Abstract
A bioinformatic covariation analysis of a collection of 119 novel variants of the antigenomic, self-cleaving hepatitis delta virus (HDV) RNA motif supported the formation of all of the Watson–Crick base pairs (bp) of the catalytic centre except the C19–G81 pair located at the bottom of the P2 stem. In fact, a novel Watson–Crick bp between C19 and G80 is suggested by the data. Both chemical and enzymatic probing demonstrated that initially the C19–G81 pair is formed in the ribozyme (Rz), but upon substrate (S) binding and the formation of the P1.1 pseudoknot C19 switches its base-pairing partner from G81 to G80. As a result of this finding, the secondary structure of this ribozyme has been redrawn. The formation of the C19–G80 bp results in a J4/2 junction composed of four nucleotides, similar to that seen in the genomic counterpart, thereby increasing the similarities between these two catalytic RNAs. Additional mutagenesis, cleavage activity and probing experiments yield an original characterization of the structural features involving the residues of the J4/2 junction.
INTRODUCTION
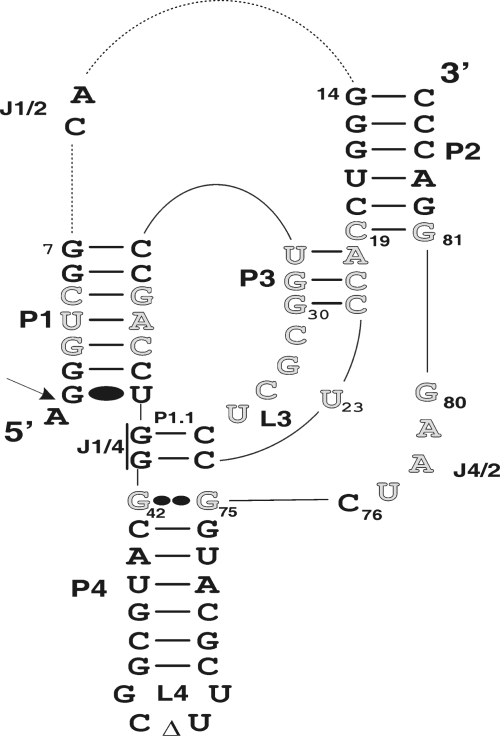
Both the genomic and antigenomic hepatitis delta virus (HDV) RNA strands include a self-cleaving RNA motif that produces 2′–3′-cyclic phosphate and 5′-hydroxyl termini (1,2). These self-cleaving RNAs have been separated into two molecules in order to develop trans-acting systems in which one molecule, the ribozyme (Rz), possesses the catalytic properties required to successively cleave several molecules of substrate (S). The HDV ribozyme folds into a double-pseudoknot secondary structure composed of one stem (P1), two pseudoknots (P1.1 and P2), two stem-loops (P3–L3 and P4–L4) and three single-stranded junctions (J1/2, J1/4 and J4/2) (Figure 1). Crystallographic studies of the HDV genomic ribozyme have provided high-resolution details of the compact tertiary structure of this catalytic RNA (3,4).
Figure 1.
Secondary structure of the antigenomic self-cleaving HDV RNA. The numbering system used is from Shih and Been (34). The empty letters indicate the 25 position randomized in the previous study (12). The Wobble bp is represented by a single large dot (G•U), while the homopurine bp at the top of the P4 stem is represented by two large dots (G••G). The triangle in the L4 loop indicates the P4 deletion (as compared to the natural variants). The arrow indicates the cleavage site. The trans-acting ribozyme was created by deleting the CA forming the J1/2 junction.
The J1/4 junction and the L3 loop are both initially single-stranded, but subsequently are involved in the formation of the P1.1 pseudoknot (5–7). The formation of this pseudoknot, which requires the presence of both the substrate and magnesium, is critical for the folding of the ribozyme into an active ternary structure (8). It permits the stacking of the P1–P1.1–P4 stem into one helix that becomes coaxial to the second stacked P2–P3 helix (3,4). Moreover, it significantly contributes to the bringing together of the scissile phosphate and the catalytic cytosine that is located within the J4/2 junction (i.e. C76) (9,10). The nucleotides of this single-stranded region are also involved in two distinct structural motifs: a ribose zipper formed between the two adenosines of the J4/2 junction (A78 and A79) and the two cytosines of the P3 stem (C21 and C22), and a trefoil turn structure formed by the catalytic cytosine (C76) and the adjacent guanosine (G77) (3). The formation of this trefoil turn positions C76 deep within the catalytic core near the scissile bond (3,11).
In a recent study, the primary results of an unbiased in vitro selection of antigenomic HDV ribozymes randomized at 25 positions within the catalytic centre (Figure 1) showed that nucleotide variation was found at all of the randomized positions, even those where the specific base in question was believed to be essential for catalytic activity (12). Analysis of the random nucleotide covariation, obtained using a database composed of 45 different sequences, supported the formation of most of ribozyme base pairs that form both the P1 and the P3 stems. Moreover, these results support the presence of the homopurine bp located at the top of the P4 stem. Altogether, these observations led us to conclude that the selection performed was unbiased and yielded many new variants. However, neither new base pairs, nor any tertiary interactions, were discovered. This might be due to the fact that the number of different variants was relatively small (only 45).
As a result, we decided to emphasize both the sequencing and the analysis of the sequence variations. This work permitted the identification of a new structural rearrangement involving the J4/2 junction and the bottom base pair of the of the P2 stem. This conformational transition increases the similarities of the geometry adopted by the J4/2 junctions of both the genomic and antigenomic HDV ribozymes.
MATERIALS AND METHODS
Sequence determination and covariation analysis
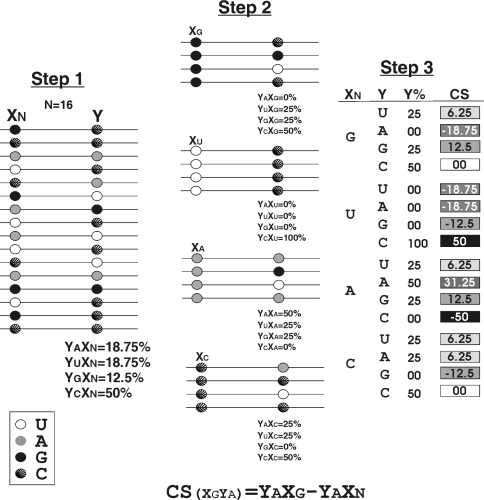
A total of 170 new, self-cleaving HDV motif clones, isolated from the previously reported in vitro selection (12), were sequenced using the T7 sequencing kit (GE Healthcare). All of the sequences from both the present and the previous studies were compiled in a database and subsequently analysed for base conservation both at each position and in terms of nucleotide covariation using homemade software written in Perl language (Figure 2). Briefly, the software analyses two positions at a time, namely X and Y. First, for a given position X, the percentages of the different nucleotides found at position Y in all sequences are determined (%YAXN,%YC XN,%YGXN and%YUXN). Second, for a specific nucleotide in position X (for example a cytosine), the percentage of each nucleotide (G, C, A and U) at position Y is determined (%YAXC,%YCXC,%YGXC,%YUXC). Third, these two percentages are compared in order to determine a relative covariation score (CS) for the specific nucleotides located in these two positions [CS = (%YN′ − %YN): CS(YAXC) = %YAXC − % YAXC]. If the CS is 0, or near 0, this suggests that the nucleotide in position X has no effect on the distribution of nucleotides in position Y. Conversely, a relatively large CS (e.g. ±0.5) suggests a significant covariation of these two bases (see Supplementary data for examples). The operation was repeated for all possible combinations of the 25 randomized positions (i.e. 9600 combinations), and the CSs were displayed in an Excel file.
