Abstract
The FemXWv aminoacyl transferase of Weissella viridescens initiates the synthesis of the side chain of peptidoglycan precursors by transferring l-Ala from Ala-tRNAAla to UDP-MurNAc-pentadepsipeptide. FemXWv is an attractive target for the development of novel antibiotics, since the side chain is essential for the last cross-linking step of peptidoglycan synthesis. Here, we show that FemXWv is highly specific for incorporation of l-Ala in vivo based on extensive analysis of the structure of peptidoglycan. Comparison of various natural and in vitro-transcribed tRNAs indicated that the specificity of FemXWv depends mainly upon the sequence of the tRNA although additional specificity determinants may include post-transcriptional modifications and recognition of the esterified amino acid. Site-directed mutagenesis identified cytosines in the G1–C72 and G2–C71 base pairs of the acceptor stem as critical for FemXWv activity in agreement with modeling of tRNAAla in the catalytic cavity of the enzyme. In contrast, semi-synthesis of Ala-tRNAAla harboring nucleotide substitutions in the G3–U70 wobble base pair showed that this main identity determinant of alanyl-tRNA synthetase is non-essential for FemXWv. The different modes of recognition of the acceptor stem indicate that specific inhibition of FemXWv could be achieved by targeting the distal portion of tRNAAla for the design of substrate analogues.
INTRODUCTION
Bacterial resistance to antibiotics will lead to therapeutic deadlocks unless new drugs are discovered. Only two families of antibiotics act on peptidoglycan polymerization, the β-lactams and the glycopeptides. Development of new generations of molecules derived from these antibiotics is not sufficient to defeat the sophisticated resistance mechanisms responsible for the gradual loss of activity against main human pathogens, such as the pneumococci and the staphylococci, responsible for life-threatening pneumonia, meningitis and sepsis. Blocking synthesis of the substrate of peptidoglycan polymerases is an attractive strategy to develop new antibiotics but this approach requires a better understanding of the substrate specificity and catalytic mechanism of the essential enzymes since screening of natural substances has led to very limited success in the discovery of novel molecules. Here, we report characterization of FemXWv, the model enzyme of a unique family of tRNA-dependent aminoacyl transferases that have not been previously explored as drug targets because of the complexity of their nucleotide substrate. Key features in the recognition of Ala-tRNAAla by FemXWv validate these enzymes as potential drug targets.
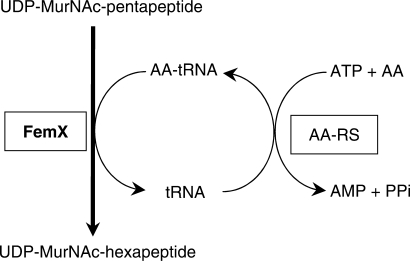
The peptidoglycan is an essential component of the bacterial cell envelope, since it provides a mechanical barrier to the internal osmotic pressure of the cytoplasm (1). The peptidoglycan subunit is assembled by a series of cytoplasmic and membrane steps (Figure 1) that lead to formation of nucleotide (UDP-linked) and lipid intermediates. The complete subunit is translocated to the cell surface by an unknown mechanism and polymerized by the glycosyltransferases, that form the β-1-4 bonds linking the repeating GlcNAc–MurNAc units (2), and by the d,d-transpeptidases, that form peptide bonds between stem peptides in order to cross-link adjacent glycan strands (3). The latter activity is the target of β-lactam antibiotics that mimic the d-Ala4-d-Ala5 extremity of peptidoglycan precursors (4) and act as ‘suicide’ substrates in an essentially irreversible acylation reaction (5). In Gram-positive bacteria, β-lactam resistance is often due to so-called low-affinity penicillin-binding proteins (PBPs) that interact poorly with β-lactams and catalyze peptidoglycan cross-linking in the presence of the drug (3).
Figure 1.
Main steps of peptidoglycan synthesis in W. viridescens. The subunit consists of β-1-4-linked N-acetyl glucosamine (GlcNAc) and N-acetyl muramic acid (MurNAc) substituted by a depsipeptide which is linked to the d-lactoyl group of MurNAc by an amide bond. The assembly of the subunit starts in the cytoplasm by the synthesis of UDP-MurNAc, the first precursor dedicated to peptidoglycan synthesis (37). In the following steps, the Mur ligases sequentially add l-Ala, d-Glu, l-Lys, attached to the γ carboxyl of d-Glu (d-iGlu) and the depsipeptide d-Ala-d-Lac to form the stem pentadepsipeptide l-Ala1-d-iGlu2-l-Lys3-d-Ala4-d-Lac5. The FemXWv aminoacyl transferase adds the first residue of the side chain onto this nucleotide precursor. Synthesis of the subunit proceeds by the transfer of the phospho-MurNAc-pentadepsipeptide moiety of UDP-MurNAc-pentadepsipeptide to the C55 lipid carrier undecaprenyl phosphate to form lipid intermediate I (undecaprenyl-PP-MurNAc-pentadepsipeptide or lipid I). The addition of GlcNAc to lipid I leads to lipid intermediate II [undecaprenyl-PP-MurNAc-(pentadepsipeptide)GlcNAc] or lipid II. The second and third residues of the l-Ala-l-Ser and l-Ala-l-Ser-l-Ala side chains are added to the lipid intermediates by unknown Fem transferases. The α carboxyl of d-iGlu2 is amidated in mature peptidoglycan (d-iGln2). The insets indicate the relative abundance of precursors and muropeptides, which was determined by the absorbance at 260 and 195 nm, respectively.
A wide variety of modifications of the disaccharide-peptide subunit has been detected in mature peptidoglycan from Gram-positive bacteria, including the addition of a side chain comprising one to seven amino acids to the ɛ amino group of l-Lys3 (6). These modifications are introduced in the subunit at different steps of peptidoglycan synthesis and can therefore involve the nucleotide precursor as well as both lipid intermediates (7). The dicarboxylic amino acid d-Asp is incorporated into the side chain by aspartate ligases that belong to the ATPGrasp superfamily and activate their substrate by ATP-dependent formation of a β-aspartyl-phosphate intermediary (8). In contrast, l-amino acids and glycine are activated as aminoacyl-tRNAs (9) and incorporated into the side chain by Fem transferases (10) that belong to the GCN5-related N-acetyltransferase (GNAT) protein superfamily (11,12). Characterized members of this family include FemXWv from Weissella viridescens that transfers l-Ala from l-Ala-tRNAAla to UDP-MurNAc-pentapeptide (13,14) (Figure 1) and FemABX from Staphylococcus aureus that sequentially add one (FemX) or two (FemA and FemB) glycines to lipid II (15).
FemXWv catalysis proceeds by an ordered bi–bi mechanism with sequential fixation of the UDP-MurNAc-pentapeptide and Ala-tRNAAla substrates and sequential release of the tRNAAla and UDP-MurNAc-hexapeptide products (14). Binding of the first substrate to FemXWv involves a complex network of hydrogen bonds and stacking interactions that constraint UDP-MurNAc-pentapeptide in a bent conformation essential for transferase activity (13). The mode of interaction of FemXWv with the second substrate, Ala-tRNAAla, as well as the catalytic mechanism of the enzyme remains speculative (14).
The side chain of peptidoglycan precursors assembled by Fem transferases carries the amine used for peptide bond formation by the transpeptidases in the last cross-linking step of peptidoglycan polymerization (Figure 1) (13,16). The specificity of Fem transferases is essential for the bacterium since incorporation of erroneous amino acids would produce precursors acting as chain terminators, thereby preventing formation of the stress-bearing peptidoglycan network. In spite of its critical role for the integrity of peptidoglycan, little is known about the amino acid specificity of Fem transferases. Surprisingly, FemXWv was reported to poorly discriminate between Ala-tRNAAla and Ser-tRNASer in vitro (14). The specificity constant kcat/Km for the two aminoacyl-tRNAs differed only 11-fold implying a likely incorporation of both amino acids into the peptidoglycan precursors (14). In this report, we have evaluated the in vivo specificity of FemXWv by mass spectrometry analyses of peptidoglycan and its precursors. The in vitro specificity of the enzyme was also evaluated by comparing the catalytic efficiency of the enzyme with natural tRNAs extracted from different bacterial species or obtained by in vitro transcription. The modes of recognition of the acceptor stem of the tRNA by FemXWv and alanyl-tRNA synthetase were compared based on saturation mutagenesis and semi-synthesis of Ala-tRNAAla analogues.
