Abstract
While much is known about abasic DNA, the biological impact of abasic RNA is largely unexplored. To test the mutagenic potential of this RNA lesion in the context of retroviruses, we synthesized a 31-mer oligoribonucleotide containing an abasic (rAS) site and used it as a template for studying DNA primer extension by HIV-1, avian myeloblastosis virus (AMV) and moloney murine leukemia virus (MMLV) reversed transcriptases (RT). We found that trans-lesion synthesis readily takes place with HIV-1 RT and to a lesser extent with AMV RT while MMLV RT aborts DNA synthesis. The preference of dNTP incorporation follows the order A∼G > C∼T and thus obeys to the ‘A-rule’. In the case of HIV-1 RT, we measured the kinetic data of dNTP incorporation and compared it to abasic DNA. We found that A-incorporation is only 2-fold slower relative to a matched (undamaged) RNA template while it is 7-fold slower in the case of DNA. Furthermore, there is less discrimination in incorporation between the four dNTPs in the case of abasic RNA compared to abasic DNA. These experiments clearly point to a higher promiscuity of lesion bypass on abasic RNA. Given their known higher chemical stability, such rAS sites can clearly contribute to (retro)viral evolution.
INTRODUCTION
Abasic sites are well-known DNA lesions that occur spontaneously via depurination at a frequency of ca. 10 000 per genome per day in mammalian cells (1). Acidic or alkylating conditions as well as oxidative stress greatly increase this number. Abasic DNA is unstable and undergoes strand cleavage 3′ to the abasic site with an average lifetime of 8 days at 37°C, pH 7.4 and physiological ionic strength (2). Due to the degradation of the genetic material and to missing coding information, such sites are highly mutagenic and are a major threat to living cells (3,4). Living organisms have therefore evolved a highly efficient and complex DNA repair machinery that maintains genome integrity (5). An important component of this machinery is the base excision repair (BER) pathway in which abasic sites are produced via removal of damaged bases by specific glycosylases, followed by endonucleolytic cleavage 5′ to the abasic site by the endonuclease APE1 to generate a nick. Polymerase β (Polβ) then removes the abasic moiety with its AP lyase activity and reinserts a correct nucleotide. In the final step, the ends of the repaired strand are ligated by DNA ligase III.
Abasic lesions are a priori not restricted to DNA but can also occur in RNA. Given the fact that not only short-lived mRNA but long-lived tRNAs and ribosomal RNAs are present at all times in a cell in much higher concentrations than DNA, and given also the fact that RNA viruses as well as retroviruses store their genetic information in form of RNA, it is not excluded that abasic RNA is of biological relevance. Literature about the chemical biology of RNA abasic sites is, however, rare and their biological impact is largely unexplored with a few exceptions. For example, RNA abasic sites are known to be the result of the action of RNA N-ribohydrolases (6) that deactivate eukaryotic ribosomes by depurination of a specific adenosine residue on 28S rRNA. Such ribosome-inactivating proteins (RIPs) are long known, but only recently it was found that class-I RIPs as e.g. the pokeweed antiviral protein (PAP) cannot only depurinate the sarcin/ricin loop of the large rRNA but also specific adenine and guanine residues throughout the sequence of capped mRNAs (7) or other viral RNAs (8). Besides this there is also evidence for a class of RNA-specific lyases that act on ribosomal RNA abasic sites, leading to complete inactivation of ribosomes (9,10). Taken together, this proves that there exist not only spontaneous but also enzymatic pathways to abasic RNA for which further metabolic processing may be required.
Given the lack of knowledge on the chemical biology of RNA abasic sites, we set out to investigate it in more detail. We (11) and others (12) developed a chemical synthesis of abasic RNA based on a precursor phosphoramidite carrying a photocleavable protecting group at the anomeric center. Using this method, we recently investigated the chemical reactivity of RNA abasic sites toward 3′-β-elimination chemistry. We found that, compared to abasic DNA, abasic RNA is ca. 150-fold more stable under basic conditions (0.1 M NaOH) and ca. 15-fold more stable under slightly acidic conditions in the presence of an aromatic amine (11). This has of course to be put into perspective with the ca. 100- to 1000-fold higher stability of the glycosidic bond in a ribonucleoside compared to a 2′-deoxyribonucleoside (13), which makes the spontaneous formation of abasic RNA a much rarer event compared to DNA. Anyway, once formed the relative longevity of abasic RNA can be expected to have a non-negligible impact on mutation in RNA viruses and retroviruses.
Reverse transcriptases (RT) are the key enzymes in retroviral replication. They synthesize in a first step a complementary DNA strand on the retroviral RNA template followed by degradation of the RNA strand by the RNaseH subunit and synthesis of the complementary DNA strand by the polymerase subunit. An abasic site on the RNA template can affect both the polymerase and the RNaseH activity of an RT. In preliminary work, we reported on the trans-lesion synthesis of HIV-1 reverse transcriptase on a RNA-template/DNA-primer system with an abasic site in the RNA template (14). Here we present a detailed investigation of the impact of abasic RNA on reverse transcription by HIV-1 RT, avian myeloblastosis virus (AMV) RT and moloney murine leukemia virus (MMLV) (H−) RT. More specifically, we investigated the insertion preferences for any of the natural dNTPs opposite the lesion in standing start (ss) and running start (rs) experiments with DNA primers. We also investigated the RNaseH activity of HIV-1 RT and AMV RT on the abasic template. In the case of HIV-1 RT, we determined the relative kinetic efficiencies of dNTP incorporation and compared it to those of a true abasic DNA template/DNA primer system of the same sequence.
