Abstract
Oxidative stress activates the transcription factor NRF2, which in turn binds cis-acting antioxidant response element (ARE) enhancers and induces expression of protective antioxidant genes. In contrast, the transcriptional repressor BACH1 binds ARE-like enhancers in cells naïve to oxidative stress and antagonizes NRF2 binding until it becomes inactivated by pro-oxidants. Here, we describe the dynamic roles of BACH1 and NRF2 in the transcription of the heme oxygenase-1 (HMOX1) gene. HMOX1 induction, elicited by arsenite-mediated oxidative stress, follows inactivation of BACH1 and precedes activation of NRF2. BACH1 repression is dominant over NRF2-mediated HMOX1 transcription and inactivation of BACH1 is a prerequisite for HMOX1 induction. In contrast, thioredoxin reductase 1 (TXNRD1) is regulated by NRF2 but not by BACH1. By comparing the expression levels of HMOX1 with TXNRD1, we show that nuclear accumulation of NRF2 is not necessary for HMOX1 induction; rather, BACH1 inactivation permits NRF2 already present in the nucleus at low basal levels to bind the HMOX1 promoter and elicit HMOX1 induction. Thus, BACH1 confers an additional level of regulation to ARE-dependent genes that reveals a new dimension to the oxidative stress response.
INTRODUCTION
Reactive oxygen species (ROS) pose a serious threat to all aerobic organisms that maintain redox homeostasis in the face of constant exposure to environmental oxidants. ROS are harmful due to their reactivity with many cellular macromolecules. To maintain cells in a state of redox balance, biochemical antioxidants and a host of antioxidant enzymatic reactions supply the needed reduction potential. Antioxidant defenses exist in a balance with endogenous oxidants, and it is the disruption of this balance that characterizes the pathogenesis of many human diseases and aging.
Arsenite, the trivalent form of inorganic arsenic, is an environmental contaminant of major concern. Arsenic is a potent electrophilic inducer of oxidative stress with many of its effects attributable to its affinity for soft nucleophiles, such as cysteine residues in glutathione (GSH) and proteins (1). Arsenite exposure results in rapid oxidation of glutathione (2) thereby disrupting intracellular redox status (3). In response to the oxidative stress mediated by arsenite, cells induce the antioxidant battery of protective enzymes, of which heme oxygenase-1 (HMOX1) and thioredoxin reductase-1 (THXRD1) are two well-recognized members.
Transactivation of HMOX1 and of other antioxidant genes is regulated by binding of the transcription factor NRF2 (Nuclear factor erythoid-derived 2 related factor 2) to a cis-acting enhancer element known as the antioxidant response element (ARE). Activation of NRF2 requires its translocation to the nucleus, formation of a transcriptionally active complex through dimerization with small MAF (sMAF) proteins and binding to ARE enhancer motifs (4). The ARE is one form of MAF response element (MARE) having the core sequence RTGAYNNNGC (reverse complement: GCNNNRTGAY) (5) and additional flanking nucleotides that increase the specificity of NRF2 recognition (6,7). Some variations of the ARE motif can be recognized by other regulatory factors, in addition to NRF2, including NRF1 (8), NRF3 (9) and BACH1 (BTB and CNC homolog 1) (10). Thus, variation of ARE motif sequences contribute to overlapping DNA binding by factors that compete with NRF2 for ARE binding (11). This arrangement appears to exist between NRF2 and BACH1.
BACH1 is a transcriptional repressor (12) that is conserved and ubiquitously expressed in tissues (13,14) though its global activity in gene regulation is poorly characterized. Like several other MAF-related transcription factors, particularly NRF2, BACH1 heterodimerizes with sMAF proteins in order to bind DNA. In the basal state, BACH1/sMAF heterodimers interact with the MARE-like enhancer sites recognized by NRF2 or NF-E2 to inhibit expression of the corresponding genes (13,15). As a repressive transcription factor, BACH1 allows gene induction upon its release from enhancer elements. Originally, BACH1 was described as a heme-regulated repressor of β-globin genes (12) and HMOX1 (14–18). More recently, BACH1 has been suggested to play a role as a sensor of oxidative stress. Human BACH1 is a thiol-rich protein possessing 34 interspersed cysteine amino acids, of which two are responsible for BACH1 inactivation by oxidants (17). Therefore, it is possible that heme and oxidants trigger gene induction simply by relieving BACH1 repression (19). In this regard, it has been demonstrated that BACH1 plays a role in redox induction of HMOX1 (17) and NQO1 (10), though the exact mechanism of this repression is not clear.
The relationship between BACH1 inactivation and NRF2 activation during the induction of antioxidant genes is unknown. NRF2 coordinates induction of genes through its interaction with ARE enhancer motifs, frequently located 5′ to the transcriptional start site (TSS) of several well-characterized antioxidant genes (5). In the absence of oxidative stress, the cytosolic protein KEAP1 (Kelch-like ECH Associated Protein 1) directs E3 ligase-dependent proteasomal degradation of newly synthesized NRF2. As a result of this continuous degradation, NRF2 is effectively maintained at very low cellular levels. KEAP1, like BACH1, is a thiol-rich protein, and oxidation of a few of KEAP1's cysteines blocks NRF2 degradation (20–22). Consequently, activation of NRF2 is dependent on nuclear accumulation of de novo synthesized protein for subsequent binding of ARE motifs (23). At a few known genes, including HMOX1, BACH1 binds ARE motifs to the exclusion of NRF2.
Here, we have tested whether oxidative stress induced by arsenite exposure can trigger gene induction by simply releasing repressive BACH1 from ARE motifs. We find that NRF2 and BACH1 bind to two distal ARE enhancer sites far upstream of the HMOX1 TSS and that BACH1 removal is necessary for NRF2-mediated HMOX1 gene induction. In contrast to HMOX1, TXNRD1, which has a single ARE motif located 9 bp upstream of the TSS, is regulated by NRF2 but not by BACH1. Comparison of TXNRD1 expression with that of HMOX1 demonstrates that BACH1 repression is dominant over NRF2-mediated transcription. The dynamic interplay between BACH1 and NRF-2 can produce distinct regulatory expression patterns at different oxidative stress-induced genes.
MATERIALS AND METHODS
Cells and chemical treatments
HaCaT (24) human keratinocytes were grown in DMEM (Mediatech, Herndon, VA, USA) supplemented with 5% fetal bovine serum (Sigma Aldrich, St Louis, MO, USA) and 1% penicillin–streptomycin solution (Invitrogen, Carlsbad, CA, USA). Cells were incubated at 37°C with 5% CO2 and grown to ∼90% confluence before treatment. Aqueous solutions of NaAsO2 (hereafter referred to as arsenite) (Sigma Aldrich) were prepared from a 1000 × stock in ddH2O.
Whole cell extracts, nuclear extracts and immunoblotting
For preparation of whole cell extracts, cells were washed and harvested in PBS containing 1 × Complete Protease Inhibitor ™ (Roche Diagnostics Corporation, Indianapolis, IN) and lyzed by sonication (Sonic Dismembrator 60, Fisher Scientific) on ice 3 × 10 s in 300 μl NETN buffer (100 mM NaCl, 20 mM Tris pH 8.0, 1 mM EDTA, 0.5% NP-40, 1 × Complete Protease Inhibitor). Nuclear and cytosolic extracts were prepared using a nuclear extraction kit from Panomics (Fremont, CA, USA) according to the manufacturer's protocol. Protein concentrations were measured using the Bradford assay and concentrations adjusted to 1 μg/μl. Proteins were separated by SDS–PAGE, transferred to PVDF and probed for NRF2 (H300; Santa Cruz Biotechnology), β-actin (Sigma), HMOX1 (C20; Santa Cruz Biotechnology), in blocking buffer containing 3% NFDM in PBST (0.1 M PBS with 0.2% Tween 20). Blots were probed for BACH1 (C20; Santa Cruz Biotechnology) in blocking buffer containing 1% BSA in PBST. After washing, the blots were incubated with species-appropriate HRP-conjugated secondary antibody (Santa Cruz), incubated with chemiluminescent reagent (PicoWest Super Signal, Pierce Rickford, IL, USA) and visualized by exposing to film (GE Healthcare, Piscataway, NJ, USA).
