aacC4), enhanced green fluorescent protein (egfp), enhanced yellow fluorescent protein (eyfp), enhanced cyan fluorescent protein (ecfp), Venus (venus), Cerulean (cerulean), mCherry (mCherry), FLAG-tag (flag), Strep-tag II (strep), tetracysteine tag (tcys). Plasmids are shown to scale, except where indicated by breaks or dotted lines.
Figure 7.
Multiple cloning sites. The sequences of the different MCS (MCS A–E) are shown in boldface. Flanking Shine-Dalgarno motifs (RBS, as present in Pvan-containing constructs) and fusion tags (exemplified by egfp) are indicated. Restriction sites that are unique in all plasmids carrying the respective MCS are highlighted in boldface, whereas those that are unique in a subset of constructs only are shown in normal print.
A second group of plasmids (Figure 6B) essentially combines the features described above with a short chromosomal fragment comprising the minimal Pvan or Pxyl promoter region and the corresponding downstream translation initiation signals. An NdeI restriction site (5′-CATATG-3′) was engineered at the respective start codons to allow expression of cloned genes using the xylX or vanA Shine-Dalgarno sequence (Figure 7, MCS B). Despite their small size, the promoter fragments used still contain all regulatory elements necessary to ensure tight transcriptional control. However, they are too short to undergo efficient homologous recombination, thus favoring integration of the plasmids at a chromosomal locus which matches the sequence inserted into the MCS. If the construct contains the 5′ part of a gene only, its incorporation into the chromosome will generate a truncated version of the native gene and a second, full-length copy under control of the plasmid-borne inducible promoter, thus offering a straightforward method to create conditional alleles. Primers suitable for sequencing inserted genes are listed in Table 1.
Table 1.
Primers for DNA sequencing and analytical PCR reactions
| Primera | Sequence | Application |
|---|---|---|
| Pvan-for | 5′-GACGTCCGTTTGATTACGATCAAGATTGG-3′ | Sequencing of the region downstream of Pvan |
| Pxyl-for | 5′-CCCACATGTTAGCGCTACCAAGTGC-3′ | Sequencing of the region downstream of Pxyl |
| M13-forb | 5′-GCCAGGGTTTTCCCAGTCACGA-3′ | Reverse sequencing primer |
| eGYC-1 | 5′-GTTTACGTCGCCGTCCAGCTCGAC-3′ | Sequencing of the region upstream of egfp, eyfp, ecfp, venus, or cerulean |
| eGYC-2 | 5′-ATGGTCCTGCTGGAGTTCGTGACC-3′ | Sequencing of the region downstream of egfp, eyfp, ecfp, venus, or cerulean |
| RFP-1 | 5′-CTTGAAGCGCATGAACTCCTTGATG-3′ | Sequencing of the region upstream of mrfp1 or mCherry |
| RFP-2 | 5′-CGAGGACTACACCATCGTGGAACAGTAC-3′ | Sequencing of the region downstream of mrfp1 or mCherry |
| RecUni-1c | 5′-ATGCCGTTTGTGATGGCTTCCATGTCG-3′ | Test of recombinants for integration of a plasmid into the vanA or xylX locus |
| RecVan-2 c | 5′-CAGCCTTGGCCACGGTTTCGGTACC-3′ | Test for integration into the vanA locus |
| RecXyl-2 c | 5′-TCTTCCGGCAGGAATTCACTCACGCC-3′ | Test for integration into the xylX locus |
aAll primers are suitable for DNA sequencing under standard conditions. They have been optimized for PCR reactions performed with an annealing temperature of 65°C in the presence of 5% DMSO.
bPrimer M13-for is not applicable for sequencing plasmids bearing the tetRA resistance cassette and plasmids encoding C-terminal protein fusions.
cRecUni-1 anneals to a region upstream of the promoter fragment in all Pvan- and Pxyl-bearing integration plasmids, whereas RecVan-2 and RecXyl-2 anneal to the 5’ region of the chromosomal vanA and xylX genes, respectively. Insertion of a construct into the vanA and xylX locus can be tested by colony PCR using the primer pairs RecUni-1/RecVan-2 and RecUni-1/RecXyl-2, respectively.