MATERIALS AND METHODS
Preparation and analysis of cytoplasmic peptidoglycan precursors
Bacteria were grown at 30°C to an optical density at 650 nm of 0.75 in Man, Rogosa and Sharpe (MRS) broth (Biorad). Bacitracin (250 µg/ml) was added to inhibit dephosphorylation of undecaprenyl diphosphate and incubation was continued for 30 min to accumulate cytoplasmic peptidoglycan precursors. Extraction of the precursors with formic acid and separation by rp-HPLC in a C18 column with a methanol gradient (0–20%) in 50 mM ammonium formiate (pH 4.0) was performed as previously described (17). The precursors were analyzed by mass spectrometry using an electrospray time-of-flight mass spectrometer operating in positive mode (Qstar Pulsar I, Applied Biosystem, Courtaboeuf, France) (18). The sequence of the precursors was determined by tandem mass spectrometry with nitrogen as the collision gas (18).
Peptidoglycan structure analysis
Bacteria were grown at 30°C to an optical density of 0.75 at 650 nm in MRS broth. Peptidoglycan was extracted with boiling SDS, treated with proteases and digested with mutanolysin and lysozyme, as previously described (16). The resulting muropeptides were treated with ammonium hydroxide to cleave the ether link in MurNAc, separated by rp-HPLC and analyzed by mass spectrometry and tandem mass spectrometry (16).
Enzyme production and purification
The same procedures were used for production and purification of the proteins unless specified otherwise. Genes were amplified by PCR and cloned into the vector pTrc-His60 (19) in order to introduce a C-terminal 6XHis tag in the recombinant proteins. Derivatives of Escherichia coli TOP10 harboring recombinant plasmids were grown to an optical density at 600 nm of 0.7 in brain heart infusion broth (Difco) containing 100 µg ml−1 of ampicillin, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.5 mM, and incubation was continued overnight at 16°C. Bacteria were harvested by centrifugation (4200×g for 30 min at 4°C), resuspended in 50 mM Tris–HCl (pH 7.5) containing 300 mM NaCl (buffer A), and disrupted by sonication with a Branson Sonifier 450 for 2 min with cooling. The extract was centrifuged at 13 800 × g for 30 min at 4°C and the supernatant was mixed with a HIS-Select™ nickel affinity gel (SIGMA, Saint Louis, Missouri, USA) for affinity purification. Proteins eluted with imidazole were dialyzed against buffer A containing 100 instead of 300 mM NaCl prior to anion exchange chromatography on a Mono Q 5/50 GL column (Amersham Biosciences, Piscataway, USA) with an NaCl gradient in Tris–HCl (pH 7.5). Size exclusion chromatography was performed on a Superdex 75 HR 10/30® column (Amersham Biosciences) equilibrated in buffer A. Protein concentration was determined by using the Bradford reagent (Bio-Rad, Munich, Germany) with bovine serum albumin as a standard. Proteins were stored at −20°C in buffer A supplemented with 50% glycerol.
The recombinant plasmid encoding the FemXWv transferase of W. viridescens has been previously described (13). The protein was purified by affinity, anion exchange and size exclusion chromatographies.
The aminoacyl-tRNA synthetases were purified in two steps by affinity and anion-exchange chromatographies. Construction of the recombinant plasmid encoding the alanyl–tRNA synthetase of Enterococcus faecalis JH2-2 (AlaRS) has been previously described (20). The same cloning approach was used to construct recombinant plasmids encoding the glycyl-tRNA synthetase of S. aureus Mu50 (GlyRS) and one (EF3292) of the two seryl–tRNA synthetases of E. faecalis JH2-2 (SerRS).
The gene encoding the T4 RNA ligase was cloned into pTrc-His60 and the enzyme was purified by affinity, anion exchange and size exclusion chromatographies. For production of the T7 RNA polymerase, we used a pQE30 derivative in E. coli M15 (a kind gift of K. Tomita). The protein was purified in a single step by nickel affinity chromatography followed by dialysis against Tris–HCl 50 mM (pH 7.5), MgCl2 10 mM, KCl 200 mM and dithiothreitol 2 mM. The protein was stored in the same buffer supplemented with 50% glycerol.
DNA templates for preparation of tRNAs by in vitro transcription
The sequence of the tRNAAla used in this study (5′-GGGGCCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGG UUCGAUCCCGCUAGGCUCCACCA-3′) corresponds to the three identical sequences annotated as tRNAAla in the genome sequence of E. faecalis strain V583 (http://www.tigr.org/) (13). The sequence of the tRNASer (5′- GGAGAGUUGUCCGAGAGGCCGAAGGAGCAUGAUUGGAAAUCAUGUAGGCGGUAAA CACUGUCUCAAGGGUUCGAAUCCCUUACUCUCCGCCA-3′) corresponds to one of the five sequences annotated as tRNASer in the genome of E. faecalis V583. The sequence of the tRNAGly (5′-GCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCG AUUCCCAUCACCCGCUCCA-3′) corresponded to two of the six sequences annotated as tRNAGly in the genome of S. aureus strain Mu50. The sequence (5′-TAATACGACTCACTATA-3′) (21) containing the T7 promoter was fused to the DNA templates encoding the tRNAs and cloned into the vectors pUC18 (tRNAAla and tRNAGly) or pCRblunt (Invitrogen) (tRNASer). The DNA templates used for in vitro transcription were obtained by digestion of these recombinant plasmids with BstNI, as previously described (21), or alternatively by PCR amplification of a portion of the same recombinant plasmids using primers 5′-AGGCTTTACACTTTATGCTTCCGG-3′ and 5′-TGGTGGAGCCTAGCGGGAT-3′ for tRNAAla, 5′-CGAATTCGATTTAATACGACTC-3′ and 5′-TGGCGGAGAGTAAGGGAT-3′ for tRNASer, and 5′-GCGGGTGTAGTTTAATGG-3′ and 5′-TGGAGCGGGTGATGGGGA-3′ for tRNAGly and DNA fragments used as templates were purified by agarose gel electrophoresis.
DNA templates for mutagenesis of tRNAAla
The plasmid templates used to generate the 35- and 36-nucleotide (nt) tRNAAla mini helices were generated by deleting internal portions of the pUC18 recombinant plasmid carrying the tRNAAla sequence (above). The plasmid templates used to generate the 24-nt micro helix was obtained by hybridizing two oligodeoxyribonucleotides (5′-AATTGCTGCAGTAATACGACTCACTATAGGGGCCTTAGCTCAGGCTCCACCA-3′ and 5′-TGGTGGAGCCTGAGCTAAGGCCCCTATAGTGAGTCGTATTACTGCAGCAATT-3′). The same approach was used generate the two strands of the acceptor stem (5′-CGAATTCGATTTAATACGACTCACTATAGGGGCCT-3′ and 5′-AGGCCCCTATAGTGAGTCGTATTAAATCGAATTCG-3′ for the 7-nt transcript; 5′-CGAATTCGATTTAATACGACTCACTATAAGGCTCCACCA-3′ and 5′-TGGTGGAGCCTTATAGTGAGTCGTATTAAATCGAATTCG-3′ for the 11-nt transcript).
For site-directed mutagenesis, a recombinant plasmid containing a portion (positions 6 to 76) of the sequence specifying tRNAAla (5′-CTTAGCTCAGCTGGGAGAGCGCCTGCTTTGCACGCAGGAGGTCAGCGGTTCGATCC CGCTAGGCTCCACCA-3′) was introduced into pUC18. This plasmid was used as a template for PCR amplification with combinations of two types of mutagenic primers. The first type of primers (5′-TTTAATACGACTCACTATAGGGGCCTTAGCTCAGCTGGGAG-3′) contained the sequence of the T7 promoter (5′-TAATACGACTCACTATA-3′), positions 1 to 5 of tRNAAla (underlined) with all possible single nucleotide substitutions at positions 1 to 4, and 17 nt complementary to the plasmid template (italics). The second type of primers (5′-TGGTGGAGCCTAGCGGGAT-3′) was complementary to the template (positions 59 to 76 of tRNAAla) with all possible single nucleotide substitutions in positions 69 to 76 (underlined). Using this approach, transcription of the wild-type tRNAAla sequence could not occur in the following step since the pUC derivative used for PCR amplification did not carry the T7 promoter and the first five bases of the tRNA.