RNA isolation and real-time RT–PCR
Total RNA was extracted using NucleoSpin RNA II columns (Macherey-Nage, Bethlehem, PA) according to the manufacturer's protocol. cDNA was synthesized by reverse transcription of 1 μg total RNA in a total volume of 20 μl containing 1 × reverse transcriptase buffer (Invitrogen, Carlsbad, CA, USA), 25 μg/ml oligo (dT)12–18 (Invitrogen), 0.5 mM dNTP mix (GeneChoice, Frederick, MD), 10 mM dithiothreitol (Invitrogen), 20 U of RNase inhibitor (RNasin®; Promega, Madison WI) and 100 U of SuperScript≈ II reverse transcriptase (Invitrogen). Resulting cDNA products were diluted in a final volume of 200 μl and a 2 μl aliquot was used as template for subsequent quantification by real-time PCR amplification. Quantitative real-time PCR was performed in a 25 μl reaction mixture containing 1 × Clonetech QTaq polymerase reaction mix (Mountain View, CA), 1 × SybrGreen (Invitrogen) as a marker of amplification, 0.1 μM of each primer and 2 μl of template cDNA. Products were amplified with human HMOX1 primers (forward: 5′-CTCAAACCTCCAAAAGCC-3′ and reverse: 5′-TCAAAAACCACCCCAACCC-3′), TXNRD1 primers (forward: 5′-CTTTTTCATTCCTGCTACTCTACC-3′ and reverse: 5′-CTCTCTCCTTTTCCCTTTTCC-3′), BACH1 primers (forward: 5′-TGCGATGTCACCATCTTTGT-3′ and reverse: 5′-CCTGGCCTACGATTCTTGAG-3′), NRF2 primers (forward: 5′-GAGAGCCCAGTCTTCATTGC-3′ and reverse: 5′-TGCTCAATGTCCTGTTGCAT-3′). Amplifications were performed using an Opticon 2 Real-Time PCR Detection System (MJ research). Cycle threshold (Ct) of each sample was automatically determined to be the first cycle at which a significant increase in optical signal above an arbitrary baseline was detected. Amplification of β-actin cDNA in the same samples was used as an internal control for all PCR amplification reactions. Relative mRNA expression was quantified using the comparative Ct (ΔCt) method and expressed as 2−ΔΔ Ct. Each assay was done in triplicate.
Chromatin immunoprecipitation (ChIP) analysis
Nuclei were isolated from formaldehyde (1% final) fixed cells by lysing cells in buffer containing 5 mM PIPES (pH 8.0), 85 mM KCl, 0.5% NP-40 and 1 × Complete Protease Inhibitor ™. DNA was fragmented by sonication (Diagenode Bioruptor) in buffer containing 50 mM Tris–HCl pH 8.1, 10 mM EDTA, 1% SDS, 1 × Complete Protease Inhibitor ™. DNA-cross-linked proteins were immunoprecipitated from precleared samples with 2–5 μg of each specific antibodies, including anti-NRF2, anti-BACH1 or rabbit IgG control (Millipore), or for use as total input chromatin. Antibodies were pulled down with protein A beads at 4°C overnight. Recovered beads were re-suspended in 1 × dialysis buffer [2 mM EDTA, 50 mM Tris–HCl (pH 8.0), 0.2% Sarkosyl and 1 × Complete Protease Inhibitor ™] and washed twice with dialysis buffer, following which pellets were washed four times with IP wash buffer [100 mM Tris–HCl (pH 9.0), 500 mM LiCl, 1% NP-40, 1% deoxycholic acid and 1 × Complete Protease Inhibitor ™]. Antibody/protein/DNA complexes were eluted in IP elution buffer (50 mM NaHCO3, 1% SDS) with vigorous shaking. Immunoprecipitated DNA and total input chromatin were diluted to 120 μl with water, brought up to 0.3 M NaCl. Cross-linking was reversed at 65°C overnight in the presence of RNase A (10 μg), followed by the addition of RNase A followed by protease K digestion at 45°C for 2 h. DNA was purified using a Qiaquick PCR purification kit per manufacturer protocols. Samples were evaluated for enrichment by quantitative real-time PCR (qRT–PCR) in a 25 μl reaction mixture containing 1 × BD QTaq polymerase reaction mix (BD Biosciences), 1 × SybrGreen (Invitrogen) as a marker of DNA amplification, and 0.1 μM of each primer (Supplementary Table 1). ChIP enrichment was evaluated using primers to regions upstream of the human HMOX1 or TXNRD1 genes (Table S1). Relative efficiency of each PCR primer was determined using input DNA and adjusted accordingly. The DNA in each ChIPed sample was normalized to the corresponding input chromatin (ΔCt) and enrichment was defined as the change in Ct in treated samples relative to untreated controls (ΔΔCt), relative to the IgG negative control. Exponential ΔΔCt values were converted to linear values (2−ΔΔCt) for graphical presentation as either as fold change or percent change, where indicated.
RESULTS
BACH1 inactivation precedes NRF2 activation
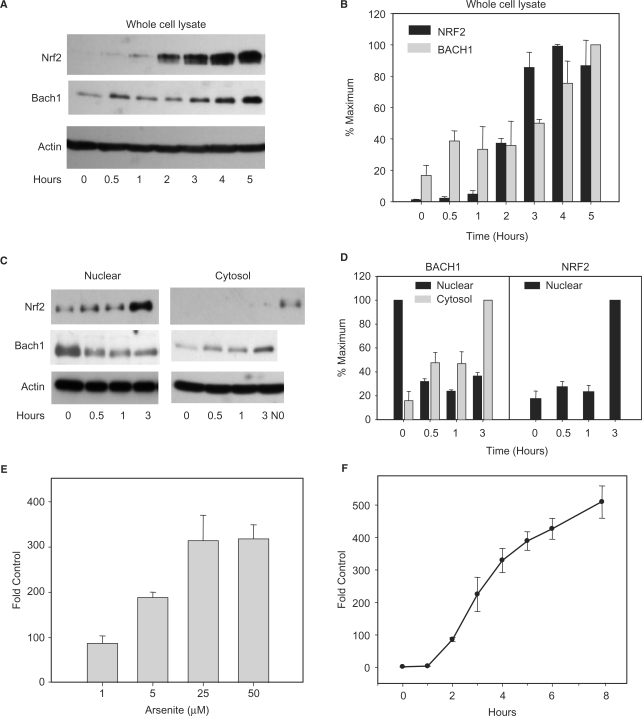
To characterize activation of the antioxidant response following arsenite exposure, we first characterized cellular and subcellular changes in NRF2 and BACH1 disposition. In control cells, NRF2 is present at very low levels in whole cell extracts attributable to ongoing proteasomal degradation mediated by KEAP1 (25). After treatment with 25 μM arsenite, NRF2 accumulates to high levels in whole cell extracts consistent with oxidative inactivation of KEAP1 (23) (Figure 1A and B). Importantly, NRF2 accumulation is negligible during the first hour after arsenite treatment, and does not reach maximum levels until 3 h after treatment. This initial lag period represents the time necessary for translational synthesis of new NRF2 protein (26), which precedes nuclear translocation and subsequent ARE-mediated gene induction. In comparison to NRF2, BACH1 gradually accumulates in whole cell extracts over 5 h after treatment. The extent to which BACH1 accumulation is attributable to protein stabilization is unknown; however, no change in BACH1 mRNA expression was observed (data not shown).