MATERIALS AND METHODS
Synthesis of primers and templates
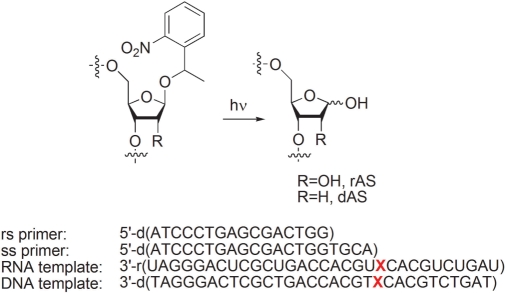
All phosphoramidites, (2′-TBDMS protection in the case of RNA building blocks) for oligonucleotide synthesis were from Glen Research. The caged amidites for the true DNA and RNA abasic sites were prepared as described (11). Synthesis of DNA primers and RNA and DNA templates as well as deprotection/detachment from solid support was performed as described (11,14). DNA primers were purified by standard reversed phase HPLC and were desalted over Sep-Pak C18 columns (Waters). Caged DNA and RNA templates were purified by preparative gel electrophoresis on a 20% (19:1) denaturing polyacrylamide gel. Oligonucleotide containing bands were excised and electroeluted with a Elutrap electroelution system (Schleicher & Schuell). Where required, DNA primers or RNA templates were 5′-end labeled with T4 polynucleotide kinase (Fermentas) and [γ−32P]ATP (Hartmann Analytics). Purified caged abasic templates were transformed into true abasic templates after annealing with the corresponding primers (template/primer ratio = 2:1, 2 min, 60°C, slow cooling to RT) in enzyme buffer (in the absence of enzyme) by irradiation for 2 min with a UV lamp (TQ 150, Heraeus) or with a tungsten slide projector lamp for 6 min (11).
Reverse transcription (RT) assays
RT assays were performed with HIV-I RT (Worthington Biochemicals, Lakewood, NJ, USA), AMV RT (USB corp., Cleveland, OH, USA) and MMLV (H−) RT (Promega Corp., Madison, WI, USA) in the following buffers: HIV-I RT and AMV RT : 50 mM Tris (pH 8.3), 50 mM NaCl, 8 mM MgCl2, 1 mM DTT; M-MLV (H−) RT : 50 mM Tris (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT. Final reaction mixtures contained abasic DNA or RNA template (100 nM), DNA primer (50 nM) and dNTP (20 μM). After addition of enzyme (0.5–32 U), the mixtures were incubated at 37°C for 1 h. The reactions were quenched with gel loading buffer (98% formamide, 0.1% xylene cyanol (FF), 0.1% bromophenol blue), heated to 90°C for 5 min and applied to a 20% denaturing polyacrylamide gel. Radioactivity was detected and quantified on a Storm 820 Phosphorimager with ImageQuant software (GE Healthcare).
Steady-state kinetics
Incorporation of natural 2′-deoxynucleotides opposite the abasic lesion on the template was performed with HIV-I RT in the ss system by measuring the initial rates at which the 32P-labeled primer was extended with varying concentration of 2′-deoxynucleoside triphosphates. All experiments were carried out in 50 mM Tris (pH 8.3), 50 mM NaCl, 8 mM MgCl2, 1 mM DTT, 100 nM template and 50 nM primer at 37°C for 2 min. The dNTP concentrations were varied from 10 to 1000 μM. Enzyme concentrations used were 0.1–1.5 U (4–59 nM). Reactions were quenched by addition of 50 µl of loading dye and the mixtures heated to 90°C for 2 min. Aliquots of the reaction mixtures were analyzed by gel electrophoresis on a 20% denaturing polyacrylamide gel. A Storm 820 Phosphorimager and ImageQuant 5.2 software (GE Healthcare) was used to quantify gel band intensities corresponding to the extended and unextended primer. The measured velocities were then plotted against the concentration of the dNTPs and fitted to the Michaelis–Menten equation in KaleidaGraph 4.0 (Synergy Software) to determine Vmax and Km. kcat was obtained from Vmax by normalizing by the total enzyme concentration. Values were determined as the average of at least three independent experiments (see Supplementary Data).