A third set of integrations vectors (Figure 6C) is intended to facilitate the one-step generation of strains producing fluorescent and affinity-tagged hybrid proteins encoded on the chromosome under control of their native promoter. Based on a common basic architecture (Figure 6A), these constructs offer a choice of four different resistance determinants, each of which can be used in conjunction with a variety of carboxy-terminal fusion partners. The tag sequences are preceded by an extensiveMCS, whose translation product has been optimized for low secondary structure content and the absence of bulky hydrophobic residues, thus ensuring high flexibility and minimal interference with protein function (Figure 7, MCS D). Moreover, they have all been inserted in the same reading frame, so that genes of interest can be shuffled between different plasmids without difficulty. Among the available fusion partners, the enhanced versions of green, yellow and cyan fluorescent protein (eGFP, eYFP and eCFP) have proved to be reliable tools for cell biological studies in Caulobacter. Recently, however, diverse mutagenic approaches have significantly improved the photophysical and folding parameters of these proteins (52). To make use of these advances, three of the newly developed variants were included in this vector set. Venus resembles eYFP in its excitation and emission spectra, but shows higher folding and maturation rates, leading to a large gain in overall fluorescence (24). Cerulean, by contrast, combines the spectral properties of eCFP with an increased quantum yield, extinction coefficient and fluorescence lifetime and thus outrivals its predecessor in net fluorescence intensity (25). The red fluorescent protein mCherry, finally, can be used complementary to eGFP in dual-color imaging experiments and is characterized by its high maturation rate and photostability (26). Furthermore, it is the only protein besides its precursor mRFP1 which has been shown to fluoresce after Sec-mediated export into the bacterial periplasm (53) and, therefore, represents an indispensable tool for studying the localization of cell envelope proteins. To account for the frequent use of affinity peptides in cell biological research, the selection of available fusion partners was extended by three short peptide tags. The FLAG-tag is specifically recognized by the M2 monoclonal antibody and can be employed for immunological detection and co-immunoprecipitation analyses (28). The same types of applications are supported by the Strep-tag II peptide (29). As a high-affinity ligand of streptavidin and its derivative StrepTactin® (54), it further allows direct immobilization of its fusion partner on surfaces coated with these proteins. Binding of the Strep-tag II to StrepTactin® is readily reversible by low concentrations of desthiobiotin, thereby offering a gentle and highly selective method for the affinity purification of proteins and protein complexes (54,55). The tetracysteine tag, by contrast, has primarily been developed for in vivo protein localization studies. It tightly interacts with the membrane-permeable biarsenical dyes FlaSH (Lumio Green®) and ReAsH, forming stable yellow and red fluorescent complexes that can be detected by light microscopy (27). However, it has also been used successfully to affinity-purify proteins with the help of agarose beads that were covalently modified with a FlaSH derivative (56).
To avoid polar effects, resulting from the integration of a plasmid at the native locus of a gene, or to allow tagging of essential proteins even if their function is compromised by the fusion partner, hybrid genes are frequently put under the control of a regulatable promoter and integrated ectopically at a neutral locus. We, therefore, created modified versions of the aforementioned gene fusion vectors which carry the xylX or vanAB promoter and translation initiation signals inserted upstream of their MCS (Figures 6D and 7, MCS E), thus allowing xylose- or vanillate-inducible expression of the fusions they bear (Figures 8A and C). Moreover, since the promoter fragments chosen for this purpose comprise more than 1.5 kb of the xylX or vanA upstream region, respectively, they are on average significantly longer than the genes of interest fused to the tag sequences. Consequently, they represent the major targets of homologous recombination, favoring integration of the plasmids at the Pxyl or Pvan locus.
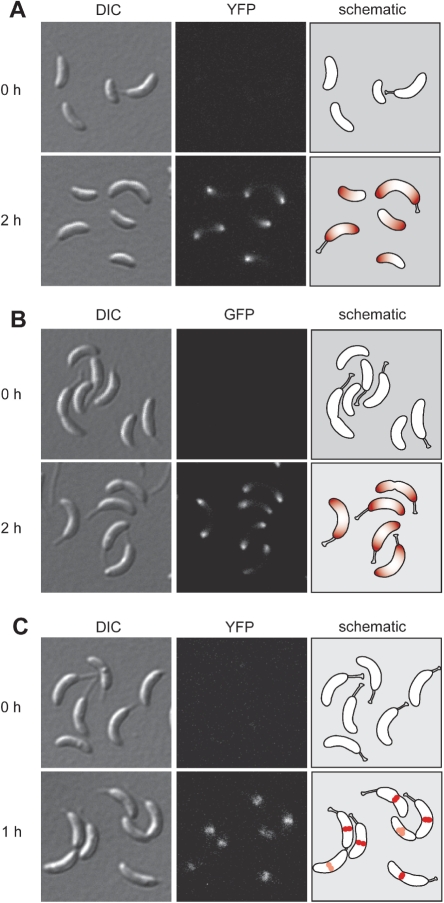
Figure 8.
