Abstract
Hairpin nucleic acid probes have been highly useful in many areas, especially for intracellular and in vitro nucleic acid detection. The success of these probes can be attributed to the ease with which their conformational change upon target binding can be coupled to a variety of signal transduction mechanisms. However, false-positive signals arise from the opening of the hairpin due mainly to thermal fluctuations and stem invasions. Stem invasions occur when the stem interacts with its complementary sequence and are especially problematic in complex biological samples. To address the problem of stem invasions in hairpin probes, we have created a modified molecular beacon that incorporates unnatural enantiomeric l-DNA in the stem and natural d-DNA or 2′-O-Me-modified RNA in the loop. l-DNA has the same physical characteristics as d-DNA except that l-DNA cannot form stable duplexes with d-DNA. Here we show that incorporating l-DNA into the stem region of a molecular beacon reduces intra- and intermolecular stem invasions, increases the melting temperature, improves selectivity to its target, and leads to enhanced bio-stability. Our results suggest that l-DNA is useful for designing functional nucleic acid probes especially for biological applications.
INTRODUCTION
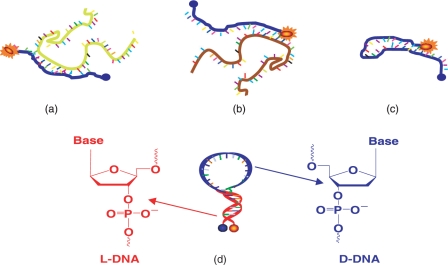
Molecular beacons (MBs) are single-stranded nucleic acid probes composed of three different functional domains: stem, loop and fluorophore/quencher pair (Figure 1d) (1). Stems function as locks that maintain the closed hairpin structures when there is no hybridization with complementary targets so that the fluorescence is quenched with high-quenching efficiency. Upon hybridization between the loop and its complementary target, the MB undergoes conformational changes from hairpin to linear structure resulting in an increased fluorescence due to the increased physical separation of the fluorophore and quencher. Various fluorophore/quencher pairs can be incorporated into experimental design so that multiple targets can be monitored simultaneously. MBs have been for real-time monitoring of RT–PCR (2,3) and mRNA expression inside of living cells (4–6). Unlike traditional mRNA analysis, MBs do not require pre- or post-treatment of cells because, theoretically, they will not give any fluorescence signal unless hybridizing with their complementary targets. Indeed, MBs are selective enough to distinguish single-mismatched targets since the principle of MB's target recognition is based on Watson–Crick base paring.
Figure 1.
Molecular beacon design and interactions. (a) MB binding to target mRNA; (b) MB opened by non-specific mRNA that binds to MB stem; (c) MB mis-folds into non-hairpin structure through the hybridization of loop sequence with stem sequence. (d) A newly designed MB using l-DNA for the stem (red) to enforce a hairpin structure,, and d-DNA for the loop (blue) for nucleic acid recognition effectively avoids the situations depicted in (b) and (c).
Although initial MB applications have demonstrated wide feasibility and great potential, challenges in probe design and applications still remain. Since stems are critical for maintaining a stable hairpin structure, major difficulties often arise from the interruption of the stem structure. There are two common causes of stem disruption which severely affect MB's sensitivity and selectivity. One problem is the undesired intermolecular interactions between stems and their complementary sequences, called stem invasion (Figure 1b). Although the ideal molecular recognition scheme is between the loops of MBs and their complementary targets (Figure 1a), there are always chances that, especially in complex cellular environments, the stems can hybridize with their matched sequences, resulting in a high background signal and a poor analytical selectivity (Figure 1b). This stem invasion by non-target sequences should not be underestimated in real biological samples since there could be high copy numbers of short sequences complementary to part of the MB stems. In theory, the expressed frequency of occurrence of any particular stretch of RNA or DNA sequence with n bases is 4−n (7). The average number of occurrences of each particular sequence in human genome is 5 × 109 × 4−n (7). Table 1 summarizes the occurrence of a nucleic acid sequence with n base pairs. This theoretical calculation indicates that if the base parings between the short sequences can take place, MBs can be vulnerable to false-positive signal coming from stem-involved recognition even though each MB is designed to target only one complementary nucleic acid sequence. Another problem arising from the stems is the thermodynamic conformational switch between hairpin and non-hairpin structures. This is a result of the unwanted intramolecular hybridization between the stem and part of the loop. In the case of MBs, this can cause a significant fluorescence background due to incomplete quenching. This problem increases the amount of effort required in designing and testing MBs for new target sequences with acceptable performance (Figure 1c). Sometimes the stem invasion cannot be avoided at all, resulting in low-performance hairpin structures in nucleic acid monitoring.