RESULTS
Three RTs differing in their intrinsic transcription fidelity were tested in their ability to bypass an abasic site on an RNA template. The human immunodeficiency virus (HIV-1) RT is a heterodimer of two related chains, a 66-kD subunit (p66) and a 51-kD subunit (p51) derived from p66 by proteolytic cleavage. It is known to be one of the most promiscuous polymerases (15–18). The AMV RT is again a heterodimer composed of a 95 kDa and a 63 kDa subunit but is 10 times less error prone than HIV-1 RT (15,19). Compared to these two enzymes, MMLV RT is a high-fidelity polymerase (19,20). The monomeric enzyme consists of a single polypeptide chain with five domains with a mass of 75 kDa. MMLV RT typically shows only weak RNaseH activity. In our experiments, we used MMLV RT (H−), a mutant enzyme with complete lack of RNaseH activity.
For the polymerase and RNaseH assays, we chose the primer/template systems depicted in Figure 1. We used two formats, namely a ss system consisting of a 31-mer abasic RNA template (X = rAS) and a 20-mer DNA primer with its 3′-end directly ending before the rAS site, and a rs system with the same RNA template and a DNA primer which was 4 nt shorter. For reference, we used a non-damaged RNA template (X = U). The nucleotide sequence of the template was chosen randomly with the annealing region to have a GC content of 60% and every base to occur at least twice in the single strand overhang to be transcribed. For determining the insertion kinetics in the case of HIV-1 RT, the ss system with both the abasic RNA and the DNA template (X = rAS and dAS, respectively) was used. Also here, the templates with X = U or T served as undamaged references. The template strands were chemically synthesized using the corresponding caged abasic site precursor phosphoramidites as described previously (11,14) and the abasic sites rAS and dAS, respectively, were revealed by UV-irradiation after primer annealing. RNaseH activity was tested in the same way on the ss primer/template system using labeled template instead of labeled primer.
Figure 1.
Primers and templates (X = dAS, rAS, T or U) used for ss primer and rs primer reverse transcription assays.
HIV-1 reverse transcriptase
Trans-lesion synthesis
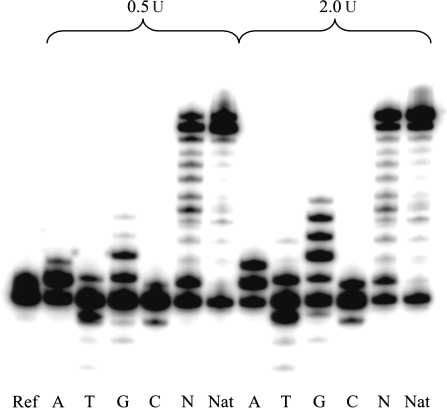
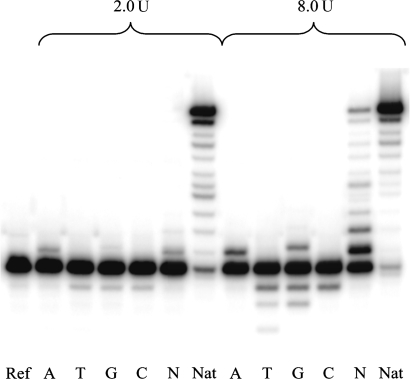
Standing start experiments with two different enzyme concentrations showed that HIV-1 reverse transcriptase incorporates all four natural dNTPs opposite an abasic RNA lesion with varying preferences (Figure 2). The efficiency of incorporation of the four dNTPs followed the order A∼G > C∼T. A relative quantification of the incorporation of the different dNTPs at the higher (2 U) enzyme concentration is given in Table 1. The amount of residual non-extended primer ranged from 30% to 60% depending on the nature of the dNTP. While insertion stopped after bypassing the abasic site in the case of the pyrimidine dTPs, additional insertions were observed in the case of the purine dTPs. For dATP, a second adenosine was misincorporated after that opposite to the abasic site. In the case of dGTP, up to five guanosines were inserted before primer extension came to an end. While positions X + 1 and X + 3 correspond to a correct G–C pair, the positions X + 2 and X + 4 correspond to G–A and G–G mispairs. In the case of dCTP and dTTP insertion, substantial residual 3′–5′-exonucleolytic activity on the primer was observed. With all four dNTPs available, full-length elongation took place readily. The size of the band at the position X + 1 thereby indicates that extension of the first nucleotide after the abasic site bypass is relatively slow in comparison with a non-damaged template (X = U).
Figure 2.
Standing start HIV-1 RT assay with abasic RNA template (X = rAS), enzyme concentrations 0.5 and 2.0 U, reaction time 1 h. Ref: without enzyme and dNTPs. A, T, G, C: reactions in presence of the according dNTP; N: reactions in presence of all four dNTPs; Nat: unmodified RNA template (X = U) and all four dNTPs.
Table 1.