Application examples. (A) Expression of a mipZ-eyfp fusion, obtained by insertion of the mipZ gene into plasmid pVYFPC-1, under the control of Pvan. Cells of strain MT232 (Pvan-mipZ-eyfp) were examined by DIC and fluorescence microscopy before and 2h after induction with 0.5 mM vanillate. Note: MipZ is a spatial regulator mediating proper positioning of the cell division apparatus in Caulobacter (10). It forms a complex with the DNA partitioning protein ParB at the chromosomal origin of replication. Before initiation of chromosome replication, the MipZ·ParB·origin complex localizes to the flagellated pole of the cell. After entry into S phase, MipZ and ParB re-associate with the newly synthesized origin regions, generating two nucleoprotein complexes which are positioned at the two opposite cell poles. (B) Expression of an egfp-mipZ fusion, created using plasmid pXGFPN-2, under the control of Pxyl. The images show cells of strain MT236 (Pxyl-egfp-mipZ) before and two hours after induction with 0.03% xylose. (C) Expression of an ftsZ-eyfp fusion, constructed with the help of plasmid pVCHYC-3, under the control of Pvan. Cells were withdrawn from a culture of strain MT240 (Pvan-ftsZ-eyfp) before and 1h after induction with 0.5 mM vanillate and analyzed by microscopy. Note: The tubulin homologue FtsZ is a fundamental component of the bacterial cell division machinery. Upon initiation of chromosome replication and bipolar positioning of MipZ, it forms a ring-like structure at the cell center which recruits the other constituents of the cytokinetic ring and plays an essential role in the constriction process (10,60).
The list of integration vectors is completed by a similar series of constructs which are intended for the generation of N-terminally tagged hybrid proteins (Figures 6E and 8B). In this case, the tag sequences were cloned immediately downstream of the xylX or vanA promoter fragments, with the MCS fused to their 3′ ends (Figure 7, MCS F). Since their start codons overlap with an engineered NdeI restriction site, they can easily be removed and replaced with any gene of interest, which makes these plasmids also suitable for ectopic expression of alleles lacking a fusion partner under the control of Pxyl or Pvan.
Replicating plasmids
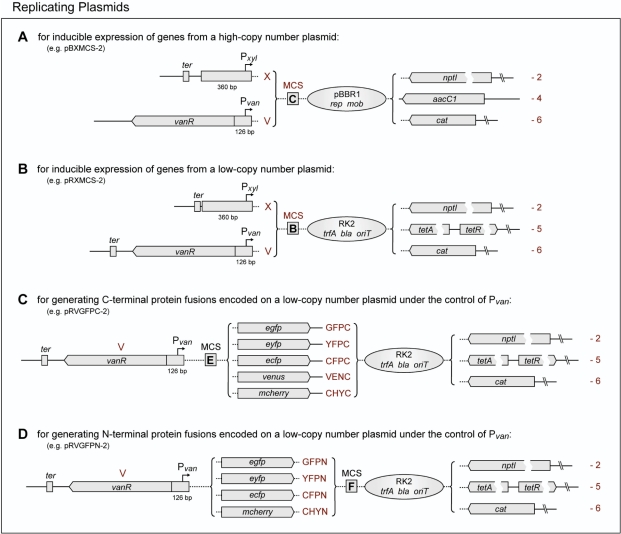
In order to facilitate inducible overproduction of proteins in Caulobacter, we inserted the xylX and vanAB promoters including the respective translation initiation signals into vectors whose replication is driven by the broad host range and mid-copy number replication origin of plasmid pBBR1 (15) (Figure 9A). The corresponding start codons are positioned in the context of an NdeI recognition sequence and are followed by an extensive MCS (Figure 7, MCS C). Together, these constructs offer a selection of three different resistance markers. They further possess all functions necessary for mobilization by the RK2 conjugative transfer machinery (50,57). Since our results showed that the amount of VanR produced from a single, chromosomal allele is not sufficient to tightly regulate multiple copies of its cognate promoter (Figure 5), the vanR gene was included in all Pvan-bearing plasmids. The regulator responsible for xylose-dependent induction of xylX has been identified recently, but it was found to impair growth at elevated levels (B. Christen, C. Stephens and U. Jenal, unpublished). This observation precluded incorporation of its gene into the Pxyl-bearing overexpression vectors. However, to keep leaky expression at a minimum, a transcriptional terminator was placed upstream of the promoter fragment in these constructs, thus reducing unwanted transcription originating in the plasmid backbone.