Table 1.
Copy numbers of each nucleic acid appearing in DNA and RNA targets
| Sequence length | Number of occurrence for DNA (5 billion bp) | Number of occurrence for RNA (∼0.3 billion bp) |
|---|---|---|
| 10 | 4768 | 286 |
| 11 | 1192 | 72 |
| 12 | 298 | 18 |
| 13 | 74 | 4 |
| 14 | 18 | 1 |
| 15 | 4 | 0 |
| 16 | 1 | 0 |
There have been developments aimed at minimizing the aforementioned problems. For example, strengthening stem stability by increasing stem length or G and C content can increase the dominance of the hairpin structure, so that the background signal of MBs remains low (8). Unfortunately, a strong stem leads to slow hybridization kinetics and a high tendency of forming sticky-end pairing, reducing the fluorescence signal enhancement upon target hybridization (9). The typical example is the locked nucleic acid (LNA), or LNA MB (10). LNA is known as an extraordinarily strong nucleic acid binder and LNA MBs show ultra-high thermal stability, resulting in a low background signal. However, LNA MBs tend to have extremely slow hybridization kinetics, which limits their usefulness. In order to minimize unnecessary intermolecular interactions, MBs have been created in which the stems have structural features that inhibit stem–loop hybridization. In one study, the sense of the stem DNA was inverted relative to the loop, i.e. 3′-stem-5′-5′-loop-3′-3′-stem-5′, to ensure that any stem–loop hybridization would be disrupted at the 5′5′ and 3′3′ junctions (11). Unfortunately, the usefulness of these MBs is limited because they required longer stems causing increased sticky-end-paring problems. These examples showcase the underlying complexity of designing MBs and other DNA hairpin probes. There has yet to be a MB design that simultaneously solves the problem of stem invasion without causing other performance limiting effects.
Ideally, the MB stem should not interact with the loop or any other complementary nucleic acid sequences. If the stem can be immune to naturally occurring nucleic acids either within the probe or in the sample matrix, there would be no stem invasion, resulting in highly stable hairpin nucleic acid probes for analysis. This thought has led us to consider non-standard bases like l-DNA which do not hybridize with natural nucleic acids. Our hypothesis is that stems made of such bases will hybridize with each other and not to the loop or any other natural nucleic acids in the sample. In this way, a stable hairpin structure is formed in which the stem and loop have no innate ability to form a stable structure. This idea has been explored using HOMO DNA stem MBs (12). Unfortunately, the synthesis of such MBs is not easy and requires optimization of the stem design. Here we propose to use non-natural enantiomeric DNA, termed l-DNA, in the stem, and natural d-DNA in the loop (Figure 1d) as a model system for MB design. Since l-DNA is the mirror-image form of the naturally occurring d-DNA, its duplexes have the same physical characteristics in terms of solubility and stability as d-DNA hybrids, except for forming a left-handed double helix (13–16). l-DNA was examined as a potential antisense reagent but failed to perform adequately because there is no interaction between l-DNA and d-DNA due to the chiral difference (17). By taking advantage of these features of l-DNA to build stems we can prevent intramolecular and intermolecular nonspecific interactions in any hairpin-structured DNA probes. In addition, the easy incorporation of l-DNA stems to other non-standard bases, such as 2-O-Me-modified RNA, can be attractive to design bio-compatible MBs. In the present report, we have designed, prepared and studied MBs based on this strategy, and will show that this design can effectively minimize stem-related problems in MBs.