Figure 2.
Schematic representation of the covariation software developed for the analysis of the sequence database. The three steps of the procedure are illustrated using the example described in the text. CS indicates the covariation score. The inset shows the legend of the filling sphere for each nucleotide.
RNA synthesis and 32P-5′-end labelling
Chemical synthesis
The substrates, the SdA4 analogue, the 3′ product and the RzB strands (with and without the abasic residue at position 77) were chemically synthesized using 2′-ACE chemistry by Dharmacon Research Inc. (Lafayette, Colorado). The resulting polymers were deprotected according to the manufacturer's recommendations and then purified by denaturing 10–20% polyacrylamide gels electrophoresis (PAGE, 19:1 ratio of acrylamide to bisacrylamide) using 45 mM Tris–borate, pH 7.5, 8 M urea and 1 mM EDTA solution as buffer. The products were visualized by UV shadowing, the bands corresponding to the correct sizes were cut out and the RNA eluted by incubating overnight at room temperature in a solution containing 0.1% SDS and 0.5 M ammonium acetate. The resulting polymers were ethanol precipitated and their quantities determined by absorbance at 260 nm after dissolving in water.
In vitro transcription
All of the ribozymes and the RzA strands were synthesized by run-off transcription as described previously (19). Briefly, RNA molecules were produced by annealing two strands of complementary DNA oligonucleotides that included the T7 RNA promoter followed by the sequence of the desired ribozyme. The transcriptions were then performed in the presence of 10 µg purified T7 RNA polymerase, 24 U RNA Guard (Amersham Biosciences), 0.01 U pyrophosphatase (Roche Diagnostics) and 500 pmol of template in a buffer containing 80 mM HEPES–KOH pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT and 5 mM of each NTP in a final volume of 100 µl at 37°C for 2 h. Upon completion, the reaction mixtures were treated with DNase RQ1 (Amersham Biosciences) at 37°C for 20 min. The RNA was then purified by phenol extraction and ethanol precipitation, and was fractionated by denaturing 8% PAGE. The bands corresponding to the correct sizes for the RNA species were cut out, the transcripts eluted and then ethanol precipitated.
32P-5′-end labelling
Purified substrates or ribozymes (40 pmol) were dephosphorylated in a final volume of 10 µl using 10 U of Antartic phosphatase according to the manufacturer's conditions (New England Biolabs). The reactions were stopped by heating for 5 min at 65°C. Dephosphorylated RNAs (10 pmol) were 5′-end labelled in a final volume of 10 µl containing 3.2 pmol [γ-32P]-ATP (6000 Ci/mmol, Amersham Biosciences), 10 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 50 mM KCl and 3 U of T4 polynucleotide kinase (Amersham Biosciences) at 37°C for 90 min. The reactions were stopped by the addition of formamide dye buffer and the mixtures fractionated through denaturing 10–20% PAGE gels. After autoradiography, the bands containing the appropriate 5′-end-labelled RNAs were excised and the RNA recovered as described above.
Chemical and enzymatic probing
Ribozymes were subjected to in-line probing according to a procedure reported previously (16). Briefly, 1 nM of 32P-5′-end-labelled trans-acting ribozyme was incubated for 40 h at 25°C in a buffer containing 20 mM MgCl2, 50 mM Tris–HCl pH 8.3 and 100 mM KCl either in the absence or the presence of SdA4 analogue (20 µM). In the enzymatic digestions, trace amounts of the 5′-end-labelled ribozymes (<1 nM) were dissolved in 10 µl of buffer containing 20 mM Tris–HCl, pH 7.5, 10 mM MgCl2 and 100 mM NH4Cl. The mixtures were incubated for 0.5–1.0 min at 25°C in the presence of either 0.2 U of RNase T1 (Amersham Biosciences) or of 0.001 U of RNase V1 (Pierce Molecular Biology), and were then quenched by adding 10 µl of formamide. For alkaline hydrolysis, the ribozymes (<1 nM) were dissolved in 4 µl of water and 2 µl of 2 N NaOH were added. The reaction was incubated at room temperature for 15 s, and was then quenched by the addition of 6 µl of 500 mM Tris–HCl, pH 7.5 and 5 µl of loading buffer. The resulting mixtures were ethanol precipitated, resuspended in loading buffer, separated on denaturating 10% PAGE gels and visualized by exposure of the gels to phosphor imaging screens.
Cleavage reactions and kinetic assays
The cleavage reactions were performed as described previously (19). Briefly, cleavage reactions were carried out in 20 µl reaction mixtures containing 50 mM Tris–HCl, pH 7.5 and 10 mM MgCl2 at 37°C under single-turnover conditions ([Rz] ≫ [S]). Prior to the reaction, trace amounts of 5′-end-labelled substrate (<1 nM) and non-radioactive ribozymes (100 nM) were mixed together, heated at 70°C for 1 min, snap-cooled on ice for 3 min and then incubated at 37°C for 5 min. Similarly in the case of the bimolecular constructs (RzA–RzB or−RzB–Ab77) 100 nM of each RNA species were used in each reaction. Following this pre-incubation step, the cleavage reactions were initiated by the addition of MgCl2. Aliquots (2 µl) were removed either at various times up to 1 h, or until the end point of the cleavage was reached, and were quenched by the addition of ice-cold formamide dye buffer (8 µl). The mixtures were fractionated on denaturing 20% PAGE gels and exposed to phosphor Imager screens (Molecular Dynamics). The extent of cleavage was determined from measurements of the radioactivity present both in the substrate and in the 5′ product bands at each time point using the ImageQuant software. When required, the rate of cleavage (kobs) was obtained by fitting the data to the equation At = A∞ (1 − e−kt), where At is the percentage of cleavage at time t, A∞ is the maximum percent cleavage (or the end point of cleavage) and k is the rate constant (kobs). Each rate constant was calculated from at least two independent measurements.
RESULTS
Analysis of all selected HDV variants
The different experiments reported in the previous study were performed using a database containing a total of 330 clones, but involving only 45 different sequence variants (12). In order to find novel variants, we decided to sequence 170 additional clones that were obtained via a PCR amplification strategy that avoided domination by the most active self-cleaving ribozymes. A new database containing 119 different variants was constructed (Supplementary data Table 1). Subsequently, each variant was individually synthesized by run-off transcription, and its self-cleavage activity determined (Supplementary data Table 1). All of the selected variants were active, confirming the accuracy of the selection protocol. Among the 119 variants, 21 sequences exhibited self-cleavage activities at a level smaller than 10% after 60 min of incubation. The 98 most active variants were retained for further analysis in order to study the interactions important for efficient self-cleavage.
In order to perform a covariation analysis of the randomized nucleotides, we developed a software program that analyses the covariation between the randomized positions. This program can detect not only Watson–Crick bp, but also any tertiary interactions (Figure 2). Briefly, a CS is calculated for each pair of nucleotides. A CS that tends near zero indicates there is no covariation between these two nucleotides. A significant positive CS indicates that the presence of a cytosine in position X favours the presence of an adenosine in position Y, and vice versa. Conversely, a significant negative difference is an indication that covariation is disfavoured.
