Abstract
Sequencing ribosomal RNA (rRNA) genes is currently the method of choice for phylogenetic reconstruction, nucleic acid based detection and quantification of microbial diversity. The ARB software suite with its corresponding rRNA datasets has been accepted by researchers worldwide as a standard tool for large scale rRNA analysis. However, the rapid increase of publicly available rRNA sequence data has recently hampered the maintenance of comprehensive and curated rRNA knowledge databases. A new system, SILVA (from Latin silva, forest), was implemented to provide a central comprehensive web resource for up to date, quality controlled databases of aligned rRNA sequences from the Bacteria, Archaea and Eukarya domains. All sequences are checked for anomalies, carry a rich set of sequence associated contextual information, have multiple taxonomic classifications, and the latest validly described nomenclature. Furthermore, two precompiled sequence datasets compatible with ARB are offered for download on the SILVA website: (i) the reference (Ref) datasets, comprising only high quality, nearly full length sequences suitable for in-depth phylogenetic analysis and probe design and (ii) the comprehensive Parc datasets with all publicly available rRNA sequences longer than 300 nucleotides suitable for biodiversity analyses. The latest publicly available database release 91 (August 2007) hosts 547 521 sequences split into 461 823 small subunit and 85 689 large subunit rRNAs.
INTRODUCTION
Initiated by the pioneering studies of Fox and Woese (1) 30 years ago and later on pursued by Pace, Olsen, Giovannoni, and Ward (2–5), the ribosomal RNA (rRNA) molecule has been established as the ‘gold-standard’ for the investigation of the phylogeny and ecology of microorganisms (6,7). Today the more than 500 000 publicly available small and large subunit (SSU and LSU) rRNA sequences ask for specialized quality controlled databases and appropriate software tools. In anticipation of this impending deluge of rRNA data, the development of the ARB software suite and the curation of its associated databases began more than 12 years ago (8). The software suite offers a graphical user interface and a wide variety of interacting software tools built around a common database. Furthermore, the ARB project provides structured, integrative knowledge databases for small and large subunit rRNAs. Based on regularly offered international workshops and the ARB mailing list it is currently estimated that the ARB software suite and its databases are employed worldwide by several thousand users from academia and industry. In addition to the ARB approach, there are currently three projects offering access to a set of curated ribosomal RNA sequence and alignment databases: the European rRNA databank at the University of Gent (http://www.psb.ugent.be/rRNA/) (9), the Ribosomal Database Project II (http://rdp.cme.msu.edu/) at Michigan State University in East Lansing, MI (10,11), and the greengenes project (http://greengenes.lbl.gov/) maintained by the Lawrence Berkeley National Laboratory in Berkeley, CA (12). All four projects offer at least one 16S rRNA dataset, but vary in the amount of sequences, quality checks, alignments, and update procedures. However, the ARB project is the only platform that actively incorporates homologous small (SSU) as well as large (LSU) subunit sequences from all three domains of life, the Bacteria, Archaea (16S/23S) and Eukarya (18S/28S). All projects provide web-based software tools for the alignment and classification of sequences as well as probe match functionalities. Downloading of sequences is provided in various formats including the commonly used FASTA and GenBank file formats. Additionally, greengenes provides ARB compatible datasets, but only for nearly full length sequences (>1250 bases) of Bacteria and Archaea.
An increasing awareness and motivation to catalogue and protect the biodiversity on Earth using molecular techniques demands comprehensive knowledge databases spanning all three domains of life. Furthermore, a majority of the sequences available is derived from cultivation independent biodiversity surveys, which rely on rapid pattern- or clone-based approaches that often generate partial rRNA sequences. To conserve this suboptimal information especially for diversity studies, state of the art databases need to retain partial sequences.
To compensate for the limited phylogenetic resolution of the SSU rRNA (13,14) the two fold larger LSU rRNA should now also be included in the rRNA approach (6). Especially for Eukaryotes the highly variable regions in the LSU rRNA are already commonly used for species discrimination (15). Triggered by a new capacity for cheap and rapid sequencing, there is a steady flow of approximately 10 000 rRNA sequences per month into the public databases of the International Nucleotide Sequence Database Collaboration (http://www.insdc.org). To make full use of these data for reliable phylogenetic reconstructions and biodiversity analysis careful inspection of each sequence and alignment is necessary. To support the users with this task, standardized procedures to assign a defined set of contextual information to each sequence must be established. Unified quality control mechanisms are urgently needed to intuitively flag potential problems with each sequence. The recent introduction of accelerated and less expensive sequencing technologies, such as pyrosequencing (16), and their successful application for a census of marine microbial diversity (17), further substantiates the need for comprehensive quality controlled databases for comparisons. Such databases provide a stable framework enabling biologists to transfer the copious data into reliable biological knowledge. The SILVA database project is designed to satisfy the request for comprehensive quality controlled and aligned rRNA datasets. It is intended to provide a central knowledge resource to alleviate users of the time consuming manual curation process.
