Abstract
Hepatitis C is a major public health concern, with an estimated 170 million people infected worldwide and an urgent need for new drug development. An attractive therapeutic approach is to prevent the ‘cap-independent’ translation initiation of the viral proteins by interfering with both the structure and function of the hepatitis C viral internal ribosomal entry site (HCV IRES). Towards this goal, we report the design, synthesis and purification of novel bi-functional molecules containing DNA or RNA antisenses attached to functional groups performing RNA hydrolysis. These 5′ or 3′-coupled conjugates bind the HCV IRES with affinity and specificity and elicit targeted hydrolysis of the viral genomic RNA after short (1 h) incubation at low (500 nM) concentration at 37°C in vitro. Additional secondary cleavage sites are induced and their mapping within the RNA structure indicates that functional domains IIIb-e are excised from the IRES that, based on cryo-EM studies, becomes incapable of binding the small ribosomal subunit and initiation factor 3 (eIF3). All these molecules inhibit, in a dose-dependent manner, the ‘IRES-dependent’ translation in vitro. The 5′-coupled imidazole conjugate reduces viral protein synthesis by half at a 300 nM concentration (IC50), corresponding to a 4-fold increase of activity when compared to the naked oligonucleotide. These new conjugates are now being tested for activity on infected hepatic cell lines.
INTRODUCTION
The hepatitis C virus (HCV) belongs to the Flaviviridae family, with a positive single-strand RNA genome. An estimated 170 million people are infected worldwide. Most will develop chronic hepatitis, some with severe complications including liver failure or hepatocellular carcinoma (1). Despite clinical improvements allowed by the combination of interferon-alpha and ribavirin, this therapeutic approach fails in about half the patients (2). The HCV virus possesses high genomic plasticity and drug-resistant viruses emerge under pharmacological pressure. Ongoing drug development includes novel protease and RNA polymerase inhibitors, as well as immuno-modulators but there is no vaccine against HCV yet.
Cap-dependent eukaryotic protein synthesis is a sophisticated mechanism requiring the association of numerous initiation factors at the 5′-end of the capped mRNA. Active 80S ribosomes are formed after message scanning by the 40S subunit to find an AUG initiation codon for placing the initiator tRNA (3). In the case of HCV genomic RNA, translation initiation is triggered by a ‘cap-independent’ mechanism involving its 5′ and 3′-untranslated regions (UTRs). Correct positioning of the ribosomes from infected cells onto the internal viral AUG codon requires two initiation factors, eIF2 and eIF3. For the HCV RNA translation, as for prokaryotic translation initiation, there is no scanning but the process needs a structured sequence at the genomic RNA 5′-end called an Internal Ribosome Entry Site [IRES, (3,4)].
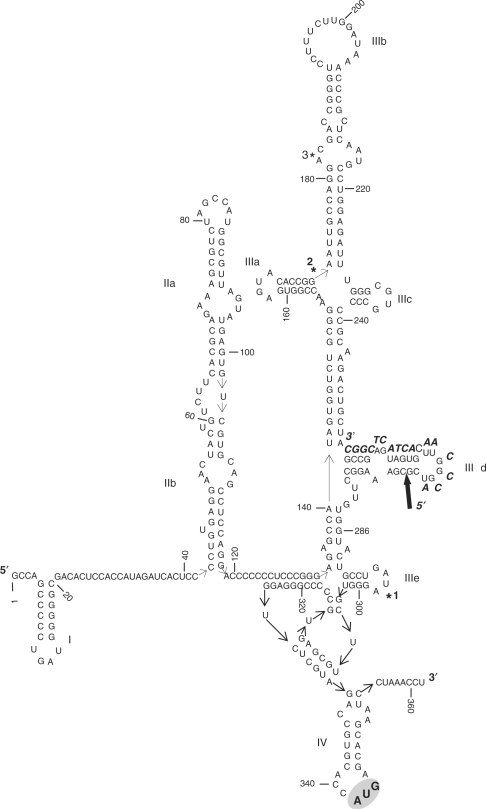
The HCV IRES is a ∼340 nt-long RNA motif encompassing four structural domains (I–IV, Figure 1). Unlike most of the primary sequence of the HCV genome, the IRES nucleotide sequence is highly conserved among all the genotypes (5), suggesting its importance for the virus life cycle (6–8). Its secondary structure was deduced from comparative sequence analysis and structural mapping (9). Domain I is a short hairpin, domain II is a long interrupted stem with internal bulges, domain III contains five stem–loops, denoted ‘a to e’ with a four-way junction, abutting to a pseudoknot. Domain IV is a short stem–loop with the AUG start codon embedded within the loop. Structures of the isolated domains II, IIId and IIIe were determined by NMR spectroscopy and/or X-ray crystallography (10–12). As revealed by ‘footprinting/toeprinting’ experiments (13) and by cryo-electron microscopy (14,15), the HCV IRES forms a binary complex with the 40S ribosomal subunit thanks to direct interactions with domains II and III. Domain II is bended and forms an angle between IIa and IIb (14). It loops away from the 40S, inducing contacts with a region next to the E-site thanks to subdomain IIb. Domain III contacts the platform of the 40S and interacts simultaneously with initiation factor eIF3 (16).
Figure 1.
Secondary structure of the IRES from the HCV genomic RNA deduced from comparative sequence analysis and structural mapping (9). Nucleotides are numbered every 20 nt and the major structural domains (I to IV) are indicated. The predicted binding site of the 17-mer antisense oligonucleotide within the IRES sequence is indicated. The arrow points to the ribose-phosphate bond that is hydrolysed by the 5′-coupled imidazole oligonucleotide conjugate 1. The three numbered stars indicate the position of the secondary cleavage sites triggered by oligonucleotide conjugates 1 and 4 within domains IIIb and IIIe. The AUG translation initiation codon of the viral genomic RNA is grey-boxed.