Table 1 presents a compilation of the CSs of random nucleotides known to interact together based on the secondary structure of the antigenomic HDV ribozyme. The covariation results are in good agreement with most of the interactions proposed in the secondary structure of the HDV ribozyme (1). For example, the base pairs of the P3 stem are well supported by the covariation results: each base pair is found to be formed in at least 98% of the selected ribozymes, with a significant preference for Watson–Crick base pairing over GU/UG Wobble bp (≥95% compared to ≤3%). This is in good agreement with the results of earlier directed mutagenesis studies which indicated that the size of the P3 stem is critical to self-cleaving RNA strands, and that the identities of its nucleotides are also important (13). Similarly, covariation analysis supported the formation of the base pairs located in the middle of the P1 stem (see Table 1 and Supplementary data Table 2). However, statistical analysis showed that these base pairs (G3C37, U4A36 and C5G35) tolerate the presence of more Wobble bp and mismatches than do those of the P3 stem (Table 1). This indicates that they are important, but not essential, at least for self-cleaving RNA strands. The fact that mismatches are observed in the middle of the P1 stem is in agreement with the results from enzymatic probing experiments that illustrated the susceptibility of this domain to a single-strand specific ribonuclease (RNase T2) (14). The presence of the homopurine bp found at the top of the P4 stem (positions 42 and 75) is well supported by the covariation analysis. AA and GG homopurines were retrieved in 96% of the sequences, while AG and GA were found in only 3%. Surprisingly, the selected sequences predominantly include an AA homopurine bp (79%). This contrasts with the exclusive presence of a GG homopurine bp in the sequence variants found in nature (15). This difference might be due to different selective pressures existing in vivo as compared to in vitro. Thus, the covariation analysis confirmed the secondary structure of the self-cleaving antigenomic sequences, including both the homopurine and all Watson–Crick bp, with one exception (see below).
Table 1.
Compilation of data from the covariation analysis
| Positions | Combinations of nucleotides (%) | Base-pairing (%) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mismatchs | Waston-Crick bp | G/U:U/G Wobble | Waston-Crick bp | G/U:U/G Wobble | |||||||||||||||
| Waston-Crick bp | G/U:U/G Wobble | ||||||||||||||||||
| CC | GG | UU | AA | AC | AG | GA | UC | CU | CA | AU | UA | GC | CG | UG | GU | ||||
| (3–37) | 2 | 11 | 2 | 0 | 1 | 4 | 0 | 1 | 5 | 2 | 4 | 0 | 17 | 18 | 3 | 30 | 39 | 33 | |
| P1 stem | (4–36) | 2 | 15 | 2 | 0 | 0 | 2 | 11 | 0 | 3 | 3 | 1 | 0 | 9 | 29 | 3 | 20 | 39 | 23 |
| (5–35) | 2 | 2 | 1 | 0 | 3 | 1 | 0 | 3 | 0 | 0 | 3 | 2 | 29 | 35 | 1 | 18 | 69 | 19 | |
| (20–32) | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 80 | 10 | 5 | 0 | 0 | 3 | 95 | 3 | |
| P3 stem | (21–31) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 11 | 5 | 82 | 0 | 0 | 99 | 0 |
| (22–30) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 18 | 21 | 56 | 1 | 0 | 98 | 1 | |
| Homopurine | (42–75) | 0 | 17 | 1 | 79 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bottom of the P2 stem | (19–80) | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 42 | 13 | 21 | 17 | 1 | 1 | 93 | 2 |
| (19–81) | 0 | 18 | 0 | 15 | 3 | 24 | 3 | 13 | 3 | 2 | 0 | 0 | 1 | 14 | 3 | 1 | 15 | 4 | |
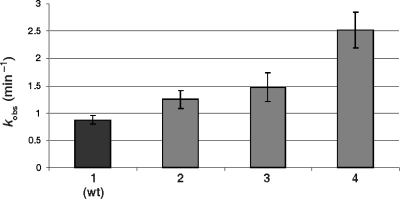
The base pairing of the nucleotides at the bottom of the P2 stem (C19–G81) was believed to form a Watson–Crick bp (Figure 1). The covariation analysis does not support the presence of this base pair (see Table 1 and Supplementary data Table 2). According to the set of selected sequences, only 19% of the selected variants possess either a Watson–Crick or Wobble bp at this position (Table 1). CSs attributed to the different nucleotides that can form Watson–Crick bp are either very close to zero, or negative (e.g. A19 − U81 = 0; U19 − A81 = − 0.2; G19 − C81 = − 0.1; C19 − G81 = 0.08). In order to add biochemical support to this observation, trans-acting mutant ribozymes, including the three possible mutations at position 81, were synthesized and their cleavage activities assessed under single-turnover conditions. The rate constants (kobs) of the mutants were determined, and are presented in histogram form (Figure 3). The least active mutant was the wild-type ribozyme harbouring a C19–G81 bp. The three mutants that did not permit the formation of a base pair were all more active. Independent experiments with several other mutants at the bottom of the P2 stem (positions 19 and 81) also led to the same conclusion. Even several ribozymes without a base pair at this position were found to be more active than the wild-type ribozyme (Ouellet,J. and Perreault,J.P, unpublished data).
Figure 3.
Histogram of the determined rate constant (kobs) for the wild-type ribozyme (1), the RzG81A (2), the RzG81C (3) and the RzG41U and (4) mutants.
The analysis of the nucleotide occupying positions 19 and 80 gave high CSs suggesting that these nucleotides were able to form a Watson–Crick bp (e.g. A19 − U80 = 0.43; U19 − A80 = 0.57; G19 − C80 = 0.69; C19 − G80 = 0.63) (see Supplementary data Table 2). In fact, 93% of the selected sequences possess nucleotides at these positions that have the ability to form a Watson–Crick bp, and 2% a Wobble bp, between these two residues (Table 1). In other words, 95% of the sequence variants seemed to include a base pair at these positions. Conversely, only 15% of the selected ribozymes were able to form a base pair between positions 19 and 81. All possible base-pairing combinations were found, but at different levels (A19–U80, U19–A80, G19–C80 and C19–G80 were found at 42%, 13%, 21% and 17%, respectively). To our knowledge, this is the first demonstration of a base pairing between these two specific nucleotides.