MATERIALS AND METHODS
Synthesis of MBs and their targets
The three oligonucleotide sequences shown in Table 2 were synthesized. MB1 sequence which does not have any biological targets is randomly designed with high signal enhancement and moderate hybridization kinetics. MB2 and MB3 were specially designed to target β-actin mRNA (GenBank Accession No. BC014861) and MnSOD mRNA (Gen-Bank Accession No. NM-000636), respectively (18). The l-deoxyphosphoramidites were obtained from ChemGenes Corporation (Wilmington, MA). The other synthesis reagents, such as 3′-dabcyl (quencher) CPG, 6-fluorescein phosphoramidite (fluorophore), 2′-O-Me-modified RNA phosphoramidite, and d-deoxyphosphoramidites, were from Glen Research Corporation (Sterling, VA). All MBs and their targets were synthesized using an ABI 3400 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA) at 1 μmol scale with the standard synthesis protocol. Dabcyl was placed at 3′ to ensure that every DNA strand contains a quencher. For high coupling yield, the coupling of 6-fluorescein phosphoramidite to 5′ end of each MB was extended to 15 min. For the complete cleavage and deprotection, overnight incubation with ammonia was used. After ethanol precipitation, the precipitates were then dissolved in 0.5 ml of 0.1 M triethylammonium acetate (TEAA, pH7.0) for further purification with high-performance liquid chromatography (HPLC). The HPLC was performed on a ProStar HPLC Station (Varian, CA) equipped with a fluorescence detector and a photodiode array detector. A C-18 reverse phase column (Alltech, C18, 5 μM, 250 × 4.6 mm) was used.
Table 2.
The sequences of hairpin probes and their targets
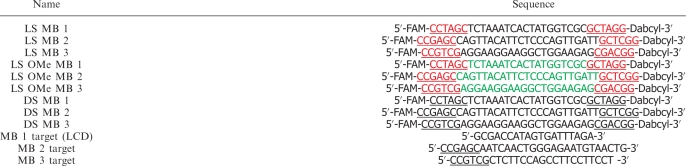 |
Black letters indicate natural DNA bases, red letters are l-DNA bases, and green letters are 2′-O-Me-modified RNA bases. Underlined regions are stems.
Hybridization experiments
Fluorescence measurements were performed with a Fluorolog-3 Model FL3-22 spectrofluorometer (JOBIN YVON-SPEX Industries, Edison, NJ) using a 100 μl quartz cuvette. All hybridizations were performed at room temperature. First, the background fluorescence from 200 μl of the buffer containing 20 mM of Tris–HCl (pH7.5), 50 mM NaCl and 5 mM MgCl2, designated as MB buffer, was monitored for about 1 min and then each stock MB solution (20 μM) was added to the hybridization buffer to reach final concentration of 65 nM and the fluorescence was monitored. After a stable fluorescence signal was obtained from the MB, an excess of target oligonucleotide (650 nM) was added. The level of fluorescence intensity was recorded until the signal reached plateau. The excitation and emission wavelengths were set to 488 nm and 520 nm, respectively. Signal enhancement was calculated using the following equation,
where Fopen are fluorescence signals from MBs with target; Fclosed the fluorescence signals from MBs without target; Fbuffer the fluorescence signal of buffer.
Melting temperature (Tm) measurement of MBs
Thermal denaturizing profiles of each MB were recorded to evaluate the stability of the stems using by a BioRad RT–PCR thermal cycler. The identical buffer to the one used for hybridization study was applied. The solution was prepared with 100 nM final concentration of each MB. The fluorescence intensity of each MB in buffer at temperatures ranging from 10°C to 95°C in the intervals of 1°C, was measured and plotted against the temperature to generate the melting temperature curve.
Nuclease Sensitivity
To test the nuclease digestion of MBs, Deoxynuclease I from Sigma–Aldrich, Inc. (St Louis, MO) was chosen as a standard nuclease. The fluorescence of 65 nM of MBs in MB buffer was measured as a function of time at room temperature. Once the fluorescence is stabilized, two units of ribonuclease-free DNase I was added, and the fluorescence change was monitored until it reached to plateau.
RNase H sensitivity
To test the vulnerability of MB-RNA duplexes to ribonuclease H from Sigma–Aldrich, Inc. (St Louis, MO) digestion, 65 nM of MBs and RNA targets were incubated in the MB buffer while the fluorescence intensity was monitored. After the hybridization reached equilibrium, two units of ribonuclease H were added, and the change in fluorescence was recorded as a function of time.