There are growing interests in developing new drugs targeting HCV viral genome, with some emphasis to the IRES that is indispensable for virus survival. Domains II and III are reasonable candidates to design molecules that interfere with initiation complex assembly, preventing viral protein synthesis. Pathogenic RNAs involved in acute and chronic infectious diseases can be selectively inactivated by various pharmacological compounds (17), including modified nucleic acids. Among those, antisense oligonucleotides, ribozymes, small interfering- or micro-RNAs do receive a particular attention at the present time (18). As recent examples, several RNA or DNA aptamers have been selected against domains II and IIId of the HCV IRES and they decrease viral translation up to 90% in vitro and/or in vivo (19–22). Also, modified oligonucleotides, peptide nucleic acids (PNAs) and siRNAs inactivate domains II or IIId of the HCV IRES in vitro and in vivo (20–28). An antisense oligonucleotide targeting domain II of HCV IRES is currently tested in a phase-one trial (29).
Among the compounds capable of RNA hydrolysis, imidazole moieties are RNase A catalytic centre mimics (30–32). The ‘imidazole-catalyzed’ RNA hydrolysis, however, requires long incubation periods and millimolar concentrations to cleave target RNAs (33,34). We showed previously that polyamine-imidazole conjugates induce non-random, site-selective RNA scission, even with a single imidazole moiety (35). However, polyamines bind RNAs with low specificity and such conjugates become active at millimolar concentrations and require long incubations (10–15 h) with the RNA substrates.
In this report, we describe the design, synthesis, purification and testing of novel nucleic acids conjugates that cleave the HCV IRES genomic RNA at specific locations and decrease, in a dose-dependent manner, the ‘IRES-mediated’ in vitro translation of reporter proteins. The antisense nucleotide sequence was inspired from an RNA aptamer identified by in vitro selection, that binds domain IIId of the IRES RNA from HCV and inhibits IRES-dependent in vitro translation (21). These conjugates contain various RNase A catalytic centre mimics, each attached to either the 5′ or the 3′ end of selected DNA oligonucleotides that can pair specific RNA sequences (domain IIId) from the HCV IRES RNA genome. This novel set of bi-functional RNA-cleaving molecules recognizes and cleaves the targeted RNA sequences with specificity and efficiency. These conjugates are active at low (500 nM) concentrations, require short (<1 h) incubation times and inhibit IRES-dependent translation in vitro in a dose-dependent manner. These novel molecules capable of directed degradation of a functional domain of the HCV genomic RNA are now being tested for antiviral activity and toxicity on infected hepatic cell lines.
MATERIAL AND METHODS
Synthesis of oligonucleotide conjugates
The chemicals were purchased by Sigma–Aldrich (St Louis, USA). Dihydrourocanic acid was obtained by catalytic hydrogenation of urocanic acid in methanol (10% Pd/C) followed by crystallisation in ethanol as previously described (36). All the oligonucleotides were assembled by Eurogentec (Liège, Belgium). The following DNA oligonucleotide was used to target the IIId domain: 5′-ACCCAACACTACTCGGC-3′ (AS), it was either non-modified or bearing a C3-amino group at the 5′- or 3′-end. Its 2′-O-methyl RNA analogue (5′-ACCCAACACUACUCGGC-3′ RNA_AS) was also synthesized with a C3-amino group at the 5′ end. A non-specific DNA oligonucleotide (NS: 5′-CAACCCTAGCCCGTCAA-3′), scrambled from the original sequence, was also synthesized.
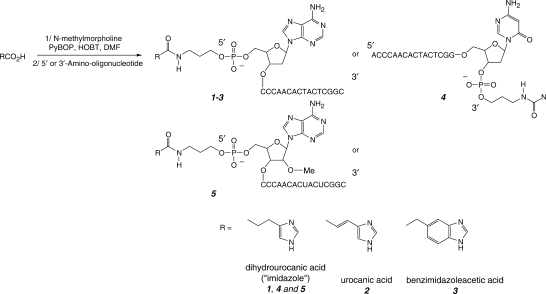
The procedure for coupling the amino-oligonucleotides (Figure 2) was adapted from (37). 30 μmol (1 eq) of acid RCO2H were activated during 150 min under argon with 1.2 eq of PyBOP, 1.5 eq of HOBT and 3 eq of freshly dried N-methylmorpholine in 1 ml of dry DMF. Fifty micro liters of this solution were added to 40 nmol of the amino-oligonucleotide in 150 μl of buffer (133 mM NaHCO3/Na2CO3, pH 9). One hour later, 50 μl of the activated ester were added to the oligonucleotide solution. This addition was repeated four times. The mixture was kept overnight at room temperature. Prior to analysis and purification, the oligonucleotides were precipitated with 0.1 vol of 3 M sodium acetate (pH 5.3) and six volumes of absolute ethanol. The pellets were dissolved in 100 μl of water. DNA concentration was assessed by UV measurement at 260 nm. The coupling protocol was the same for the DNA amino-oligonucleotide and its 2′-O-Me-RNA analogue.
Figure 2.
Chemical procedure used to couple the functional groups to the 5′ or to the 3′-ends of the DNA oligonucleotide. The activated ester was added to the amino-oligonucleotide (DNA or 2′-O-Me-RNA oligonucleotide) to form a peptide bond. The imidazole-conjugated oligonucleotides were obtained from dihydrourocanic acid. The DNA amino-oligonucleotide was coupled either at the 5′ or the 3′-end, they are respectively denoted 5′-imidazole (5′-Im, 1) and 3′-imidazole (3′-Im, 4). The imidazole-coupled 2′-O-Me-RNA oligonucleotide, denoted 5′-imidazole_RNA (5′-Im-RNA, 5), was coupled at the 5′-end. Urocanic acid and benzimidazoleacetic acid functional groups were also coupled at the 5′-end of the DNA oligonucleotide. The corresponding oligonucleotides are respectively denoted 5′-UA (2) and 5′-BI (3).
