Abstract
Telomerase adds telomeric DNA repeats to the ends of linear chromosomal DNA. 3′-Azido-3′-deoxythymidine 5′-triphosphate (AZTTP) is a known telomerase inhibitor. To obtain more selective and potent inhibitors that can be employed as tools for studying telomerase, we investigated the telomerase-inhibitory effects of purine nucleosides bearing a 3′-down azido group: 3′-azido-2′,3′-dideoxyguanosine (AZddG) 5′-triphosphate (AZddGTP), 3′-azido-2′,3′-dideoxy-6-thioguanosine (AZddSG) 5′-triphosphate (AZddSGTP), 3′-azido-2′,3′-dideoxyadenosine (AZddA) 5′-triphosphate (AZddATP) and 3′-azido-2′,3′-dideoxy-2-aminoadenosine (AZddAA) 5′-triphosphate (AZddAATP). Of these, AZddGTP showed the most potent inhibitory activity against HeLa cell telomerase. AZddGTP was significantly incorporated into the 3′-terminus of DNA by partially purified telomerase. However, AZddGTP did not exhibit significant inhibitory activity against DNA polymerases α and δ, suggesting that AZddGTP is a selective inhibitor of telomerase.
We also investigated whether long-term treatment with these nucleosides could alter telomere length and growth rates of human HL60 cells in culture. Southern hybridization analysis of genomic DNA prepared from cells cultured in the presence of AZddG and AZddAA revealed reproducible telomere shortening.
INTRODUCTION
Telomeres constitute the termini of eukaryotic chromosomes and incorporate linear chromosomal DNA ends consisting of guanine-rich sequences and associated protein components (1). Telomeres protect the ends of each chromosome from degradation and loss of essential genes, and allow the cell to distinguish between double-strand breaks and natural chromosome ends. Functional telomeres are essential for continued cell proliferation. As a result of incomplete replication of lagging-strand DNA synthesis and other end-processing events, telomeres progressively shorten in all somatic cells with each cell division (2). When telomeres become short, cells usually undergo replicative senescence (3).
Telomerase is a cellular endogenous reverse transcriptase (RT) believed to counteract this progressive shortening by directing the appropriate nucleotides onto the telomeric ends of chromosomes, and play an important role in the mechanism of tumor cell immortalization. Telomerase is expressed in embryonic cells and adult male germline cells (4), but is undetectable in normal somatic cells with the exception of proliferating cells in tissues undergoing renewal (5,6). In normal somatic cells, progressive telomere shortening occurs, eventually leading to greatly shortened telomeres and, consequently, limited replicative capacity. In contrast to normal cells, tumor cells generally have short telomeres and show no net loss of average telomere length with successive cell divisions, suggesting that telomere stability might be required for cells to escape replicative senescence and proliferate indefinitely. In normal cells, telomerase activity appears to be tightly controlled, but is reactivated in ∼90% of malignant tumor cells. Telomerase activity could therefore be a rate-limiting step for the continuing proliferation of advanced cancers (7–11). Thus, a potential therapeutic window exists in which cancer cells can be efficiently targeted by telomerase inhibitors, while normal telomerase-expressing cells, such as stem and germline cells, remain unaffected as a result of their longer telomeres and slower rates of cell division (12–14).
Numerous approaches for targeting telomeres and telomerase activity have been studied (15). Telomerase is a ribonucleoprotein in which the internal RNA serves as a template for directing the telomere DNA sequence, which in vertebrates is (TTAGGG)n (16). Therefore, telomerase is classified as a RT (17,18). Classic methods for affecting enzymatic reverse transcription have proven useful for identifying telomerase inhibitors. Strahl and Blackburn analyzed whether known inhibitors of retroviral RTs, 2′,3′-dideoxyguanosine (ddG), 3′-azido-2′,3′-dideoxythymidine (AZT), 2′,3′-dideoxyadenosine (ddA), 2′,3′-dideoxyinosine (ddI) and 2′,3′-didehydro-2′,3′-dideoxythymidine (d4T), could perturb telomere length and growth rates of two immortalized human cell lines. Of these, only ddG caused reproducible telomere shortening, but had no observable effect on cell growth rates or morphology (19). Gomez et al. (20) reported that treatment of HeLa cells with 800 µM AZT caused shortening of the telomeric DNA. In this context, guanine counterparts may be more potent inhibitors than other base analogs, since telomerase catalyzes telomere DNA elongation through addition of repeated guanine-rich sequences, (e.g. TTAGGG). Additionally, the azido group confers enhanced lipophilicity, which could be expected to contribute significantly to non-selective transport across membranes (21). We have previously demonstrated that 3′-azido-2′,3′-dideoxyguanosine (AZddG) 5′-triphosphate (AZddGTP) (Figure 1) shows more potent inhibition than 3′-azido-3′-deoxythymidine 5′-triphosphate (AZTTP) (22). The present article describes the inhibition of telomerase by purine counterparts of AZTTP and the mechanism of activity. We report the effects of AZddG, 3′-azido-2′,3′-dideoxy-2-aminoadenosine (AZddAA) and AZT on telomere length and growth properties of the immortalized cell line HL60, derived from human leukemia cells.
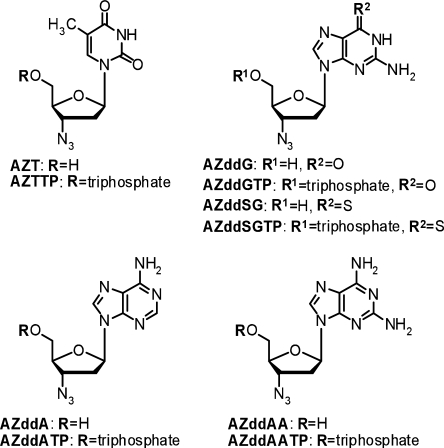
Figure 1.
Nucleoside and nucleotide analogs examined in this study.