Preparation of cell lysate
Cell lysate was prepared using CCRF-CEM (CCL-119, T cell line, human ALL) obtained from American Type Culture Collection (Manassas, VA) and Cell culture lysis buffer containing 25 mM Tris (pH 7.8 with H3PO4), 2 mM CDTA 2 mM DTT, 10% glycerol and 1% Triton®X-100 purchased from Promega (Madison, WI) with the protocol recommended by manufacture. CCRF-CEM cells were cultured in RPMI medium 1640 (American Type Culture Collection) supplemented with 10% FBS (heat-inactivated; GIBCO) and 100 U/ml penicillin–streptomycin (Cellgro).
Briefly, ∼6 million cells were centrifuged to remove cell culture media and then washed twice with 500 μL of 5 mM MgCl 2 in Dulbecco's PBS Dulbecco's PBS with calcium chloride and magnesium chloride purchased from Sigma–Aldrich, Inc. (St Louis, MO). All these steps were done at 4°C. After the buffer was removed, the cells were incubated with 200 μl of cell lysis buffer for 3 min at room temperature. Finally, the mixture was centrifuged to remove any cell debris. To determine biostability of each MB, 2 μl of cell lysate was applied.
RESULT AND DISCUSSION
Stability and sensitivity of l-DNA stem MBs (LS MBs)
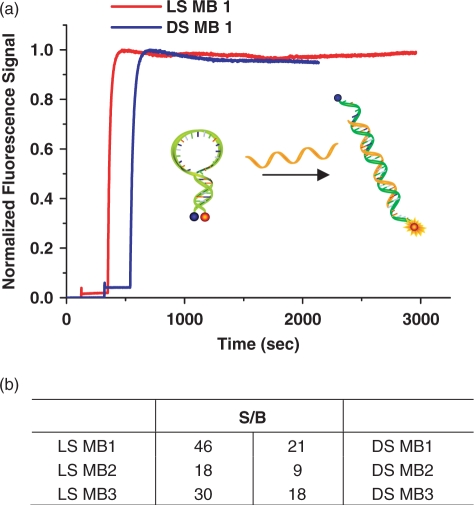
To show the feasibility of an l-DNA stem, we synthesized three l-DNA stem MBs using sequence MB1, MB2 and MB3 shown in Table 2. The same sequences were used to prepare control MBs, called DS MBs entirely made of d-DNA bases. Hybridization of each MB to its corresponding natural DNA target was performed under the same conditions. In order to ensure all MBs are opened, we used 10 times more target DNA. The signal enhancement was calculated with the S/B equation given in Materials and Methods section. As shown in Figure 2a, the l-DNA stem duplex was able to maintain the hairpin conformation and dehybridizes when the loop binds to its target with moderate hybridization kinetics. More interestingly, compared to DS MB 1, LS MB 1 produced a lower fluorescence signal in the absence of its target, which leads to a signal enhancement ratio of more than twice that of the DS MB 1, 46 compared to 21, respectively (Figure 2b). High signal enhancements were consistently observed from all three LS MBs. LS MB 2 and LS MB 3 had 18 and 30 times enhancement compared to their counterparts MB 2 and MB 3, which had 9 and 18 times, respectively (Figure 2b). Thus, the better S/B of l-DNA stem MBs over regular MBs can be generalized regardless of oligo sequences. The improved sensitivity is believed to be a result of the enhanced stability of the hairpin conformation in l-DNA MBs due to the lack of stem–loop interactions which could otherwise contribute to a significant background. The better structural stability causing higher S/B was supported by the higher Tm of stems for LS MBs compared to that of DS MBs.
Figure 2.
The responses of LS MBs and DS MBs to the targets. (a) The hybridization of LS MB1 and DS MB1 took place with 10 times excess target DNA, and their fluorescence change was recorded. (b) Signal enhancement (S/B) of each MB was calculated and compared.