The oligonucleotide conjugates were analysed and purified by reverse phase HPLC on Uptisphere ODB C18-5μ columns (Interchim, France) using an ISS200 LC system (Perkin Elmer, USA). The flow rate was 1 ml/min for analytical purpose (250 × 4.6 column) and 3 ml/min. for quantitative purification (250 × 10 column). The following sequence was used: after 5 min elution with 25 mM TEAA (pH 7.0), a linear gradient from 0 to 15% of acetonitrile in 25 mM TEAA was applied for 15 min. It was followed by elution with 15% acetonitrile in TEAA for 10 min. Retention times of the antisense oligonucleotides were, respectively, 20.2 min for the amino-oligonucleotides (5′ or 3′) and 20.5 min for the corresponding conjugate. No significant difference could be observed between the retention times of the various imidazole-coupled oligonucleotides (Figure 2). For the 2′-O-Me-RNA antisense, the following sequence was used: after 5 min elution with 25 mM TEAA (pH 7.0), a linear gradient from 0 to 30% of acetonitrile in 25 mM TEAA was applied for 30 min. It was followed by elution with 30% acetonitrile in TEAA for 10 min. Retention times of the antisense RNA oligonucleotides were respectively 27.5 min for the amino-2′-O-Me-RNA oligonucleotide and 27.8 min for the corresponding conjugate (5). Monitoring was performed at 260 nm. After HPLC purification and precipitation, the final yield of conjugate formation is close to 50%. The purified oligonucleotide conjugates were analysed by MALDI-TOF mass spectrometry using an Ultra Flex Mass Spectrometer (Bruker Daltonics, Germany). For each molecule synthesized, the measured mass was in accordance to the theoretically calculated value.
Plasmids preparation
Bicistronic pIRF HCV (5) and pIRF ECMV (EnCephaloMyocarditis Virus) were both digested by BamH1 and Kpn1 restriction enzymes to extract the Firefly luciferase coding sequence. After ligation of the 5′ and 3′ ends by the Klenow fragment of DNA Polymerase 1 from Escherichia coli, we obtained the monocistronic plasmids pIRF_HCV-RLuc and pIRF_ECMV-RLuc coding for the reporter gene of Renilla luciferase under the control of each of the IRES sequences. The integrity of the coding genes sequences was verified by sequencing the plasmid.
In vitro transcription
HCV_IRES-RLuc and ECMV_IRES-RLuc DNA templates used for the in vitro translation assays were produced after the linearization of plasmids pIRF by BstX1. The HCV_IRES DNA template was produced from the pUC19 plasmid linearized by BamH1. All these DNA sequences were purified by phenol/ether extraction and ethanol precipitated. RNAs were synthesized for 4 h using the T7 Megascript Kit (Ambion, USA) and purified with MegaClear columns (Ambion, USA).
RNAs and oligonucleotides labelling
The HCV IRES RNAs were labelled at their 3′-ends using T4 RNA ligase (New England Biolabs, USA) and Cytidine 3′,5′-Bis32Phosphate (32pCp). The radiolabelled RNAs were loaded onto a 6% polyacrylamide/bisacrylamide (19:1) 8 M urea denaturing gel in TBE buffer [90 mM Tris–borate, 2 mM EDTA (pH 8) at 20°C]. The electrophoresis was performed at 30 V/cm for 3 h. The band corresponding to the intact RNA was excised and eluted for 8 h at 37°C in 20 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA, 1% SDS. The RNA was ethanol precipitated and the pellets were dried and aliquoted at –20°C. The RNA was stored in 10 mM HEPES (pH 7.5) prior to use. Both the naked oligonucleotides and the 3′-coupled oligonucleotides were labelled at their 5′ ends using T4 Polynucleotide kinase (New England Biolabs, USA) and [γ-32P] ATP. The 5′-coupled oligonucleotides were labelled at their 3′-ends using Terminal Deoxynucleotidyl-Transferase (Promega, France) and [α-32P]-UTP.
IRES refolding
Before all the experiments, the IRES RNAs were heated for 2 min at 80°C in folding buffer (10 mM HEPES pH 7.5, 20 mM NH4Cl and 3 mM MgCl2) and slowly cooled down at room temperature for 30 min.
Native gel retardation assays
One picomole of each of the labeled antisense oligonucleotides was incubated for 15 min at 37°C in 10 μl of 10 mM spermidine, 80 mM HEPES (pH 7.5), 200 mM NH4Cl, 3 mM MgCl2 with increasing concentrations of refolded IRES RNAs (from 10 to 2000 nM). Samples were loaded onto a 5% acrylamide/bisacrylamide (37.5:1) gel in TBM buffer [45 mM Tris–borate (pH 8.3), 0.2 mM MgCl2] at 300 V (8 V/cm, 3 W) for 2 h 30 min. The percentage of oligonucleotides bound to the IRES RNAs was determined as the ratio of radioactivity measured in the RNA/oligonucleotide complex to the total amount of radioactivity measured in the lane. Bands corresponding to the RNA–conjugate duplexes and to the unbound oligonucleotide were quantified using ‘ImageJ’ (38).
IRES cleavage reactions
Refolded 3′-end labelled IRES transcript measuring 100 000 c.p.m. (∼0.1 pmol) were incubated for 1 h at 37°C with increasing concentrations (from 0.5 to 50 μM) of either naked or conjugated oligonucleotides in 10 μL of cleavage buffer (1.5 mM MgCl2, 150 mM NaCl, 50 mM HEPES pH 7.5, 10 μM ZnSO4). Reactions were quenched by RNA precipitation in 20 μL of 0.3 M sodium acetate (pH 5.3) and 400 μL of 2% lithium perchlorate (diluted in acetone). The pellets were dissolved in loading buffer (80% formamide, 10 mM EDTA pH 8.0, 0.1% xylene cyanol, 0.1% bromophenol blue) and the samples loaded onto denaturing gels (7% acrylamide/bisacrylamide (19:1), 8 M urea, TBE 1×). Both the digestions with RNase T1 (Amersham, USA), RNase U2 (Pierce Nucleic Acids Technologies, USA) and the alkaline hydrolysis were performed in parallel in order to map the cleavage sites.
RNase H cleavage assays
Radiolabelled IRES RNA 100 000 c.p.m. (∼0.1 pmol) was annealed with 50 pmol of antisense oligonucleotide. RNase H cleavage assays of the labelled oligonucleotides/IRES complexes were then performed by incubating 1 U of RNase H (USB Corporation, USA) for 45 min at 37°C in 10 μl of a solution containing 1 mM DTT, 10 mM MgCl2, 160 mM KCl, 64 mM H (pH 7.5) and 8 mM spermidine. The samples were loaded onto a 7% polyacrylamide denaturing gel. Both the concentration and the incubation time of the oligonucleotide conjugates were selected, on purpose, sufficiently low so that they do not trigger RNA hydrolysis.