Quantification of the primer elongation/shortening with HIV-1 RT (2 U) in%
| dNTP | − 3 | − 2 | − 1 | Primer | + 1 | + 2 | + 3 | + 4 | + 5 |
|---|---|---|---|---|---|---|---|---|---|
| dATP | – | – | – | 30.9 | 42.4 | 26.6 | – | – | – |
| dTTP | 0.7 | 1.2 | 25.5 | 46.6 | 26.1 | – | – | – | – |
| dGTP | – | 0.6 | 3.8 | 29.2 | 12.6 | 24.3 | 14.6 | 12.2 | 2.8 |
| dCTP | – | – | 10.8 | 60.4 | 28.8 | – | – | – | – |
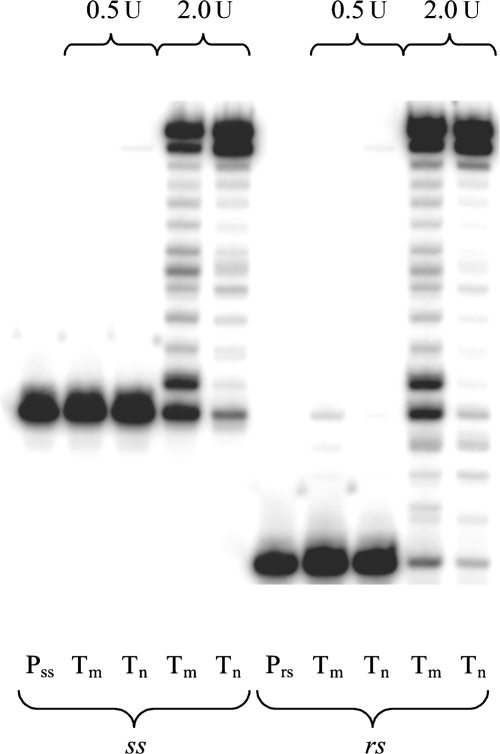
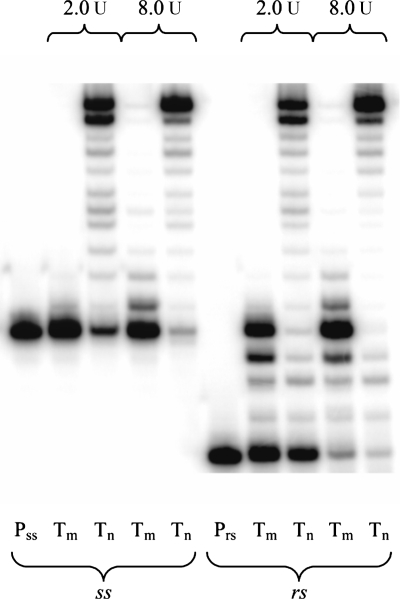
To study the insertion steps preceding the abasic lesion, we performed a rs experiment (Figure 3). At the higher enzyme concentration, the ss as well as the rs experiments showed efficient trans-lesion synthesis. The rs assay confirmed that the slowest nucleotide insertions occur at the positions opposite X and X + 1. The subsequent incorporations then proceed readily, although typically a higher number of truncated fragments are observed compared with the non-damaged template system (X = U).
Figure 3.
Comparison of ss (left) and rs (right) elongation experiments with HIV-1 RT, reaction time 1 h Pss: primer ss, Prs: primer rs, Tm: abasic RNA template (X = rAS), Tn: non-damaged RNA template (X = U).
As a general conclusion at this point, it appears that preferential purine (especially A) insertion opposite an abasic site also pertains to RNA templates. This fact, often referred to as the ‘A-rule’, is well known for a many RTs and other DNA polymerases on abasic DNA templates (21).
Kinetics of insertion at the abasic site of an RNA template compared to a DNA template
Given the fact that reverse transcriptases have evolved to use both RNA and DNA as templates for polymerase activity, it is of interest to know the differences in the insertion kinetics of a dNTP opposite an abasic site as a function of the nature of the template backbone (DNA or RNA). While the kinetics of insertion on DNA templates containing abasic site analogs (open chain and THF analog of an abasic site) by HIV-1 RT have already been described (22), no such data is available for nucleotide insertion opposite a true abasic site on either a DNA or an RNA template.
It is well known that abasic nucleic acids are chemically labile and tend to β-elimination and strand cleavage 3′-to the abasic site. To test for chemical stability, we subjected the corresponding abasic templates to assay conditions in the absence of enzyme and found no significant cleavage (data not shown). With the confidence of chemical stability under the assay conditions, we analyzed the rate of insertion of each of the four dNTPs opposite the abasic site on both the RNA and DNA template. More specifically we determined the apparent binding of the dNTPs (KM) as well as the first order rate constants (kcat) of the resulting ternary complex. The ratio kcat/KM is the second order rate constant and a measure of the catalytic efficiency. The kinetic data are summarized in Table 2. The corresponding gels and plots are in the Supplementary Data.
Table 2.