In vitro transcription
The assay was performed at 37°C for 4 h in a total volume of 1 ml containing NTPs (4 mM each, Amersham), GMP (20 mM), a DNA template (ca. 100 pmol), T7 RNA polymerase (100 µg), Triton ×100 (0.05% V/V), MgCl2 (15 mM), spermidine (2 mM), dithiothreitol (10 mM) and Tris–HCl (40 mM, pH 7.5). The reaction mixture was incubated 20 min at 65°C, DNAse was added (30 U; RQ1 RNase-Free DNAse, Promega) and incubation was continued for 1 h at 37°C. RNA was extracted with one volume of phenol (pH 4.5), followed by one volume of phenol/chloroform (1/1 V/V), and precipitated with 2 volumes of ethanol at −20°C overnight. RNA was centrifuged at 4°C, the pellet was washed three times with 70% ethanol, dried under vacuum and dissolved in 100 µl of water. The RNA was incubated for 1 min at 85°C and slowly (ca. 45 min) cooled to room temperature by switching off the heating of the water bath. The RNA was purified by size exclusion chromatography (Superdex® 75 HR 10/30, Pharmacia) in Tris–HCl (25 mM, pH 7.5) containing 100 mM NaCl and 5 mM MgCl2. The concentration of tRNA was determined by aminoacylation as described subsequently. A simplified procedure was used for the analysis of RNA molecules obtained by site-directed mutagenesis of tRNAAla. Briefly, size exclusion chromatography was omitted and the concentration of tRNA was estimated following ethidium bromide staining of denaturing polyacrylamide gels containing appropriate dilution of purified tRNAAla used as a standard.
Purification of natural tRNAs
The method of Meinnel and Blanquet (22) was used to extract tRNA from four bacterial species representative of the polymorphism at the third position of peptidoglycan precursors including W. viridescens CIP 102810 T (formerly Lactobacillus viridescens ATCC 12706), E. faecalis JH2-2 (23), Enterococcus faecium D344S (17) and S. aureus COL. Briefly, bacteria were grown at 37°C to mid log phase (optical density of 0.7 at 600 nm) in MRS broth for W. viridescens or in brain heart infusion broth (Difco) for the other strains. Bacteria were concentrated 100-fold by centrifugation, resuspended in Tris–HCl 10 mM (pH 7.5) containing magnesium acetate 10 mM, and the tRNAs were extracted from bacterial suspensions with one volume of phenol (pH 4.5). NaCl was added to obtain a concentration of 0.5 M and the tRNAs were precipitated with 0.7 volume of isopropanol at −80°C for 30 min. Deacylation was performed in Tris–HCl (pH 8.0) for 2 h at 37°C. The tRNAs were dialysed against water, lyophilized and dissolved in water. The concentration of the tRNAs was determined by the absorbance at 260 nm.
Acylation of tRNAs
The concentration of tRNA was determined based on full aminoacylation with a high concentration of aminoacyl-tRNA synthetases (800 nM) that was shown to be sufficient for full acylation in less than 1 min. The assay (30 µl) contained 50 mM Tris–HCl (pH 7.5), 12.5 mM MgCl2, 2 mM β-mercaptoethanol, aminoacyl-tRNA synthetases (800 nM) and radiolabeled amino acids 50 µM (l-[14C]Ala, 6.3 GBq mmol−1; l-[14C]Ser, 5.8 GBq mmol−1; [14C]Gly, 3.7 GBq mmol−1; Perkin Elmer). The reaction was incubated 8 min at 37°C and aminoacyl-tRNAs were precipitated with 5% TCA and 0.5% casamino acids, filtered on glass-fiber filters and determined by liquid scintillation counting.
FemXWv coupled assay
The transferase activity of FemXWv was determined in a coupled assay containing an auxiliary system to generate aminoacyl-tRNAs as described by Maillard et al. (13) with minor modifications. The assay contained 50 mM Tris–HCl (pH 7.5), 12.5 mM MgCl2, 2 mM β-mercaptoethanol, 7.5 mM ATP, 800 nM aminoacyl-tRNA synthetase, 50 μM UDP-MurNAc-pentapeptide, 50 µM of [14C] amino acids and tRNAs at the concentrations specified in the text. The reaction was performed at 30°C with a preincubation of 10 min prior to the addition of FemXWv for synthesis of the aminoacyl-tRNAs by the auxiliary system. The concentration of FemXWv (0.5–500 nM) was adjusted to obtain initial velocities in conditions where transferase activity was rate-limiting. Under such conditions, the tRNAs were completely acylated at the onset of the FemXWv reaction and remained fully acylated during the entire reaction, as judged by direct determination of aminoacyl-tRNAs. The reaction was stopped at 95°C for 10 min and analysed by descending paper chromatography (Whatman 4MM, Elancourt, France) with isobutyric acid : ammonia 1 M (5 : 3 per vol). Radioactive spots were identified by autoradiography, cut-out and counted by liquid scintillation.
Synthesis of N-(4-pentenoyl)-l-alanine cyanomethyl ester
To a solution of pentenoic acid (500 µl, 4.9 mmol) in DME (18 ml) cooled down to −10°C were added successively N-methylmorpholine (539 µl, 4.9 mmol) and isobutyl chloroformate (640 µl, 4.9 mmol). In another flask, a mixture of alanine methyl ester hydrochloride (684 mg, 4.9 mmol) and N-methylmorpholine (539 µl, 4.9 mmol) in DME (2 ml) was stirred for 10 min at room temperature before being added dropwise to the first solution. The reaction was stirred overnight at room temperature, quenched with H2O and DME was evaporated under reduced pressure. The aqueous layer was extracted twice with DCM and organic layers were gathered and washed successively with saturated aqueous NaHCO3 solution, HCl (0.5 M) and brine. After drying over MgSO4 and concentrated under reduced pressure, the N-(4-pentenoyl)-l-alanine methyl ester was obtained as a yellowish oil (92% yield) which was used without any further purification. To a solution of this compound (834 mg, 4.5 mmol) in THF (20 ml) was added a solution of LiOH (0.5 M, 2 equiv.) and the mixture was stirred for 30 min at room temperature. The reaction mixture was washed twice with Et2O. The aqueous layer was acidified to pH 1 with HCl and extracted twice with Et2O. The last two organic layers were dried over MgSO4 and concentrated under reduced pressure to give the N-(4-pentenoyl)-l-alanine as a yellowish oil (48% yield) which was used without further purification. To a solution of this compound (325 mg, 1.9 mmol) in CH3CN (5 ml) were added trietylamine (1.14 ml, 8.17 mmol) and chloroacetonitrile (300 µl, 4.75 mmol); the mixture was stirred overnight at room temperature and concentrated to dryness under reduced pressure. The residue was dissolved in AcOEt and H2O, the organic layer was dried over MgSO4 and concentrated. A purification by flash chromatography (cyclohexane/EtOAc : 2/1 to 1/1) afforded the N-(4-pentenoyl)-l-alanine cyanomethyl ester as a yellow oil in 72% yield. 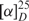 −42 (c 1, CHCl3); 1H NMR (250 MHz, CDCl3) : δ1.40 (d, 3H, J 7.4), 2.3 (bs, 4H), 4.5 (dq, 1H, J 8.0), 4.74 (d, J 15), 4.9–5.1 (m, 2H), 5.77 (m, 1H), 6.36 (bd, 1H, J 6.8); 13C NMR (63 MHz, CDCl3): d 17.6, 29.3, 35.2, 47.7, 49.0, 114.1, 115.7, 136.8, 171.7, 172.4.
−42 (c 1, CHCl3); 1H NMR (250 MHz, CDCl3) : δ1.40 (d, 3H, J 7.4), 2.3 (bs, 4H), 4.5 (dq, 1H, J 8.0), 4.74 (d, J 15), 4.9–5.1 (m, 2H), 5.77 (m, 1H), 6.36 (bd, 1H, J 6.8); 13C NMR (63 MHz, CDCl3): d 17.6, 29.3, 35.2, 47.7, 49.0, 114.1, 115.7, 136.8, 171.7, 172.4.