MATERIALS AND METHODS
Compounds
AZddG, 3′-azido-2′,3′-dideoxy-6-thioguanosine (AZddSG), AZT and AZddAA were synthesized according to the procedures reported by Imazawa and Eckstein (23) and Marchand et al. (24), with slight modifications. Their triphosphate derivatives were also synthesized as described, with slight modifications (25,26). The purity of the triphosphate derivatives was confirmed to be higher than 95% as detected by UV absorption at the λmax of each compound during HPLC analysis. HPLC analysis was conducted using a TSK-GEL DEAE-2SW (TOSOH, Tokyo, Japan) anion-exchange column as the solid phase and 0.21 M potassium phosphate buffer (pH 6.9) containing 20% CH3CN as the mobile phase (1 ml/min) at room temperature. 9-β-d-Arabinofuranosylguanine 5′-triphosphate (araGTP) was synthesized as reported previously (27), and 3′-azido-2′,3′-dideoxyadenosine (AZddA) 5′-triphosphate (AZddATP) and ddGTP were purchased from TriLink BioTechnologies (San Diego, CA, USA).
Cells and enzymes
HeLa cells and HL60 cells were obtained from the Riken Gene Bank (Tsukuba, Japan). Recombinant HIV-1 RT was purchased from Seikagaku Kogyo (Tokyo, Japan). Calf DNA polymerase (pol) α (28) and human pol δ were provided by Dr M. Takemura of Tokyo University of Science (Tokyo, Japan) and Dr K. Sakaguchi of Tokyo University of Science (Chiba, Japan), respectively. Human pol δ was purified by the nuclear fractionation of human peripheral blood cancer cells (Molt-4) using the second subunit of pol δ-conjugated to an affinity column chromatography (29). Partially purified cherry salmon telomerase was prepared from immature cherry salmon testis as described previously (30).
Semi-quantitative telomerase assay based on the ‘stretch PCR’ method
Standard telomerase assay
The reaction mixture (20 µl) for telomere DNA extension was comprised of 50 mM Tris–acetate/50 mM potassium acetate (pH 8.5), 1 mM MgCl2, 1 mM dithiothreitol, 1 µM TAG-U primer (5′-GTA AAA CGA CGG CCA GTT TGG GGT TGG GGT TGG GGT TG-3′), 10–200 µM dGTP, 10–200 µM dTTP, 10–200 µM dATP, inhibitors and a HeLa cell S-100 extract containing telomerase (1–2 µg of protein). The reaction mixture was incubated for 10 min at 30°C followed by the addition of 13 mM EDTA (pH 8.0) (40 µl). Isolation of product DNA, stretch PCR using CTA-R reverse primer (5′-CAG GAA ACA GCT ATG ACC CCT AAC CCT AAC CCT AAC CCT-3′), separation by polyacrylamide gel electrophoresis, DNA detection by staining with SYBR green I (Cambrex BioScience, Rockland, ME, USA) and estimation of the product concentrations were carried out as described previously (31). A standard curve correlating the amount of enzyme and PCR product was prepared, allowing the linearity of the reaction to be confirmed in each experiment.
Inhibition study and determination of kinetic constants
Inhibition studies were performed by the addition of various concentrations of nucleotide analogs to the standard telomerase assay mixture. Inhibition mode and kinetic constants were analyzed by Lineweaver–Burk plot analyses.
Primer extension assay
The assay was performed essentially as described by Tendian et al. (30,32). The reaction mixture (40 µl) was comprised of 50 mM Tris–HOAc (pH 8.5), 50 mM KOAc, 1 mM MgCl2, 1 mM dithiothreitol, 2 µM (TTAGGG)3, 5 µM [α-32P]dTTP (GE Healthcare Bio-Science, Buckinghamshire, UK), 5 µM dATP, 5 or 0 µM dGTP, several concentrations of dGTP analogs and 20 µl of partially purified cherry salmon telomerase. Incubation was performed for 1 h at 25°C. The reaction was terminated by adding 0.5 M EDTA (1 µl) and 2 µg/µl RNase A (2 µl) and incubating at 37°C for 15 min. The reaction mix was further treated by adding 10% SDS (1 µl) and 20 µg/µl proteinase K (1 µl), then incubated at 37°C for 15 min. Unincorporated radioactivity was removed by gel filtration using a MicroSpin G-25 (GE Healthcare Bio-Science). DNA products were treated by phenol extraction and isolated by ethanol precipitation, then analyzed by electrophoresis on a 20% polyacrylamide–7 M urea sequencing gel. Autoradiography was carried out by one-day exposure to imaging plates (Fuji Film, Tokyo, Japan). Signals were detected using a fluorescence image analyzer FLA 3000 (Fuji Film).
Assay of DNA polymerases α and δ
The reaction mixture (25 µl) was comprised of 50 mM Tris–HCl (pH 7.5), 4 mM MgCl2, 1 mM dithiothreitol, 100 µg/ml activated DNA, 20 µM dGTP, 50 µM each dATP, dCTP and [3H] dTTP (GE Healthcare Bio-Science), 200 µg/ml bovine serum albumin, 10% glycerol and the enzyme preparation. Incubation was carried out at 37°C for 15 min. The radioactive DNA product was collected on a paper disc (DE 81, Whatman, Middlesex, UK) and the radioactivity was measured (33).
Assay of HIV-1 RT
The reaction mixture (25 µl) was comprised of 50 mM Tris–HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 1 µg/ml polyU-oligo(dA)16 (10: 1), 50 µM dATP, inhibitors and HIV-1 RT (0.25 U). Incubation was performed for 20 min at 37°C followed by addition of SYBR green I (125 µl) which was diluted 5000-fold with 60 mM Tris, 60 mM boric acid and 1.2 mM EDTA. The fluorescence was analyzed at 485 nm for excitation and 535 nm for emission using a micro-plate reader (Wallac 1420 ARVOSX, Perkin Elmer, Waltham, MA, USA). Under these reaction conditions, the relationship between the amount of HIV-1 RT and fluorescence intensity was linear in the range 0–0.5 U of the enzyme.