Rates of incorporation of dNTPs opposite an abasic lesion on either the DNA or RNA template with the standing start primer (Figure 1) by HIV-1 RT
| Template backbone | X | dTP | Vmax (nM·min−1) | KM (μM) | kcat (min−1) | kcat/KM (μM−1·min−1) | krela |
|---|---|---|---|---|---|---|---|
| RNA | U | A | 1.75 ± 0.08 | 13.47 ± 5.99 | 0.089 ± 0.004 | 6.57×10−3 | 13.5 |
| rAS | A | 3.61 ± 0.40 | 54.84 ± 10.64 | 0.183 ± 0.020 | 3.33 × 10−3 | 6.7 | |
| rAS | G | 7.19 ± 0.92 | 80.62 ± 15.87 | 0.182 ± 0.023 | 2.26 × 10−3 | 4.5 | |
| rAS | C | 4.83 ± 0.15 | 148.75 ± 12.05 | 0.082 ± 0.003 | 0.55 × 10−3 | 1.1 | |
| rAS | T | 8.05 ± 2.17 | 275.21 ± 75.95 | 0.136 ± 0.037 | 0.50 × 10−3 | 1 | |
| DNA | T | A | 5.47 ± 1.04 | 2.88 ± 0.67 | 1.385 ± 0.263 | 4.81 × 10−1 | 1551 |
| dAS | A | 6.98 ± 0.24 | 25.86 ± 1.90 | 1.768 ± 0.060 | 6.84 × 10−2 | 221 | |
| dAS | G | 7.16 ± 0.81 | 68.46 ± 14.88 | 0.725 ± 0.083 | 1.06 × 10−2 | 34 | |
| dAS | C | 3.39 ± 0.31 | 185.77 ± 18.89 | 0.057 ± 0.005 | 0.31 × 10−3 | 1 | |
| dAS | T | 5.93 ± 1.76 | 349.88 ± 150.86 | 0.150 ± 0.044 | 0.43 × 10−3 | 1.4 |
akrel = (kcat/KM)N/(kcat/KM)min.
The data clearly show that on both the RNA and DNA template, purine nucleotides are more efficiently inserted opposite the abasic site compared to the pyrimidines. There are, however, backbone intrinsic differences. In the case of the RNA backbone, the efficiency of A and G insertion is similar while a clear preference for A is observed in the case of the DNA template. Furthermore, the relative discrimination of the four dNTPs opposite the abasic site is much smaller on the RNA template compared to the DNA template as can be easily inferred from krel. It varies only 6.7-fold in the former but 221-fold in the latter case. Last but not least, incorporation efficiency opposite an abasic site relative to a matched insertion (X = U or T, dTP = A) is much higher in the case of the RNA backbone compared to the DNA backbone. This is again apparent from krel which shows that insertion of A opposite U is only 2-fold more efficient compared to the best nucleotide incorporated opposite the abasic lesion in the case of the RNA template while it is 7-fold more efficient in the case of the DNA template with X = T. This clearly shows that HIV-1 RT is more promiscuous in bypassing an abasic lesion in RNA compared to DNA.
RNaseH activity
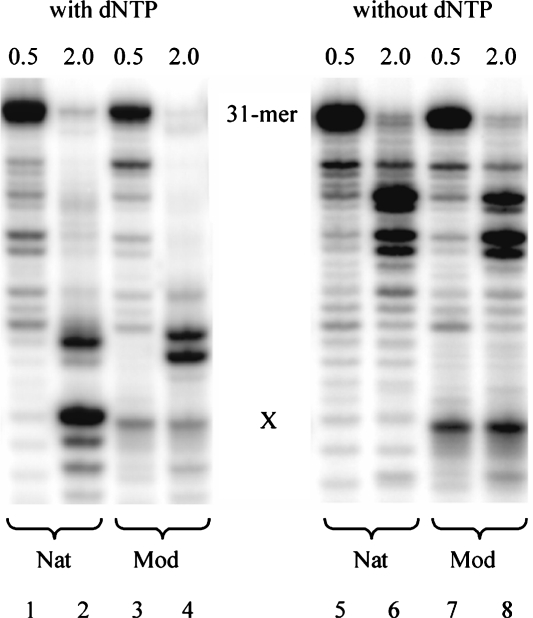
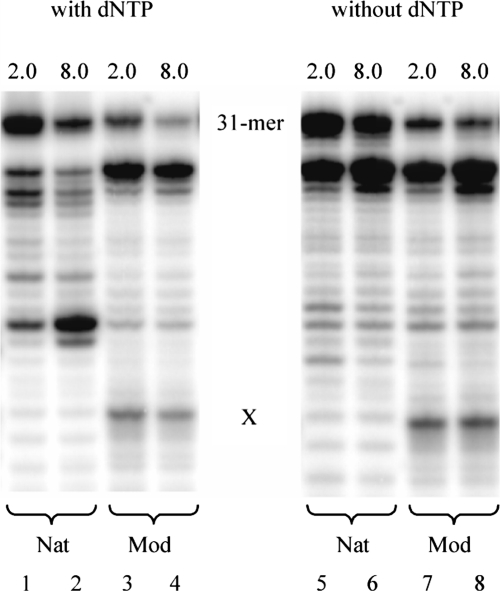
We also determined the RNaseH activity of HIV-1 RT (23) on the abasic RNA template in the presence and absence of dNTPs. For this, 5′-32P-labeled template RNA and unlabeled DNA ss primer were used. At low enzyme concentration, only weak RNaseH activity in the case of both damaged and non-damaged RNA template, independent on the presence or absence of dNTPs, was observed (lanes 1, 3, 5 and 7 in Figure 4). Increasing the enzyme concentration resulted in complete disappearance of the full-length RNA template in all cases (lanes 2, 4, 6 and 8). Interestingly, in the presence of dNTPs (where primer extension is possible) there are only small differences in the RNA degradation pattern as a function of the presence or absence of the RNA abasic site. In the unmodified RNA template, cleavage occurs predominantly at the positions X and X − 4 (direction of DNA synthesis) whereas in the abasic template, cleavage is effected predominantly at positions X − 3 and X − 4. The same experiment performed in the absence of dNTPs (no primer extension possible) shows again similar degradation characteristics for the natural and the abasic template. In these cases, however, the major cleavage sites are deep in the double helical primer/template region and not near to the X site. This differential behavior is an indication that primer extension in the presence of dNTPs is faster than RNA degradation also in the case of the damaged template.