Figure 1.
Activation of antioxidant response and induction of HMOX1. (A) Representative immunoblots of total cellular proteins (20 μg) illustrating the effect of 25 μM arsenite on NRF2 and BACH1 protein levels. (B) Graphical representation of three separate experiments showing changes in expression of NRF2 and BACH1 proteins normalized to β-actin levels. (C) Representative immunoblots of nuclear (10 μg) and cytosolic proteins (20 μg) illustrating changes in subcellular localization of NRF2 and BACH1 following arsenite treatment (25 μM). (D) Graphical representation of three separate experiments showing changes in nuclear and cytosolic NRF2 and BACH1 proteins levels normalized to β-actin levels. The presence of NRF2 in nuclear extract at 0 h (N0) is provided as a positive immunoblot control for cytosolic NRF2. Quantification represents the mean ± SEM of three independent experiments. (E) Dose response of HMOX1 mRNA expression in HaCaT cells following a 6-h continuous treatment with the indicated concentration of arsenite. (F) Time course of HMOX1 mRNA expression in HaCaT cells following treatment with 25 μM arsenite. HMOX1 expression was determined using quantitative real-time PCR (qRT–PCR). HMOX1 mRNA concentrations were normalized to β-actin mRNA and expressed as fold change relative to untreated controls. Values represent at least three independent experiments quantified in triplicate.
Nuclear localization of both NRF2 and BACH1 is essential for their transcriptional activity and can be used as an indicator of their activation. To investigate their temporal activation following arsenite treatment, changes in nuclear and cytosolic distribution were examined (Figure 1C and D). Nuclear NRF2 accumulation parallels that observed in whole cell extracts. Prior to treatment, very low levels of NRF2 are present in nuclear fractions. Consistent with NRF2 levels in whole cell extracts, nuclear NRF2 accumulation is negligible during the first hour after arsenite treatment but becomes highly elevated by 3 h after treatment. Throughout the time course, and despite the accumulation of NRF2 in whole cell and nuclear fractions, there is little or no NRF2 accumulation in the cytoplasmic fraction. Rather, whenever detected, NRF2 is always associated with nuclear fractions, consistent with the rapid transport of NRF2 into the nucleus after its synthesis. Detection of NRF2 in nuclear fractions of control cells shows that even in the absence of oxidant exposure a low level of NRF2 resides in the nucleus.
In contrast to NRF2, BACH1 is rapidly exported from the nucleus following arsenite treatment, reaching minimal nuclear levels by 30 min after treatment, corresponding with an increase in cytosolic levels. BACH1 levels in the cytosol increase by 2-fold during the first hour after arsenite treatment and reach maximum observed levels by 3 h after treatment (Figure 1C and D).
Oxidative stress and arsenite are known to strongly induce HMOX1. Time-course analyses following 25 μM arsenite treatment shows a rapid induction of HMOX1 mRNA following an initial 1-h lag period (Figure 1F), coinciding with the time required for nuclear localization of de novo synthesized NRF2 protein. This induction continues in a linear fashion through 8 h after treatment without achieving a steady-state plateau. Together, these results show that HMOX1 transcription is preceded by BACH1 inactivation and occurs in parallel with, rather than following, NRF2 activation, suggesting that BACH1 inactivation is the antecedent event corresponding with transcriptional initiation of HMOX1.
BACH1 and NRF2 bind to the same ARE motifs in the HMOX1 promoter
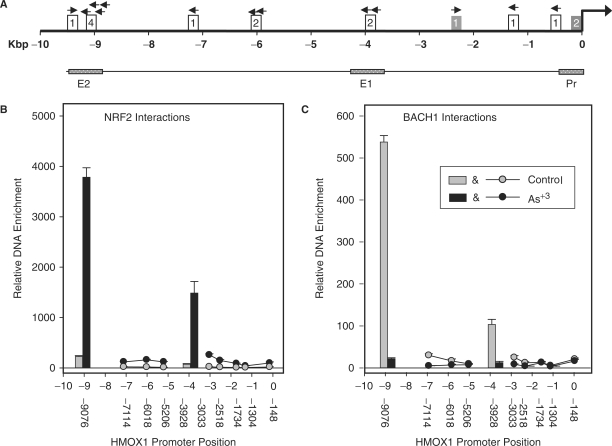
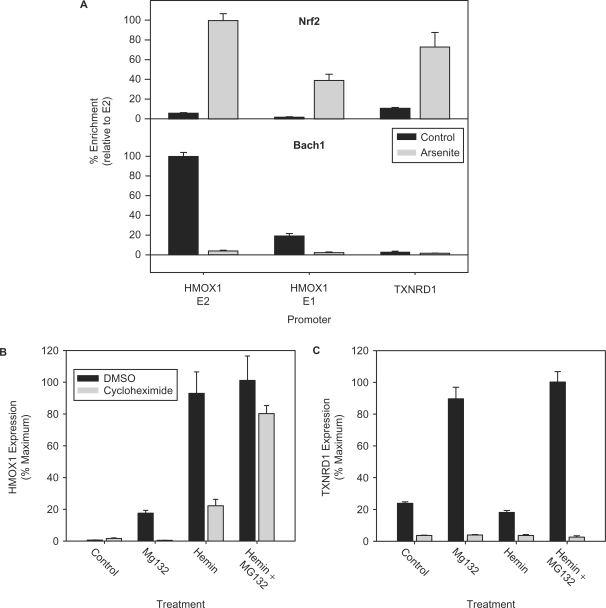
To identify the maximum possible BACH1- and NRF2-binding sites, we searched 10 kb upstream of the HMOX1 TSS for core ARE motifs conforming to the sequence RTGAYNNNGC or its reverse complement (5). Twelve consensus elements were identified (Table 1) and each of these sites were investigated for NRF2 and BACH1 interactions by ChIP analysis (Figure 2A). Of the 12 ARE motifs, NRF2 and BACH1 interact with the same two sites containing multiple ARE motifs; one, a more proximal site located at −3928 bp upstream of the TSS (E1) and the other a more distal site at −8979 bp upstream (E2). NRF2 binds both of these sites after arsenite treatment (Figure 2B) while BACH1 binds both of them in arsenic naïve cells (Figure 2C). The proximal E1 element is composed of two slightly different ARE core motifs having the sequences GCtgcGTCAT and GCtgaGTCAC and separated by 54 nt. The more distal E2 site consists of four identical repeats of the core ARE motif having the sequence GCtraGTCAC, each separated by 19 nt. An additional single motif is located at −9491 bp, 415 bp further upstream from the end of this ARE tetrad for a total of five elements in this DNA region. In control cells, BACH1 binding at the E2 site is 5-fold greater than at E1. Following arsenite treatment, NRF2 binding at the distal E2 tetrad is greater than at the E1 site by ∼2.5-fold. None of the other five remaining ARE motifs were bound by either NRF2 or BACH1. These data show that BACH1 and NRF2 undergo reciprocal binding at the E1 and E2 enhancers following oxidative stress initiated by arsenite treatment.