Cells and cell culture
HL60 cells were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing l-glutamine, supplemented with 0.07 µg/ml kanamycin and 10% heat-inactivated fetal calf serum at 37°C under 5% carbon dioxide. Cultures were grown in 25 cm2 flasks, 6 ml per flask, in duplicate, with AZddG (in the presence of 0.005% DMSO), AZddAA or AZT added to the medium to a final concentration of 25 or 50 µM, before culture. Cells were counted with a hemacytometer and transferred every four days (three to four mean population doublings), with 0.3 × 105 cells per flask seeded into fresh medium containing analog or control medium. The remaining cells were collected by centrifugation (800g), washed with PBS(−) and stored at −80°C until use.
DNA extraction, restriction digestion and Southern analysis
Cell pellets (∼2 × 106 cells) were suspended in 50 µl of TE buffer [10 mM Tris–HCl (pH 8.0), 1 mM EDTA], mixed with 0.5 ml of lysis buffer [10 mM Tris–HCl (pH 8.0), 125 mM EDTA, 10% sodium dodecyl sulfate (SDS), 20 µg/ml RNase A], incubated at 37°C for 1 h, and then mixed with 20 mg/ml proteinase K (5 µl). Incubation was then continued at 50°C overnight, followed by phenol extraction and ethanol precipitation. Genomic DNA (∼15 µg) was completely digested in 50 µl of mixture [50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 100 mM NaCl, 0.1 mg/ml bovine serum albumin and 40 U of Hinf I] at 37°C for 4 h. The mixture was further treated by adding 10% SDS (1 µl) and 20 µg/µl proteinase K (1 µl), then incubated at 37°C for 15 min. The resulting digested DNA was treated by phenol extraction and isolated by ethanol precipitation. Approximately 2 µg of digested DNA per lane was electrophoresed on a 1% agarose gel. The DNA was transferred to a nylon support membrane and then UV-cross-linked to the membrane. Hybridization using a digoxigenin-labeled telomere DNA probe, and detection of telomere DNA by treatment with AttoPhos (Promega, Madison, WI, USA) were performed as described previously (30).
RESULTS
Inhibitory effects of 3′-azido-2′,3′-dideoxynucleoside 5′-triphosphates on HeLa cell telomerase activity
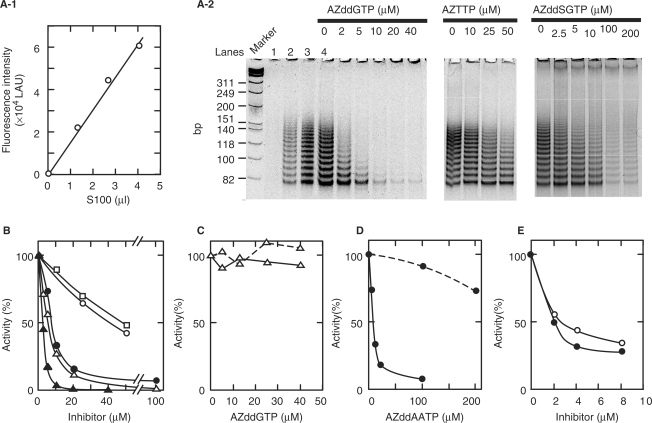
A semi-quantitative telomerase assay based on the ‘stretch PCR’ method was performed to investigate the inhibitory effects of several nucleoside triphosphate derivatives on HeLa cell telomerase (34). As shown in Figure 2A-2, the telomerase products amplified by the stretch PCR method were detected as a ladder consisting of bands spaced six bases apart. The ladders were abolished by incubation (30°C, 3 min) with 1 µg of RNase A or by heating (90°C, 5 min) (data not shown). The inhibitory effects of AZddGTP, AZddSGTP, AZddAATP, AZddATP and AZTTP on human telomerase were examined (Figure 2B). Although the inhibitory effects of AZTTP and AZddATP were weak, those of the guanine analogs AZddGTP and AZddSGTP were potent. As the concentration of AZddGTP was increased in the presence of 10 µM dGTP, the products became shorter and their total amount decreased (Figure 2A-2). On the other hand, AZddGTP showed very weak or no inhibition against replicative DNA polymerases α and δ (Figure 2C). An inhibition study also showed that AZddAATP moderately inhibited telomerase activity in the presence of 10 µM dATP and each of dGTP and dTTP at 200 µM, but slightly in the presence of 10 µM dGTP and each of dATP and dTTP at 200 µM (Figure 2D). As shown in Figure 2E, AZddAATP and AZddATP showed similar inhibitory activity against HIV-1 RT. HIV-1 RT activity is generally assayed on the basis of incorporation of radioisotope-labeled nucleotide substrates into DNA, as described for the assay of DNA polymerases α and δ in Materials and Methods section. However, in the present study HIV-1 RT activity was estimated from the amount of elongated DNA using SYBR green I staining as a convenient assay method. This reagent, which has recently been used for monitoring amplified DNA in real-time PCR (35), was also useful for assaying HIV-1 RT using poly U-oligo(dA), poly A-oligo(dT) and poly C-oligo(dG) as the template–primers (data not shown).
Figure 2.