Figure 4.
Comparison of the RNaseH activity of HIV-1 RT with (lanes 1–4) and without (lanes 5–8) dNTPs at different enzyme concentrations: 0.5 U (lanes 1, 3, 5, 7) and 2.0 U (lanes 2, 4, 6, 8). Nat = unmodified RNA template (X = U), Mod = abasic RNA template (X = rAS).
AMV reverse transcriptase
Trans-lesion synthesis
Standing start experiments were again performed at two different enzyme concentrations. (Figure 5). At higher enzyme concentrations, faint bands are visible for single insertions in the case of dATP and dGTP. No multiple, consecutive incorporations as in the case of the HIV-1 RT could be found. Also, no primer extension takes place in the case of dTTP and dCTP. Instead some 3′–5′-exonuclease activity is present leading to 1–2 nt shortening of the primer. In the case of higher enzyme concentration and in the presence of all four dNTPs, full-length synthesis occurs to a moderate extent in the case of the abasic RNA template. This contrasts to the efficient full-length primer extension in the case of the non-damaged template (X = U) at both enzyme concentrations.
Figure 5.
Standing start AMV RT assay with abasic RNA template (X = rAS). Enzyme concentrations 2.0 and 8.0 U, reaction time 1 h. Ref: without enzyme and dNTPs. A, T, G, C: reactions in presence of the respective dNTP; N: reactions in presence of all four dNTPs; Nat: unmodified RNA template (X = U) and all four dNTPs.
A comparison of ss and rs experiments in the presence of all dNTPs is shown in Figure 6. Again only a very weak band for the full-length transcript is visible in both cases. The rs series shows that the enzyme slows down mostly directly at the abasic site as well as at positions X − 1 and X + 1. This contrasts with the findings on HIV-1 RT, where essentially no slowdown at position X − 1 was observed and where positions X and X + 1 are extended with similarly low efficiency. It therefore appears that the two polymerases react differently on local structural perturbations at the abasic site.
Figure 6.
Comparison of ss (left) and rs (right) primer elongation experiments with AMV RT, reaction time 1 h. Pss: primer ss, Prs: primer rs, Tm: abasic RNA template (X = rAS), Tn: natural RNA template (X = U).
RNaseH activity
RNaseH activity was again assayed in the presence and absence of dNTPs and at two different enzyme concentrations. From the gels, it becomes clear that RNaseH activity is generally slightly higher on the damaged template as compared to the non-damaged template irrespective of the enzyme concentration and the presence or absence of dNTPs (Figure 7). In the presence of the dNTPs, there is a clear difference in the preferred sites of cleavage. In the non-damaged template, the preferred site of cleavage is near to the 3′-end of the primer (positions X − 5 and X − 6, direction of DNA synthesis) while on the abasic template, the preferred site of cleavage is deep into the non-extended primer/template region. In the absence of dNTPs, the cleavage pattern is essentially the same in both cases with the preferred cleavage site deep in the non-extended primer/template part. The similar behavior in the presence or absence of dNTPs in the case of the damaged template can easily be explained by the very inefficient primer extension, excluding cleavage near to the 3′-end of the primer due to lack of elongation. The slightly higher cleavage at the abasic site in the damaged template is most likely not due to enzymatic activity but of β-elimination chemistry as a consequence of the denaturation of the sample after reaction and before application onto the gel.
Figure 7.
Comparison of the RNaseH activity of AMV reverse transcriptase with (lanes 1–4) and without (lanes 5–8) dNTPs for different enzyme concentrations: 2.0 U (lanes 1, 3, 5, 7) and 8.0 U (lanes 2, 4, 6, 8). Nat = unmodified RNA template (X = U); Mod = abasic RNA template (X = rAS).
MMLV reverse transcriptase
Trans-lesion synthesis
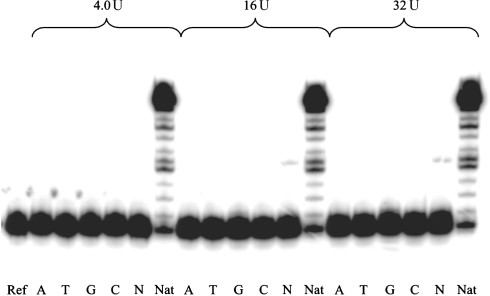
MMLV RT is known to be one of the less active and less promiscuous reverse transcriptases. Therefore, generally higher enzyme concentrations were required to obtain full-length transcripts compared to HIV-1 or AMV RT. Standing start experiments at three different enzyme concentrations showed efficient elongation for the natural template (Figure 8). To the contrary, no nucleotide insertion at the abasic site in the case of the damaged template/primer system appeared, regardless of the nature of the dNTP and the enzyme concentration. Also no 3′–5′-exonuclease activity was observed as in the case of the other RTs investigated. As expected, no RNaseH activity was found (data not shown). It thus appeared that for MMLV RT, an abasic site on the RNA is a very efficient block for reverse transcription.