Semi-synthesis of Ala-tRNAAla analogues
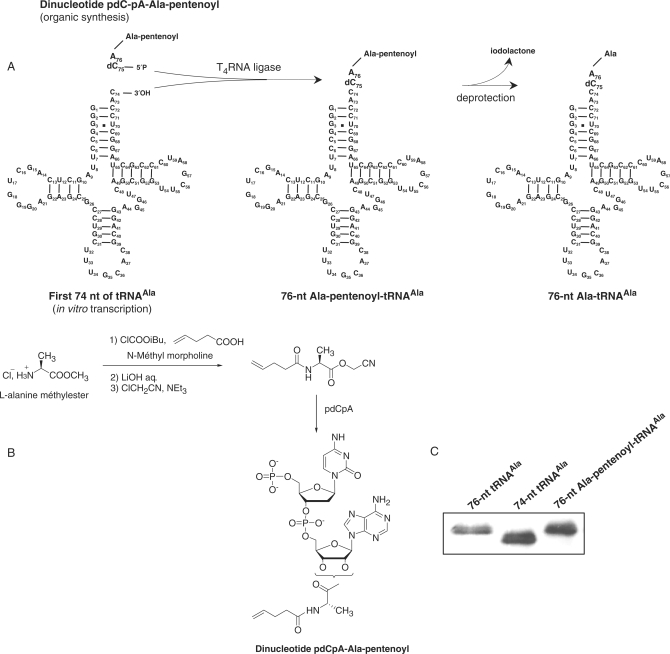
Chemical aminoacylation of the hybrid dinucleotide 5′-phospho-2′-deoxyribocytidylylriboadenosine (pdCpA) by N-(4-pentenoyl)-l-alanine cyanomethyl ester was performed as previously described (24,25). Ligation of pdCpA-Ala-pentenoyl (400 µM) to the 74-nt in vitro transcript (4 µM) lacking the last 3′ nucleotide C75–A76 of tRNAAla was performed with the purified T4 RNA ligase (above) in 50 µl of 50 mM HEPES buffer (pH 7.5) containing 0.75 mM ATP, 15 mM MgCl2, and 10% DMSO (per volume) for 2 h at 37°C. The Ala-tRNA substrate (ca. 200 pmol) was deprotected with 5 mM I2 in THF/H2O (1/1) (25), purified by ethanol precipitation, and immediately incubated with FemXWv (5 pmol) and UDP-MurNAc-pentapeptide (200 pmol) in 10 µl of 50 mM Tris–HCl (pH 7.5) containing 12.5 mM MgCl2 and 2 mM β-mercaptoethanol. The reaction was incubated for 2 h at 30°C, desalted using a micro column (ZipTipC18, Millipore), and analyzed by nanoelectrospray mass spectrometry in the positive mode in order to detect formation of UDP-MurNAc-hexapeptide.
Docking model of the FemXWv-tRNAAla complex
The model of tRNAAla was built from tRNAAsp in complex with E. coli aspartyl tRNA-synthetase (26) by replacing nucleotides of E. coli tRNAAsp by the E. faecalis tRNAAla counterparts. The tRNAAla was manually docked in the FemXWv active site (PDB code 1P4N) (11) using the position of puromycin in the active site of the leucyl/phenylalanyl-tRNA-protein transferase to position the 3′-end A76 of the tRNAAla (PDB code 2DPT) (27). The structure was energy minimized in 100 steps with a dielectric constant of 1, using the CNS program (28). The puromycin: leucyl/phenylalanyl-tRNA synthetase complex was used to position the 3′-end of the acceptor arm of tRNAAla since this transferase and FemXWv share a common fold indicating that their tRNA-binding sites are likely to be structurally related (11,27).
RESULTS
In vivo specificity of the FemXWv transferase
Cytoplasmic peptidoglycan precursors were extracted from W. viridescens to identify the amino acid incorporated by FemXWv in vivo (Figure 1). The main precursor had a monoisotopic mass of 1221.40. Sequencing by tandem mass spectrometry indicated that this precursor consisted of UDP-MurNAc, a linear pentadepsipeptide terminating in d-lactate (l-Ala1-d-iGlu2-l-Lys3-d-Ala4-d-Lac5) and an l-Ala residue linked to the side chain of l-Lys3. Production of d-Lac-ending precursors is in agreement with phenotypic expression of glycopeptide resistance by W. viridescens (data not shown) since these precursors bind the drugs with reduced affinity (29). Minor amounts of UDP-MurNAc-l-Ala1-d-iGlu2-l-Lys3-d-Ala4 (UDP-MurNAc-tetrapeptide) with a monoisotopic mass of 1078.31 and of UDP-MurNAc-tetrapeptide with a side chain consisting of a single l-Ala (1149.35) were also detected (Figure 1). Incorporation of amino acid other than l-Ala into the side chain was not detected indicating that FemXWv is highly specific for l-Ala in vivo.
Synthesis of the side chain of peptidoglycan precursors was also assessed by analyzing the structure of mature peptidoglycan by mass spectrometry. Identification of 120 peptidoglycan fragments obtained by digestion of the cell wall by muramidases revealed variations in the length of stem peptides and of side chains in combination with various degree of oligomerization (Figure 1 and data not shown). Briefly, the stem of monomers consisted of a tripeptide (l-Ala1-d-iGln2-l-Lys3) or a tetrapeptide (l-Ala1-d-iGln2-l-Lys3-d-Ala4) in similar amounts indicating that the l-Lys3-d-Ala4 and d-Ala4-d-Lac5 peptide bonds were cleaved by l,d and d,d-carboxypeptidases, respectively (18). The side chains of monomers mainly consisted of the sequence l-Ala-l-Ser and l-Ala-l-Ser-l-Ala. Side chains consisting of a single l-Ala were less abundant and unsubstituted monomers were not detected. These observations indicate that FemXWv efficiently and specifically adds l-Ala to the precursors in agreement with the analysis of the cytoplasmic pool of UDP-MurNAc-peptides. The second (l-Ser) and third (l-Ala) residues of the side chain were added by unknown Fem transferases, since FemXWv adds a single residue in the in vitro coupled assay containing the accessory systems for Ala-tRNAAla and Ser-tRNASer synthesis (Figure 2 and data not shown). The second and third residues were added after the transfer of the phospho-MurNAc-peptide moiety of the precursor to the lipid carrier since a single l-Ala residue was detected in the nucleotide precursors (Figure 1). The peptidoglycan of W. viridescens contained multimeric structure ranging from dimers to hexamers. The cross-bridges contained the sequence l-Ala-l-Ser and l-Ala-l-Ser-l-Ala indicating that the d,d-transpeptidases used acceptor containing at least two residues in their side chains.
Figure 2.
Coupled assay for UDP-MurNAc-hexapeptide synthesis. The auxiliary system used to generate aminoacyl-tRNA (AA-tRNA) involves acylation of tRNA with a radiolabeled amino acid by aminoacyl-tRNA synthetases (AA-RS). Purified alanyl-tRNA synthetase (AlaRS), seryl-tRNA synthetase (SerRS) and glycyl-tRNA synthetase (GlyRS) were used in this study. FemXWv transfers the amino acids from the aminoacyl-tRNAs to the ɛ -amino group of l-Lys at the third position of UDP-MurNAc-pentapeptide to form the corresponding hexapeptide which was determined by liquid scintillation.
In conclusion, analyses of the structure of peptidoglycan and its precursors indicated that FemXWv only adds l-Ala at the first position of l-Ala-l-Ser and l-Ala-l-Ser-l-Ala side chains. Incorporation of l-Ala substitutes in more than 1% of the precursors would have been easily detected since peptidoglycan fragments containing Gly, d-Asp and l-Ser are readily detected by electrospray mass spectrometry (16,17).