Inhibition of human telomerase, DNA polymerases α and δ, and HIV-1 RT activities by nucleotide analogs. (A-1) Relationship between telomerase in HeLa cell extract (S-100) and amount of amplified products obtained by stretch PCR method (lanes 1–4 of A-2). Lane 1, 0 µl (0 µg); lane 2, 1.3 µl (0.7 µg); lane 3, 2.7 µl (1.3 µg); lane 4, 4 µl (2 µg). The amount of product was determined with a fluorescence image analyzer and plotted against the amount of S-100 extract. Values on the Y-axis are presented as linear arbitrary units (LAU). A standard curve correlating the amount of enzyme and PCR product was prepared, allowing the linearity of the reaction to be confirmed in each experiment. (A-2) Telomerase activity in the presence of various concentrations of nucleotide analogs. Telomerase activity was measured as described in Materials and Methods section in the presence of the indicated concentrations of inhibitors, 10 µM dNTP bearing a similar base moiety to the inhibitor, and two other dNTPs (200 µM each). Typical results are shown. (B) Remaining activity in the presence of AZddGTP (filled triangles), AZddSGTP (open triangles), AZddAATP (filled circles), AZddATP (open circles) and AZTTP (open squares) are shown. Activity in the absence of inhibitor was taken as 100%. The amount of PCR product comprising a DNA ladder in each lane, as shown in panel A-2, was quantified using a fluorescence image analyzer and the percent activity was estimated. (C) Inhibitory effects of AZddGTP on calf DNA polymerase α (broken line) and human DNA polymerase δ (solid line). The remaining activity of DNA polymerase in the presence of various concentrations of AZddGTP, 20 µM dGTP, 50 µM dATP, 50 µM dCTP and 50 µM [3H]dTTP is shown. Activity in the absence of inhibitor was taken as 100%. (D) Influence of dNTP concentration on the inhibitory effects of AZddAATP on human telomerase. Remaining activity in the presence of 10 µM dATP, 200 µM dGTP and 200 µM dTTP (solid line) or 200 µM dATP, 10 µM dGTP and 200 µM dTTP (broken line) is shown. Activity without inhibitor was taken as 100%. (E) Inhibitory effects of AZddAATP (filled circles) and AZddATP (clear circles) on HIV-1 RT. The remaining activity of HIV-1 RT is shown in the presence of the indicated concentrations of inhibitor, 50 µM dATP as a dNTP substrate and 1 µg poly U-oligo(dA)18 as a template–primer. Activity in the absence of inhibitor was taken as 100%.
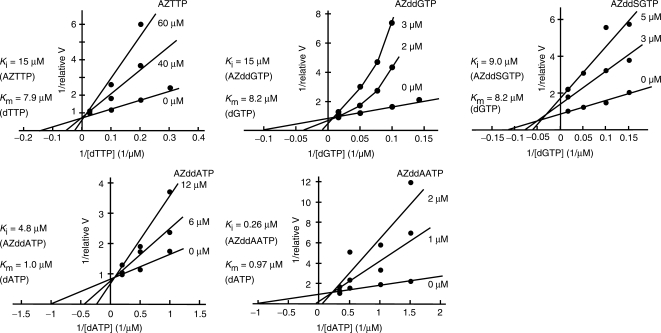
Next, the inhibitory effects of nucleotide analogs on telomerase were analyzed using Lineweaver–Burk plots (Figure 3). The inhibition modes of AZTTP and AZddGTP were shown to be nearly competitive with dTTP or dGTP, respectively. However, the inhibition mode of AZddGTP, shown in Figure 3, was not competitive in a simple manner with respect to dGTP, but rather in a complex manner, as indicated reproducibly by the concave curves obtained at lower concentrations of dGTP. The inhibition mode of AZddAATP was also not simple. On the other hand, the inhibition mode of AZddSGTP was shown to be due to a mixed-type mechanism. The Ki values of AZTTP, AZddGTP, AZddSGTP, AZddATP and AZddAATP were estimated to be 15, 1.5, 9.0, 4.8 and 0.26 µM, respectively, and the Km values of dTTP, dGTP and dATP were estimated to be 7.9 ± 1.8, 8.2 ± 2.1 and 0.97 ± 0.31 µM, respectively.
Figure 3.
Lineweaver–Burk plot analyses of the inhibitory effects of AZTTP, AZddGTP, AZddSGTP, AZddATP and AZddAATP. Relative reaction velocity (relative V) was calculated relative to the highest activity without inhibition, which was taken as 1.
Incorporation of analogs into DNA
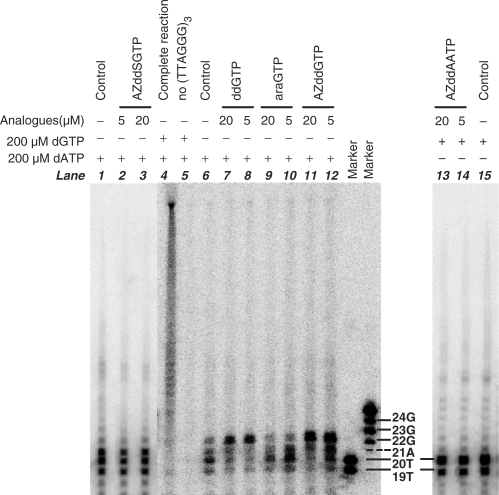
We analyzed the primer extension products synthesized by cherry salmon telomerase in the presence of ddGTP, araGTP, AZddSGTP or AZddGTP instead of dGTP, and in the presence of AZddAATP instead of dATP. The results are shown in Figure 4. These experiments were performed using an 18-mer of (5′-TTAGGG-3′)3 capable of incorporating two 32P-labeled dTMP molecules at the initial two residues, unlabled dAMP at the third and unlabled dGMP at the fourth position (22-mer) from the 3′-end of the primer. The primer extension experiment in the presence of [α-32P]dTTP, dATP and dGTP (complete reaction, lane 4 in Figure 4) gave long products of various lengths, as expected. Omission of dGTP (control reactions, lanes 1 and 6) gave products that migrated as 19-, 20- and 21-mer bands. Similarly, omission of dATP (control reaction, lane 15) gave products that migrated as 19- and 20-mer bands. When examined in the presence of [α-32P]dTTP, dATP with AZddGTP (lanes 11 and 12) or ddGTP (lanes 7 and 8) instead of dGTP gave a distinct 22-mer product, whereas AZddSGTP (lanes 2 and 3) did not give a clear 22-mer product. This means that AZddGTP, but not AZddSGTP, is effectively utilized by telomerase as a substrate and incorporated into the 3′-terminus of the primer strand, and that chain termination is elicited. When examined in the presence of [α-32P]dTTP, dGTP and AZddAATP (lanes 13 and 14) instead of dATP gave only 19- and 20-mer products, similar to the control reaction (lane 15). In this experiment, the telomerase employed was partially purified from a crude extract of immature testes of cherry salmon by diethylaminoethyl cellulose column chromatography and 70% ammonium sulfate fractionation, followed by dialysis against buffer. One explanation for the faint 22-mer bands in lanes 1, 2, 3 and 6 of Figure 4 is misincorporation of dGMP. However, we did not investigate this issue further.