Figure 8.
Standing start MMLV RT (H−) assay with abasic RNA template (X = rAS), enzyme concentrations 4.0, 16 and 32 U, reaction time 1 h. Ref: without enzyme and dNTPs. A, T, G, C: reactions in presence of the according dNTP; N: reactions in presence of all four dNTPs; Nat: non-damaged template (X = U) and all four dNTPs.
DISCUSSION
Abasic sites as a consequence of base damage on genomic DNA are highly mutagenic if unrepaired. Given the fact that any diploid cell has only two copies of its genetic material, it becomes intuitively clear that the installation of a complex repair machinery is vital for maintaining cellular integrity. The question whether there is need for a similar repair system for damaged RNA is less clear and needs differential consideration. Arguments against a necessity of repairing RNA abasic sites include the typically short lifetime and the high copy numbers as in the case of mRNA. On the other hand, there exist long-lived tRNAs and ribosomal RNAs as well as genomic RNA in RNA viruses and retroviruses and their damage may have fatal consequences to their survival. The influence of such a lesion on the RNA backbone on polymerase activity has not been explored so far. This deficit in information is mostly due to the unavailability of RNA with a site-directed lesion.
In 1993, Cai et al. reported on HIV-1 reverse transcriptase incorporating dNTPs opposite an abasic DNA template (22,24). They found that in rs reactions, Vmax/Km for insertion of A was about 5, 15 and 20 times larger than for insertion of G, T and C opposite X supporting the ‘A rule’. In our own experiments, we directly compared polymerase efficiency on both a RNA and DNA template containing a true abasic lesion in the ss format. While the general paradigm of the ‘A rule’ is fulfilled in both backbone systems, there are subtle differences in the insertion efficiencies. From our experiments it follows that the incorporation of dATP versus rAS in the RNA template is slower by a factor of only 2, whereas it is slower by a factor of 7 in the DNA template, compared to the respective match (dATP versus U/T). Furthermore, the range of relative incorporation efficiencies of dNTPs is much narrower in the case of RNA (1- to 6-fold) as compared to DNA (1- to 221-fold). This suggests that RNA abasic sites, once formed, are more mutagenic compared to DNA abasic sites, and the preference for incorporation of A relative to the other bases is less pronounced in the case of abasic RNA as compared to abasic DNA. The qualitative analysis of trans-lesion synthesis with AMV RT shows that incorporation efficiencies are generally much poorer as for HIV-1 RT, and mostly stop after X + 1. Nevertheless at higher enzyme concentration, some full-length product could be observed. These findings are in line with earlier observations on abasic DNA (22). Not surprisingly, MMLV RT that is known to be a high-fidelity RT does not process abasic RNA sites at all in a reasonable enzyme concentration range.
Previous investigations on the RNaseH activity showed that HIV-1 RT and MMLV RT generated smaller RNA products while AMV RT typically produced larger products. In all cases, some of the template RNA remained undigested (16,25–27). In the case of HIV-1 RT, ca. ∼20% of the template RNA remained annealed, while with AMV RT, ∼80% of the template RNA remained annealed after one round of processive DNA synthesis (23). We investigated the RNaseH activity in the case of HIV-1 and AMV RT in the presence and absence of dNTPs. In the case of AMV RT, essentially no difference could be seen on the damaged RNA template irrespective whether dNTPs were present or not. The major cleavage site was invariantly located deep in the primer/template double-helical range, indicating that forward polymerase activity is relatively slow compared to RNaseH digestion of the damaged RNA. The picture is different in the case of HIV-1 RT, where also in the case of the damaged template, trans-lesion synthesis is faster than RNA degradation. This fact has again implications on the level of mutagenicity of abasic RNA. Whereas the RNaseH function of AMV RT will most likely act as a control mechanism and abort DNA synthesis at an abasic lesion, HIV-1 RT will most likely continue by introducing mutations in the reverse transcript.