Probing of the bottom of the P2 stem
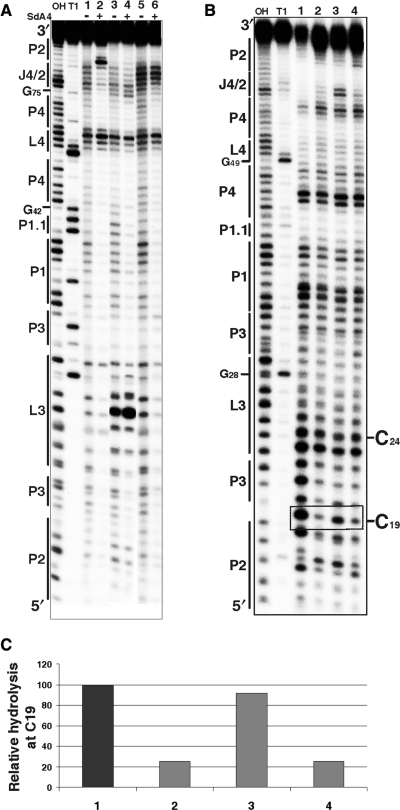
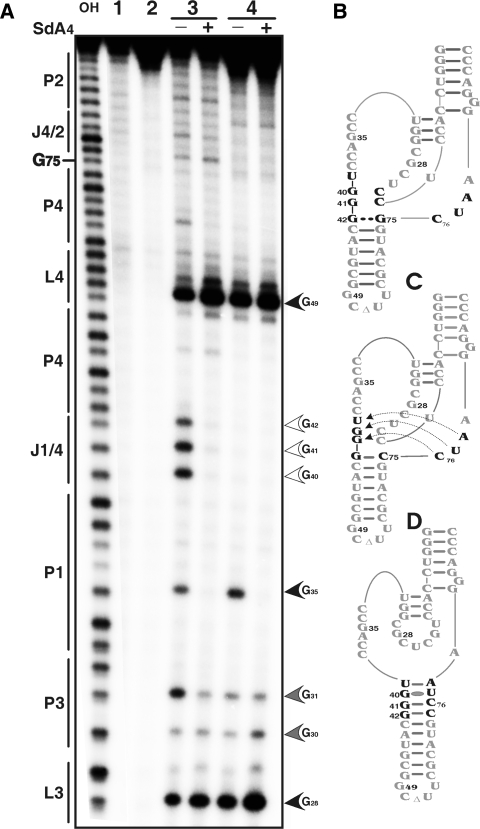
In-line probing is a method that relies on the fact that there is a natural rate of spontaneous cleavage within RNAs (16). Cleavage occurs when a phosphodiester linkage is subjected to an internal nucleophilic attack by the 2′ oxygen adjacent to and in-line with it. Structured regions of RNA, such as those in the base-paired stems, are less susceptible to spontaneous cleavage than are non-structured regions (17). In-line probing experiments were performed with the trans-acting version of the HDV ribozyme for which the kinetic behaviour has been extensively characterized under both single- and multiple-turnover conditions (18). The use of a trans-acting version permits the monitoring of the binding of the substrate to the ribozyme, a step which has been shown to be essential for many conformational transitions to take place (5,19,20). Probing experiments were performed using a trace amount (<1 nM) of 32P 5′-end-labelled ribozyme in either the presence, or the absence, of an excess of uncleavable substrate (SdA4; [S] ≫ [Rz]). The use of a substrate analogue that includes a deoxyriboadenine adjacent to the cleavage site prevents the cleavage from occurring. In the absence of SdA4, RNA degradation was observed in all single-stranded regions, including the nucleotides of both the P1 region (positions 33–39) and the J1/4 junction (G40 and G41) that are not base paired under these conditions (Figure 4A, lane 1; see also Supplementary Figure S1A). The nucleotides of the J4/2 junction, including G80, appear to be single-stranded. Because the residue G81 does not appear to be hydrolysed, this indicates that it is base paired with C19. Upon the addition of SdA4, several modifications in the banding pattern were observed (Figure 4A, lane 2; see also Supplementary Figure 1A). In general, the intensities of the majority of the bands were weaker, indicating that the presence of the substrate led to a more compact structure. Specifically, the nucleotides of the P1, P1.1 and P4 stems appeared to be less susceptible to hydrolysis, supporting the stacking of these stems into one helix (3). Moreover, the nucleotides of both the L3 loop and J4/2 junction were less hydrolysed, indicating the rearrangement of these single-stranded domains into a more compact catalytic core. However, the most obvious difference in the presence of the substrate was observed at the bottom of the P2 stem: the residue G81 became highly susceptible to the in-line attack, indicating that if it is indeed base paired in the absence of the substrate, it switches into a single-stranded conformation in its presence. Similar observations were made when the in-line probing was performed in the presence of the 3′ cleavage product that bound to the P1 region (data not shown). Moreover, terbium-mediated footprinting results also support the occurrence of a rearrangement of the bottom of the P2 stem (21).
Figure 4.
Chemical and enzymatic probing of the bottom of the P2 stem. (A) Autoradiogram of a 10% PAGE gel of in-line probing performed on 5′-end-labelled wild-type and mutated trans-acting ribozymes. Both alkaline and RNase T1 hydrolyses of the wild-type ribozyme were performed in order to determine the location of each position (lanes OH and T1, respectively). In-line probing of the wild-type ribozyme (1, 2), the RzC24U,C25U,G40U,G41U (3, 4) and the RzA78U,A79U (5, 6) mutants are shown. The experiments were performed either in the absence (−) or the presence (+) of the SdA4 analogue. The secondary structure motifs are identified on the left. (B) Autoradiogram of a 10% PAGE gel of RNase V1 probing performed on 5′-end-labelled wild-type and mutated cis-acting ribozymes. Both alkaline and RNase T1 hydrolyses of the wild-type sequence were performed in order to determine the location of each position (lanes OH and T1, respectively). Lanes 1 to 4 correspond to the wild-type sequence (C19G81G80) and the mutants RzC19,G81A,G80A, RzC19,G81AG80 and RzC19G81G80A, respectively. The positions of the C19 and C24 (used to establish the relative level of hydrolysis) are indicated on the right. (C) Histogram of the relative levels of RNase V1 hydrolysis of C19 for each ribozyme.
It is well established that substrate binding triggers many folding rearrangements, the most important of which is the formation of the P1.1 stem between the L3 loop and the J1/4 junction (3,19). In order to verify if the formation of the C19–G80 bp occurred either immediately after the annealing step, or later in the folding pathway, in-line probing of a mutant ribozyme known to prevent the formation of the P1.1 pseudoknot was performed. In this mutant the two G–C bp forming the P1.1. pseudoknot (i.e. G40–C25 and G41–C24) are replaced by four uridines (i.e. U24, U25, U40 and U41). This mutant binds the substrate, but is completely deprived of cleavage activity (7). The banding pattern obtained upon in-line probing was virtually identical, regardless of the absence or the presence of the SdA4 analogue, with the exception of within the substrate binding region (Figure 4A, lanes 3 and 4, respectively; see also Supplementary Figure 1B). With this mutant, the presence of the SdA4 did not trigger a significant folding rearrangement, and G81 remained insensitive to the in-line attack. This result indicates that G81 is most likely base paired to C19 prior the formation of the P1.1 pseudoknot, and that only subsequently does it become single-stranded.