Table 1.
DNA sequences consistent with the consensus ARE motifs located within 10 Kb upstream of the HMOX1 TSS and 1 Kb upstream of TXNRD1 TSS
| Postulated antioxidant response elements of the HMOX1 and TXNRD1 genes | ||
|---|---|---|
| PCR primer location for ChIP (bp) | Motif sequence | Motif location (bp) |
| Consensus motif: GCnnnRTCAY or CGnnnYAGTR | ||
| HMOX1 | ||
| 9069 | GTGACagaGC | 9491 |
| GCtgaGTCAC | 9066 | |
| GCtaaGTCAC | 9037 | |
| GCtgaGTCAC | 9008 | |
| GCtgaGTCAC | 8979 | |
| 7232 | GCcttGTCAC | 7104 |
| 6060 | GCagaATCAT | 6008 |
| GCtgaATCAT | 5967 | |
| 3928 | GCtgcGTCAT | 3992 |
| GCtgaGTCAC | 3928 | |
| 1319 | GCgtgGTCAC | 107 |
| 148 | GCaaaATCAC | 200 |
| TXNRD1 | ||
| 91 & 36 | GCtttGTCAT | 19 |
PCR primer and motif positions correspond to the most distant 5′ nucleotide from the TSS on the forward strand. Sequences were considered candidate ARE motifs regardless of orientation or strand.
Figure 2.
Identification of NRF2 and BACH1 interactions with core ARE motifs of HMOX1. (A) The position of all 12 putative ARE motifs, relative to the HMOX1 transcription start site as annotated by the NCBI Homo sapiens Genome Map Viewer, Build 36.2. Open boxes = motif conforming to the consensus ARE sequence, shaded boxes = imperfect ARE motif. Boxed numerals indicate the number of motif repeats in that region. Arrows indicate the plus-strand orientation of each ARE motif relative to the consensus ARE sequence (RTGAYnnnGC). Relative DNA enrichment associated with NRF2 (B) and BACH1 (C) ChIP at each of the HMOX1 ARE-containing sites. Immunoprecipitated DNA was analyzed for enrichment by qRT–PCR using primers flanking ARE motifs at the positions indicated. Vertical bars and circle symbols graphically depict the relative magnitude of DNA enrichment at each position. Vertical bars indicate enrichment at positions −3928 bp (E1) and −9069 bp (E2) while circles correspond with regions where negligible DNA enrichment was observed. Values oriented vertically along the abscissa indicate the position of each DNA region amplified by qRT–PCR. Amplification was expressed in terms of ΔΔCt as calculated by normalizing the Ct for each primer in chromatin immunoprecipitated samples to the Ct obtained from the respective input DNA and expressed as a percentage relative to the PCR primer having the maximum enrichment (–9069 bp). Values represent the mean ± SEM of at least three independent experiments performed in triplicate.
Differential regulation of BACH1 and NRF2 activity
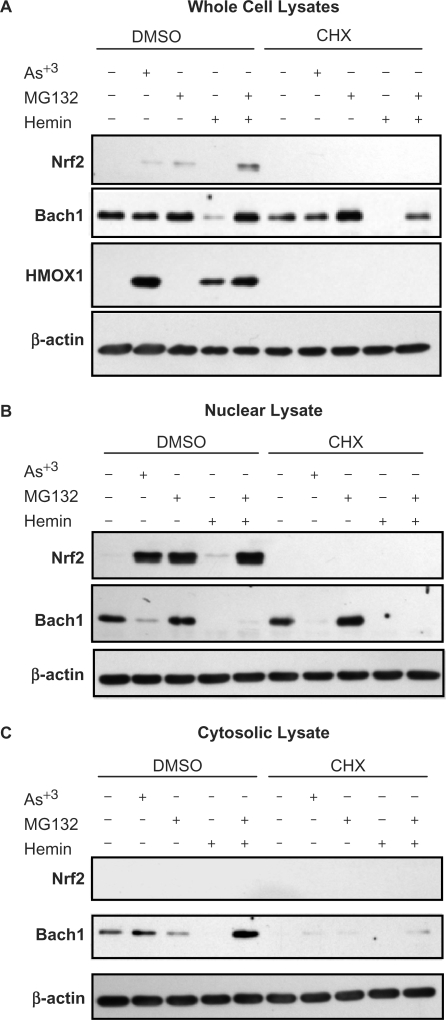
To investigate the relative contributions of BACH1 and NRF2 to HMOX1 expression, we differentially regulated their activities by treating cells with the proteasome inhibitor MG132 or with hemin. MG132 indirectly causes NRF2 activation by inhibiting KEAP1-dependent proteasomal degradation (26). Hemin inactivates BACH1 through interaction with its multiple heme-binding motifs leading to a conformational change and nuclear export (27,28). Treatment of HaCaT cells with MG132 for 3 h induces extensive NRF2 accumulation in whole cell lysates relative to untreated control cells (Figure 3A). This increase in NRF2 is almost entirely localized to the nuclear fraction (Figure 3B) with negligible levels detected in the cytosolic fraction (Figure 3C). In contrast to NRF2, BACH1 levels are only slightly affected by MG132 treatment in whole cell lysates or nuclear distribution (Figure 3). Hemin treatment contrasts with MG132 treatment in that it prominently triggers BACH1 inactivation, which manifests as a significant decrease in whole cell BACH1 protein levels (Figure 3A) and as its disappearance from the nuclear fraction (Figure 3B). Interestingly, this effect is not associated so much with nuclear efflux to the cytosolic compartment as occurs following arsenite treatment, but rather is associated with a net loss of total BACH1 (Figure 3A). Co-treatment of MG132 and hemin blocks this net loss of BACH1 (Figure 3A) and results in its cytoplasmic accumulation due to efflux from the nucleus (Figure 3C). Thus, inactivation of BACH1 by hemin triggers both its nuclear export and subsequent proteasomal degradation. In contrast to hemin, arsenite only mediates subcellular redistribution of BACH1, as indicated by nuclear efflux, without notable protein loss in whole cell lysates (Figure 3). Importantly, hemin treatment has only minimal effects on NRF2 activation, as indicated by the relative absence of NRF2 accumulation in either whole cell extracts or nuclear fractions. Co-treatment with hemin plus MG132 produces a combined pattern of NRF2 nuclear translocation and BACH1 efflux similar to that elicited by arsenite. Consistent with these findings, the expression of HMOX1 protein (Figure 3A) is associated only with treatments that result in BACH1 efflux from the nucleus, but not with NRF2 activation alone.
Figure 3.
Differential regulation of NRF2 and BACH1 activation. Immunoblots illustrating differential expression of NRF2 and BACH1 protein following treatment of HaCaT cells with 25 μM arsenite, 5 μM MG132, 25 μM hemin or MG132 + hemin combined. HaCaT cells were treated as indicated for 3 h or co-treated with 5 μM cycloheximide (CHX) following a 30-min CHX pretreatment. (A) Total cellular proteins (20 μg) from whole cell lysates. (B) Proteins extracts (10 μg) from nuclear lysates. (C) Proteins extracts (20 μg) from cytosolic lysates. Blots are representative of 2–3 separate experiments.