MATERIALS AND METHODS
Sequence data
The SILVA release cycle and numbering corresponds to that of the EMBL database, a member of the International Nucleotide Sequence Database Collaboration (http://www.insdc.org). Thus, the ribosomal RNA sequences used to build version 91 of the SILVA databases, which is referred to in this paper, were retrieved from release 91 (June 2007) of EMBL. A complex combination of keywords including all permutations of 16S/18S, 23S/28S, SSU, LSU, ribosomal and RNA was used to retrieve a comprehensive subset of all available small and large subunit ribosomal RNA sequences. All candidate rRNA sequences extracted from the EMBL database were stored locally in a relational database system (MySQL). The specificity of the SILVA databases for rRNA is assured by the subsequent processing of the primary sequence information.
The source database providing the seed alignment, required for the incremental alignment process, included a representative set of 51 601 aligned rRNA sequences from Bacteria, Archaea and Eukarya with 46 000 alignment positions. The SSU alignment positions are currently kept identical with the ssu_jan04.arb database which has officially been released by the ARB project (http://www.arb-home.de) in 2004. For the large subunit RNA databases, an in-house, aligned database was used as the seed. It encompasses a representative set of 2868 sequences from all three domains (150 000 alignment positions). Since the quality of the final datasets critically depends on the quality of the seed alignments both datasets were iteratively cross-checked by expert curators during database build-up. Within this process, all sequences that could not be unambiguously aligned were removed from the seed.
Quality checks
Every imported SSU and LSU sequence had to pass a multi-stage quality inspection. Sequences were rejected if they were shorter than 300 unaligned nucleotides, if they were composed of more than 2% of ambiguities or more than 2% homopolymeric stretches longer than four bases, which means only bases exceeding homotetramers are counted, or if they had more than 5% identity to vector sequences. The identity was checked by querying a database of commonly used vector sequences, based on the EMVEC (http://www.ebi.ac.uk/blastall/vectors.html) and UniVec (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html) databases using the blastn tool. All thresholds to reject sequences were defined based on statistical analysis of the retrieved SSU and LSU sequences. Each sequence in the SILVA databases carries the percentages of ambiguities, homopolymers, and vector contamination. Additionally, a summary ‘sequence quality’ score is calculated according to the following formula, where Sq = sequence quality, A = % ambiguities, H = % homopolymers and V = % vector identity:
This score represents the mean of the three individual parameters, such that 100 is the best possible value. All sequences that passed the quality thresholds were automatically aligned against the seed alignment using the new SILVA INcremental Aligner (SINA).
Aligner
To cope with the huge amount of sequence information and to minimize the workload for manual curation, a new dynamic incremental profile sequence aligner (SINA) was developed. In the first step the aligner uses the suffix tree concept of ARB (8) to search for up to 40 closely related sequences in the seed alignment. The reference sequences from the seed are transferred into a partial order graph as used in (18), while preserving the positional identity from the reference alignment. The sequence under investigation is then aligned to this graph using a variant of the Needleman-Wunsch algorithm (19) with affine gap penalties and cost free overhang. The graph concept allows ‘jumping’ between the different references to find an optimal alignment for the different sequence regions. This technique enables the algorithm to correctly place bases that were e.g. deleted from the closest relative, but are present in the candidate sequence and other relatives. It also eliminates the need for synthetic full length sequences in the reference alignment as introduced for the NAST aligner (20). To further improve the alignment quality a variability statistic is used to give more weight to conserved positions. Results of each step of the aligner are reported to the database and shown in the corresponding fields of the exported ARB file (Tables 1–3). The ‘alignment quality’ score is a measure of the similarity with the seed sequences that are taken into account for the alignment process. The score is derived from the dynamic programming score by removing the effects of sequence length and positional weighting. High values (>90) mean that nearly identical sequences have been found within the seed alignment, resulting in a high likeliness for the alignment to be accurate. Low values indicate a high distance as perceived by the aligner, making the alignment task more difficult and lowering the average accuracy. Due to the size of the seed alignment, low values are rather rare and ask for manual inspection of the alignment. The ‘basepair’ score is calculated from the number of bases involved in helix binding according to the secondary structure model of Gutell et al. (21) as already implemented in the ARB package. Canonical and non-canonical base pairings are evaluated, weighted according to the cost model implemented in the Probe_Match (‘weighted mismatches’) tool in ARB (8). To fit our unified scoring scheme, the alignment quality and the base pair score were normalized to values between 0 and 100, such that 100 represents the maximum score. After aligning, the number of successfully aligned bases was again counted and sequences with less than 300 bases within the boundaries of the respective SSU or LSU rRNA genes were discarded.