Figure 9.
Replicating plasmids. Shown are four sets of replicating shuttle vectors designed for the inducible expression of genes from (A) a high-copy number or (B) a low-copy number plasmid and for the construction of (C) C-terminal and (D) N-terminal fluorescent protein fusions, expressed under the control of Pvan from a low-copy number plasmid. The nomenclature of these plasmids follows the same rules as described in Figure 6, but the prefix is ‘pB’ for high-copy number plasmids and ‘pR’ for low-copy number plasmids. The gene names used are as follows: pBBR1 replication protein (rep), pBBR1 mobilization protein (mob), RK2 replication initiatior protein (trfA), β-lactamase (AmpR, bla). All other gene designations are detailed in Figure 6.
A comparable set of vectors was also created based on a minimal version of the low-copy number and broad host range plasmid RK2 (17) (Figure 9B). In addition to their specific resistance determinants, these constructs all carry a β-lactamase (bla) gene, allowing the use of standard ampicillin plates for cloning purposes. They are stably maintained under selective conditions. However, since they lack a partitioning system, the plasmids are easily lost in the absence of antibiotics (data not shown), which makes them a versatile tool for gene function and complementation analyses. All vectors contain the RK2 origin of transfer and can be mobilized into Caulobacter by conjugation using a suitable host strain (50).
Finally, in order to cope with conditions that necessitate the synthesis of fluorescent protein fusions at levels higher than afforded by chromosomally integrated constructs, we created two sets of replicating plasmids (Figures 9C and D) which allow the inducible expression of C- and N-terminally tagged proteins under control of the vanAB promoter and translation initiation signals. Based on the Pvan-bearing RK2 derivatives described above (Figure 9B), they offer a choice of three different resistance markers, an extensive MCS (Figure 7, MCS E and F), and a selection of fluorescent protein tags that covers all common cell biological applications.
DISCUSSION
This work extends the molecular toolset for Caulobacter by a second inducible promoter and a variety of different gene fusion and expression vectors, thereby considerably facilitating the study of this model organism. Moreover, many of the constructs are also suitable for use as replicating or integrating vectors in other bacteria, complementing their genetic systems by adding additional resistance genes and a number of different protein fusion tags.
Our results demonstrate that Caulobacter is able to utilize the aromatic compound vanillate as the sole carbon source. The first step of vanillate degradation involves removal of the O-methyl group by a two-subunit vanillate demethylase complex, formed by the products of open reading frames CC2393 (vanA) and CC2394 (vanB). The reaction product, protocatechuate, is then funneled into the citric acid cycle via the β-ketoadipate pathway. Expression of the vanAB operon is regulated by the GntR-type repressor CC2392 (vanR) and induced by vanillate as well as, to a smaller degree, by its reduced derivatives vanillin and vanillyl alcohol. Interestingly, while the enzymes constituting the β-ketoadipate pathway are highly conserved among α-proteobacteria, the closest homologs of CC2392, CC2393 and CC2394 are found in terrestrial and aquatic representatives of the β- (Ralstonia, Burkholderia) and γ-proteobacteria (Pseudomonas, Acinetobacter). This observation indicates that Caulobacter obtained the ability to degrade vanillate by horizontal gene transfer from distantly related bacteria sharing the same environmental niche. Pvan is tightly repressed in the absence of inducer, while vanillate concentrations in the nanomolar range are sufficient to stimulate its activity. In addition, expression of vanAB—like that of xylose-regulated genes (7)—is not subject to catabolite repression. These characteristics might reflect an adaptation of Caulobacter to the nutrient-poor environments it inhabits, facilitating the simultaneous utilization of multiple carbon sources, even if present in trace amounts only.