The elimination rate of NRF2 exceeds its rate of synthesis (26); hence, NRF2 activation requires both inhibition of proteasomal degradation and continuous NRF2 synthesis. To characterize the roles of NRF2 in basal and inducible expression of HMOX1 and TXHRD1, NRF2 activation was prevented by pretreating cells for 30 min with cycloheximide (CHX) to block protein synthesis prior to treatment with MG132. Blocking ongoing protein synthesis prior to inhibition of NRF2 degradation by MG132 prevents accumulation of de novo synthesized NRF2 while eliciting depletion of basal nuclear NRF2. The right half of Figure 3A shows that pretreatment of HaCaT cells with 5 μM CHX blocks synthesis of new NRF2, thereby preventing its activation when followed by MG132 or arsenite treatment. The fact that CHX pretreatment results in the elimination of NRF2 (Figure 3) establishes that under these conditions NRF2 cannot be the mediator of gene transcription. In parallel treatments, CHX has no effect on hemin- or arsenite-induced export of BACH1 from the nucleus, though total BACH1 levels increase in the presence of MG132 relative to untreated and CHX-treated controls. These data support a role for proteasomal degradation in the turnover of BACH1 and indicate that BACH1 has a much longer half-life than NRF2 in cells naïve to oxidative stress or hemin.
The effect of differentially modulating BACH1 or NRF2 activities was examined in relation to HMOX1 expression. HMOX1 is expressed in control cells at nearly undetectable levels and its induction coincides with the disappearance of nuclear BACH1 but not nuclear translocation of NRF2. Treatment with MG132 triggers prominent nuclear accumulation of NRF2 but fails to induce HMOX1 expression (Figure 3A). In contrast, treatment with either arsenite or hemin leads to nuclear export of BACH1 and to a prominent increase in HMOX1 expression. Combined treatment with MG132 and hemin augments HMOX1 protein levels, suggesting that activation of NRF2 contributes to HMOX1 expression in a manner that is at best additive to the effect of BACH1. These data suggest that BACH1 inactivation plays a larger role in the regulation of antioxidant gene expression than NRF2 activation.
Temporal induction of HMOX1 transcription depends on BACH1 removal from the enhancers
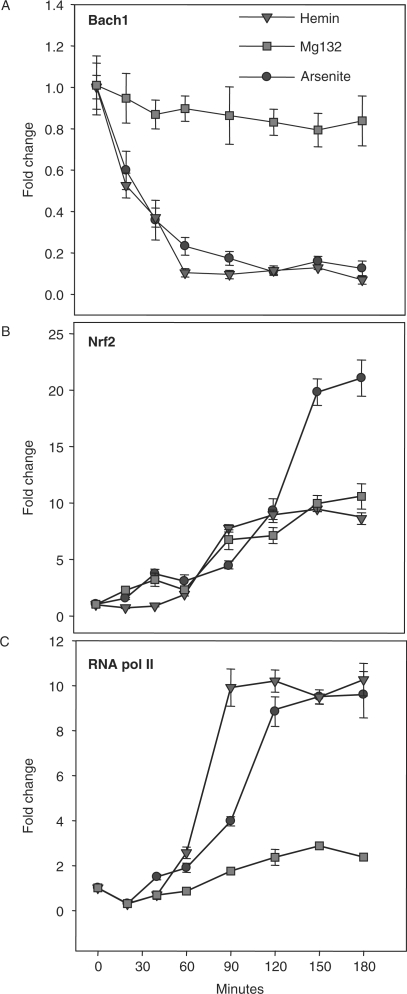
The observation that expression of HMOX1 is associated with nuclear efflux of BACH1 rather than with NRF2 activation, suggests that BACH1 removal is the dominant event regulating HMOX1 induction. To test this hypothesis we followed the temporal dynamics of HMOX1 transcriptional initiation by ChIP analysis. The time course of BACH1 and NRF2 binding to the E1 (Figure 4) and E2 (data not shown) enhancer regions of HMOX1, as well as RNA pol II binding at the proximal promoter (−148), were quantified after treatments with arsenite, hemin or MG132. These treatments induced very similar interactions of NRF2 and BACH1 at both of these enhancer regions, of which the observed interactions at E1 are representative. As anticipated, arsenite treatment produces a significant loss of BACH1 binding that is clearly detected 30 min after treatment and reaches minimal level by 60 min (Figure 4A). Conversely, arsenite triggers a prominent increase in NRF2 binding (Figure 4B), but this increase is delayed by at least 60 min relative to the BACH1 decrease, and is maximal at 150 min after treatment. Arsenite also produces a large increase in RNA pol II binding (Figure 4C) that temporally follows the loss of BACH1 but precedes NRF2 binding by at least 60 min and this coincides with the relatively low level of NRF2 binding during the period between 60 and 120 min.
Figure 4.
Association of NRF2 and BACH1 DNA binding with transcriptional activation. Time course of DNA binding by BACH1 (A) NRF2 (B) and RNA polymerase II (C) following treatment with 25 μM arsenite, 5 μM MG132 or 25 μM hemin. ChIP-enriched DNA was quantified using qRT–PCR with primers flanking the HMOX1 ARE motifs at positions −3992 (NRF2 and BACH1) and −148 (RNA Pol II) and is expressed as the value for each treatment normalized to its corresponding input and expressed as fold enrichment relative to untreated control. Figures represent the results of two independent experiments performed in triplicate ± SEM.
To investigate the individual contributions of BACH1 and NRF2 to HMOX1 induction, cells were treated with either hemin or MG132 in parallel with arsenite. Hemin triggers a rapid decrease in BACH1 DNA binding, mirroring the effect of arsenite and producing nearly complete BACH1 loss from the promoter 60 min after treatment (Figure 4A). In comparison to arsenite-mediated NRF2 binding, hemin treatment is associated with relatively weak NRF2 binding at time points earlier than 90 min (Figure 4B) and significantly less than that observed with arsenite after 120 min. Interestingly, hemin induces RNA pol II binding that proceeds in a manner similar in magnitude and time course to that associated with arsenite treatment (Figure 4C).
In stark contrast to both arsenite and hemin, MG132 has little effect on promoter binding by BACH1 (Figure 4A) and limited binding of NRF2, even after 120 min of treatment (Figure 4B). The level of NRF2 binding elicited by MG132 is of the order of that observed with hemin and approximately one-half of the level found with arsenite. Furthermore, MG132 elicits weak RNA pol II DNA binding relative to the levels associated with hemin and arsenite treatments, implying much lower levels of transcription initiation (Figure 4C). Together, these results strongly suggest that loss of BACH1 from the HMOX1 promoter is associated with a high level of RNA polymerase II binding, while NRF2 activation in the absence of BACH1 removal is associated with weak levels of gene induction, as inferred from the extent of RNA pol II binding. Consistent with whole cell lysate and nuclear extract immunoblots, it is evident that RNA pol II HMOX1 binding precedes NRF2 activation, further supporting the conclusion that loss of BACH1-mediated transcriptional repression is more important than nuclear accumulation of NRF2 for HMOX1 induction.
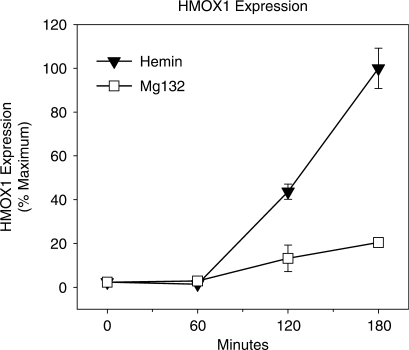
To confirm that RNA pol II binding is associated with gene induction by hemin but not by MG132, relative steady-state levels of HMOX1 mRNA accumulation were measured by qRT–PCR over the time course of HMOX1 protein induction. Induction of HMOX1 mRNA expression was differentially regulated by treatment with either hemin or MG132. Despite the absence of NRF2 activation, hemin triggers induction of much greater levels of HMOX1 mRNA than MG132 (Figure 5). Given that NRF2 activation is generally considered a critical event for antioxidant gene induction, it is surprising that inactivation of BACH1 by hemin produced 5-fold greater levels of HMOX1 induction than were elicited by MG132-mediated NRF2 activation.