Specificity of the FemXWv transferase for tRNAs obtained by in vitro transcription
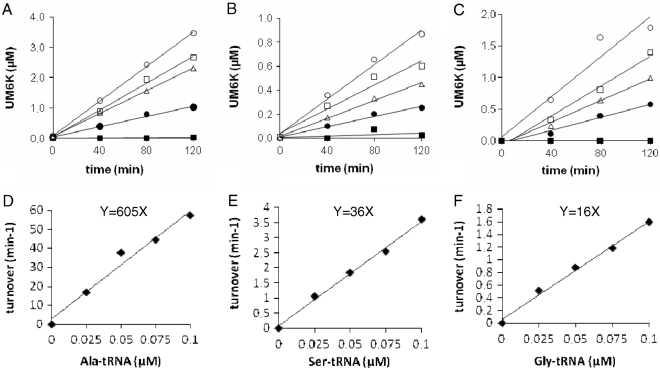
In order to compare the catalytic efficiency of FemXWv with different aminoacyl-tRNAs, kinetics of synthesis of UDP-MurNAc-hexapeptides containing Ala (Figure 3A), Ser (Figure 3B), and Gly (Figure 3C) were performed at different aminoacyl-tRNA concentrations. The turnover number of FemXWv was deduced from the individual kinetics and plotted against the aminoacyl-tRNA concentrations (Figure 3D–F). The linear relationship indicates that the reaction conditions meet the first order conditions with respect to aminoacyl-tRNAs in agreement with the Km value of 1.7 µM previously reported for Ala-tRNAAla (13). Ser-tRNASer and Gly-tRNAGly were used 17- and 38-fold less efficiently than Ala-tRNAAla by FemXWv (36 and 16 versus 605 min−1 µM−1, respectively; Table 1).
Figure 3.
Catalytic efficiency of FemXWv with different tRNAs obtained by in vitro transcription. Kinetics of transfer of Ala (A), Ser (B) and Gly (C) from aminoacyl-tRNAs to UDP-MurNAc-pentapeptide was determined with four concentrations of tRNAs (filled square, 0 µM; filled circle, 0.025 µM; open circle, 0.05 µM; open square, 0.075 µM; open circle, 0.1 µM). The concentrations of the two enzymes in the coupled assay were adjusted to obtain initial velocities in conditions where the transferase activity of FemXWv was rate-limiting and the tRNAs were completely acylated by the aminoacyl-tRNA synthetases, (A) FemXWv (0.5 nM), alanyl-tRNA synthetase (0.8 µM); (B) FemXWv (2 nM), seryl-tRNA synthetase (0.8 µM); (C) FemXWv (10 nM), glycyl-tRNA synthetase (0.8 µM). The FemXWv turnover numbers deduced from the kinetics were plotted as a function of the concentrations of Ala-tRNAAla (D), Ser-tRNASer (E), Gly-tRNAGly (F). The slopes (y) provide an estimate of the relative catalytic efficiencies of FemXWv for the transfer of Ala, Ser and Gly at non-saturating concentrations of aminoacyl-tRNAs (turnover per min and per µM of tRNAs).
Table 1.
Relative turnover numbers of FemXWv with different tRNAs
| In vitro transcript | Natural tRNAs | ||||
|---|---|---|---|---|---|
| W. viridescens (l-Ala-l-Ser -l-Ala) | S. aureus (Gly5) | E. faecalis (l-Ala2) | E. faecium (d-iAsn) | ||
| tRNAAla | 605 ± 54 | 2890 ± 350 | 410 ± 45 | 570 ± 57 | 830 ± 78 |
| tRNASer | 36 ± 1 | 29 ± 3 | 8 ± 1 | 32 ± 4 | 37 ± 2 |
| tRNAGly | 16 ± 1 | 0.75 ± 0.07 | 0.65 ± 0.06 | 1.44 ± 0.10 | 1.99 ± 0.20 |
The relative catalytic efficiency of FemXWv for the transfer of Ala, Ser and Gly at non-saturating concentrations of aminoacyl-tRNAs (turnover per min and per µM of tRNAs) was determined for tRNA obtained by in vitro transcription and for natural tRNAs. The in vitro transcripts had the sequence of tRNAs from E. faecalis (tRNAAla and tRNASer) or S. aureus (tRNAGly; see Materials and methods section). The natural tRNAs were extracted from four bacterial species representative of the diversity of the side chain of peptidoglycan precursors (indicated in parenthesis).
In vitro specificity of the FemXWv transferase for natural tRNAs
The relative efficiency of FemXWv with natural tRNAs from W. viridescens, S. aureus, E. faecalis and E. faecium was evaluated based on determination of the turnover number of the enzyme at four aminoacyl-tRNA concentrations as performed for in vitro transcripts (data not shown and Table 1). The highest FemXWv turnover number was observed with Ala-tRNAAla extracted from W. viridescens. The Ser-tRNASer and Gly-tRNAGly were used 100- and 4000-fold less efficiently than Ala-tRNAAla (29 and 0.75 versus 2890 min−1 µM−1, respectively). FemXWv was less specific with heterologous tRNAs from S. aureus, E. faecalis and E. faecium. This was mainly due to the fact that heterologous tRNAAla were not efficiently used by FemXWv. The heterologous tRNAs were extracted from bacterial species representative of the diversity in the sequence and the mode of assembly of the side chain of peptidoglycan precursors. E. faecium produces an ATP-dependent ligase for incorporation of d-Asp whereas S. aureus and E. faecalis produce Fem transferases for synthesis of Gly5 and l-Ala2 side chains (6). FemXWv displayed similar catalytic efficiencies with the heterologous tRNAs irrespective of the mode of synthesis of the side chain in the three species.
The catalytic efficiencies of FemXWv with tRNAAla obtained by in vitro transcription or present in the tRNA pool extracted from E. faecalis were very similar (605 versus 570 min−1 µM−1). Since the genome of E. faecalis contains a single tRNAAla sequence present in triplicate, this result indicates that post-transcriptional modification of the tRNA does not modulate the activity of FemXWv. Similar catalytic efficiencies were also observed for tRNASer obtained in vitro or present in the tRNA pool of E. faecalis (36 versus 32 min−1 µM−1). For tRNAGly, the in vitro transcript was used more efficiently than the natural tRNAs from the four species (16 versus 0.65–1.99 min−1 µM−1). This suggests that unknown post-transcriptional modifications of tRNAGly may impair FemXWv activity.
Identification of a minimal RNA substrate in the AlaRS-FemXWv coupled assay
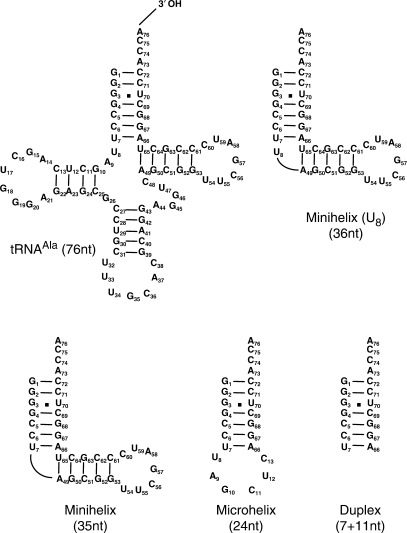
Various derivatives of the E. faecalis tRNAAla (Figure 4) were tested as substrates of AlaRS and FemXWv in the coupled assay (Figure 2) for UDP-MurNAc-hexapeptide synthesis. Deletion of the anticodon and pseudouridine arms of tRNAAla resulted in two minihelices, with or without bulged U8, which were both functional in the coupled assay. Formation of UDP-MurNAc-hexapeptide was also obtained with a 24-nt microhelix consisting of the acceptor stem of tRNAAla and a loop of 6 nt. Finally, an RNA duplex consisting only of the acceptor stem was also functional. These results indicate that the bases essential for FemXWv and AlaRS activities are all located within the acceptor stem of tRNAAla. We therefore focused the following mutagenesis studies on the acceptor stem of tRNAAla although we have not excluded the possibility that additional region of the tRNA might modulate enzyme activity.
Figure 4.
Functional RNA molecules in the synthesis of UDP-MurNAc-hexapeptide by AlaRS and FemXWv in the coupled assay. The 76-nt RNA corresponds to the only tRNAAla of E. faecalis. The minimal RNA substrate in the AlaRS-FemXWv coupled assay was a duplex (7 + 11 nt) mimicking the acceptor stem of the tRNAAla.