IRES translation assays in the presence of the conjugates
In vitro translation assays were performed using cell-free translation assay [Rabbit Reticulocyte Lysates, Nuclease-treated (Promega, France)]. Reactions were performed by adding first each oligonucleotide conjugate (from 200 nM to 5 μM final concentration) to 0.2 μg of Renilla Luciferase-driven IRES RNA. Reticulocyte lysate was then added straight ahead to start the reaction. The reaction proceeds in 10 μl according to the manufacturer's recommendations, in the presence of 0.3 mM MgCl2, 0.1 mM KCl. The naked oligonucleotide with free imidazole (1:1 ratio) was used as a control. After 45 min at 30°C, the translation levels were evaluated by Renilla luciferase assays (Promega, France) using a microplate luminometer (Centro XS3 LB960, Berthold, Germany). The effect of the ribonucleases was evaluated by comparison of the Renilla luciferase activity in the absence and presence of the conjugates. All the experiments were performed in triplicates.
RESULTS
Influence of the cleaving group on the oligonucleotide binding to the HCV IRES RNA
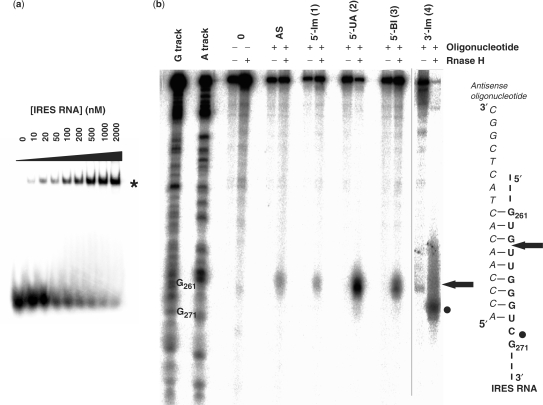
The binding affinities of the four labelled conjugates for increasing amounts of the complete IRES from HCV were assayed by native gel retardation assays and compared to that of the naked oligonucleotide sequence (Figure 3a). A retarded band (Figure 3a, star), not detected in the control lane in the absence of the RNA, corresponds to the RNA–conjugate complex. Two additional negative controls were performed in the same conditions. First, we incubated the HCV IRES with a non-specific oligonucleotide (NS). This mismatched sequence does not bind the IRES RNA, up to the higher concentration used for the antisense oligonucleotide (data not shown). Then, the ECMV IRES was incubated with the antisense (AS). No retarded bands were detected (data not shown). The apparent dissociation constants (Kd) of the HCV IRES-oligonucleotide conjugates complex were calculated. The uncoupled antisense oligonucleotide has a ∼70 nM apparent Kd. Adding any of the three imidazole groups to the 3′ or 5′ end of the antisense oligonucleotide does not modify significantly this binding affinity (data not shown).
Figure 3.
All the DNA conjugates bind to the HCV IRES at the predicted location within the RNA sequence. (a) Representative native gel retardation assay between the radiolabelled 3′-imidazole conjugate and increasing concentrations (from 0 to 2000 nM) of purified unlabelled IRES RNA. The slower migrating band corresponding to the IRES-conjugate duplex is indicated by a star. Apparent dissociation constants between each conjugate and the IRES were calculated from three independent experiments. (b) RNase H cleavage assays between each of the unlabelled conjugates and labelled IRES RNA. Partial RNA sequencing using RNases T1 and U2 under denaturing conditions demonstrates that the complex between each of the antisense oligonucleotides and the IRES takes place at the predicted location (indicated by the arrow and the black circle) deduced by nucleotide pairing, at hairpin IIId.
According to its sequence, the antisense oligonucleotide should bind to nucleotides 253 to 269 from domain IIId of HCV IRES (Figure 1). RNase H experiments were carried out to verify that the naked antisense oligonucleotide as well as the four antisense conjugates bind at the expected location, and not at additional ectopic sites within the HCV IRES sequence. For all the five antisense derivatives, RNase H-mediated cleavages are observed at or around the expected location of pairing onto the IRES RNA (Figure 3b, arrow and circles). Interestingly, the 3′-coupled imidazole conjugate 4 induces a cleavage of the IRES next to the pairing area, whereas the other three molecules trigger a cut within the pairing, which is stronger for the 5′-coupled urocanic acid 2 (Figure 3b). Altogether, these results confirm the specificity of the antisense conjugates for their predicted binding site. The 5′ or the 3′ coupling of various chemicals do not affect or reduce binding specificity.
Targeted degradation of the HCV IRES by the DNA conjugates
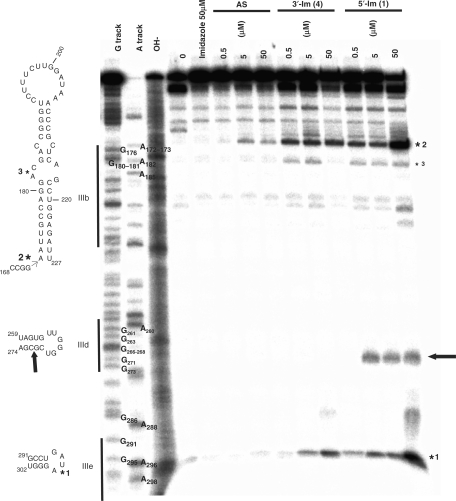
We designed and performed additional experiments, in the absence of RNase H, in which the refolded IRES RNA (Figure 1) is incubated from 5 to 60 min with increasing concentrations (from 0.5 to 50 μM) of either the naked DNA antisense AS or the oligonucleotides conjugates 1 or 4. As negative controls, the naked antisense and the free, uncoupled imidazole were used at concentrations and incubation times identical to those used for the conjugates (Material and Methods section). Alkaline hydrolysis and RNase T1 and U2 digestions of the RNA allow the positioning of the cleavage sites. One-hour incubation at the lower concentration (0.5 μM) is required to observe a significant cleavage at the expected location (Figure 4, arrow) only for the 5′-coupled imidazole conjugate 1. Three strong additional cleavages are observed for conjugates 1 and 4 (stars in Figure 4). With the naked oligonucleotide AS, these cleavages are very weak but can also be detected.
Figure 4.