Figure 5.
Time course of HMOX1 expression elicited by hemin and MG132 treatment. The time course of HMOX1 mRNA expression following treatment with 25 μM hemin or 5 μM MG132. HMOX1 mRNA was determined by quantitative real-time PCR (qRT–PCR), normalized to β-actin mRNA and expressed as percent maximum expression. Values represent at least three independent experiments quantified in triplicate ± SEM.
Differential gene induction regulated by NRF2 and BACH1
Although NRF2 and BACH1 interact with ARE motifs, several lines of evidence support that these interactions with DNA are not identical, thereby suggesting that genes could be identified that are regulated by one factor but not by the other (12,29,30). TXNRD1 was identified as one such gene. ChIP analysis was used to test for NRF2 and BACH1 binding to the single ARE motif located 9 bp upstream of the TSS in the TXNRD1 5′-flanking region. As illustrated in the upper panel of Figure 6A, NRF2 binds the ARE motif of the TXNRD1 in ‘control cells’ with an affinity significantly greater than either at HMOX1 E1 or E2. After arsenite-induced oxidative stress, this site becomes more strongly bound by NRF2, reaching a level that is intermediate between NRF2 binding at the HMOX1 E1 and E2 sites after activation, and ∼6.5-fold greater than control levels. In contrast, the lower panel of Figure 6A shows negligible binding of BACH1 to the TXNRD1 ARE in either control or arsenite-treated cells. As a consequence of differential transcription factor binding, we anticipated that MG132, but not hemin, would elicit NRF2-dependent TXNRD1 induction. To test this prediction, we treated HaCaT cells with MG132 or hemin and quantified mRNA levels of HMOX1 and TXNRD1 by qRT–PCR. In untreated control cells, basal TXNRD1 mRNA levels are 35-fold greater than the levels of HMOX1 mRNA. Inhibition of NRF2 activation with CHX significantly inhibited basal TXNRD1 expression but left HMOX1 expression unchanged (Figure 6B and C). This pattern of expression is consistent with functionally active NRF2 residing basally in the nucleus and rules out the possibility that this resident nuclear NRF2 contributes to basal HMOX1 expression. The role of activated NRF2 is demonstrated by the MG132 treatments that trigger maximal TXNRD1 induction but mediate only weak HMOX1 expression (Figure 6B and C). Pretreatment with CHX prior to MG132 blocks NRF2 synthesis, returning HMOX1 mRNA to control levels and reducing TXNRD1 mRNA to the levels of CHX-treated control cells. These expression data reflect the primary role of NRF2 in basal and inducible expression of TXNRD1 but not HMOX1. Hemin treatment, which inactivates BACH1, has no effect on TXNRD1 expression (Figure 6C), reflecting the fact that TXNRD1 is regulated independently of BACH1. In contrast, hemin elicits a significant and nearly maximal increase in HMOX1 expression (Figure 6B). CHX pretreatment predominantly blocks this induction, demonstrating a requisite role for resident nuclear NRF2 in HMOX1 induction without necessitating nuclear accumulation of activated NRF2. Combined treatment with hemin plus MG132 results in induction of TXNRD1 to levels similar to those in cells treated with MG132 alone while the level of HMOX1 induction is similar to that elicited by hemin (Figure 6B and C). These results show that TXNRD1 induction is attributable solely to activation of NRF2 by proteasome inhibition but that HMOX1 induction requires inactivation of BACH1. Thus, BACH1 inactivation is predominantly responsible for HMOX1 induction and that NRF2 activation contributes nominally to this process. Interestingly, the combination of MG132 plus hemin also shows that, when preceded by CHX treatment, HMOX1 expression remains significantly induced despite the lack of NRF2 and hints at the possible role for BACH1 in repressing the activity of transcriptional activators in addition to NRF2. These data establish that inactivation of BACH1 is more important than activation of NRF2 for induction of HMOX1 expression and that not all NRF2-regulated genes respond in this manner, as is the case of TXNRD1.
Figure 6.
Differential regulation of HMOX1 and TXNRD1 by BACH1 and NRF2. (A) ChIP analysis of NRF2 and BACH1 interactions with ARE sites of HMOX1 and TXNRD1 before and after 25 μM arsenite treatment for 3 h. Binding of NRF2 (top panel) and BACH1 (bottom panel) to HMOX1 E1 and TXNRD1 is expressed relative to binding at the HMOX1 E2 enhancer element. Expression of HMOX1 (B) and TXNRD1 (C) mRNA was measured in HaCaT cells following treatment with 25 μM hemin, 5 μM MG132 or both in the presence (shaded bars) or absence (filled bars) of 5 μM CHX. Relative mRNA was determined by quantitative real-time PCR (qRT–PCR), normalized to β-actin mRNA and expressed as percent maximum expression. Values represent at least three independent experiments quantified in triplicate ± SEM.
DISCUSSION
In the present article, we report for the first time the dynamics associated with inactivation of BACH1 and NRF2 activation underlying the initiation of endogenous gene targets. BACH1, which is predominantly localized to the nucleus of control cells, is exported to the cytosol within 30 min following arsenite or hemin treatment. Temporally, inactivation of BACH1 precedes NRF2 activation by at least 30 min, correlating with the period necessary for de novo synthesis of NRF2, as supported by the absence of immunoreactive NRF2 when CHX treatment precedes proteasome inhibition. Consequently, biological coupling of these transcriptional regulators insures that the response to stressors is rapid and not dependent on the delay required for synthesis of high NRF2 levels.
Of the 12 putative HMOX1 ARE motifs, only two sites, at −3928 bp (E1) and −8979 bp (E2), are reciprocally bound by BACH1 and NRF2. Both sites contain multiple ARE motifs and have been previously recognized as HMOX1 transcriptional regulators (15). Our data confirm that these are the only two HMOX1 elements recognized by NRF2 and BACH1 in vivo. This pattern of binding contrasts markedly with binding at the TXNRD1 promoter, where NRF2 is capable of significantly binding the ARE motif but BACH1 is not. Distinct recognition of ARE motifs by BACH1, compared to NRF2, conveys an additional level of transcriptional regulation to HMOX1 that is lacking in the regulation of TXNRD1. The fact that BACH1 and NRF2 differently recognize ARE motifs presents the possibility that BACH1 regulates an overlapping but separate gene battery. NRF2 and BACH1 differ in their affinities for ARE motifs that otherwise appear to conform equally well to the consensus ARE core motif; however, this short 10 bp sequence is not the sole determinant of factor binding. Nucleotides flanking the ARE core as well as motif multiplicity also contribute to differential motif recognition by BACH1 and NRF2. Several reports indicate that extended sequences flanking the ARE core are vital for recognition by NRF2 (5,31–33), though the precise sequence requirements of the extended ARE motif are not well defined leaving a great deal of uncertainty when attempting to predict high-affinity NRF2-binding elements. Similar difficulties exist in predicting the sequence requirements for high-affinity BACH1-binding sites. Unlike NRF2 however, detailed description of BACH1-binding elements is hampered by the fact that only a few genes are known to be under its regulatory control (13,34), including NQO1 (10) and HMOX1 (14). The sequences that BACH1 interacts with at these genes seems to conform to the core ARE motif recognized by NRF2; however, only one report has experimentally examined sequence requirements for BACH1 binding, the results of which generally support motif homology with the ARE (29). Binding element multiplicity may also contribute to differences in DNA-binding affinity between NRF2 and BACH1. While Nrf2 is capable of binding an individual ARE motif, BACH1 appears to bind poorly to single motifs, preferring to interact at sites containing multiple motifs. The BTB/POZ domain of BACH1 is thought to contribute to DNA binding through oligomerization with neighboring BACH1 heterodimers to stabilize their interactions with DNA (12). In this respect, both HMOX1 enhancers recognized by BACH1 consist of multiple elements while the TXNRD1 element is a single consensus motif. Thus, the multiple ARE motifs of HMOX1 might permit BACH1 oligomerization while TXNRD1, with its single motif, would not allow formation of stable BACH1 oligomers. Together, nucleotide sequence and motif multiplicity may account for the observation that the TXNRD1 promoter is poorly recognized by BACH1.