Development and validation of a novel method for saturation mutagenesis of the acceptor stem of tRNAAla
To facilitate site-directed mutagenesis of the DNA template for in vitro transcription of tRNAAla by T7 RNA polymerase, we have developed a method based on PCR amplification using mutagenic primers. In the classical method (21), the 5′-end of the DNA template that determines the 3′-end of the run off transcript is generated by digestion of a plasmid by the restriction endonuclease BstNI. In this study, the 5′-end of a primer used for PCR amplification of the DNA template provided an alternate way to define the 3′-end of the tRNA transcript. Twenty-four mutagenic primers were used to introduce all possible single nucleotide substitutions in positions C69 to A76 of the acceptor stem. The 5′-end of the in vitro transcript was defined by the localization of the T7 promoter, which was incorporated at the 5′-end of the second primer used for PCR amplification. The mutagenic primers also contained the sequence corresponding to bases 1 to 22 of tRNAAla, thereby allowing the introduction of all possible single nucleotide substitutions in positions 1 to 4. The plasmid template used for PCR amplification did not contain the T7 promoter sequence. Thus, the wild-type tRNAAla sequence could not originate from transcription of the plasmid template. In preliminary experiments, we compared tRNAAla with the wild-type sequence obtained by in vitro transcription of DNA matrices generated by PCR or by the classical digestion of a plasmid template with BstNI. The two types of tRNAAla preparations were equivalent in conditions where the tRNA substrate and FemXWv were rate limiting (data not shown). We also checked that the fidelity of the DNA polymerase used for the PCR was sufficient to avoid false positives. This was established by showing that several substitutions abolished formation of the UDP-MurNAc-hexapeptide product of FemXWv in the coupled assay (Figure 5, see subsequently).
Figure 5.
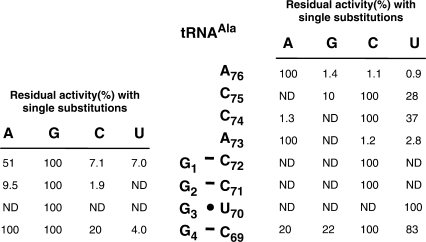
Residual (%) UDP-MurNAc-hexapeptide synthesis in the AlaRS-FemXWv coupled assay with derivatives of tRNAAla (76-nt) harboring single base substitutions in the acceptor stem. The standard assay was performed with AlaRS (800 nM), FemXWv (500 nM), l-[14C]Ala (50 µM) and the different tRNAs (ca. 5.0 µM). The reaction was incubated for 2 h at 30°C. Under these conditions, total l-[14C]Ala (0.5 nmol) was incorporated into UDP-MurNAc-hexapeptide when RNA with the wild-type sequence was used (100%). ND, not detected.
Impact of single nucleotide substitutions in tRNAAla
Nucleotide substitutions were introduced at positions 1 to 4 of tRNAAla and residual activity was determined by the coupled AlaRS-FemXWv assay (Figure 5). All substitutions at position 3 abolished formation of the UDP-MurNAc-hexapeptide indicating that G3 is essential. At position 2, low residual activity was detected only for the G2A and G2C substitutions. All substitutions were tolerated at positions 1 and 4 with various impacts on residual activities. Analysis of the complementary strand by the same method identified three essential bases (C72, C71 and U70). Relatively high residual activities were detected for position C69. Several substitutions were tolerated in the single-strand ACCA extremity of the acceptor stem and none of the four bases was essential. In conclusion, analysis of single substitutions identified four bases, C72, C71 and G3–U70 that were essential for synthesis of UDP-MurNAc-hexapeptide by AlaRS and FemXWv.
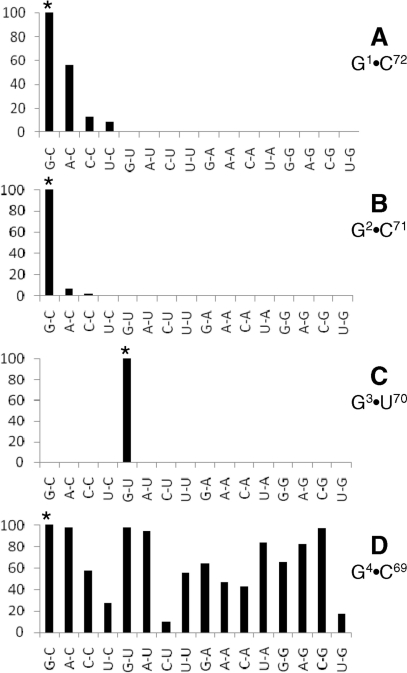
Impact of double nucleotide substitutions in tRNAAla
All 15 possible combinations of nucleotides were introduced in base pairs G1–C72, G2–C71, G3–U70 and G4–C69 using pairs of mutagenic primers and residual activity was determined in the coupled AlaRS-FemXWv (Figure 6). For the G1–C72 base pair, C at position 72, rather than a Watson–Crick base pair, was essential for enzyme activity since tRNAs containing C1–G72, A1–U72, U1–A72 did not support UDP-MurNAc-hexapeptide synthesis. In the following base pair, residual activity was only detected for two of the 15 combinations, A2–C71 and C2–C71, revealing again that destabilization of the acceptor stem is not the main factor that determines loss of activity. At position G3–U70, none of the 15 combinations were tolerated. Finally, synthesis of UDP-MurNAc-hexapeptide was obtained with the set of 16 tRNAs containing all base combinations at positions 4 and 69. These results confirmed that four specific bases, C72, C71 and the G3–U70 pair, are essential for UDP-MurNAc-hexapeptide synthesis. None of the double substitutions that restored Watson–Crick base pairs compensated for substitution at C72 and C71. Thus, modification of the sequence rather than destabilization of the acceptor stem was the primary impact of nucleotide substitutions at these two positions.
Figure 6.
Histogram showing residual (%) UDP-MurNAc-hexapeptide synthesis in the AlaRS-FemXWv coupled assay with derivatives of tRNAAla (76-nt) harboring all possible combinations in base pairs G1–C72 (A), G2–C71 (B), G3–U70 (C) and G4–C69 (D). The standard assay was performed with AlaRS (800 nM), FemXWv (500 nM), l-[14C]Ala (50 µM) and the different tRNAs (ca. 5 µM). The reaction was incubated for 2 h at 30°C. Under these conditions, total l-[14C]Ala (0.5 nmol) was incorporated into UDP-MurNAc-hexapeptide when RNA with the wild type sequence was used (100%, indicated by a star).
Semi-synthesis of analogues of Ala-tRNAAla
Detection of UDP-MurNAc-hexapeptide synthesis in the coupled assay used above implies that nucleotide substitutions are tolerated both by the AlaRS and by FemXWv. Thus, the assay provides a rapid method to determine whether nucleotide substitutions are tolerated by FemXWv provided that the modified tRNAAla molecules are substrate of the AlaRS. The assay is highly sensitive to qualitatively detect residual activity since each tRNA molecule can participate to several catalytic cycles (Figure 2). However, the absence of UDP-MurNAc-hexapeptide synthesis in the coupled assay, as observed for the substitutions at positions C72, C71 and G3–U70, may indicate that the modified tRNAs are not substrate of AlaRS although they can be used by FemXWv. Semi-synthesis of Ala-tRNAAla analogues harboring these substitutions was therefore developed to obtain esterified tRNAAla independently from the AlaRS (Figure 7A). This approach was originally developed to introduce non-natural amino acids into proteins by using in vitro coupled transcription-translation systems (30). Briefly, a dinucleotide esterified by an amino acid analogue is obtained by organic synthesis (Figure 7B) and added to an incomplete tRNA lacking the 3′-end CA dinucleotide using the T4 RNA ligase (30). The ligation restores the complete tRNA sequence with a 3′ esterified residue. A 2′ deoxycytidine is incorporated into the dinucleotide to facilitate its synthesis. In preliminary experiments, we generated a FemXWv substrate by this approach which contained the wild-type tRNAAla sequence and l-Ala as the 3′ amino acyl group (Figure 7C). Transfer of l-Ala from this Ala-tRNAAla analogue to UDP-MurNAc-pentapeptide was detected by mass spectrometry indicating that FemXWv tolerates substitution of a ribose by a 2′deoxyribose at position 75.
Figure 7.