Next, in order to gain more confidence for the conformational rearrangement of the bottom of the P2 stem, enzymatic probing was performed using RNase V1, an enzyme that specifically cleaves double-stranded nucleotides. This experiment was performed using a 5′-end-labelled 3′-product derived from the cis-acting self-cleaving strand of either the wild type or the mutated sequences (Figure 4B and C). The banding pattern produced by the wild-type ribozyme is in agreement with the proposed secondary structure (Figure 4B, lane 1). C19 was hydrolysed, indicating that it is indeed double-stranded. The three mutants tested exhibited essentially identical banding patterns, with only minor differences being observed. For example, the band pattern of the mutant RzC19,G81AG80 included additional bands corresponding to positions 76 and 77. These two bands might be resulting from an alternative conformation adopted by the J4/2 region or the J4/2 stacked slightly more on the P4 stem, enough for RNase V1 to be active. The most important differences were observed for C19 (Figure 4B and C, lanes 2–4). For the C19,G81A,G80A mutant, which is unable to form any base pair involving C19, RNase V1 poorly hydrolysed this residue. Conversely, it efficiently hydrolysed the C19 of the C19,G81A,G80 mutant, suggesting the formation of the C19–G80 bp (Figure 4B and C, lane 3). Finally, the RNase V1 did not hydrolyse the C19 of the C19,G81,G80A mutant (Figure 4B, lane 4). The fact that C19 remains primarily single-stranded in this mutant, even though base pairing with G81 is possible, is an indication that, upon substrate binding, and especially after the formation of the P1.1 stem, G81 is extruded out of the catalytic site even if C19 is not allowed to base pair with another nucleotide. Together, these results confirm that C19 base-paires with G80, and not with G81.
Redrawing the secondary structure of the antigenomic ribozyme
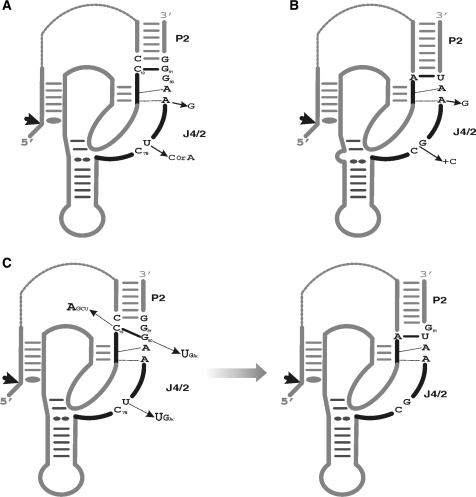
Prior to this study the J4/2 junctions of both the genomic and antigenomic HDV ribozymes were considered to be different (22). The J4/2 junction of the genomic self-cleaving sequences is composed of four nucleotides, C75G76A/G76A78 (Figure 5). This contrasts with that of the antigenomic self-cleaving sequences which is composed of five nucleotides, specifically C76U/C/A77A/G78A79G80 (Figure 5). However, when the new C19–G80 bp is taken into account, the secondary structure of the antigenomic self-cleaving sequence can be redrawn so as to include a J4/2 junction composed of only four nucleotides (Figure 5C). The similarities are not limited to their length, but also include the nucleotide composition. Among the selected sequences, the base pairing between positions 19 and 80 favours an A19–U80 bp (42% AU, 13% UA, 21% GC, 17% CG, 2% GU/UG Wobble bp and 7% mismatches, Table 1), as is observed in all natural genomic variants. Moreover, the identity of the nucleotides located in positions 76–79 in both versions is similar, specifically C76N77A78A79 (Figure 5B and C). Although the selection experiment was designed with the antigenomic version of the HDV ribozyme, the selected sequences corresponded to a consensus including both genomic and antigenomic ribozymes.
Figure 5.
Proposed secondary structures for the HDV self-cleaving RNA motifs. (A) and (B) are the structures proposed for the antigenomic and genomic motifs prior to the present study. The natural sequence variants reported to date for the J4/2 junction are indicated. (C) The novel proposed secondary structures for the antigenomic RNA motif including the C19–G80 bp. The differences between both structures are the representations of the C19–G80 bp. The sequence variations of the J4/2 junction retrieved via the in vitro selection are illustrated. The arrows indicate the cleavage sites.
The switch from C19G81 to C19G80 occurs late in the folding pathway
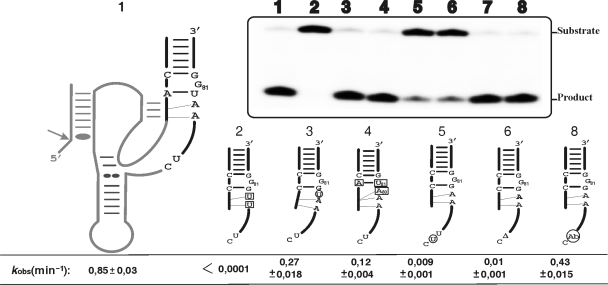
The results described above demonstrate that initially C19 is base paired to G81 and either simultaneously with, or after the formation of, the P1.1 pseudoknot this base pair is disrupted in order to permit formation of the C19–G80 bp (Figure 4A). The formation of the P1.1 pseudoknot has been demonstrated to be the limiting step of the HDV folding pathway, in addition to being critical for the folding of the complete J4/2 junction (3,8). The folding of the J4/2 junction involves the formation of a ribose zipper, a trefoil turn and the positioning of the catalytic cytosine within the catalytic core (C76), three features that received physical support from X-ray diffraction, nuclear magnetic resonance (NMR) and fluorescence spectroscopy studies (3,4,23–25). In order to acquire additional biochemical understanding of the J4/2 junction's folding, we analysed our sequence database in terms of these structural features and then synthesized mutated trans-acting ribozymes in order to evaluate their cleavage activities (Figure 6).
Figure 6.
Cleavage activity assays of various mutants of the J4/2 junction. The inset shows a typical autoradiogram of a PAGE gel for a 30 min reaction for each ribozyme tested. The secondary structure of the wild-type ribozyme is illustrated entirely on the left, while only the P2 stem, J4/2 junction and the most relevant nucleotides are illustrated for the mutants. The mutations and insertion are denoted by squared and circled nucleotides, respectively, while deletions are indicated by a triangle (▵). The lane numbers at the top of the autoradiogram correspond to the number of each ribozyme: 1, wild-type; 2, RzA78U,A79U; 3, Rz–U+79–80; 4, Rz–C19A,G81U,G80A; 5, Rz–U+76–77; 6, Rz–▵77; 7 is the original bimolecular construct (RzA–RzB); and, 8, the bimolecular construct including an abasic residue (Ab) in position 77 (RzB–Ab77). The positions of the substrate and product are indicated adjacent to the gel. The rate constants (kobs) for each ribozyme are indicated below the gel.
The ribose zipper is formed by several tertiary interactions between the residues of the G–C bp of the P3 stem and the adenosines, A78 and A79, of the J4/2 junction (3,26). According to the selected self-cleaving sequences, A79 was perfectly conserved, while A78 was highly conserved and could be substituted for by G78 in a small number of variants (see Supplementary data Table 2). This mutation has been observed in several natural variants (Figure 5A and B). An important point to bear in mind is that both adenosine and guanosine moieties possess a nitrogen group in position 3 which is involved in the interactions that form the ribose zipper (3). The substitution of both adenosines by two uridines completely abolishes the cleavage activity of the trans-acting ribozyme (Figure 6, mutant Rz–A78U,A79U), confirming the importance of the ribose zipper. In order to learn more, the in-line probing of this mutant was performed (Figure 4A, lanes 7 and 8). Upon the addition of the substrate, the P1.1 pseudoknot is formed as shown by the absence of hydrolysis of the corresponding residues (i.e. C24, C25, G40 and G41). Interestingly, G81 remained intact while G80 was hydrolysed, regardless of the presence or not of the substrate. This indicates that the formation of the C19–G80 bp requires the formation of the ribose zipper. In order to investigate whether or not the distance between the C19–G80 bp and the ribose zipper is important, a mutant with a uridine inserted between these two structural features was synthesized. The resulting ribozyme exhibited a cleavage activity only three-fold less than that of the wild type, suggesting that the distance is not significant (Figure 6, 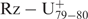 ).