In vitro cleavage assays between the conjugates and the IRES from HCV. Purified 3′-labelled IRES is incubated with either 50 μM free imidazole or increasing concentrations of the naked or the 3′/5′ imidazole-coupled antisense oligonucleotides for one hour at 37°C. Partial RNA sequencing using RNases T1 and U2 under denaturing conditions as well as partial alkaline RNA hydrolysis allows the mapping of the cleavage sites within the IRES RNA sequence. The arrow points to the cleavage site, between G271 and C272, induced by conjugate 1. The stars point to three additional cleavage sites induced by conjugates 1 and 4, mapped between U297 and A298 in domain IIIe (*1), between G171 and A172 between domains IIIa and IIIb (*2) and between G181 and A182 in domain IIIb (*3). Note that the two highest concentrations of the naked oligonucleotide elicit a weak cleavage between G171 and A172 (*2).
The cut triggered by the 5′-coupled imidazole conjugate 1 occurs in stem IIId, between G271 and C272. These two nucleotides are at the second and third positions after the last nucleotide hybridized to the antisense sequence. Despite a similar apparent binding constant for the IRES, the 3′-coupled imidazole conjugate 4 does not cleave at the location predicted from base-pairing, even at the highest concentration.
The three additional cleavages (Figure 4, stars), located away from the binding sites of the complementary sequence of the antisense within the IRES sequence, are observed for all the conjugates, but not with free imidazole. Of these three, two are heavy cuts, one in domain IIIe between U297 and A298 (Figures 1 and 4, *1) and the other is between domains IIIa and IIIb, between G171 and A172 (Figure 4, *2). The third weaker cut is situated in domain IIIb between G181 and A182 (Figure 4, *3). Strikingly, whereas the 3′-coupled imidazole conjugate 4 does not induce a cleavage around its primary recognition site, the binding to its target sequence within the IRES structure is sufficient to induce the three secondary cuts (Figure 4). This suggests that binding of the conjugates is sufficient to modify the IRES structure on a large scale that, in turn, reveals novel degradation sites at specific locations.
Next, the influence of various conjugates on both the site of cleavage and the efficiency of the RNA hydrolysis was addressed. 5′-coupling of either an urocanic acid or a benzimidazoleacetic acid tests the influence of both the pKa (benzimidazoleacetic acid, urocanic acid and dihydrourocanic acid groups have pKas of 4.6, 5.3 and 6.95, respectively) and the flexibility (benzimidazoleacetic acid is the more rigid of the three because of a phenyl group, urocanic acid is intermediate with one double bond and dihydrourocanic acid is the more flexible, Figure 2). Neither the benzimidazoleacetic acid 3 nor the urocanic acid 2 conjugates elicit cleavage at the location predicted from base-pairing, probably because of their lower pKas and flexibilities compared to the dihydrourocanic acid conjugate 1 (data not shown). Very weak secondary cuts *1 and *2 are induced by conjugates 2 and 3 (data not shown).
Inhibition of HCV IRES-dependent translation by the oligonucleotide conjugates
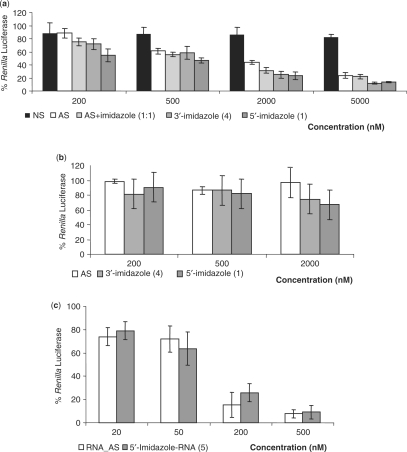
Translation assays were performed using a Renilla luciferase reporter gene under the control of the HCV IRES (Figures 5a and c) or the ECMV IRES (Figure 5b). The IRES-Renilla luciferase RNA (IRES-RLuc) transcripts were first incubated with increasing concentrations of the conjugates and then with the reticulocyte lysate containing the ribosomes (see Material and Methods section). The translation reactions were performed for 45 min, a time at which no RNA degradation could be observed in the absence of the antisense oligonucleotides (data not shown). The Renilla luciferase protein levels were measured in the presence of the conjugates and compared to those obtained in the absence of the putative inhibitors. Direct comparisons between the conjugates were inferred from the determination of the concentration of the drug required to inhibit reporter gene translation by half (IC50). As a control, the effect of a mismatched oligonucleotide (NS) was tested and it doesn't inhibit the translation of the HCV IRES RNA (Figure 5a).
Figure 5.
Dose-dependent inhibition of viral protein synthesis by the DNA conjugates in vitro. The translation assays were performed at 30°C for 45 min. The Renilla luciferase protein levels measured in the presence of each conjugates were all divided by the protein level obtained in the absence of translation inhibitors. (a) Concentration-dependent viral translation inhibition of the HCV-IRES RNA by the non-specific oligonucleotide NS, the naked DNA antisense AS, the ‘naked-free imidazole’ in a one-to-one ratio or the 3′- or the 5′-imidazole-coupled oligonucleotides 4 and 1. (b) translation inhibition of the ECMV-IRES RNA by the naked (AS), the 3′- or the 5′-imidazole-coupled oligonucleotides 4 and 1. (c) Concentration-dependent translation inhibition of the HCV-IRES RNA by the naked 2′-O-Me RNA antisense RNA_AS or the 5′-imidazole-coupled 2′-O-Me-RNA oligonucleotides 5.
Long incubations of the IRES from HCV in a molar imidazole buffer do not induce any RNA hydrolysis or any detectable translation inhibition (not shown). Naked oligonucleotide AS, from a 200 nM to a 5 μM concentration, reduces the rate of translation from 90 to 25%, respectively, with a 1200 ± 200 nM IC50. This suggests that the uncoupled oligonucleotide already has substantial inhibitory activity (Figure 5). Adding exogenous free imidazole in a one-to-one ratio to the naked oligonucleotide reduces the IC50 down to 900 ± 200 nM.