Figure 4.
Utilization of dGTP and dATP analogs as substrates by cherry salmon telomerase. An 18-mer (TTAGGG)3 was extended by partially purified cherry salmon telomerase in the presence of 5 µM [α-32P]dTTP, 200 µM dATP, 0 or 5 µM dGTP and 0, 5 or 20 µM dGTP analogs for dGTP analog analysis, and 5 µM [α-32P]dTTP, 200 µM dGTP, 0 or 5 µM dATP and 0, 5 or 20 µM dATP analogs for dATP analog analysis.
Growth rates and telomere length of HL60 cells grown in the presence of AZddG, AZddAA and AZT
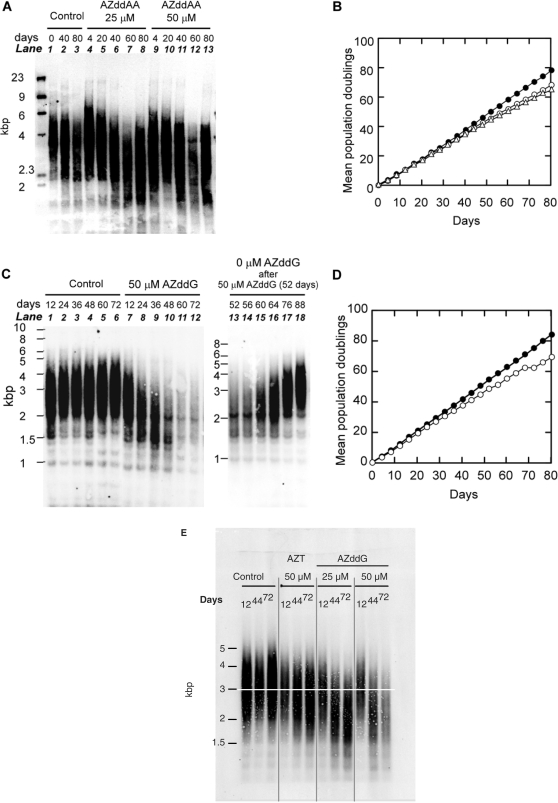
Cultures of HL60 cells were tested with AZddG in the presence of 0.005% dimethylsulfoxide (DMSO) and AZddAA (Figure 5). At the concentrations chosen, 50 µM AZddAA and 50 µM AZddG did not cause any significant change in cell growth rate for first four days. For each culture, every four days, cells were counted to determine cell growth rates, and passaged quantitatively to maintain them in logarithmic growth conditions. We tried to detect the critical point for significant cessation of cell growth or cell death after telomere shortening by treatment with AZddAA or AZddG. However, as shown in Figure 5B and D, the lines representing the rates of cell division were almost linear, and their slopes were not changed after treatment with these compounds. These results suggest that the slight growth inhibition evident in Figure 5B and D was not due to telomere shortening.
Figure 5.
Telomere lengths and growth rates of HL60 cells grown in the presence or absence of AZddAA (A and B) and AZddG (C and D), and comparison of telomere shortening activity of AZT and AZddG (E). DNAs from serial passages of HL60 cells grown in the presence or absence of AZddAA (A), AZddG (C and E) and AZT (E) were digested with Hinf I, run on a 1% agarose gel, and Southern blotted with a digoxigenin-labeled telomere DNA probe. Lanes 13–18 of Figure 5C show the telomere lengths of HL60 cells cultured after removal of AZddG from the culture medium, using cells previously treated with 50 µM AZddG for 52 days. MPDs of cultures treated with 0 µM nucleoside analogs [control cultures, filled circles in (B), and (D)], 25 µM [open circles in (B)] and 50 µM [open triangles in (B)] AZddAA and 50 µM AZddG [open circles in (D)], are plotted against days cultured.
Genomic DNAs from HL60 cells cultured with AZddG and AZddAA were restriction digested with Hinf I, and their telomere length distributions were determined by Southern hybridization analysis with a digoxigenin-labeled telomeric DNA probe (30). The results are shown in Figure 5. Lanes 1–3 in Figure 5A and lanes 1–6 in Figure 5C show the results of control experiments carried out in the absence of nucleoside analogs. Lanes 4–8 and 9–13 in Figure 5A show the effects of 25 and 50 µM AZddAA, respectively, and lanes 7–12 in Figure 5B show the effects of 50 µM AZddG. In control experiments, telomere length showed a broad distribution ranging from ∼2 to ∼6 kb (Figure 5A) and ∼2 to ∼5 kb (Figure 5C), and did not change significantly during all passages. However, after 60 days in the presence of AZddAA, slight telomere shortening was detected (Figure 5A), and after 24 days in the presence of 50 µM AZddG, marked telomere shortening was seen in comparison with the control experiments (Figure 5C). However, these shortened telomeres returned to the length of telomeres in the control cells within three weeks following removal of AZddG from the culture medium (Figure 5C, lanes 13–18).
Telomere shortening activities of AZT and AZddG were compared (Figure 5E). When HL60 cells were treated, AZddG showed more potent activity than AZT. The telomere shortening activities of these two compounds appeared to be correlated with telomerase-inhibitory activities of their triphosphate derivatives.