).
We noted that only 10% of the selected sequences included an A80. The presence of an adenosine in position 80 decreases the cleavage activity of the ribozyme in question (Supplementary data Table 2), an effect that was more dramatic when base pairing was possible between positions 19 and 81. For example, the mutant ribozyme C19A,G81U,G80A exhibited a cleavage activity that was seven-fold less than that of the wild-type ribozyme (Figure 6, wt/C19A,G81U,G80A, kobs of 0.85 and 0.12 min−1, respectively). One possible explanation for this effect is that the presence of a single-stranded A80 leads to the formation of a ribose zipper that includes an A80A79 pair instead of the A79A78 one. If this is indeed the case, then the catalytic cytosine would be displaced and the J4/2 junction would contain an additional nucleotide in the trefoil turn domain. In order to verify this possibility, a mutant with an additional uridine inserted between the ribose zipper and C76 was synthesized (Figure 6, 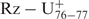 ). This mutant exhibited a cleavage activity that was dramatically reduced, in the order of 100-fold as compared to that of the wild-type ribozyme (0.009 min−1 compared to 0.85 min−1). This clearly shows that the distance between the ribose zipper and the catalytic C76 is very important. In other words, the trefoil turn cannot be altered so as to accommodate an additional nucleotide. The deletion of the residue (U77) caused an even more dramatic effect (Figure 6, Rz–▵U77, kobs of 0.01 min−1), showing that this residue is crucial for the formation of the trefoil turn. The database of selected self-cleaving sequences showed that the identity of the nucleotide at position 77 is not important as all four bases were found at this position (see Supplementary data Table 2). To further support this idea U77 was replaced by an abasic nucleotide. To do so we adopted a bimolecular ribozyme designed by opening the wild-type ribozyme within the L4 loop, thus generating two pieces: RzA that includes the nucleotides 14–48; and, RzB, which corresponds to nucleotides 67–88 (Figure 1), (27). The ribozymes formed by an RzB strand with either a U77 (Figure 6, RzA–RzB) or an abasic residue (RzB–Ab77) exhibited essentially identical cleavage activities (Figure 6; kobs of 0.63 min−1 ±0.02 and 0.43 min−1 ±0.015 for the RzA–RzB and RzB–Ab77, respectively), confirming that the presence of the base moieties in position 77 is essential, but its identity is not. Only the presence of the phosphodiester backbone at this position is important for the formation of the trefoil turn motif. Altogether, these results show the importance of the base pair switching, and provide additional information on the structural environment required for the formation of the ribose zipper, the trefoil turn and the positioning of the catalytic cytosine within the core centre.
). This mutant exhibited a cleavage activity that was dramatically reduced, in the order of 100-fold as compared to that of the wild-type ribozyme (0.009 min−1 compared to 0.85 min−1). This clearly shows that the distance between the ribose zipper and the catalytic C76 is very important. In other words, the trefoil turn cannot be altered so as to accommodate an additional nucleotide. The deletion of the residue (U77) caused an even more dramatic effect (Figure 6, Rz–▵U77, kobs of 0.01 min−1), showing that this residue is crucial for the formation of the trefoil turn. The database of selected self-cleaving sequences showed that the identity of the nucleotide at position 77 is not important as all four bases were found at this position (see Supplementary data Table 2). To further support this idea U77 was replaced by an abasic nucleotide. To do so we adopted a bimolecular ribozyme designed by opening the wild-type ribozyme within the L4 loop, thus generating two pieces: RzA that includes the nucleotides 14–48; and, RzB, which corresponds to nucleotides 67–88 (Figure 1), (27). The ribozymes formed by an RzB strand with either a U77 (Figure 6, RzA–RzB) or an abasic residue (RzB–Ab77) exhibited essentially identical cleavage activities (Figure 6; kobs of 0.63 min−1 ±0.02 and 0.43 min−1 ±0.015 for the RzA–RzB and RzB–Ab77, respectively), confirming that the presence of the base moieties in position 77 is essential, but its identity is not. Only the presence of the phosphodiester backbone at this position is important for the formation of the trefoil turn motif. Altogether, these results show the importance of the base pair switching, and provide additional information on the structural environment required for the formation of the ribose zipper, the trefoil turn and the positioning of the catalytic cytosine within the core centre.
Contribution of the homopurine bp
The homopurine bp is located at the top of the P4 stem and is therefore adjacent to the catalytic cytosine (C76) (see Figure 1, positions 42 and 75). As mentioned previously, this structural feature is perfectly conserved in both the natural and the selected variants. However, it is an A42A75 bp in most of the selected variants (72%) and a G42G75 bp in the natural variants. A similar observation was detected when analysing the sequence variants from another in vitro selection of HDV ribozyme (28). This homopurine bp is important to the ribozyme's activity; its replacement by a Watson–Crick bp has been shown to drastically reduce the cleavage activity (15). However, its contribution to the molecular mechanism of the HDV ribozyme remains elusive.
In order to verify if the adoption of the homopurine bp is influenced by the binding of the substrate to the ribozyme, RNase T1 probing was performed. RNase T1 hydrolyses all guanosines located in single-stranded regions. In the ribozyme alone, the three consecutive guanosines, G40, G41 and G42, appeared as being single-stranded (Figure 7A). Upon addition of the SdA4 analogue these guanosines became inaccessible. The P1.1 pseudoknot, which includes G40 and G41, is formed in addition to the homopurine bp (Figure 7A). Thus, the binding of the substrate is required for the formation of these structural features. Several mutants were designed in order to probe the effect of homopurine bp formation on the secondary structure of the ribozyme, especially on the positioning of the catalytic C76. RNase T1 probing of a mutant ribozyme (RzG75C) including a G42–C75 Watson–Crick bp instead of a G42–G75 homopurine bp was performed (Figure 7A–C). This mutant is completely devoid of any cleavage activity (15). The resulting banding pattern showed several differences (Figure 7A, lanes 3 and 5), the most striking being at the level of the J1/4 junction: the three consecutives guanosines were not hydrolysed regardless of the presence or absence of the SdA4 analogue. This suggests that even in the absence of the substrate analogue, these residues were already engaged in the formation of base pairs. This observation received additional support from in-line probing data showing that the residues of both the J1/4 junction and the bottom of the J4/2 junction (i.e. C75, C76, U77 and A78) were not hydrolysed (data not shown). Together, these results suggest that the residues of these regions form a double-stranded structure that extends the P4 stem (Figure 7C and D). This suggests to us that a potential contribution of the homopurine bp might be to prevent the formation of such a non-productive structure (i.e. alternative inactive folding). The homopurine bp seems to interrupt an elongation of the P4 stem, and contributes to the conservation of the catalytic cytosine as a single-stranded residue. Results from mutagenesis of the homopurine bp have demonstrated that an A42A75 homopurine bp is more active than a G42G75 bp, as has been observed previously (15). The presence of an AA homopurine bp appears to limit the number of potential alternative structures that can be formed, which may explain the higher occurrence of A42A75, rather than G42G75, in the selection performed.
Figure 7.