We have shown previously that the intrinsic chemical stability of an RNA abasic site is higher compared to a DNA abasic site (11). Although they are less spontaneously formed, there also exist enzymes that can produce such lesions on mRNA or viral RNA (7,8). Once formed, there is good reason to believe that they can play an important biological role. In the context of mRNA, it will therefore be of interest to find out how the ribosome processes abasic mRNA during translation and how abasic mRNA will eventually be metabolized. In the context of RNA viruses or retroviruses abasic sites can act in concert with the intrinsically low transcription fidelity of their encoded polymerases and add to the fact that viral evolution very often occurs at a rate near to the critical threshold for loss of genetic information (28).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Funding of this work as well as to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation (Grant. No. 200020-107692).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T, Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 3.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 4.Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Schärer OD. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. 2003;42:2946–2974. doi: 10.1002/anie.200200523. [DOI] [PubMed] [Google Scholar]
- 6.Schramm VL. Enzymatic N-riboside scission in RNA and RNA precursors. Curr. Opin. Chem. Biol. 1997;1:323–331. doi: 10.1016/s1367-5931(97)80069-5. [DOI] [PubMed] [Google Scholar]
- 7.Hudak KA, Bauman JD, Tumer NE. Pokeweed antiviral protein binds to the cap structure of eukaryotic mRNA and depurinates the mRNA downstream of the cap. RNA. 2002;8:1148–1159. doi: 10.1017/s1355838202026638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh BA, Tumer NE. Antiviral activity of ribosome inactivating proteins in medicine. Mini Rev. Med. Chem. 2004;4:523–543. doi: 10.2174/1389557043403800. [DOI] [PubMed] [Google Scholar]
- 9.Ogasawara T, Sawasaki T, Morishita R, Ozawa A, Madin K, Endo Y. A new class of enzyme acting on damaged ribosomes: ribosomal RNA apurinic site specific lyase found in wheat germ. EMBO J. 1999;18:6522–6531. doi: 10.1093/emboj/18.22.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozawa A, Sawasaki T, Takai K, Uchiumi T, Hori H, Endo Y. RALyase; a terminator of elongation function of depurinated ribosomes. FEBS Lett. 2003;555:455–458. doi: 10.1016/s0014-5793(03)01304-8. [DOI] [PubMed] [Google Scholar]
- 11.Küpfer PA, Leumann CJ. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trzupek JD, Sheppard TL. Photochemical generation of ribose abasic sites in RNA oligonucleotides. Org. Lett. 2005;7:1493–1496. doi: 10.1021/ol050120h. [DOI] [PubMed] [Google Scholar]
- 13.Kochetkov NK, Budovskii EI. Hydrolysis of N-glycosidic bonds in nucleosides, nucleotides and their derivatives. In: Kochetkov NK, Budovskii EI, editors. Organic Chemistry of Nucleic Acids. NY: Plenum; 1972. pp. 425–448. [Google Scholar]
- 14.Küpfer PA, Leumann CJ. RNA abasic sites: preparation and trans-lesion synthesis by HIV-1 reverse transcriptase. ChemBioChem. 2005;6:1970–1973. doi: 10.1002/cbic.200500204. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Goodman MF. Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J. Biol. Chem. 1992;267:10888–10896. [PubMed] [Google Scholar]
- 16.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 17.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 18.Menendez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- 19.Roberts JD, Preston BD, Johnston LA, Soni A, Loeb LA, Kunkel TA. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol. Cell. Biol. 1989;9:469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth MJ, Tanese N, Goff SP. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J. Biol. Chem. 1985;260:9326–9335. [PubMed] [Google Scholar]
- 21.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 22.Cai H, Bloom LB, Eritja R, Goodman MF. Kinetics of deoxyribonucleotide insertion and extension at abasic template lesions in different sequence contexts using HIV-1 reverse transcriptase. J. Biol. Chem. 1993;268:23567–23572. [PubMed] [Google Scholar]
- 23.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Quantitative analysis of RNA cleavage during RNA-directed DNA synthesis by human immunodeficiency and avian myeloblastosis virus reverse transcriptases. Nucleic Acids Res. 1994;22:3793–3800. doi: 10.1093/nar/22.18.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman MF, Cai H, Bloom LB, Eritja R. Nucleotide insertion and primer extension at abasic template sites in different sequence contexts. Ann. N.Y. Acad. Sci. 1994;726:132–143. doi: 10.1111/j.1749-6632.1994.tb52804.x. [DOI] [PubMed] [Google Scholar]
- 25.DeStefano JJ, Buiser RG, Mallaber LM, Bambara RA, Fay PJ. Human immunodeficiency virus reverse transcriptase displays a partially processive 3′ to 5′ endonuclease activity. J. Biol. Chem. 1991;266:24295–24301. [PubMed] [Google Scholar]
- 26.Dudding LR, Nkabinde NC, Mizrahi V. Analysis of the RNA- and DNA-dependent DNA polymerase activities of point mutants of HIV-1 reverse transcriptase lacking ribonuclease H activity. Biochemistry. 1991;30:10498–10506. doi: 10.1021/bi00107a019. [DOI] [PubMed] [Google Scholar]
- 27.Fuentes GM, Fay PJ, Bambara RA. Relationship between plus strand DNA synthesis removal of downstream segments of RNA by human immunodeficiency virus, murine leukemia virus and avian myeloblastoma virus reverse transcriptases. Nucleic Acids Res. 1996;24:1719–1726. doi: 10.1093/nar/24.9.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domingo E, Biebricher CK, Eigen M, Holland JJ. Quasispecies and RNA virus evolution: principles and consequences. Georgetown TX: Landes Bioscience; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.