Table 1.
Description of database fields in ARB files exported from SILVA for ARB specific fields and entries
| ARB field name | Owned by | Description |
|---|---|---|
| aligned | User | User-defined entry, e.g. name and date of the person who aligned the sequence |
| ambig | ARB | Ambiguities calculated in ARB using ‘count ambiguities’ |
| ARB_color | ARB | Stores the information about sequence colors |
| name | ARB | Internal ARB database ID, do not change! |
| nuc | ARB | Number of nucleotides; calculated by ARB using ‘count nucleotides’ |
| nuc_term | ARB | Number of nucleotides coding for the respective rRNA gene; calculated by ‘count nucleotides gene’ |
| remark | User | Field for remarks |
| tmp | ARB | Used by several ARB modules |
Table 2.
Description of database fields in ARB files exported from SILVA for Fields and entries imported from EMBL
| ARB field name | EMBL field | Description |
|---|---|---|
| acc | AC | Accession number |
| ali_xx/data | sequence | Sequence information |
| author | RA | Reference author(s) |
| clone | FT/clone | Clone from which the sequence was obtained |
| collected by | FT/collected_by | Name of the person who collected the specimen |
| collection_date | FT/collection_date | Date that the specimen was collected |
| country | FT/country | Geographical origin of sequenced sample |
| date | DT | Entry creation and update date separated by; |
| description | DE | Description |
| full_name | OS | Organism species |
| gene | FT/gene | Symbol of the gene corresponding to a sequence region |
| insdc | PR | The International Nucleotide Sequence Database Collaboration (INSDC) Project Identifier that has been assigned to the entry |
| isolate | FT/isolate | Individual isolate from which the sequence was obtained |
| isolation_source | FT/isolation_source | Describes the physical, environmental and/or local geographical source of the biological sample from which the sequence was derived |
| journal | RL | Reference location |
| lat_lon | FT/lat_lon | Geographical coordinates of the location where the specimen was collected |
| nuc_region | FT source | Identifies the biological source of the specified span of the sequence |
| nuc_rp | RP | Reference positions |
| product | FT/product | Name of the product associated with the feature |
| publication_doi | RX | Cross-reference DOI number |
| pubmed_id | RX | Cross-reference Pubmed ID |
| specific_host | FT/specific_host | Natural host from which the sequence was obtained |
| specimen_voucher | FT/specimen_Voucher | An identifier of the individual or collection of the source organism and the place where it is currently stored, usually an institution |
| start | FT rRNA | Start of the ribosomal RNA gene |
| stop | FT rRNA | Stop of the ribosomal RNA gene |
| strain | FT/strain | Strain from which the sequence was obtained |
| submit_author | RL | Submission authors from reference location |
| submit_date | RL | Submission date from reference location |
| tax_embl | OC | Organism classification according to EMBL |
| tax_embl_name | OC | Organism name taken from the classification field |
| tax_xref_embl | FT/db_xref | Database cross-reference: pointer to related information in another database |
| title | RT | Reference title |
| version | ID SV | Subversion from identification line |
Table 3.