RNase T1 mapping of the wild type and homopurine bp mutant ribozymes. (A) Autoradiogram of a 10% PAGE gel of T1 probing performed with 5′-end-labelled wild-type and mutated trans-acting ribozymes. Alkaline hydrolysis of the wild-type ribozyme was performed in order to determine the location of each position (lane OH). Lanes 1 and 2 are negative controls (no reaction and no substrate) performed with the wild-type and mutant RzG75C ribozymes, respectively. RNase T1 hydrolysis was performed on both the wild-type ribozyme (lane 3) and the RzG75C (lane 4) either in the absence or the presence of the SdA4 analogue as indicated by the symbols (−) and (+), respectively. The sites of RNase T1 hydrolyses are identified, and intensities of the hydrolyses correlate with the intensities of the arrow heads. The positions of the guanosines are indicated on the left. (B) to (D) are schematic representation of the nucleotide sequences and secondary structures of the ribozymes. (B) is the wild-type ribozyme. (C) and (D) are two secondary structures for the RzG75C mutant that differs for the J1/4 and J4/2 junctions.
DISCUSSION
The development of an unbiased in vitro selection protocol for the isolation of self-cleaving HDV RNA strands, in conjunction with a sequencing effort, permitted the construction of a database containing 119 novel variants. Subsequently, a software programme that evaluates the covariation of nucleotides was developed. The finding that the covariation data supports the base pairs composing both the P1 and P3 stems, as well as that of the homopurine bp, illustrates the usefulness of this software. High CSs were also obtained for some pairs of nucleotides of both the P1 and the P3 stems, suggesting that some interactions must exist between the residues of these two helical regions (e.g. the U36 favours the presence of a G22–C30 bp in the P3 stem with a CS of +0.47). However, current knowledge does not permit the elucidation of the nature of these putative interactions. NMR studies have already suggested the presence of a magnesium ion located between these two helices (25). Other biochemical experiments will be required in order to establish and characterize these interactions. More importantly, the C19–G81 bp at the bottom of the P2 stem was not supported by the covariation analysis. Conversely, the covariation data indicated the presence of a novel base pair formed between C19 and G80. In fact, chemical and enzymatic probing revealed that, upon formation of the P1.1 pseudoknot, the C19–G81 bp is disrupted in order to favour the formation of the C19–G80 bp. This structural rearrangement occurred as a molecular switch, leaving G81 without any contribution to the catalysis.
The P2 stem has been shown to be crucial in the HDV trans-acting ribozyme while dispensable for self-cleaving under certain conditions (29, 30). Specifically, an experiment of determination of the 3′ boundary of the self-cleaving antigenomic RNA strand showed that even the ribozyme ending at G80 was active (30). However, the level of activity of such a self-catalytic RNA strand remains undefined. It is most likely that the covalent linking between the substrate and catalytic domain is sufficient to permit folding of a significant proportion of the RNA strands into the active conformation. The fact that a cis-acting ribozyme ending with the G80 at the 3′ end is active supports the importance of the G80–C19 base pairing forming the base of P2 stem. According to the older representation of the antigenomic HDV ribozyme, a ribozyme ending with the G80 would be deprived of the P2 stem. The identity of the residue at position G80 was conserved in all natural variants. Our data showed that the identity of the nucleotide at position 80 affects the level of the cleavage activity; however, the most important feature for this residue is to form a base pair with the residue in position 19; 95% of the self-cleaving sequence included N19–N′80 bp (Table 1). It was reported previously that a self-cleaving sequence that did not support the formation of this base pair was not necessarily deprived of all cleavage activity (31). Moreover, we reported recently that the replacement of the G80 by a 4-thiouridine residue in a trans-acting HDV ribozyme that possess a C19 was not detrimental (19). In fact, the cleavage activity was evaluated to be seven-fold smaller (19). In other words, there is some flexibility at position 80. This may be an indication that the switch performed by the G80 is probably a consequence of the J4/2 rearrangement instead of being an essential conformation transition. Importantly, the new C19–G80 bp sheds light on the contribution of G80 that was previously proposed to stabilize the catalytic centre through interaction with the sequence of the P3 stem (30). Rather than being with the P3 stem, this interaction take place with the nucleotide just before the last one of the P2 stem (i.e. C19).
With the formation of the new C19–G80 bp, the secondary structure of the antigenomic ribozyme becomes reminiscent of that of the genomic version, that is to say it includes a J4/2 junction composed of only four nucleotides. It has already been suggested that the self-cleaving HDV motifs of both the genomic and the antigenomic polarities adopt a similar global architecture (32), a conclusion supported by our results. The notable differences between the two polarities are primarily limited to the unique single-stranded CAA located within the J1/2 junction of the genomic ribozyme. The reduced number of differences between the two ribozymes strongly suggests that both evolved from a unique ancestral sequence. The various selective pressures exerted along the life cycle of the HDV virus, including its self-cleaving sequences, are most likely responsible for these differences in a manner analogous to that observed between the in vitro selected sequences and the natural variants (e.g. AA and GG homopurine bp as well as the C19–G80 and A19–U80 bp). Interestingly a genome-wide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene (33). The sequence retrieved within the human genome includes neither a G80 as found in the genomic version of the self-cleaving HDV strand, nor the single-stranded CAA located within the J1/2 genomic version, this is reminiscent of the antigenomic HDV sequence. Thereby, it looks like a consensus sequence between the genomic and antigenomic sequence variants, supporting the hypothesis that the HDV arose from the human transcriptome (33).
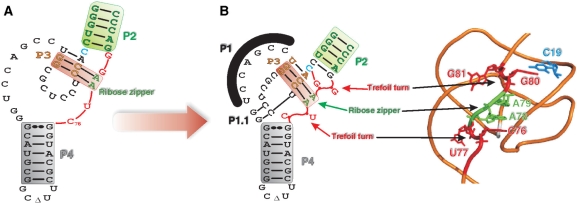
The molecular switch leading to the formation of the C19–G80 bp appears to be one feature of a complex structural rearrangement that either occurred simultaneously with, or rapidly following, the formation of the P1.1 pseudoknot. The latter step, which was shown to take place after both the annealing of the substrate to the ribozyme and the docking of the P1 stem within the catalytic centre, has been proposed to be the limiting step of the HDV ribozyme catalysis (8,24). The folding of the P1.1 pseudoknot appears to be the central event that triggers all of the conformational changes leading to the cleavage reaction. This results in the complete folding of the J4/2 junction, including the formation of the C19–G80 bp, the ribose zipper (including the A78A79) and the trefoil turn (C76U77) (Figure 8). Together, the formation of these features leads to the correct positioning of the catalytic cytosine (C76). Furthermore, G81 is most likely extruded out of the structure, as is U77. The extrusion of U77 is associated with the positioning of C76 deeply within the catalytic centre (11). This structural rearrangement might be viewed as two opposing forces causing the exclusion of one nucleotide. In the case of G81, it is tempting to suggest that this is reminiscent of a trefoil turn. Prior to this study, the presence of the structural motifs that form the J4/2 junction, with the exception of the base pair switch C19–G80, were all discovered by high-resolution approaches (3,4,23–25). The present work provides an original biochemical study of these motifs, as well as attempting to determine their order of formation. Moreover, it shows that G81 is optional for the ribozyme's catalysis. A mutant lacking G81 and including either a C19–G80 or U19–A80 bp exhibited virtually the same cleavage activity as did the wild-type ribozyme. Therefore, the conservation of this guanosine in all natural antigenomic variants is intriguing. Similarly, the GG homopurine bp is perfectly conserved in all of the natural variants, while in vitro selection revealed that the most efficient homopurine bp is AA. In addition, we observed several guanosine residues outside of the catalytic centre of the ribozyme (e.g. in the P4 stem-loop) that were conserved in the natural variants, but are not critical to the self-cleavage activity. According to the secondary structure predicted for several HDV RNA genome variants, most, if not all, of these guanosines appear to form base pairs. Therefore, it is tempting to speculate that a potential contribution of these guanosines would explain their conservation in the natural variants and they favour both, the unfolding of the ribozyme after cleavage and the subsequent formation of the rod-like structure that prevents the self-cleavage of the newly circular HDV RNA strands. Clearly, verifying this hypothesis requires additional experimentation.