Next, the ability of various concentrations of the oligonucleotide conjugates to inhibit reporter gene translation was measured. The calculated IC50 of all the oligonucleotides are listed in Table 1. Binding of each of the conjugates to the IRES domain of this RNA construct, followed by subsequent RNA hydrolysis, reduces the expression of the reporter protein in a dose-dependent manner (Figure 5a). Compared to the naked oligonucleotide, the presence of a 3′-coupled imidazole slightly improves translation inhibition as when free imidazole is added in the reaction buffer with the naked antisense. Interestingly, the 5′-coupled imidazole conjugate 1 has 3-fold higher inhibition activity of the IRES-driven reporter gene translation than the 3′-coupled imidazole 4 (Figure 5). The result is in agreement with the fact that only the 5′-conjugate 1 cleaves the IRES sequence near its binding site in vitro and induces the strongest specific degradation of the target (Figure 4). The 5′-coupled imidazole conjugate 1 is the most powerful inhibitor of HCV IRES-dependent translation, decreasing the IC50 to 300 ± 100 nM, a 4-fold difference compared to the naked oligonucleotide AS. When modifying the cleaving group, neither the urocanic acid conjugate 2 nor the benzimidazoleacetic acid conjugate 3 improved the IC50 of the 5′-coupled imidazole 1 (Figure 5). The 5′-coupled benzimidazoleacetic acid conjugate 3 is nonetheless an interesting conjugate, lowering the IC50 3-fold compared to the naked oligonucleotide AS. To confirm the specificity of translation inhibition of the conjugates on the IRES from HCV, similar experiments were carried out on the IRES from ECMV by using all the conjugates and the naked oligonucleotide (Figure 5b). Within the same concentration range, no significant inhibitory effect on translation could be detected.
Table 1.
Concentration at which translation is reduced by half (IC50)
| Oligonucleotide | IC50 (μM) |
| AS | 1.2 ± 0.2 |
| Imidazole + AS (1:1) | 0.9 ± 0.2 |
| 5′-Imidazole (1) | 0.3 ± 0.1 |
| 5′-UA (2) | 0.8 ± 0.2 |
| 5′-BI (3) | 0.4 ± 0.1 |
| 3′-Imidazole (4) | 0.9 ± 0.2 |
| RNA_AS | 0.10 ± 0.02 |
| 5′-Imidazole RNA (5) | 0.09 ± 0.03 |
The IC50 has been calculated for either the naked antisense or the conjugates the DNA-conjugates 1–4 and for the 2′-O-Me-RNA oligonucleotides, naked RNA_AS or conjugate 5. These data were derived from at least three independent experiments. Concerning the inhibition of the ECMV-IRES RNA by the DNA conjugates, the IC50 could not be determined.
Due to the results obtained with the 5′-imidazole coupled DNA antisense oligonucleotide 1, we coupled an imidazole moiety at the 5′-end of the 2′-O-Me-RNA antisense. Despite the significant decrease of the IC50 that could be observed when adding an imidazole at the 5′-end of the DNA antisense, no decrease of the IC50 could be observed when adding an imidazole at the 5′-end of the antisense RNA.
DISCUSSION
In this report, novel oligonucleotide conjugates containing imidazole groups were designed to target and cleave the IIId domain of the HCV Internal Ribosome Entry Site (IRES) with specificity, affinity and efficiency. The antisense oligonucleotide sequence is complementary to nucleotides 253 to 269 from hairpin IIId of the IRES from HCV; it binds to the HCV IRES at its expected location and inhibits cap-independent IRES-mediated translation (21). This oligonucleotide sequence is specific of the HCV-IRES RNA and, as a negative control, it could not bind or inhibit the ECMV IRES.
Oligomers that pair with domain IIId of the IRES from HCV have been selected by various methods and all are potent inhibitors of viral translation in vitro (20,21,28,39). Interestingly, all contain a 5′ACCCAA3′sequence that can pair with the 5′264UUGGGU2693′ sequence from loop IIId, probably important to initiate pairing between the oligonucleotide and its RNA target. To improve the ability of the oligonucleotide to inhibit IRES-mediated translation, imidazole derivatives were coupled first to the DNA sequence, then to its 2′-O-Me-RNA analogue, reasoning that if specific cleavages of the IRES RNA can be triggered at the vicinity of the antisense binding site, it may increase further the inhibition of translation initiation.
The RNA-cleaving group could be coupled at either the 5′ or the 3′ end of the oligonucleotide sequence. The 5′-imidazole conjugate 1 induces cleavage of the IRES between nts G271 and C272, two nucleotides away from the last 5′-paired nt (Figure 4) and decreases translation 4-fold compared to the naked oligonucleotide (the IC50 goes from 1200 ± 200 nM for the naked oligonucleotide AS to 300 ± 100 nM for the conjugate 1 (Table 1). The conjugate 1 has the highest efficiency to cleave the IRES RNA in vitro and is also the most potent inhibitor of viral translation. The IC50 of the naked antisense into an imidazole buffer is three-times higher than that of the 5′-imidazole conjugate 1, suggesting that the higher activity of the latter comes from the higher local concentration of the cleaving group in the vicinity of the RNA backbone. The 3′-imidazole conjugate 4 is not able to cleave around its binding site, even at a 50 μM concentration (Figure 4). Both the 5′ and the 3′ conjugates place their RNA-cleaving groups next to paired regions, C270–A274 for the 5′-conjugate and A244–A252 for the 3′-conjugate, that are less favourable for RNA hydrolysis than RNA single-strands. The short (5 bp) stem near the imidazole moiety of 5′-conjugate probably allows sufficient flexibility of the ribose-phosphate chain for imidazole-induced RNA hydrolysis, whereas the nine base-paired helix next to the imidazole moiety of the 3′-conjugate is probably too rigid to allow acid-base catalysis to occur (Figure 1). The cleavage of the phosphodiester backbone of an RNA by an imidazole moiety generally involves two imidazoles, one acting as a base and the other one as an acid. However, the 5′-coupled imidazole oligonucleotide 1 that specifically cleaves the HCV-IRES RNA bears only one imidazole. One explanation could involve the divalent metal ions contained in the cleavage buffer. Indeed, divalent metal cations such as Mg2+ and Zn2+ can act as a Lewis acid (40), thus contributing to the cleavage mechanism of the ribonucleic chain by a single imidazole. In a second time, the 5′-imidazole cleaving group was replaced by a benzimidazoleacetic acid 3, or by an urocanic acid 2. These two functional groups are more rigid and have lower pKas than an imidazole. None of these two conjugates induce specific cleavage of the HCV-IRES RNA at the vicinity of its binding site within the IRES. Therefore, we can conclude that lowering both the pKas and the flexibility of the cleaving group inhibit RNA hydrolysis. The best results were obtained with an imidazole coupled at the 5′-end of the AS sequence. Note that the use of a 2′-O-Me RNA antisense sequence increases the overall inhibitory effect of the naked compound, but is not further improved by the addition of an imidazole. The affinity of the 2′-O-Me RNA antisense for the IRES RNA is about 2 nM (21). It is 35-fold higher than the affinity of the DNA antisense AS (apparent Kd of ∼70 nM). The high affinity of the antisense RNA for the IRES makes an important contribution for translation inhibition. The lack of improvement that we observe when we couple an imidazole at the 5′-end of a 2′-O-Me RNA antisense is probably due to this increased affinity that takes over the influence of the imidazole.