DISCUSSION
Because of the structural and mechanistic similarity between telomerase and HIV-1 RT, it has been hypothesized that known RT inhibitors may potently inhibit human telomerase. AZT is used clinically against human immunodeficiency virus type 1 (HIV-1). It can be phosphorylated to its 5′-phosphate derivative (AZTTP), which inhibits HIV-1 RT and exerts antiviral activity. Inhibitory activity of AZTTP on telomerase has been reported (19,36). In order to obtain more selective and potent inhibitors of telomerase, we investigated the inhibitory effects of purine analogs of 3′-azido-2′,3′-dideoxynucleotides on telomerase in the hope that conversion of AZTTP to its guanine or adenine counterpart might change its affinity for telomerase. It should be noted that telomerase incorporates as many as three dGTP residues during the extension reaction of the six residue sequence TTAGGG. We are especially interested in the inhibitory effect of a guanine counterpart of AZT, AZddGTP, on telomerase. As shown in Figure 2B, the guanine analogs AZddGTP and AZddSGTP showed potent inhibitory effects. On the other hand, the thymine and adenine counterparts, AZTTP and AZddATP, showed moderate or weak inhibition. We previously observed similar phenomena when comparing thymine analogs and guanine counterparts. Arabinofuranosylthymine 5′-triphosphate (araTTP), 2′-deoxy-2′-fluoro-5-methylarabinofuranosyl-uracil 5′-triphosphate (FMAU-TP) and 1-[(2-hydroxyethoxy)methyl]thymine 5′-triphosphate (acycloTTP) showed weaker telomerase-inhibitory activity than the corresponding guanine analogs (22).
Potent inhibitory activity of AZddGTP is not due only to a frequent incorporation of dGMP during elongation. A possible explanation is as follows. As shown in Figure 2B, remaining telomerase activity in the presence of 10 μM AZddATP and 10 μM dATP was estimated to be 85%. The inhibition mode of AZddATP was shown to be approximately competitive (Figure 3). Therefore, under the present assay conditions, incorporation of dGMP and dTMP by telomerase was hardly inhibited by 10 μM AZddATP because of the high concentrations of dGTP and dTTP (200 μM each). This means that one-on-one competition by AZddATP with dATP during the extension of six residues (TTAGGG) resulted in a residual telomerase activity of 85%. From a statistical viewpoint, if AZddATP can compete with dATP three times during the extension of these six residues in the presence of 10 μM dATP, total inhibition can be calculated as (85%)3 = 61%. On the other hand, with 10 μM AZddGTP, remaining telomerase activity was only 3% in the presence of 10 μM dGTP (Figure 2B). Thus, the difference in inhibition potency between AZddGTP and AZddATP may depend not only on a high frequency of appearance of G-residues during elongation but also other factors such as affinity for the enzyme.
Although AZddATP and AZddAATP showed similar inhibitory activity against HIV-1 RT (Figure 2E), as shown in Figure 2B the IC50 value of AZddAATP (8 μM) for human telomerase was five times lower than that of AZddATP (40 μM), suggesting that the two-amino group of AZddAATP is effective at increasing the telomerase inhibitory activity. HIV-1 RT replicates the HIV-1 genome and shows both RNA- and DNA-dependent DNA polymerase activities. However, telomerase elongates only the TTAGGG repeat at the very end of genomic DNA, and is composed of a protein subunit and a RNA subunit that includes a template sequence for DNA synthesis (37). Thus, the two reverse transcriptases fulfill different functions. The two-amino group of the guanine ring is a distinctive feature in the structure of dGTP. Telomerase also showed higher affinity for AZddAATP bearing a two-amino group in the adenine ring than it did for AZddAA (Figure 3). Thus, telomerase appears to be an enzyme specialized for the frequent incorporation of the dGMP moiety.
We were also interested in the 1,6-position tautomerism equilibration of the 2,6-diaminopurine moiety of 3′-azido-2′,3′-dideoxy-2-aminoadenosine (AZddAA). Therefore, 5′-triphosphate (AZddAATP) was designed with the anticipation that it would function as a competitor of both dGTP and dATP. Contrary to our expectation, AZddAATP behaved like a competitor of dATP only (Figure 2D).
The Ki value of AZddGTP was estimated to be 1.5 µM. This value is somewhat higher than that of ddGTP (0.65 µM), when compared with that of araGTP (1.2 µM) (31), and ∼5 times smaller than the Km of dGTP (8.2 ± 2.1 µM). Human telomerase seems to prefer AZddGTP as well as ddGTP and araGTP to dGTP. Likewise, the Ki value of AZddSGTP was estimated to be 9 µM, similar to the Km value of dGTP (8.2 ± 2.1 µM). In the case of AZddAATP, the Ki value was low (0.26 µM), about a quarter of the Km value of dATP (0.97 ± 0.31 µM). Only the Km for dATP was about eight times smaller than the Km values of dGTP and dTTP. The Km values of dGTP and dTTP (8.2 ± 2.1 and 7.9 ± 1.8 µM) obtained here were similar to those (8–12 µM) of telomerase from human leukemia cells and HeLa cells (38).
As shown in Figure 4, the amounts of a 22-mer chain-terminated product in lanes 11 and 12 in the presence of AZddGTP were somewhat higher than those in lanes 7 and 8 in the presence of ddGTP, respectively, under similar experimental conditions. Therefore, it is suggested that AZddGTP can be incorporated into the primer more efficiently than ddGTP.
Strahl and Blackburn demonstrated that long-term treatment of immortalized human B- and T-cell lines with 10 µM ddG caused reproducible progressive telomere DNA shortening. However, they also reported that ddG did not have observable effects on cell population doubling rates or morphology (19). Similarly, Gomez et al. demonstrated progressive telomere DNA shortening in HeLa cells upon treatment with 800 µM AZT (20), but no evidence of cellular senescence could be detected. Interestingly, Ji et al. recently reported that cyclic treatment of human breast cancer MCF-7 cells with AZT suppressed telomerase, followed by suppression of telomerase RNA, and that further treatment with AZT accelerated both telomere loss and apoptosis (39). They confirmed that AZT-induced telomere shortening is irreversible and cumulative.
Treatment of HL60 with 50 µM AZddG for 24 days or more caused significant telomere DNA shortening. Additionally, shortened telomeres began to elongate when AZddG was removed from the culture medium (Figure 5C). These observations indicate that the telomere shortening in AZddG-treated cells is attributed to AZddGTP because AZddG showed only low cytotoxicity for cultured cells (21), and AZddGTP is a potent and selective inhibitor for telomerase (Figure 2B and C). Treatment of HL60 cells with both AZddAA and AZddG caused telomeres to shorten significantly during early passages (up to about 40–50 days) but then stabilized during later passages. This finding is consistent with the previous analyses demonstrating that reduced telomerase activity can maintain telomeres at shorter equilibrium length (40). It appears that even reduced telomerase activity, caused by inhibitors, is sufficient for telomere maintenance when telomeres are very short.