Description of database fields in ARB files exported from SILVA for SILVA specific fields and entries
| ARB field name | Description |
|---|---|
| align_bp_score_slv | Calculates the number of bases in helices in the aligned sequence taken into account canonical and non canonical basepairing. The cost matrix is taken from ARB Probe_Match (8) |
| align_cutoff_head_slv | Unaligned bases at the beginning of the sequence |
| align_cutoff_tail_slv | Unaligned bases at the end of the sequence |
| align_family_slv | Names and scores of reference sequences in the alignment process |
| align_log_slv | Detailed aligner comments |
| align_quality_slv | Maximal similarity to reference sequence in the seed |
| aligned_slv | Data and time of alignment by Silva |
| ambig_slv | Calculated percent ambiguities in the sequences, a maximum of 2% is allowed |
| homop_slv | Calculated percentages repetitive bases with more than four bases, a maximum of 2% is allowed |
| homop_events_slv | Absolute number of repetitive elements with more than four bases |
| nuc_gene_slv | Aligned bases within gene boundaries |
| pintail_slv | Information about potential sequence anomalies detected by Pintail (22); 100 means no anomalies found. |
| alternative_name_slv | Synonyms or basonyms of the species according to the DSMZ ‘nomenclature up to date’ catalogue |
| seq_quality_slv | Summary sequence quality value calculated based on values from vector, ambiguities and homopolymers, 100 means very good |
| tax_gg | Taxonomy mapped from greengenes |
| tax_gg_name | Organism name in greengenes |
| tax_rdp | Nomenclatural taxonomy mapped from RDP II |
| tax_rdp_name | Organism name in RDP II |
| vector_slv | Percent vector contamination, a maximum of 5% is allowed |
Anomaly check
To check for sequence anomalies, a custom version of the Pintail software (22) was used. The software was specifically adapted for batch processing by the RDP II team. By design, Pintail can only detect anomalies between two sequences. To circumvent this limitation, a pairwise comparison of all sequences in the seed against a group of 20 sequences was performed. If a majority of the comparisons was deemed anomalous, the sequences were iteratively eliminated from the seed alignment until no such sequences remained. Subsequently, all aligned sequences of the SSU database were tested against their five closest relatives within this pruned seed. The number of ‘yes’, ‘likely’ and ‘no’ reported by Pintail was counted for each sequence and transferred into the ‘Pintail quality’ value. This score was normalized between 0 and 100, such that 100 indicates the best quality and a low probability that the sequence is anomalous or chimeric. Only SSU sequences were checked for anomalies because the Pintail software is currently not designed to handle LSU sequences.
Taxonomy
Every sequence in the SILVA databases carries the EMBL taxonomy assignment. Where available, the greengenes and RDP taxonomies where added for comparison. The EMBL taxonomy was retrieved simultaneously with the sequence, whereas the other taxonomies have been assigned to the sequences based on accession numbers. The greengenes taxonomic outline was acquired in June 2007 from the greengenes website (http://greengenes.lbl.gov/) and the RDP Nomenclatural Taxonomy was acquired from RDP II release 9.51. At the moment, no other up to date databases containing aligned LSU sequences are available. Therefore, the only taxonomy provided with the LSU database is the taxonomy used by EMBL. Type strain information has been added to the field ‘strain’ and is indicated by [T]. Mapping was done based on the RDP II dataset and is therefore only available for Bacteria.
Nomenclature
All organism names have been synchronized with the ‘Nomenclature up to date’ website of the Deutsche Sammlung für Mikroorganismen und Zellkulturen” DSMZ (released June 2007, http://www.dsmz.de/download/bactnom/names.txt) in order to stay consistent with the constant renaming of validly described species according to the recommendations published in the ‘International Journal of Systematic and Evolutionary Microbiology’. All former names are stored in the database and are visible on the web page, as well as in the corresponding field of the ARB databases (Tables 1–3).
SSU and LSU rRNA databases for ARB
Two types of precompiled databases for both small and large subunit ribosomal RNA sequences are available in the ARB format: the high-quality Ref databases and the comprehensive Parc databases. The Ref databases are subsets of Parc, which are exclusively comprised of nearly full length 16S/18S and 23S/28S rRNA sequences. A sequence is accepted if it is at least 1200 bases long. Additionally, sequences as short as 900 bases are included if they belong to the domain Archaea. Applying a strict cut-off at 1200 bases would result in the loss of the majority of sequences of this domain. Sequences in the LSU Ref database have to be at least 1900 bases long. For quality control, all sequences that could not be unambiguously aligned (alignment quality score <50 and <30 for SSU and LSU, respectively) were removed from the Ref databases. Both Ref databases are supplemented with a guide tree based on the full length sequence tree of the ARB Jan 04 SSU and the Ludwig LSU databases, respectively. The trees were built using the ARB parsimony tool with filters to remove highly variable positions. Common filters like the positional variability filters were recalculated for the Ref databases. Sequences with long branches in combination with low alignment qualities (<80) were removed from the Ref databases.