In control cells, we have noted that the cytosol is essentially devoid of detectable NRF2 consistent with reports that NRF2 activity is governed by regulation of its stability (23,35,36) rather than by being sequestered in the cytosol (25,37). In the absence of oxidative stress, KEAP1 mediates Cul3-dependent ubiquitylation of NRF2 that targets it for rapid proteasomal destruction (36). During oxidative stress several sensitive KEAP1 cysteines are oxidized resulting a loss of KEAP1-directed proteasomal degradation of NRF2 and increased proteasomal degradation of KEAP1 itself (21,38). In the absence of ongoing KEAP1-directed degradation of NRF2, the half-life of newly synthesized protein is prolonged permitting rapid nuclear translocation and accumulation with subsequent antioxidant gene induction (35). Consistent with a short residence time of NRF2 in the cytosol, we observe that when NRF2 escapes proteasomal degradation it accumulates in the nucleus while remaining nearly undetectable in the cytoplasm. In this respect, we have observed the persistence of nuclear NRF2 in untreated control cells indicating that even in the absence of oxidative stress some NRF2 escapes KEAP1-mediated degradation and is capable of binding ARE elements and eliciting gene induction. We propose that the persistence of resident nuclear NRF2 plays a crucial role in both the induction of HMOX1 as well as basal TXNRD1 expression. BACH1 inactivation elicits maximal HMOX1 expression without requiring concurrent NRF2 activation by permitting resident nuclear NRF2 to bind HMOX1 ARE motifs. In contrast, BACH1 does not inhibit basal nuclear NRF2 binding to the TXNRD1 promoter and therefore basal TXNRD1 expression is 20% of maximal expression, and 35-fold higher than basal HMOX1 expression. Loss of basal nuclear NRF2 following CHX treatment precipitously decreases both hemin-mediated HMOX1 induction and basal TXNRD1. It is noteworthy that some HMOX1 expression elicited by hemin inactivation of BACH1 persists despite the absence of NRF2 protein, suggesting BACH1 may also play a role in the repression of factor(s) in addition to NRF2. In this regard, HMOX1 expression is even more pronounced when CHX treatment precedes co-treatment with hemin plus MG132, suggesting that proteasomal inhibition may stabilize other putative transcriptional activator(s).
Without prior inactivation of BACH1, activated NRF2 binds inefficiently to HMOX1 enhancers compared to the binding elicited by arsenite. Although the level of NRF2 binding associated with MG132 treatment is similar in extent to that initiated by hemin treatment, NRF2 activation in the absence of BACH1 inactivation results in significantly lower levels of HMOX1 transcription. This contrasts profoundly with TXNRD1 expression, where MG132 strongly stimulates induction and hemin treatment has no effect. Since transcription factor binding to cognate sites is a stochastic event (39), the massive increase in nuclear NRF2 concentration associated with its activation could increase competition with BACH1 for ARE motifs or sMAF-binding partners. By mass action, NRF2 activation could establish a new equilibrium favoring DNA-bound NRF2/sMAF, thereby increasing the stoichiometric DNA residence of NRF2. Since BACH1 appears to bind at sites containing multiple AREs, NRF2 may not displace a large enough fraction of DNA-bound BACH1 to overcome transcriptional repression. CHX treatment decreased hemin-mediated HMOX1 induction and basal TXNRD1 expression. When interpreted in the light of our ChIP enhancer binding data, these findings support the conclusion that inactivation of BACH1 by hemin decreases its DNA-binding affinity resulting in its generalized removal from ARE motifs and concomitant loss of transcriptional repression, leaving the AREs available for binding by NRF2 and possibly other transcription that mediate gene induction.
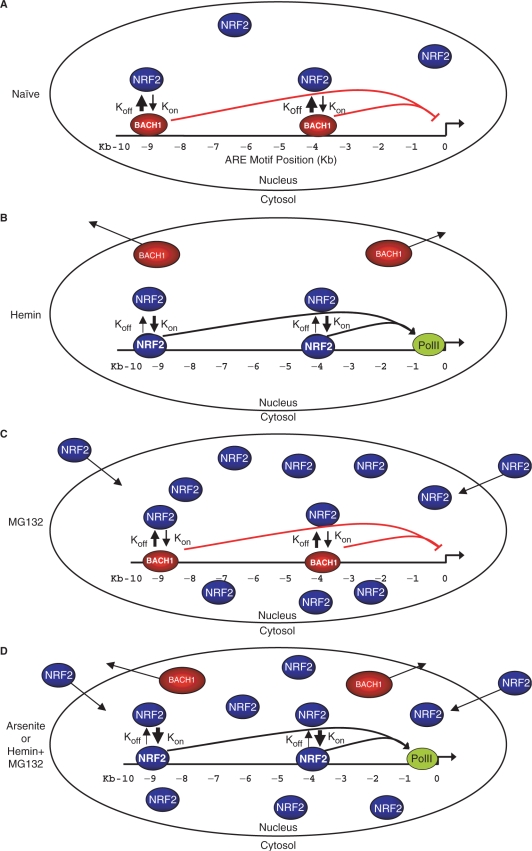
Based on the dynamic exchange of BACH1 and NRF2 we propose that in cells naïve to oxidative stress, BACH1 is bound to the ARE enhancer motifs preventing NRF2 from binding and thereby repressing transcription (Figure 7A). Hemin treatment triggers removal of BACH1 from HMOX1 enhancers thereby allowing NRF2 that is already present in the nucleus to interact with ARE motifs and elicit gene induction (Figure 7B). On the other hand, MG132 triggers NRF2 translocation to the nucleus but DNA-bound BACH1 blocks NRF2–ARE interactions to prevent gene induction (Figure 7C). The vast increase in the presence of nuclear NRF2 permits some increase in NRF2 binding to HMOX1 AREs but the presence of BACH1 maintains gene repression. Treatment with either arsenite or hemin plus MG132 triggers both the removal of BACH1 and the activation of NRF2, which can now freely bind vacant enhancer motifs (Figure 7D).
Figure 7.