Although the number of mean population doublings (MPDs) of HL60 cells in control culture medium was estimated to be ∼4.1 per passage that of treated cells cultured in the presence of AZddG for 24 days or more was estimated to be ∼3.6. Thus, AZddG treatment caused a slight but steady decrease in cell growth rate during 80 days of culture. However, HL60 cells maintained for ∼200 days in the presence of 50 µM AZddG did not show significant growth cessation or cell death (data not shown). These results suggest that AZddG alone cannot critically shorten telomeres to a degree that would lead to cell death (41), even though AZddG caused efficient telomere shortening in our experiments.
Among the compounds tested in this study, the Ki value for AZddAATP was the lowest (0.26 µM). However, during inhibition, AZddAATP behaved as a dATP analog (Figure 2D). Although the Ki value of AZddAATP was lower than that of AZddGTP (Ki = 1.5 µM), the value (0.27) of Ki, AZddAATP/Km, dATP was slightly larger than the value (0.18) of Ki, AZddGTP/Km, dGTP. These results suggest that the inhibitory activity of AZddGTP is comparable to that of AZddAATP. Furthermore, cellular deoxyribonucleotide pool levels may also influence the inhibitory activity of nucleotide analogs. It has been reported that the levels of dATP and dGTP pools in HL60 cells are ∼15 and ∼8 μM, respectively (42). If AZddG and AZddAA were equally activated to the corresponding 5′-triphosphates by cellular kinases, then the inhibitory activity of AZddAATP would be decreased by the higher dATP pool level. Indeed, AZddAA caused less telomere shortening (Figure 5).
It was reported that the antitumor effects of cisplatin on proliferation of the human endometrial cancer cell line HEC-1 are enhanced in the presence of RT inhibitors (43). Therefore, it will be of interest to search for potent telomerase inhibitors like AZddG, which show considerable promise for potentiating the activity of some antitumor agents when combined with conventional chemotherapeutics.
CONCLUSION
AZddGTP is a potent and selective inhibitor of human telomerase. Displacement of the 2-H of AZddATP with an amino group is important for its inhibitory activity on telomerase, although this modification did not affect its inhibitory activity towards HIV-1 RT. Treatment of HL60 cells with AZddG resulted in more potent effects on telomere shortening than treatment with AZT. Thus, AZddG was shown to be a useful inhibitor for reproducible preparation of telomere-shortened HL60 cells; however, AZddG alone could not critically shorten telomeres to a degree that would lead to cell death. Characterization of the inhibitor-treated cells is now underway in our laboratory.
ACKNOWLEDGEMENTS
We thank Dr Teruo Azuma, Nikko Branch, National Research Institute of Aquaculture, Fisheries Research Agency, for kindly providing immature male cherry salmon. We thank Mr J. Takashima, Mr K. Kazama, Mr K. Sato, Ms M. Yamamura and Ms M. Takeuchi, of our laboratory, for their helpful support. This work was supported in part by a Grant-in-Aid for Scientific Research on Bioscience/Biotechnology Areas from the Ministry of Education, Culture, Sports, Science and Technology in Japan. Funding to pay the Open Access publication charges for this article was provided by High-Tech Research Project for private universities: matching fund from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 3.Wright W.E., Pereira-Smith O.M., Shay J.W. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol. Cell. Biol. 1989;9:3088–3092. doi: 10.1128/mcb.9.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W., Shay J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Aisner D.L., Wright W.E., Shay J.W. Telomerase regulation: not just flipping the switch. Curr. Opin. Genet. Dev. 2002;12:80–85. doi: 10.1016/s0959-437x(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 6.Forsyth N.R., Wright W.E., Shay J.W. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 7.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 8.Broccoli D., Young J.W., de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc. Natl Acad. Sci. USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Counter C.M., Gupta J., Harley C.B., Leber B., Bacchetti S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood. 1995;85:2315–2320. [PubMed] [Google Scholar]
- 10.Prowse K.R., Greider C.W. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl Acad. Sci. USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shay J.W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 12.Morin G.B. Is telomerase a universal cancer target? J. Natl Cancer Inst. 1995;87:859–861. doi: 10.1093/jnci/87.12.859. [DOI] [PubMed] [Google Scholar]
- 13.Parkinson E.K. Do telomerase antagonists represent a novel anti-cancer strategy? Brit. J. Cancer. 1996;73:1–4. doi: 10.1038/bjc.1996.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg R.A., Chin L., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 15.Mergny J.L., Riou J.F., Mailliet P., Teulade-Fichou M.P., Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin G.B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn E.H. Telomerases. Annu. Rev. Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 18.Lingner J., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 19.Strahl C., Blackburn E.H. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol. Cell. Biol. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez D.E., Tejera A.M., Olivero O.A. Irreversible telomere shortening by 3′-azido-2′,3′-dideoxythymidine (AZT) treatment. Biochem. Biophys. Res. Commun. 1998;246:107–110. doi: 10.1006/bbrc.1998.8555. [DOI] [PubMed] [Google Scholar]