Semi-synthesis of Ala-tRNAAla analogues. (A) The protected dinucleotide pdCpA-Ala-pentenoyl was ligated to a 74-nt in vitro transcript by the T4 RNA ligase. The product of the reaction was deprotected leading to an acylated-RNA molecule identical to Ala-tRNAAla except for the presence of a 3′deoxycytidine instead of a cytidine at position 75. Nucleotide substitutions were introduced in the 74-nt in vitro transcript using the PCR approach described above except that one of the two mutagenic primers lacked the two 5′ bases specifying bases 75 and 76 of the transcript. (B) Schematic representation of the main steps for synthesis of the dinucleotide pdCpA-Ala-pentenoyl. (C) Analysis of the T4 ligation products by denaturing polyacrylamide electrophoresis. Lane 1, 76-nt transcript with the wild-type tRNAAla sequence; lane 2, 74-nt in vitro transcript; lane 3, ligation of the 74-nt transcript with pdCpA-Ala-pentenoyl.
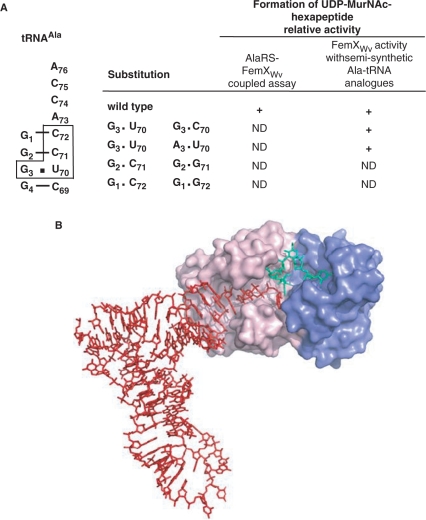
Semi-synthetic Ala-tRNAAla analogues containing Watson–Crick G3–C70 or A3–U70 base pairs instead of G3–U70 were substrates of FemXWv. Thus, neither G at position 3 nor U at position 70 was necessary for transferase activity. In addition, FemXWv did not require a wobble base pair at positions 3–70. None of the 15 base combinations were tolerated at these positions in the AlaRS-FemXWv coupled assay (Figure 6, above). These negative results can be attributed to the lack of amino acylation by the AlaRS since the semi-synthetic Ala-tRNAAla analogues containing G3–C70 and A3–U70 were substrate of FemXWv. In contrast, C72 and C71, identified as essential in the coupled assay (Figure 5, above), were also found to be essential for FemXWv activity since introduction of the C72G and C71G substitutions by semi-synthesis abolished formation of UDP-MurNAc-hexapeptide. In conclusion, C72 and C71, but not G3–U70, were essential for FemXWv indicating that the mode recognition of the tRNA body by this enzyme and AlaRS are different.
DISCUSSION
Synthesis of the side chain of peptidoglycan precursors by Fem transferases in Gram-positive bacteria implies that translation and peptidoglycan synthesis share common aminoacyl-tRNA synthetases and tRNAs. Fem transferases have evolved to redirect a portion of the aminoacyl-tRNAs toward peptidoglycan synthesis. The specificity of these enzymes is essential since the side chain is used by the transpeptidases in the last cross-linking step of peptidoglycan synthesis (Introduction section and Figure 1A). In this study, we have investigated tRNA recognition by a model transferase, FemXWv from W. viridescens, to identify and compare the identity determinants used by aminoacyl-tRNA synthetases and Fem transferases.
In vivo evaluation of the specificity of FemXWv based on extensive analyses of the structure of mature peptidoglycan and its precursors (Figure 1) revealed that this enzyme exclusively uses Ala-tRNAAla for aminoacyl transfer. Evaluation of homologous tRNAs extracted from W. viridescens indicated that FemXWv is also highly specific in vitro since Ser-tRNASer and Gly-tRNAGly were used 100- and 4000-fold less efficiently than Ala-tRNAAla, respectively (Table 1). Comparison of in vitro transcripts and natural tRNAs revealed similar catalytic efficiencies indicating that post-transcriptional modifications do not contribute to discrimination between Ala-tRNAAla and Ser-tRNASer (Table 1). In contrast, unknown post-transcriptional modifications might act as negative (anti-determinant) elements for Gly-tRNAGly since incorporation of Gly was 25-fold less efficient with natural tRNAs than with in vitro transcripts (Table 1). FemXWv retained preferential incorporation of alanine with heterologous tRNAs extracted from related Gram-positive species, although the capacity of the enzyme to discriminate between the different tRNAs was reduced mainly because heterologous Ala-tRNAAla were used less efficiently than Ala-tRNAAla from W. viridescens. The use of heterologous Ala-tRNAAla and Ser-tRNASer from E. coli in a previous study may therefore account for the reported poor discrimination of the two aminoacyl-tRNAs by FemXWv (14).
To identify the tRNA elements essential for FemXWv activity, deletions were introduced into tRNAAla revealing that the acceptor stem is sufficient for the alanine transferase activity of FemXWv (Figure 4). Site-directed mutagenesis identified four bases (C72, C71 and G3–U70) essential for UDP-MurNAc-hexapeptide synthesis in the coupled AlaRS-FemXWv assay (Figure 5). Introduction of double nucleotide substitutions showed that C72 and C71 were both essential since destabilization of the acceptor stem was not the primary impact of nucleotide substitutions at these two positions (Figure 6). Site-directed mutagenesis (Figures 5 and 6) combined to semi-synthesis (Figure 7) showed that the identity determinants of the E. faecalis AlaRS and FemXWv are overlapping but distinct (Figure 8A). In particular, the G3–U70 base pair of tRNAAla, which is the main identity determinant of AlaRS from bacteria, archaea and eukarya (31,32), occasionally found at positions 2–71 (33,34), was non-essential to the alanyl transferase activity of FemXWv since nucleotide substitutions at positions 3 and 70 were both tolerated by the enzyme (Figure 8A). Modeling of tRNAAla in the FemXWv active site (Figure 8B) indicated that the first two base pairs (G1–C72 and G2–C71) of the acceptor stem and the ACCA end are likely to be in close contact with the protein. The lack of direct interaction between the 3–70 base pair and FemXWv is in agreement with the non-essential nature of G3 and U70 for the alanyl transferase activity of this enzyme. Substitutions that restored Watson–Crick base pairing at position 3–70 did not abolish FemXWv activity. Thus, indirect recognition of the wobble base pair through a structural perturbation in the acceptor stem (34,35) appears also to be excluded for FemXWv.
Figure 8.
Recognition of tRNAs by alanyl-tRNA synthetases and FemXWv. (A) Sequence of the tRNAAla acceptor stem. The bases essential for formation of UDP-MurNAc-hexapeptide in the coupled AlaRS-FemXWv assay are boxed. Among these four bases, only C72 and C71 are essential for FemXWv activity as shown by comparing the impact of substitutions in the coupled assay and the direct determination of FemXWv activity using semi-synthetic substrates (right panel). (B) Model of FemXWv-tRNAAla interaction showing domains I (blue) and II (pink) of FemXWv and the tRNAAla (red) and UDP-MurNAc-pentapeptide (green) substrates.
The capacity of FemXWv to discriminate between different aminoacyl-tRNA is likely to depend both upon the tRNA body and the esterified amino acid as demonstrated for elongation factor EF-Tu that delivers aminoacyl-tRNAs to the ribosome during translation (36). For this protein, the affinities for the tRNA and the amino acid have compensatory role to ensure roughly equivalent affinities for all elongator aminoacyl-tRNAs. The sequence-dependent recognition of the tRNA body involves hydrogen bonding between a Glu residue of the protein and the amino group of a guanine located in the minor groove of the T-stem (base pair 51–63). Aminoacyl-tRNA recognition by the eubacterial leucyl/phenylalanyl-tRNA-protein transferase (L/F-transferase) involves a different mechanism (27). This enzyme conjugates leucine or phenylalanine to the N-terminus of proteins triggered to proteolytic degradation using Leu-tRNALeu or Phe-tRNAPhe as a substrate. The C-terminal domain of L/F-transferase belongs to the GCN5-related N-transferase fold as the two domains of FemXWv (11). The enzyme contains a hydrophobic pocket that accommodates Leu and Phe (and Met and Trp less efficiently) ensuring amino acid specificity (27). Recognition of the tRNA body occurs in a sequence-independent manner. Based on modeling, recognition involves the phosphate backbone of the bottom region of the D-stem and of the 3′ region of the acceptor stem. The recognition of amino acyl-tRNAs by L/F-transferase and EF-Tu is competitive. It has been proposed that L/F-transferase might overcome this competition by using a different site in the aminoacyl-tRNAs, presumably in the D-loop, that does not overlap with the EF-Tu binding site. A similar mechanism may operate for FemXWv which is expected to compete with EF-Tu for uptake of aminoacyl-tRNAs.