Figure 8.
Hypothetical representation of the folding of the J4/2 junction region before and after the formation of the P1.1 pseudoknot. (A) Nucleotide sequence and secondary structure of the antigenomic ribozyme characterized in this work. The substrate is represented by the thin line in order to simplify the representation. (B) 3D model representation of the antigenomic ribozyme drawn based on the backbone structure obtained from the crystal structure of the genomic version (3). The structural motifs are identified.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Cedric Reymond for his technical assistance. This work was supported by grants from the Canadian Institute of Health Research (CIHR; grant number MOP-44002) to J.P.P. The RNA group is supported by a grant from the Université de Sherbrooke. A.N. was the recipient of a pre-doctoral fellowship from Ministère de l’Enseignement Supérieur et Recherche Scientifique de Tunisie. J.P.P. holds the Canada Research Chair in Genomics and Catalytic RNA. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Been MD. HDV ribozymes. Curr. Top. Microbiol. Immunol. 2006;307:47–65. doi: 10.1007/3-540-29802-9_3. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron LJ, Ouellet J, Perreault JP. Ribozyme-based gene-inactivation systems require a fine comprehension of their substrates specificities; the case of delta ribozyme. Curr. Med. Chem. 2003;10:2589–2597. doi: 10.2174/0929867033456486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FerréD’Amaré AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 4.Ke A, Zhou K, Ding F, Cate J, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 5.Wadkins TS, Perrotta AT, FerréD’Amaré AR, Doudna JA, Been MD. A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA. 1999;6:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishikawa F, Nishikawa S. Requirement of the canonical base pairing in the short pseudoknot structure of genomic hepatitis delta virus ribozyme. Nucleic Acids Res. 2000;28:925–931. doi: 10.1093/nar/28.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deschênes P, Ouellet J, Perreault J, Perreault JP. Formation of the P1.1 pseudoknot is critical for both the cleavage activity and substrate specificity of an antigenomic trans-acting hepatitis delta ribozyme. Nucleic Acids Res. 2003;31:2087–2096. doi: 10.1093/nar/gkg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ananvoranich S, Perreault JP. The kinetic and magnesium requirements for the folding of antigenomic δ ribozymes. Biochem. Biophys. Res. Comm. 2000;270:600–607. doi: 10.1006/bbrc.2000.2465. [DOI] [PubMed] [Google Scholar]
- 9.Bevilacqua PC, Brown TS, Nakano S, Yajima R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- 10.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 11.Sefcikova J, Krasovska MV, Spackova N, Sponer J, Walter NG. Impact of an extruded nucleotide on cleavage activity and dynamic catalytic core conformation of the HDV ribozyme. Biopolymers. 2007;85:392–406. doi: 10.1002/bip.20693. [DOI] [PubMed] [Google Scholar]
- 12.Nehdi A, Perreault JP. Unbiased in vitro selection reveals the unique character of the self-cleaving antigenomic HDV RNA sequence. Nucleic Acids Res. 2006;34:584–592. doi: 10.1093/nar/gkj463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu HN, Lee JY, Huang HW, Huang YS, Hsueh TG. Mutagenesis analysis of a hepatitis delta virus genomic ribozyme. Nucleic Acids Res. 1993;21:4193–4199. doi: 10.1093/nar/21.18.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafontaine DA, Ananvoranich S, Perreault JP. Presence of a coordinated metal ion in a trans-acting antigenomic delta ribozyme. Nucleic Acids Res. 1999;27:3236–3243. doi: 10.1093/nar/27.15.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Been MD, Perrotta AT. Optimal self-cleavage activity of the hepatitis delta virus RNA is dependent on a homopurine base pair in the ribozyme core. RNA. 1995;1:1061–1070. [PMC free article] [PubMed] [Google Scholar]
- 16.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 18.Ananvoranich S, Perreault JP. Substrate specificity of delta ribozyme cleavage. J. Biol. Chem. 1998;273:13182–13188. doi: 10.1074/jbc.273.21.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reymond C, Ouellet J, Bisaillon M, Perreault JP. Examination of the folding pathway of the antigenomic hepatitis delta virus ribozyme reveals key interactions of the L3 loop. RNA. 2007;13:44–54. doi: 10.1261/rna.263407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouellet J, Perreault JP. Cross-linking experiments reveal the presence of novel structural features between a hepatitis delta virus ribozyme and its substrate. RNA. 2004;10:1059–1072. doi: 10.1261/rna.7230604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris DA, Tinsley RA, Walter NG. Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 2004;341:389–403. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 22.Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 23.Harris DA, Rueda D, Walters NG. Local conformational changes in the catalytic core of the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 24.Pereira MJB, Harris DA, Rueda D, Walters NG. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka Y, Hori T, Tagaya M, Sakamoto T, Kurihara M, Uesugi S. Imino proton NMR analysis of HDV ribozyme: nested double pseudoknot structure and Mg2+ ion-binding site close to the catalytic core in solution. Nucleic Acids Res. 2002;30:766–774. doi: 10.1093/nar/30.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiola K, Perreault JP. Kinetic and binding analysis of the catalytic involvement of ribose moieties of a trans-acting delta ribozyme. J. Biol. Chem. 2002;277:26508–26516. doi: 10.1074/jbc.M203468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ananvoranich S, Fiola K, Ouellet J, Deschênes P, Perreault JP. Kinetic analysis of bimolecular hepatitis delta virus ribozyme. Methods Enzymol. 2001;341:553–566. doi: 10.1016/s0076-6879(01)41176-1. [DOI] [PubMed] [Google Scholar]
- 28.Legiewicz M, Wichlacz A, Brzezicha B, Ciesiolka J. Antigenomic delta ribozyme variants with mutations in the catalytic core obtained by the in vitro selection method. Nucleic Acids Res. 2006;34:1270–1280. doi: 10.1093/nar/gkl018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotta AT, Been MD. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 30.Lee CB, Lai YC, Ping YH, Huang ZS, Lin JY, Wu HN. The importance of the helix 2 region for the cis-cleaving and trans-cleaving activities of hepatitis delta virus ribozymes. Biochemistry. 1996;35:12303–12312. doi: 10.1021/bi961219m. [DOI] [PubMed] [Google Scholar]
- 31.Perrotta AT, Been MD. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenstein SR, Been MD. Hepatitis delta virus ribozymes fold to generate a solvent inaccessible core with essential nucleotides near the cleavage-site phosphate. Biochemistry. 1996;35:11403–11413. doi: 10.1021/bi9609984. [DOI] [PubMed] [Google Scholar]
- 33.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 34.Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]