Improved structural stability
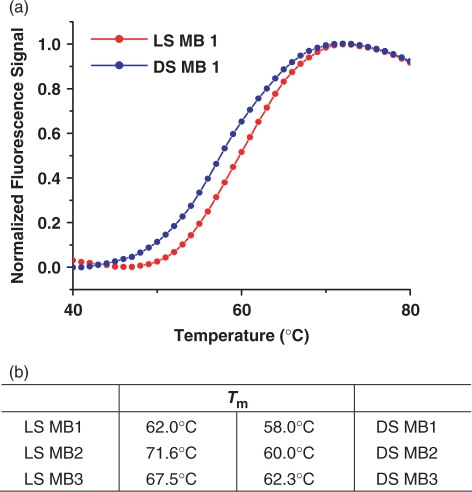
To evaluate the stem stability of MBs, the Tm of each probe was determined and compared. For all MB sequences prepared, LS MBs showed higher melting temperatures than their counterparts. For example, the LS MB 1 has a Tm of about 62°C while that for the DS MB 1 is about 58°C (Figure 3a). This difference is well above the errors in Tm measurement in our instrument (<0.5°C). The other sequences, MB 2 and MB 3, also had consistent higher Tm, ∼5–10°C, in the case of l-DNA stem design (Figure 3b). As l-DNA:l-DNA base pairs have comparable stability to that of their d-DNA counterparts, such an increase in Tm is probably due to a more stable hairpin conformation of the LS MB than that of the DS MB rather than stronger base paring between l-DNA bases (19). The improved stability of the l-DNA stem is consistent with the enhanced sensitivity of the probe observed in our hybridization experiments. In an earlier report, the partial replacement of d-DNA bases with l-DNA bases in DNA duplexes caused lower Tm (19). Our results do not contradict this work since the double-stranded stem of the LS MBs is made entirely of l-DNA and therefore does not have to accommodate both helicities simultaneously. Thus, the l-DNA stems are strong enough to maintain the stable hairpin structure, and, actually, show better stability than the pure d-DNA probes.
Figure 3.
Melting temperature of DS and LS MBs. (a) Melting temperature profiles of DS and LS MB 1 and (b) comparisons of stem melting temperatures of MBs. LS MBs generally showed much higher melting temperature, about 5–10°C, compared to their counterparts.
Elimination of intramolecular interaction
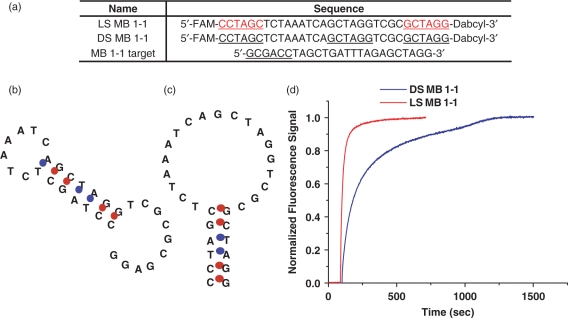
As discussed above, intramolecular interactions between stem and loop domains can cause a high background due to thermal fluctuation between the hairpin structure and other conformations. In the case of FRET-based linear nucleic acid probes, conformational changes due to thermal fluctuations are not critical since linear probes do not require any proper conformation to minimize FRET background prior to target binding. However, in the case of hairpin structures like MBs, the minor contribution of non-hairpin conformations can affect S/B more dramatically due to incomplete quenching caused by non-hairpin conformations, leading to a high background signal. Thus, the addition of l-DNA stem eliminates unwanted intramolecular interactions between the stem and loop, favoring the hairpin conformation and therefore improving sensitivity of MBs as well as easing restrictions in stem design. This situation can be understood thermodynamically in the following way. Free of target, the unwanted intramolecular interactions give the DS MB higher entropy than the LS MB. This causes the difference in the free energy between open or random coil states and closed states to be more negative for the LS MB. This argument correlates well with the higher Tm of the LS MB compared to DS MB from the previous section. Since the LS MB structure is dominated by the hairpin state, more heat energy must be added to dehybridize the stem and open the beacon. Conversely, the DS MB requires less heat energy to open since the hairpin structure is not as dominant and the less stable intramolecular configurations melt at lower temperatures. To prove the hypothesis that the low affinity of l-DNA to d-DNA can eliminate the stem–loop interaction and stabilize hairpin structure more efficiently, a specific MB was designed, called MB 1-1. This sequence was designed such that the non-hairpin structure is dominant for DS MB 1-1 as shown in Figure 4b. Thus, DS MB 1-1 can have two dominant conformations, hairpin and non-hairpin structures (Figure 4b and c), and their distributions are dependent on thermodynamic stability. On the other hand, replacing the MB stem with an l-DNA stem can ensure a hairpin structure as a dominant structure as shown in Figure 4c since the l-DNA cannot hybridize with d-DNA. The major difference between the two MBs was observed in the hybridization kinetics with their targets. The DS MB 1-1 showed noticeably slow hybridization rate (Figure 4d). We believed that there are two major reasons. One is the reduction of exposed loop sequences to its target, causing minimized chances of interactions between a probe and its target. The other is the extra energy required for dehybridization of the partially hybridized loop region before the probe binds to the target. In contrast, the LS MB 1-1 hybridized with the target much faster. This is because the fully open loop region can be easily accessed for interaction to its target, resulting in a fast dynamic response. Our results confirm that using l-DNA stem in designing MBs can effectively eliminate undesired stem–loop interactions in hairpin structured nucleic acid probes.