The rRNA Parc databases are a collection of all quality checked and automatically aligned rRNA sequences longer than 300 bases of the aligned rRNA gene (field ‘nuc_gene_slv’, Tables 1–3). The name Parc has been chosen according to the UniProt concept (23), where Parc stands for the comprehensive protein sequence archive. All sequences in the SILVA databases are associated with a rich set of sequence and process parameters. Included is information from the initial quality checks to the alignment process, as well as information taken directly from the EMBL entry (Tables 1–3). Together with the search and query functionalities on the web site and in ARB, one can quickly search for problematic sequences or generate individual high or low quality sequence subsets for further processing or curation. The ARB package can be used to export sequences in various formats like EMBL, GenBank, or aligned and unaligned FASTA.
Availability/Webpage
The SILVA databases are available via a web-based interface at http://www.arb-silva.de. The web interface is divided into six sections: the browser, search, list, download, background, and FAQs pages. Download of the complete Parc and Ref datasets in ARB format is available in the download section. It is also possible to download files that gain additional sequences from new releases. Subsets of aligned sequences from the Parc dataset can be retrieved from the website. This is currently possible via two entry points: a taxonomic browser and advanced search functions. After selecting a database and the desired taxonomy in the browser, the user can navigate through the taxonomy by clicking on the respective nodes. A cart system is used to easily select subsets of single sequences, complete groups or even phyla. The cart system keeps the selections for the SSU and LSU databases distinct. This allows the user to select sequences from both databases simultaneously without mixing the two sequence types. However, it must be noted that any misclassification or erroneous information provided by INSDC is currently propagated on the SILVA webpage.
Additionally, the advanced search functions of the SILVA website can be used to build custom subsets of sequences. In addition to simple searches e.g. for accession numbers, organism names, taxonomic entities, or publication DOI/PubMed IDs, complex queries over several database fields using constraints such as sequence length or quality values are possible. The results can be sorted according to accession numbers, organism names, sequence length, sequence and alignment quality and Pintail values. Before download, the search results must be added to the ‘List’. This can be done by either manually selecting the sequences by mouse click or by clicking on ‘Add complete result to List’ to mark and transfer all results.
The coloured bars on the search page and in the short and detailed sequence views of the browser given a fast overview of the different quality aspects assigned to every sequence. The length of the bars is a graphical representation of the respective quality value. The colours classify the information into four categories: A green bar represents a value equal to or greater than 75. Yellow bars stand for values equal to or greater than 50 but less than 75. Values less than 50 are expressed by an orange bar. Red bars are only used for scores of 0. Since ‘problematic’ sequences, sequences of inadequate quality, as well as insufficiently aligned sequences were discarded from the databases only the Pintail scores can have 0.
In the ‘List’ section of the website, the entries can be inspected, items can be deleted, and the download files can be created. By clicking on the ‘generate download’ button the user will be asked whether he would like to download the sequences as a multi-FASTA or ARB file from the download section of the web page. All generated files will be available for download on the download page for up to 24 h. The background section of the website provides additional information about the current status of the databases, and the FAQ section describes the main steps necessary to download subsets of sequences and how to merge the retrieved ARB databases with the user's personal ARB database.
Operating systems and programming languages
The SILVA core system was written in C++ and runs on an Ubuntu GNU/Linux 6.06 LTS based 64bit Dual Dual-Core Opteron cluster with at least 16 GB of main memory on each node. The database server runs MySQL 5.0 and features 32 GB of main memory. The Sun-grid engine (N1GE 6.0) is used to distribute jobs, such as importing, quality check, and aligning on the cluster. The web server is a LAMP system running Ubuntu GNU/Linux 6.06 LTS, Apache 2, MySQL 5.0, and PHP 5. It is connected to the internet via a synchronous 34 Mb connection. The website was written in PHP and Ajax and it is managed using the typo3 content management system in version 4.1. Due to the complexity of the system and the high hardware requirements the system is currently not intended for local installation.
RESULTS AND DISCUSSION
Data retrieval and processing
The total numbers of retrieved sequences and the number of and reasons for rejected sequences are listed in Table 4. Cross checks with RDP II and greengenes indicated a sensitivity of our search procedure of >99%. For ambiguities, homopolymers and vector contamination the numbers are non-additive, since some of the sequences may be affected by two or three parameters. Cut-off values have been determined based on a statistical evaluation with relaxed parameters (data not shown), and are intended to balance the quality of the databases with any loss of information. Manual inspection of the sequences rejected by the aligner showed that most of these sequences were not ribosomal RNA sequences.