- 21.Robins M.J., Wood S.G., Dalley N.K., Herdewijn P., Balzarini J., de Clercq E. Nucleic acid related compounds. 57. Synthesis, x-ray crystal structure, lipophilic partition properties, and antiretroviral activities of anomeric 3′-azido-2′,3′-dideoxy-2,6-diaminopurine ribosides. J. Med. Chem. 1989;32:1763–1768. doi: 10.1021/jm00128a017. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi T., Takahashi H., Jinmei H., Takayama Y., Saneyoshi M. Inhibition of vertebrate telomerases by the triphosphate derivatives of some biologically active nucleosides. Nucleos. Nucleot. Nucl. 2003;22:1575–1577. doi: 10.1081/NCN-120023037. [DOI] [PubMed] [Google Scholar]
- 23.Imazawa M., Eckstein F. Synthesis of 3′-azido-2′,3′-dideoxyribofuranosylpurines. J. Org. Chem. 1978;43:3044–3048. [Google Scholar]
- 24.Marchand A., Mathe C., Imbach J.L., Gosselin G. Synthesis and antiviral evaluation of unnatural β-L-enantiomers of 3′-fluoro- and 3′-azido-2′,3′-dideoxyguanosine derivatives. Nucleos. Nucleot. Nucl. 2000;19:205–217. doi: 10.1080/15257770008033004. [DOI] [PubMed] [Google Scholar]
- 25.Yoshikawa M., Kato T., Takenishi T. Studies of phosphorylation. III. Selective phosphorylation of unprotected nucleosides. B. Chem. Soc. Jpn. 1969;42:3505–3508. [Google Scholar]
- 26.Maeda M., Patel A.D., Hampton A. Formation of ribonucleotide 2′,3′-cyclic carbonates during conversion of ribonucleoside 5′-phosphates to diphosphates and triphosphates by the phosphorimidazolidate procedure. Nucleic Acids Res. 1977;4:2843–2853. doi: 10.1093/nar/4.8.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono K., Ohashi A., Yamamoto A., Matsukage A., Takahasi T., Saneyoshi M., Ueda T. Inhibitory effects of 9-β-D-arabinofuranosylguanine 5′-triphosphate and 9-β-D-arabinofuranosyladenine 5′-triphosphate on DNA polymerases from murine cells and oncornavirus. Cancer Res. 1979;39:4673–4680. [PubMed] [Google Scholar]
- 28.Mizushina Y., Xu X., Asahara H., Takeuchi R., Oshige M., Shimazaki N., Takemura M., Yamaguchi T., Kuroda K., et al. A sulphoquinovosyl diacylglycerol is a DNA polymerase epsilon inhibitor. Biochem. J. 2003;370:299–305. doi: 10.1042/BJ20021737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshige M., Takeuchi R., Ruike T., Kuroda K., Sakaguchi K. Subunit protein-affinity isolation of Drosophila DNA polymerase catalytic subunit. Protein Expres. Purif. 2004;35:248–256. doi: 10.1016/j.pep.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi T., Saneyoshi M., Takahashi H., Hirokawa S., Amano R., Liu X., Inomata M., Maruyama T. Synthetic nucleosides and nucleotides. 43. Inhibition of vertebrate telomerases by carbocyclic oxetanocin G (C.OXT-G) triphosphate analogues and influence of C.OXT-G treatment on telomere length in human HL60 cells. Nucleos. Nucleot. Nucl. 2006;25:539–551. doi: 10.1080/15257770600684217. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi T., Yamada R., Tomikawa A., Shudo K., Saito M., Ishikawa F., Saneyoshi M. Recognition of 2′-deoxy-L-ribonucleoside 5′-triphosphates by human telomerase. Biochem. Biophys. Res. Commun. 2000;279:475–481. doi: 10.1006/bbrc.2000.3982. [DOI] [PubMed] [Google Scholar]
- 32.Tendian S.W., Parker W.B. Interaction of deoxyguanosine nucleotide analogs with human telomerase. Mol. Pharmacol. 2000;57:695–699. doi: 10.1124/mol.57.4.695. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi M., Matsukage A., Takahashi T. Chick embryo DNA polymerase γ. Purification and structural analysis of nearly homogeneous enzyme. J. Biol. Chem. 1980;255:7002–7009. [PubMed] [Google Scholar]
- 34.Tatematsu K., Nakayama J., Danbara M., Shionoya S., Sato H., Omine M., Ishikawa F. A novel quantitative ‘stretch PCR assay’, that detects a dramatic increase in telomerase activity during the progression of myeloid leukemias. Oncogene. 1996;13:2265–2274. [PubMed] [Google Scholar]
- 35.Lutfalla G., Uze G. Performing quantitative reverse-transcribed polymerase chain reaction experiments. Meth. Enzymol. 2006;410:386–400. doi: 10.1016/S0076-6879(06)10019-1. [DOI] [PubMed] [Google Scholar]
- 36.Strahl C., Blackburn E.H. The effects of nucleoside analogs on telomerase and telomeres in Tetrahymena. Nucleic Acids Res. 1994;22:893–900. doi: 10.1093/nar/22.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 38.Pai R.B., Pai S.B., Kukhanova M., Dutschman G.E., Guo X., Cheng Y.C. Telomerase from human leukemia cells: properties and its interaction with deoxynucleoside analogues. Cancer Res. 1998;58:1909–1913. [PubMed] [Google Scholar]
- 39.Ji H.J., Rha S.Y., Jeung H.C., Yang S.H., An S.W., Chung H.C. Cyclic induction of senescence with intermittent AZT treatment accelerates both apoptosis and telomere loss. Breast Cancer Res. Treat. 2005;93:227–236. doi: 10.1007/s10549-005-5156-0. [DOI] [PubMed] [Google Scholar]
- 40.Erdmann N., Liu Y., Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc. Natl Acad. Sci. USA. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armbruster B.N., Etheridge K.T., Broccoli D., Counter C.M. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol. Cell. Biol. 2003;23:3237–3246. doi: 10.1128/MCB.23.9.3237-3246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansson E., Spasokoukotskaja T., Sallstrom J., Eriksson S., Albertioni F. Molecular and biochemical mechanisms of fludarabine and cladribine resistance in a human promyelocytic cell line. Cancer Res. 1999;59:5956–5963. [PubMed] [Google Scholar]
- 43.Murakami J., Nagai N., Shigemasa K., Ohama K. Inhibition of telomerase activity and cell proliferation by a reverse transcriptase inhibitor in gynaecological cancer cell lines. Eur. J. Cancer. 1999;35:1027–1034. doi: 10.1016/s0959-8049(99)00037-4. [DOI] [PubMed] [Google Scholar]