Figure 4.
Elimination of stem and loop interaction using l-DNA stem. (a) MB 1-1 and its target sequences. The underlines at the end for sequences represent stems and the one at the middle is the complementary to one of the stem. (b–c) Possible conformations of MB1-1 sequence as predicted by DNA/RNA folding program mfold. DS MB 1-1 folds into non-hairpin structure because of the stem-loop interaction (b). Use of l-DNA in the stem of LS MB 1-1 removes stem-loop interaction, forcing the probe to form a hairpin structure (c). (d) Hybridization curves showing faster hybridization kinetics for LS MB 1-1 compared to DS MB 1-1.
Elimination of false-positive signal
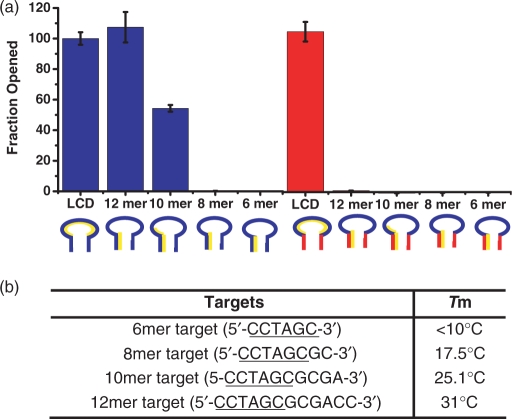
The other major advantage of using l-DNA stem in MB construction is to prevent false reporting coming from the stem and non-target sequence interactions (Figure 1b). This is an intrinsic problem of all DS MBs. Presumably, a short sequence with only 5–6 bp might not have significant contribution in stem invasion, but the one with the extended sequences matched to partial loop can have high potential to cause non-selective opening of MB and false-positive signal. In order to evaluate the extent of such non-specific interactions, we designed oligo- d-DNA with different lengths that had six bases complementary to the stem of the MB while the remaining sequence matched the loop region: 5′-CCTAGC-3′, 5′-CCTAGCGC-3′, 5′-CCTAGCGCGA-3′ and 5′-CCTAGCGCGACC-3′ (underline is complementary to the 3′ stem sequence of the MB). The calculated thermodynamic stabilities of each target with its complementary sequence are shown in Figure 5b. The loop complementary DNA target, 5′-GCGACCATAGTGATTTAGA-3′ was also prepared as a reference. These sequences were separately incubated with both LS and DS MB 1 for 1 hr. The responses of the MBs were recorded and the fluorescence signal of each sample was compared based on the fluorescence signal of MB with loop-target DNA mixture (Figure 5a). For DS MB 1, incubation with 10-fold excess of the 6mer DNA target failed to open the hairpin structure. This is expected since the intramolecular-binding constant between the stem sequences is far greater than the intermolecular interaction between one arm of the stem and the 6mer DNA target. Similar results were obtained for the 8mer sequence. On the contrary, in the presence of the 10mer DNA, which was complementary to one arm of the stem and four adjacent bases in the loop, about 55% of the DS MB 1 opened. The 12-mer sequence, in 10-fold excess, was able to fully open the DS MB 1 (Figure 5a). This result clearly demonstrates how severely the stem invasion sequence can compromise the MB's function and its selectivity in biological applications. There are tremendously diversified sequences in a real sample matrix, and the copy number of this type of short complementary sequences could be very high. The chance of such a stem invasion is thus equally high and this is one of the reasons that limit the usage of MBs in quantitative analysis. In contrast, l-DNA MB does not have such a problem. None of the mock target sequences except the full-length target sequence was able to open up the LS MB 1, indicating superior selectivity and stability. This excellent selectivity coming from the l-DNA stem will be useful when performing quantitative analysis on raw biological samples as well as for intracellular studies.