Induction of HMOX1 through the interplay of BACH1 and NRF2 with enhancer elements. (A) Binding of BACH1 (red ovals) at the E1 and E2 enhancer elements of HMOX1 in untreated control cells blocks NRF2 (blue ovals) binding and HMOX1 induction. Competition for ARE-binding elements between activated NRF2 and DNA-bound BACH1 is indicated by hypothetical rate constants representing stochastic binding (kon) and dissociation (koff) of NRF2. (B) Inactivation of BACH1 by hemin results in its removal from ARE motifs and elimination from the nucleus. Consequently, NRF2 can interact with exposed ARE enhancers to recruit RNA pol II (green oval) leading to high-level HMOX1 induction. (C) DNA-bound BACH1 blocks DNA binding of NRF2 despite its nuclear accumulation and prevents efficient HMOX1 induction. (D) Treatment with arsenite or co-treatment with hemin + MG132 results in BACH1 inactivation, nuclear accumulation and ARE binding of NRF2, and high-level HMOX1 induction.
Though not all ARE-containing genes are regulated by BACH1, whenever BACH1 interacts with ARE motifs it contributes an important regulatory dimension that provides a more complex response to environmental stress. Reciprocal ARE binding of BACH1 for NRF2 imparts an increased level of complexity to the ARE-regulated genes producing distinct patterns of gene expression patterns, as shown here for HMOX1 and THXRD1. The differential interaction of BACH1 and NRF2 with various forms of the ARE motif suggests the possibility that BACH1 may regulate a distinct but overlapping battery of genes. In this case, genes specifically repressed by BACH1 could be globally induced in response to redox stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr N. Fusenig (Division of Differentiation and Carcinogenesis in Vitro, German Cancer Research Center, Heidelberg, Germany) for a gift of HaCaT keratinocytes. This research was supported by NIEHS grants R01 ES10807, The NIEHS Center for Environmental Genetics grant P30 ES06096 and the NIEHS Superfund Basic Research Program grant P42 ES04908. J.F.R. is a Postdoctoral Trainee partly supported by NIEHS T32 ES07250, Environmental Carcinogenesis and Mutagenesis Training Grant. Funding to pay the Open Access publication charges for this article was provided by NIEHS grants RO1 ES10807.
Conflict of interest statement. None declared.
REFERENCES
- 1.Spuches A.M., Kruszyna H.G., Rich A.M., Wilcox D.E. Thermodynamics of the As(III)-thiol interaction: arsenite and monomethylarsenite complexes with glutathione, dihydrolipoic acid, and other thiol ligands. Inorg. Chem. 2005;44:2964–2972. doi: 10.1021/ic048694q. [DOI] [PubMed] [Google Scholar]
- 2.Shi H., Shi X., Liu K.J. Oxidative mechanism of arsenic toxicity and carcinogenesis. Mol. Cell Biochem. 2004;255:67–78. doi: 10.1023/b:mcbi.0000007262.26044.e8. [DOI] [PubMed] [Google Scholar]
- 3.Kann S., Estes C., Reichard J.F., Huang M.Y., Sartor M.A., Schwemberger S., Chen Y., Dalton T.P., Shertzer H.G., et al. Butylhydroquinone protects cells genetically deficient in glutathione biosynthesis from arsenite-induced apoptosis without significantly changing their prooxidant status. Toxicol. Sci. 2005;87:365–384. doi: 10.1093/toxsci/kfi253. [DOI] [PubMed] [Google Scholar]
- 4.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 5.Wasserman W.W., Fahl W.E. Functional antioxidant responsive elements. Proc. Natl Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deppmann C.D., Alvania R.S., Taparowsky E.J. Cross-species annotation of basic leucine zipper factor interactions: insight into the evolution of closed interaction networks. Mol. Biol. Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- 7.Motohashi H., O’Connor T., Katsuoka F., Engel J.D., Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 8.Venugopal R., Jaiswal A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl Acad. Sci. USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankaranarayanan K., Jaiswal A.K. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J. Biol. Chem. 2004;279:50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 10.Dhakshinamoorthy S., Jain A.K., Bloom D.A., Jaiswal A.K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 11.Amoutzias G., Veron A., Weiner A., Robinson-Rechavi M., Bornberg-Bauer E., Oliver S., Robertson D. One billion years of bZIP transcription factor evolution: conservation and change in dimerization, and DNA-binding site specificity. Mol. Biol. Evol. 2006;24:827–835. doi: 10.1093/molbev/msl211. [DOI] [PubMed] [Google Scholar]
- 12.Igarashi K., Hoshino H., Muto A., Suwabe N., Nishikawa S., Nakauchi H., Yamamoto M. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J. Biol. Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 13.Oyake T., Itoh K., Motohashi H., Hayashi N., Hoshino H., Nishizawa M., Yamamoto M., Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J., Hoshino H., Takaku K., Nakajima O., Muto A., Suzuki H., Tashiro S., Takahashi S., Shibahara S., et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J., Brand M., Zenke Y., Tashiro S., Groudine M., Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl Acad. Sci. USA. 2004;101:1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan Y., Lambrecht R.W., Ghaziani T., Donohue S.E., Bonkovsky H.L. Role of Bach-1 in regulation of heme oxygenase-1 in human liver cells: insights from studies with small interfering RNAS. J. Biol. Chem. 2004;279:51769–51774. doi: 10.1074/jbc.M409463200. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa M., Numazawa S., Yoshida T. Redox regulation of the transcriptional repressor Bach1. Free Radic. Biol. Med. 2005;38:1344–1352. doi: 10.1016/j.freeradbiomed.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Dohi Y., Alam J., Yoshizumi M., Sun J., Igarashi K. Heme Oxygenase-1 Gene Enhancer Manifests Silencing Activity in a Chromatin Environment Prior to Oxidative Stress. Antioxid. Redox. Signal. 2006;8:60–67. doi: 10.1089/ars.2006.8.60. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi K., Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox. Signal. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 20.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D.D., Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong F., Freeman M.L., Liebler D.C. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem. Res. Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 23.He X., Chen M.G., Lin G.X., Ma Q. Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2 x Keap1 x Cul3 complex and recruiting Nrf2 x Maf to the antioxidant response element enhancer. J. Biol. Chem. 2006;281:23620–23631. doi: 10.1074/jbc.M604120200. [DOI] [PubMed] [Google Scholar]
- 24.Boukamp P., Petrussevska R.T., Breitkreutz D., Hornung J., Markham A., Fusenig N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen T., Sherratt P.J., Huang H.C., Yang C.S., Pickett C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H., Tashiro S., Hira S., Sun J., Yamazaki C., Zenke Y., Ikeda-Saito M., Yoshida M., Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23:2544–2553. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanezaki R., Toki T., Yokoyama M., Yomogida K., Sugiyama K., Yamamoto M., Igarashi K., Ito E. Transcription factor BACH1 is recruited to the nucleus by its novel alternative spliced isoform. J. Biol. Chem. 2001;276:7278–7284. doi: 10.1074/jbc.M004227200. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai A., Nishimoto M., Himeno S., Imura N., Tsujimoto M., Kunimoto M., Hara S. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J. Cell Physiol. 2005;203:529–537. doi: 10.1002/jcp.20246. [DOI] [PubMed] [Google Scholar]
- 31.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 32.Erickson A.M., Nevarea Z., Gipp J.J., Mulcahy R.T. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. Revision of the ARE consensus sequence. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- 33.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tahara T., Sun J., Igarashi K., Taketani S. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem. Biophys. Res. Commun. 2004;324:77–85. doi: 10.1016/j.bbrc.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 35.McMahon M., Itoh K., Yamamoto M., Hayes J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 36.Furukawa M., Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zipper L.M., Mulcahy R.T. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D.D., Lo S.C., Sun Z., Habib G.M., Lieberman M.W., Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 39.Cranz S., Berger C., Baici A., Jelesarov I., Bosshard H.R. Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry. 2004;43:718–727. doi: 10.1021/bi0355793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.