In conclusion, modeling and mutagenesis of tRNAAla has provided key information for rational design of FemXWv inhibitors since the bases essential for FemXWv activity were mapped in a relatively small portion of the tRNAAla acceptor stem comprising a total of eight bases including its single-stranded ACCA end and the distal base pairs G1–C72 and G2–C71. Design of substrate analogues should therefore target this region of the aminoacyl-tRNA substrate. In addition, the different modes of recognition of the acceptor stem by FemXWv and the AlaRS suggests that it will be feasible to achieve inhibitor selectivity for the peptidoglycan biosynthetic enzyme.
ACKNOWLEDGEMENTS
We thank M. Uzan for the gift of T4 DNA and J. Urbonavicius for critical reading of the article. European Community (EUR-INTAFAR, Project N°LSHM-CT-2004-512138, 6th PCRD); Fondation Recherche Médicale (fin de thèse to R.V.). Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- 3.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 4.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc. Natl Acad. Sci. USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goffin C, Ghuysen JM. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. (table of contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billot-Klein D, Shlaes D, Bryant D, Bell D, Legrand R, Gutmann L, van Heijenoort J. Presence of UDP-N-acetylmuramyl-hexapeptides and -heptapeptides in enterococci and staphylococci after treatment with ramoplanin, tunicamycin, or vancomycin. J. Bacteriol. 1997;179:4684–4688. doi: 10.1128/jb.179.15.4684-4688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellais S, Arthur M, Dubost L, Hugonnet JE, Gutmann L, van Heijenoort J, Legrand R, Brouard JP, Rice L, et al. Aslfm, the d-aspartate ligase responsible for the addition of d-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J. Biol. Chem. 2006;281:11586–11594. doi: 10.1074/jbc.M600114200. [DOI] [PubMed] [Google Scholar]
- 9.Plapp R, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. XVII. Biosynthesis of peptidoglycan and of interpeptide bridges in Lactobacillus viridescens. J. Biol. Chem. 1970;245:3667–3674. [PubMed] [Google Scholar]
- 10.Rohrer S, Berger-Bachi B. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob. Agents Chemother. 2003;47:837–846. doi: 10.1128/AAC.47.3.837-846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biarrotte-Sorin S, Maillard AP, Delettre J, Sougakoff W, Arthur M, Mayer C. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition. Structure. 2004;12:257–267. doi: 10.1016/j.str.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Benson TE, Prince DB, Mutchler VT, Curry KA, Ho AM, Sarver RW, Hagadorn JC, Choi GH, Garlick RL. X-ray crystal structure of Staphylococcus aureus FemA. Structure. 2002;10:1107–1115. doi: 10.1016/s0969-2126(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 13.Maillard AP, Biarrotte-Sorin S, Villet R, Mesnage S, Bouhss A, Sougakoff W, Mayer C, Arthur M. Structure-based site-directed mutagenesis of the UDP-MurNAc-pentapeptide-binding cavity of the FemX alanyl transferase from Weissella viridescens. J. Bacteriol. 2005;187:3833–3838. doi: 10.1128/JB.187.11.3833-3838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde SS, Blanchard JS. Kinetic and mechanistic characterization of recombinant Lactobacillus viridescens FemX (UDP-N-acetylmuramoyl pentapeptide-lysine Ne-alanyltransferase) J. Biol. Chem. 2003;278:22861–22867. doi: 10.1074/jbc.M301565200. [DOI] [PubMed] [Google Scholar]
- 15.Schneider T, Senn MM, Berger-Bachi B, Tossi A, Sahl HG, Wiedemann I. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 2004;53:675–685. doi: 10.1111/j.1365-2958.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- 16.Arbeloa A, Hugonnet JE, Sentilhes AC, Josseaume N, Dubost L, Monsempes C, Blanot D, Brouard JP, Arthur M. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 2004;279:41546–41556. doi: 10.1074/jbc.M407149200. [DOI] [PubMed] [Google Scholar]
- 17.Cremniter J, Mainardi JL, Josseaume N, Quincampoix JC, Dubost L, Hugonnet JE, Marie A, Gutmann L, Rice LB, et al. Novel mechanism of resistance to glycopeptide antibiotics in Enterococcus faecium. J. Biol. Chem. 2006;281:32254–32262. doi: 10.1074/jbc.M606920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouhss A, Josseaume N, Severin A, Tabei K, Hugonnet JE, Shlaes D, Mengin-Lecreulx D, Van Heijenoort J, Arthur M. Synthesis of the l-alanyl-l-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J. Biol. Chem. 2002;277:45935–45941. doi: 10.1074/jbc.M207449200. [DOI] [PubMed] [Google Scholar]
- 19.Pompeo F, van Heijenoort J, Mengin-Lecreulx D. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J. Bacteriol. 1998;180:4799–4803. doi: 10.1128/jb.180.18.4799-4803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouhss A, Josseaume N, Allanic D, Crouvoisier M, Gutmann L, Mainardi JL, Mengin-Lecreulx D, van Heijenoort J, Arthur M. Identification of the UDP-MurNAc-pentapeptide:l-alanine ligase for synthesis of branched peptidoglycan precursors in Enterococcus faecalis. J. Bacteriol. 2001;183:5122–5127. doi: 10.1128/JB.183.17.5122-5127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinnel T, Blanquet S. Maturation of pre-tRNA(fMet) by Escherichia coli RNase P is specified by a guanosine of the 5'-flanking sequence. J. Biol. Chem. 1995;270:15908–15914. doi: 10.1074/jbc.270.26.15908. [DOI] [PubMed] [Google Scholar]
- 23.Jacob AE, Hobbs SJ. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. The use of 5′-phospho-2 deoxyribocytidylylriboadenosine as a facile route to chemical aminoacylation of tRNA. Nucleic Acids Res. 1989;17:9649–9660. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodder M, Golovine S, Laikhter AL, Karginov VA, Hecht SM. Misacylated transfer RNAs having a chemically removable protecting group. J. Org. Chem. 1998;63:794–803. doi: 10.1021/jo971692l. [DOI] [PubMed] [Google Scholar]
- 26.Eiler S, Dock-Bregeon A, Moulinier L, Thierry JC, Moras D. Synthesis of aspartyl-tRNA(Asp) in Escherichia coli—a snapshot of the second step. EMBO J. 1999;18:6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, Tomita K. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 2006;25:5942–5950. doi: 10.1038/sj.emboj.7601433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 30.Robertson SA, Ellman JA, Schultz PG. A general and efficient route for chemical aminoacylation of transfer RNAs. J. Am. Chem. Soc. 1991;113:2722–2729. [Google Scholar]
- 31.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 33.Lovato MA, Swairjo MA, Schimmel P. Positional recognition of a tRNA determinant dependent on a peptide insertion. Mol. Cell. 2004;13:843–851. doi: 10.1016/s1097-2765(04)00125-x. [DOI] [PubMed] [Google Scholar]
- 34.Swairjo MA, Otero FJ, Yang XL, Lovato MA, Skene RJ, McRee DE, Ribas de Pouplana L, Schimmel P. Alanyl-tRNA synthetase crystal structure and design for acceptor-stem recognition. Mol. Cell. 2004;13:829–841. doi: 10.1016/s1097-2765(04)00126-1. [DOI] [PubMed] [Google Scholar]
- 35.Mueller U, Schubel H, Sprinzl M, Heinemann U. Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution. RNA. 1999;5:670–677. doi: 10.1017/s1355838299982304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanderson LE, Uhlenbeck OC. The 51-63 base pair of tRNA confers specificity for binding by EF-Tu. RNA. 2007;13:835–840. doi: 10.1261/rna.485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]