Due to its tight regulation, its titratability, and its low sensitivity to the presence of sugars, Pvan is well suited as a general purpose gene expression system for Caulobacter which can be employed in parallel to the established xylose-inducible promoter (7). Its use is facilitated by a newly constructed series of integrating and replicating shuttle vectors, which cover all cell biological and physiological standard applications, such as the ectopic expression of native genes and gene fusions, the overproduction of proteins, and the construction of depletion strains. Generating a similar set of Pxyl-based plasmids, we created a standardized and versatile framework for inducible gene expression in Caulobacter, which considerably accelerates the process of plasmid construction and the swapping of promoters, resistance markers, and protein fusion tags. The performance of the new vectors is demonstrated by several application examples given in this (Figure 8) and previous (10,58,59) work. These studies indicate that, when used for the expression of fluorescent protein gene fusions, Pvan generally yields results that are qualitatively similar to those achieved with Pxyl (Figures 8A and B). However, because of its lower transcriptional activity (data not shown), it might be superior for the control of weakly transcribed genes, avoiding negative side-effects resulting from overexpression. Due to its tight regulation, it is further particularly valuable for protein depletion studies. Our data show that vanillate is consumed within a few hours after addition to uninduced cells (Figure 1E). Thus, in cases where extended induction times or tighly controlled expression levels are required, the vanA-deficient CB15N derivative MT231 is recommended as a host strain to avoid exhaustion of the inducer pool. We further observed that elevated levels of vanillate reduce the growth rate of Caulobacter in a medium-dependent manner. Based on our results, we therefore suggest inducer concentrations lower than 0.5 mM in minimal medium and 1 mM in rich medium, which still allow near-maximal induction of Pvan without adversely affecting the physiology of the cell.
ACKNOWLEDGEMENTS
Annoted sequence files in GenBank format are available for all vectors and may be obtained upon request. The integrity of the constructs has been verified by DNA sequence analysis. Material containing patented components will only be distributed to individuals holding a valid user licence. We are grateful to Esteban Toro for help with the initial characterization of the vanAB promoter. We further thank Svein Valla, Atsushi Miyawaki, David W. Piston, Roger Y. Tsien, David O. Wood, Gerben J. Zylstra, and Patrick H. Viollier for providing plasmids, and the members of the Shapiro lab for testing the performance of the different constructs. This work was supported by an EMBO long-term fellowship and startup funds from the Max Planck Society to M.T. and National Institutes of Health grants GM51426 and GM32506 to L.S. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Skerker JM, Laub MT. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2004;2:325–337. doi: 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- 2.Jensen RB, Wang SC, Shapiro L. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat. Rev. Mol. Cell Biol. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- 3.Viollier PH, Shapiro L. Spatial complexity of mechanisms controlling a bacterial cell cycle. Curr. Opin. Microbiol. 2004;7:572–578. doi: 10.1016/j.mib.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Breier AM, Cozzarelli NR. Linear ordering and dynamic segregation of the bacterial chromosome. Proc. Natl Acad. Sci. USA. 2004;101:9175–9176. doi: 10.1073/pnas.0403722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 6.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 7.Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 10.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 11.Miller JH. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 1972. Experiments in molecular genetics; pp. 352–355. [Google Scholar]
- 12.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, Albright LM, Coen D, et al., editors. USA: John Wiley & Sons, Inc; 2000. Current protocols in molecular biology. [Google Scholar]
- 13.Santos PM, Di Bartolo I, Blatny JM, Zennaro E, Valla S. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 2001;195:91–96. doi: 10.1111/j.1574-6968.2001.tb10503.x. [DOI] [PubMed] [Google Scholar]
- 14.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 15.Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 16.Orosz A, Boros I, Venetianer P. Analysis of the complex transcription termination region of the Escherichia coli rrnB gene. Eur. J. Biochem. 1991;201:653–659. doi: 10.1111/j.1432-1033.1991.tb16326.x. [DOI] [PubMed] [Google Scholar]