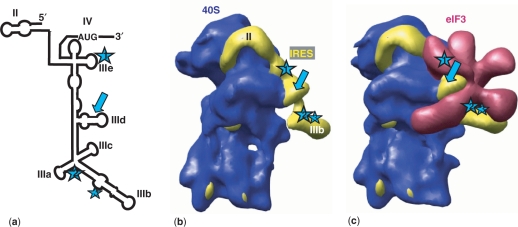
Interestingly, additional cleavages, away from the oligonucleotide recognition sequence and also distant one another onto the IRES secondary structure (Figure 1, stars), were consistently observed. These cleavages can be weakly observed with the naked antisense oligonucleotide AS. They are significantly increased with both the 5′- and the 3′-imidazole conjugates (Figure 4). These three sites were mapped by partial RNA sequencing between U297 and A298 in domain IIIe (1*) between G171 and A172 (2*) between domains IIIa and IIIb and between G181 and A182 (3*) in domain IIIb. These additional cuts remove the essential domains IIIb to IIIe from the IRES (Figure 1), most likely affecting the recognition of the IRES by eIF3 (Figure 6c) and possibly also its interaction with the 40S subunit (Figure 6b). These data are a reasonable explanation as to why a dose-dependent IRES-mediated translation inhibition is detected in vitro. A cryo-EM structural model of the IRES locked on a translation initiation state in complex with eIF3 and the 40S subunit (13) shows that the additional cleavage sites are far away when looking at the primary sequence but are clustered around the nucleotide recognition sequence on the IRES three-dimensional structure derived from cryo-EM (Figure 6c, blue stars). In the context of the naked IRES tertiary structure, those three cleavages may even be closer one another. Upon binding of the 5′ or the 3′ complementary oligonucleotide, the IRES structure may adopt alternate conformation(s) that, in turn, become(s) hydrolysed at additional secondary sites. This conformational change is specific since only three additional cleavages sites are observed. The respective contribution of the RNA hydrolysis versus the trigger of an inactive conformation of the IRES in the overall translation inhibition observed in vitro is unknown. Both the sequence and structure of domain III from the IRES are essential for the binding of eIF3 (3,16,41,42) and of ribosomal protein S9 (43). Interestingly, this is particularly true for domains IIIb and IIId (44), which are both cleaved by conjugate 1.
Figure 6.
Location of the cleavages induced by the DNA conjugates onto the HCV IRES secondary and tertiary structures in complex with the 40S subunit and eIF3 derived from cryo-EM data (16). (a) Schematic secondary structure of domains II to IV from the HCV IRES with the location of the cleavage sites (arrow and numbered stars). The four cleavage sites are visualized in the context of the binary and the ternary complexes containing ‘HCV IRES-40S subunit’ (b) or ‘HCV IRES-40S subunit-eIF3’ (c), respectively. Note that the cleavages of the IRES triggered by the conjugates remove an RNA segment (IIIb-e) essential for the recognition by eIF3.
The interaction of domain IIId with the 40S ribosomal subunit and eIF3 makes this RNA module an attracting target for blocking translation initiation of the viral proteins. We believe that this work is original and promising because the functional effects on IRES-dependent translation of each of the molecules synthesized could be analysed according to their potential for IRES RNA hydrolysis. These novel conjugates are active at low concentrations, require short (<1 h) incubation times and inhibit IRES-dependent translation in vitro in a dose-dependent manner. These novel molecules capable of directed degradation of a functional domain of HCV genomic RNA are now being tested for antiviral activity and toxicity in in vitro-infected cells.
ACKNOWLEDGEMENTS
This work was supported by grants from the ‘Région Bretagne’ (PRIR Grant n°691 and CRB 2004-1483), ACI BCMS 136 and ANR programme MIME. V.G. is supported by a post-doctoral fellowship from the ANRS (‘Agence Nationale de Recherche sur le Sida et les hépatites virales’). We are grateful to Dr Jacques Le Seyec for insightful comments on this work, to Dr Annie Cahour for providing the pIRF plasmid and to Dr C. Fraser from the Doudna lab for helping us in preparing Figure 6.
Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 2.Pearlman BL. Hepatitis C treatment update. Am. J. Med. 2004;117:344–352. doi: 10.1016/j.amjmed.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 4.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 5.Laporte J, Malet I, Andrieu T, Thibault V, Toulme JJ, Wychowski C, Pawlotsky JM, Huraux JM, Agut H, et al. Comparative analysis of translation efficiencies of hepatitis C virus 5′ untranslated regions among intraindividual quasispecies present in chronic infection: opposite behaviors depending on cell type. J. Virol. 2000;74:10827–10833. doi: 10.1128/jvi.74.22.10827-10833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukh J, Purcell R, Miller R. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc. Natl Acad. Sci. USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buratti E, Gerotto M, Pontisso P, Alberti A, Tisminetzky SG, Baralle FE. In vivo translational efficiency of different hepatitis C virus 5′-UTRs. FEBS Lett. 1997;411:275–280. doi: 10.1016/s0014-5793(97)00715-1. [DOI] [PubMed] [Google Scholar]
- 8.Honda M, Rijnbrand R, Abell G, Kim D, Lemon SM. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J. Virol. 1999;73:4941–4951. doi: 10.1128/jvi.73.6.4941-4951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 11.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 12.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the cystal and in solution. Angew. Chem. Int. Ed. Engl. 2006;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 13.Kolupaeva VG, Pestova TV, Hellen CUT. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the Internal Ribosomal Entry Site of Hepatitis C Virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 15.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure (Camb.) 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 17.Gallego J, Varani G. Targeting RNA with small-molecule drugs: Therapeutic promise and chemical challenges. Acc. Chem. Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 18.De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- 19.Martinand-Mari C, Lebleu B, Robbins I. Oligonucleotide-based strategies to inhibit human hepatitis C virus. Oligonucleotides. 2003;13:539–548. doi: 10.1089/154545703322860834. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi K, Umehara T, Fukuda K, Kuno A, Hasegawa T, Nishikawa S. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 2005;33:683–692. doi: 10.1093/nar/gki215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tallet-Lopez B, Aldaz-Carroll L, Chabas S, Dausse E, Staedel C, Toulme JJ. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 2003;31:734–742. doi: 10.1093/nar/gkg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi K, Umehara T, Fukuda K, Hwang J, Kuno A, Hasegawa T, Nishikawa S. RNA aptamers targeted to domain II of Hepatitis C Virus IRES that bind to its apical loop region. J. Biochem. (Tokyo) 2003;133:263–270. doi: 10.1093/jb/mvg036. [DOI] [PubMed] [Google Scholar]
- 23.Da Rocha Gomes S, Dausse E, Toulme J.-J. Determinants of apical loop-internal loop RNA-RNA interactions involving the HCV IRES. Biochem. Biophys. Res. Commun. 2004;322:820–826. doi: 10.1016/j.bbrc.2004.07.185. [DOI] [PubMed] [Google Scholar]
- 24.Michel T, Martinand-Mari C, Debart F, Lebleu B, Robbins I, Vasseur JJ. Cationic phosphoramidate alpha-oligonucleotides efficiently target single-stranded DNA and RNA and inhibit hepatitis C virus IRES-mediated translation. Nucleic Acids Res. 2003;31:5282–5290. doi: 10.1093/nar/gkg733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Hanecak R, Brown-Driver V, Azad R, Conklin B, Fox MC, Anderson KP. Antisense oligonucleotide inhibition of hepatitis C Virus (HCV) gene expression in livers of mice infected with an HCV-vaccinia virus recombinant. Antimicrob. Agents Chemother. 1999;43:347–353. doi: 10.1128/aac.43.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabhu R, Garry RF, Dash S. Small interfering RNA targeted to stem-loop II of the 5′ untranslated region effectively inhibits expression of six HCV genotypes. Virol. J. 2006;3:100. doi: 10.1186/1743-422X-3-100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Nulf CJ, Corey D. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) Nucleic Acids Res. 2004;32:3792–3798. doi: 10.1093/nar/gkh706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanecak R, Brown-Driver V, Fox M, Azad R, Furusako S, Nozaki C, Ford C, Sasmor H, Anderson K. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J. Virol. 1996;70:5203–5212. doi: 10.1128/jvi.70.8.5203-5212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHutchison JG, Patel K, Pockros P, Nyberg L, Pianko S, Yu RZ, Dorr FA, Kwoh TJ. A phase I trial of an antisense inhibitor of hepatitis C virus (ISIS 14803), administered to chronic hepatitis C patients. J. Hepatol. 2006;44:88–96. doi: 10.1016/j.jhep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Niittymaki T, Lonnberg H. Artificial ribonucleases. Org. Biomol. Chem. 2006;4:15–25. doi: 10.1039/b509022a. [DOI] [PubMed] [Google Scholar]
- 31.Trawick BN, Daniher AT, Bashkin JK. Inorganic mimics of ribonucleases and ribozymes: from random cleavage to sequence-specific chemistry to catalytic antisense drugs. Chem. Rev. 1998;98:939–960. doi: 10.1021/cr960422k. [DOI] [PubMed] [Google Scholar]
- 32.Konevetz DA, Beck IE, Beloglazova NG, Sulimenkov IV, Sil’nikov VN, Zenkova MA, Shishkin GV, Vlassov VV. Artificial ribonucleases: synthesis and RNA cleaving properties of cationic conjugates bearing imidazole residues. Tetrahedron. 1999;55:503–512. [Google Scholar]
- 33.Ushijima K, Gouzu H, Hosono K, Shirakawa M, Kagosima K, Takai K, Takaku H. Site-specific cleavage of tRNA by imidazole and/or primary amine groups bound at the 5′-end of oligodeoxyribonucleotides. Biochim. Biophys. Acta. 1998;1379:217–223. doi: 10.1016/s0304-4165(97)00101-3. [DOI] [PubMed] [Google Scholar]
- 34.Beloglazova NG, Fabani MM, Zenkova MA, Bichenkova EV, Polushin NN, Sil’nikov VV, Douglas KT, Vlassov VV. Sequence-specific artificial ribonucleases. I. Bis-imidazole-containing oligonucleotide conjugates prepared using precursor-based strategy. Nucleic Acids Res. 2004;32:3887–3897. doi: 10.1093/nar/gkh702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fouace S, Gaudin C, Picard S, Corvaisier S, Renault J, Carboni B, Felden B. Polyamine derivatives as selective RNaseA mimics. Nucleic Acids Res. 2004;32:151–157. doi: 10.1093/nar/gkh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baures PW, Kaliyan K, Desper J. Nα-Urocanylhistamine: A natural histamine derivative. Molecules. 2002;7:813–816. [Google Scholar]
- 37.Mestre B, Pratviel G, Meunier B. Preparation and nuclease activity of hybrid “metallotris(methylpyridinium)porphyrin oligonucleotide” molecules having a 3′-loop for protection against 3′-exonucleases. Bioconjug. Chem. 1995;6:466–472. doi: 10.1021/bc00034a017. [DOI] [PubMed] [Google Scholar]
- 38.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 39.Romero-Lopez C, Barroso-del Jesus A, Puerta-Fernandez E, Berzal-Herranz A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005;386:183–190. doi: 10.1515/BC.2005.023. [DOI] [PubMed] [Google Scholar]
- 40.Santoro SW, Joyce GF. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- 41.Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5′UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odreman-Macchioli FE, Tisminetzky SG, Zotti M, Baralle FE, Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875–885. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Mol. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]