Table 4.
Sequence retrieval and processing for SILVA 91
| SSU Parc | LSU Parc | |
|---|---|---|
| Candidates | 900 573 | 417 217 |
| <300 Bases | 320 327 | 297 218 |
| >2% Ambiguities | 8018 | 2193 |
| >2% Homopolymers | 19 240 | 4772 |
| >5% Vector contamination | 14 973 | 2573 |
| Insufficient relatives | 49 063 | 13 081 |
| <300 Gene bases | 25 961 | 7510 |
| <30 Alignment quality or base pair score | 6583 | 3390 |
| Total sequences in Parcs | 461 823 | 85 689 |
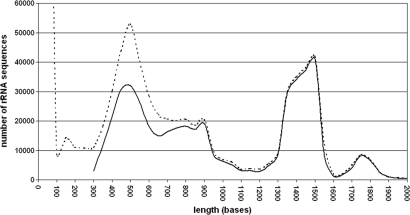
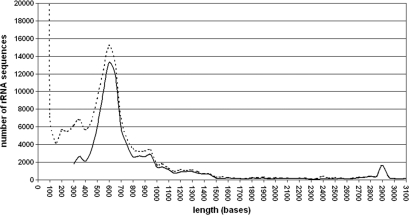
A comparison of the length distribution immediately after importing the SSU sequences with the length distribution of aligned sequences confirmed that no unexpected loss of sequences in certain length classes occurred (Figure 1). Partial sequences between 300 and 800 bases were more frequently rejected than longer ones. A closer comparison of sequence quality versus sequence length confirmed that sequences below 700 bases tend to be of low quality. These ‘problematic’ sequences may be generated in diversity studies based on single strand sequencing. The high number of rejected sequences with less than 300 bases is evidence for the increase in short length tag sequencing using e.g. pyrosequencing machines. The LSU database shows a similar distribution for rejected sequences as the SSU database (Figure 2).
Figure 1.
Sequence length distribution of rRNA genes in the SILVA 91 SSU database. The dotted line represents the sequence distribution directly after importing, the solid line after quality checks and alignment. The huge amount of sequences around 100 bases reflect the first impact of tag sequencing approaches.
Figure 2.
Sequence length distribution in the SILVA 91 LSU database. The dotted line represents the sequence distribution directly after importing, the solid line after quality checks and alignment. The huge amount of sequences around 100 bases reflect the first impact of tag sequencing approaches.
As expected, the SSU sequence length distribution follows the prominent primer sets used for sequencing specific conserved regions on the 16S/18S rRNA gene (24). A distinct peak exists around 500 bases, a small one at 900 bases, and a peak between 1300 and 1500 bases. The large number of sequences with 300 and 600 bases is typical for diversity studies that use single reads or fingerprint techniques like DGGE (25). A text search for ‘DGGE’ across all fields of the SSU Parc database using ARB showed that 8241 (93%) out of 8889 ‘DGGE’ sequences found belong to the 300–600 nucleotide length class. A taxonomic breakdown for the 300 to 600, 600 to 1000, and 1300 to 1600 bases length classes revealed that 80 to 90% of all sequences per class were of bacterial origin. Nevertheless, from the shortest to the longest length class, the relative numbers for Eukarya decreases, whereas Archaea and Bacteria peaked in the 600–1000 and 1300–1600 length classes, respectively. This again reflects the application of the typical universal primers for Bacteria (24) and Archaea (26).
A comparison of the number of sequences hosted by the SILVA, greengenes, and RDP II projects revealed that the SILVA SSU Ref database contains roughly the same amount of bacterial and archaeal sequences as greengenes (12) [SILVA: 165 928, greengenes: 165 759 (July 2007)] Furthermore, SILVA contains 2423 more nearly full length sequences for Bacteria than RDP II (163 505, release 9.52) (11). This is surprising considering SILVA's less frequent release cycle (currently synchronized with major EMBL releases); one would thus anticipate SILVA to contain fewer sequences. This may have been due to a higher sensitivity in SILVA's search and alignment protocol. Different quality control mechanisms should not have a significant influence, since only nearly full length sequences have been taken into account for this comparison.