- 17.Blatny JM, Brautaset T, Winther-Larsen HC, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiney DG, Yakobson E. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc. Natl Acad. Sci. USA. 1983;80:3595–3598. doi: 10.1073/pnas.80.12.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl Acad. Sci. USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin A, Tucker AM, Hines A, Wood DO. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 2004;70:2816–2822. doi: 10.1128/AEM.70.5.2816-2822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis JJ, Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweizer HD. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 23.Blondelet-Rouault MH, Weiser J, Lebrihi A, Branny P, Pernodet JL. Antibiotic resistance gene cassettes derived from the Ω interposon for use in E. coli and streptomyces. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 24.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 25.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 26.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 27.Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 28.Chubet RG, Brizzard BL. Vectors for expression and secretion of FLAG epitope-tagged proteins in mammalian cells. Biotechniques. 1996;20:136–141. doi: 10.2144/96201pf01. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt TG, Koepke J, Frank R, Skerra A. Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J. Mol. Biol. 1996;255:753–766. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- 30.Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benner R, Lay J, K’nees E, Hodson RE. Carbon conversion efficiency for bacterial growth on lignocellulose: implications for detritus-based food webs. Limnol. Oceanogr. 1988;33:1514–1526. [Google Scholar]
- 32.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl Acad. Sci. USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hottes AK, Meewan M, Yang D, Arana N, Romero P, McAdams HH, Stephens C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 2004;186:1448–1461. doi: 10.1128/JB.186.5.1448-1461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 35.Martinez AT, Speranza M, Ruiz-Duenas FJ, Ferreira P, Camarero S, Guillen F, Martinez MJ, Gutierrez A, del Rio JC. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005;8:195–204. [PubMed] [Google Scholar]
- 36.Harwood CS, Parales RE. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 37.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J. Bacteriol. 1988;170:4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segura A, Bunz PV, D’Argenio DA, Ornston LN. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J. Bacteriol. 1999;181:3494–3504. doi: 10.1128/jb.181.11.3494-3504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merkens H, Beckers G, Wirtz A, Burkovski A. Vanillate metabolism in Corynebacterium glutamicum. Curr. Microbiol. 2005;51:59–65. doi: 10.1007/s00284-005-4531-8. [DOI] [PubMed] [Google Scholar]
- 40.Nishimura M, Ishiyama D, Davies J. Molecular cloning of Streptomyces genes encoding vanillate demethylase. Biosci. Biotechnol. Biochem. 2006;70:2316–2319. doi: 10.1271/bbb.60180. [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee DK, Bourquin AW. Metabolism of aromatic compounds by Caulobacter crescentus. J. Bacteriol. 1987;169:1993–1996. doi: 10.1128/jb.169.5.1993-1996.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakatsu CH, Straus NA, Wyndham RC. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141(Pt 2):485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 43.Barker JL, Levitan H. Mitochondrial uncoupling agents. Effects on membrane permeability of molluscan neurons. J. Membr. Biol. 1975;25:361–380. doi: 10.1007/BF01868584. [DOI] [PubMed] [Google Scholar]
- 44.Malakooti J, Ely B. Principal sigma subunit of the Caulobacter crescentus RNA polymerase. J. Bacteriol. 1995;177:6854–6860. doi: 10.1128/jb.177.23.6854-6860.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malakooti J, Wang SP, Ely B. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthetic and housekeeping functions. J. Bacteriol. 1995;177:4372–4376. doi: 10.1128/jb.177.15.4372-4376.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tropel D, van der Meer JR. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 2004;68:474–500. doi: 10.1128/MMBR.68.3.474-500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Priefert H, Rabenhorst J, Steinbuchel A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 1997;179:2595–2607. doi: 10.1128/jb.179.8.2595-2607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor BF. Aerobic and anaerobic catabolism of vanillic acid and some other methoxy-aromatic compounds by Pseudomonas sp. Strain PN-1. Appl. Environ. Microbiol. 1983;46:1286–1292. doi: 10.1128/aem.46.6.1286-1292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morawski B, Segura A, Ornston LN. Repression of Acinetobacter vanillate demethylase synthesis by VanR, a member of the GntR family of transcriptional regulators. FEMS Microbiol. Lett. 2000;187:65–68. doi: 10.1111/j.1574-6968.2000.tb09138.x. [DOI] [PubMed] [Google Scholar]
- 50.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biol/Technology. 1983;1:784–791. [Google Scholar]
- 51.Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 52.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 53.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 54.Voss S, Skerra A. Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng. 1997;10:975–982. doi: 10.1093/protein/10.8.975. [DOI] [PubMed] [Google Scholar]
- 55.Maier T, Drapal N, Thanbichler M, Bock A. Strep-tag II affinity purification: an approach to study intermediates of metalloenzyme biosynthesis. Anal. Biochem. 1998;259:68–73. doi: 10.1006/abio.1998.2649. [DOI] [PubMed] [Google Scholar]
- 56.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 57.Szpirer CY, Faelen M, Couturier M. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 2001;183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 59.Wang SC, West L, Shapiro L. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J. Bacteriol. 2006;188:1497–1508. doi: 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]