With this respect it has to be emphasised that the primary intention of the SILVA project is not to offer the biggest database by size but more importantly to provide reliable rRNA datasets with a robust set of processing and quality values assigned to each sequence. Such quality values enable users to easily evaluate sequences in order to create subsets of sequences for specific applications, or to extract the sequences that need further attention with respect to sequence and/or alignment quality or anomalies. The alternative taxonomies and type strain information, as well as the latest nomenclature, will facilitate the daily work flow of diversity analysis using classical clone based and high throughput sequencing approaches. Additionally, SILVA provides two LSU databases to support the increasing use of molecular markers with a higher resolution than the SSU rRNA (13). A taxonomic breakdown of the LSU Parc database contents showed that already 91% of the sequences are of eukaryotic origin.
Alignment and aligner
The current SILVA alignment is based on 46 000 and 150 000 alignment positions for the small and large subunit rRNA, respectively. The reasons for the large amount of alignment positions are: (i) large insertions often present in Eukarya and (ii) sequencing errors, such as additional artificial bases often found in homopolymeric sequence stretches. Such errors are common and require placement to be filtered before phylogenetic tree reconstruction, without corrupting the rest of the alignment.
In the ‘align-to-seed’ approach implemented in the SILVA system, well aligned sequences from seed datasets are used as references for new, unaligned sequences. Therefore, the quality of the final alignment strongly depends on the accuracy of the seed alignment. To further improve the quality of the SSU and LSU seed databases a manual curation process was performed on the latest officially released ARB dataset from January 2004.
The SSU seed hosts currently over 1000 unpublished sequences that primarily cover the domain Archaea. These sequences further improve the alignment in regions of the original SSU January 2004 dataset with sparse sequence coverage. In summary, the quality and consistency of all of the seed alignments is excellent. Only minor inconsistencies could not be resolved in the Eukarya. Nevertheless, the Parc datasets exceed the corresponding SSU and LSU seeds by a factor of 8 to 25. This probably indicates that not every phylum is equally represented in the seed. Hence, before using the alignments for in-depth phylogenetic analysis, the alignment of the selected sequence should be carefully checked. Problematic sequences can be easily filtered out by the quality values followed by manual curation. The SILVA team highly appreciates the return of manually inspected and corrected alignments of sequence subsets for inclusion in the SILVA seed. This will allow us to further increase the quality of future alignments.
To manage the deluge of data currently available in the public databases, a new aligner (SINA) has been developed. Similar to existing aligners, such as the Fast Aligner implemented in ARB (8) or the NAST aligner (20), the tool uses related sequences from the reference alignment as a template. For benchmarking the performance of SINA, standard tools, such as BAliBASE (27), could not be used since they are restricted to protein sequences. Benchmark results were obtained by removing and realigning each sequence from the seed. The results were internally compared to the original alignment by counting the number of matching and non-matching columns. Overall, SINA correctly placed 99.8% of all bases in the alignment. Furthermore, 33% and 80% of all sequences tested had no, or less than 1%, alignment errors, respectively. The high accuracy was gained in extensive test runs by changing parameter sets for gap insertions/extension parameters and family sizes combined with subsequent manual inspection of the results by expert curators. The design and implementation of SINA as individually running processes allows distributed aligning on cluster nodes. More than one sequence per second can be aligned per CPU.
Future developments
To account for the growing awareness in ecology that sequence information must be treated in the proper environmental context (28), emphasis was put on the retrieval of contextual (meta)information from public databases. For easy visualisation, the ‘Environment’ subsection is available in the detailed view of the browser. Additionally, basic environmental parameters, such as exact location and time of sampling as well as physical, chemical, and biological information about the sampling site, will be added in collaboration with the International Census of Marine Microbes (ICoMM), where similar efforts are ongoing (http://icomm.mbl.edu/). In upcoming releases of the SILVA databases a crosslink to the genomes mapserver at http://www.megx.net (29) will allow the geographic visualization of the sequence information as long as the location is provided. The direct addition of tag sequences below 300 nucleotides as typically produced by pyrosequencing, is not currently planned for SILVA, since it is already a main objective of the ICoMM agenda (17). Sequence based search options and alignment of user provided sequences are under development for the SILVA webpage. Finally, it must be stressed that an appropriate and stable phylogenetic classification of all rRNA sequences is urgently needed. Efforts in collaboration with Bergey's trust are ongoing and the information will be incorporated as soon as it becomes electronically available.
CONCLUSIONS
The new SILVA system provides comprehensive, quality controlled, richly annotated and aligned, reference rRNA databases to support the molecular assessment of biodiversity, as well as investigations of the evolution of organisms. Applications of the databases range from basic research in microbiology and molecular ecology to the detection of contaminants and pathogens in biotechnology and medicine. Molecular taxonomy and diagnostics have already revolutionized our view on microbial diversity on Earth (17,30,31), and the added value of molecular techniques for the determination of eukaryotic diversity has recently been documented by Tautz et al. (32). The SILVA databases combined with the ARB software suite provide a stable and easy to use workbench for researchers worldwide to perform in depth sequence analysis and phylogenetic reconstructions. It is designed as a knowledge database to assist in the daily effort to keep pace with the increasing amount of data flooding our general-purpose primary databases.
ACKNOWLEDGMENTS
We would like to thank Ralf Westram for expert assistance with the ARB software suite, the company Pixelmotor for designing and implementing the webpage and all colleagues and students who helped with the manual curation of the databases. We would also thank James Cole, George Garrity and the RDP II team for help with Pintail and fruitful discussions. We are grateful for funding from the Max Planck Society. Funding to pay the Open Access publication charges for this article was provided by the Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fox GE, Pechman KR, Woese CR. Comparative cataloging of 16S ribosomal ribonucleic acid: molecular approach to procaryotic systematics. Int. J. Bacteriol. 1977;27:44–57. [Google Scholar]
- 2.Pace NR, Stahl DA, Olsen GJ, Lane DJ. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 3.Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 4.Giovannoni SJ, DeLong EF, Olsen GJ, Pace NR. Phylogenetic groupspecific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 6.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acid Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wuyts J, Perriere G, de Peer YV. The European ribosomal RNA database. Nucleic Acid Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acid Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acid Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig W, Schleifer KH. Molecular phylogeny of bacteria based on comparative sequence analysis of conserved genes. In: Sapp J, editor. Microbial Phylogeny and Evolution, Concepts and Controversies. New York: Oxford university press; 2005. pp. 70–98. [Google Scholar]
- 14.Peplies J, Glöckner FO, Amann R, Ludwig W. Comparative sequence analysis and oligonucleotide probe design based on 23S rRNA genes of Alphaproteobacteria from North Sea bacterioplankton. Syst. Appl. Microbiol. 2004;27:573–580. doi: 10.1078/0723202041748172. [DOI] [PubMed] [Google Scholar]
- 15.Wuyts J, De Rijk P, Van de Peer Y, Winkelmans T, De Wachter R. The European Large Subunit Ribosomal RNA Database. Nucleic Acid Res. 2001;29:175–177. doi: 10.1093/nar/29.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Genome sequencing in microfabricated high-density picolitre reactors (vol 437, pg 376, 2005) Nature. 2006;441:120–120. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 19.Needleman SB, Wunsch CD. A general method applicable to search for similarities in amino acid sequence of 2 proteins. J. Mol. Biol. 1970;48:443. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 20.DeSantis TZ, Dubosarskiy I, Murray SR, Andersen GL. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics. 2003;19:1461–1468. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 21.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang HZ, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acid Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLong EF. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: Latest developments of the multiple sequence alignment benchmark. Proteins Struct. Funct. Bioinform. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 28.Field D, Garrity G, Gray T, Selengut J, Sterk P, Thomson N, Tatusova T, Cochrane G, Glöckner FO, et al. Meeting Report: “eGenomics: Cataloguing our Complete Genome Collection III”. Comp. Funct. Genom. 2007;2007:1–7. doi: 10.1002/cfg.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombardot T, Kottmann R, Pfeffer H, Richter M, Teeling H, Quast C, Glöckner FO. Megx.net - database resource for marine ecological genomics. Nucleic Acid Res. 2006;34:D390–D393. doi: 10.1093/nar/gkj070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong SH, Bunge J, Jeon SO, Epstein SS. Predicting microbial species richness. Proc. Natl Acad Sci. USA. 2006;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedros-Alio C. Marine microbial diversity: can it be determined? Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP. DNA points the way ahead of taxonomy - In assessing new approaches, it's time for DNA's unique contribution to take a central role. Nature. 2002;418:479–479. doi: 10.1038/418479a. [DOI] [PubMed] [Google Scholar]