Figure 5.
Comparisons of selectivity of LS MB 1 and DS MB 1. (a) A final concentration of 100 nM MB was incubated with 1 μM of each target for 1 h and the fluorescence signal was measured. The experiment was repeated five times and the average value and SD were calculated. (b) The calculated melting temperature of each target with its complementary sequence.
Biostablility of LS MBs
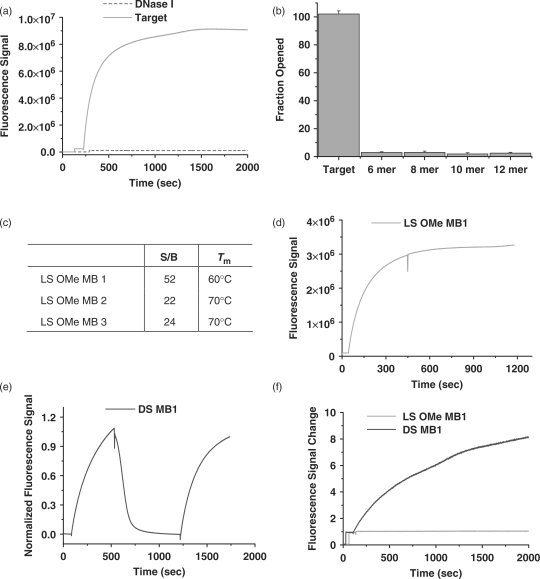
In general, degradation of d-DNA MBs is one of the most important problems in intracellular applications (20). In order to solve this problem, non-standard nucleic acid bases have been explored to design MBs, such as peptide nucleic acid (PNA) (21–23), LNA (10,24–27), and 2′-O-Me-modified RNAs (28–32). Most of these bases showed greatly improved biostability. In addition, they form stronger duplexes with their targets which lead to more stable signal production. The incorporation of l-DNA stems into such MBs can be highly beneficial to intracellular monitoring since these hybridized MBs will have high selectivity as well as stability to resist enzymatic degradation. We have chosen 2′-O-Me-modified RNA bases for the loop design using the three MB sequences as a model system, designated as LS OMe MBs. 2′-O-Me-modified RNAs have methoxyl groups at 2′ of the ribose sugar and can recognize natural nucleic acid targets but are not bio-degradable. For these advantageous features, 2′-O-Me-modified RNAs have often been used as MB building blocks. In addition, the well-characterized properties of 2′-O-Me-modified RNA bases are also beneficial to design MBs.
In order to demonstrate the feasibility of MBs composed of l-DNA stems and 2′-O-Me-modified RNA loops, called LS OMe MBs, in intracellular measurement, we investigated the biostability, sensitivity and selectivity of these probes. We prepared three different sequences of LS OMe MBs and the characterization was performed as described previously. As shown in Figure 6a–c, such a replacement of the loops did not affect structural stability and selectivity regardless of sequences. LS OMe MBs still showed higher S/B and Tm compared to DS MBs. In addition, LS OMe MBs had much better resistance to nuclease digestion (Figure 6a). DS MBs showed instant signal increase with the addition of DNase I (data not shown), whereas the LS OMe MBs did not show such an increase with DNase I (Figure 6a). The nuclease resistance of LS OMe MBs can be more beneficial to detect low copy number of mRNAs since the false-positive signal coming from MB degradation will not be the case for LS OMe MBs. Next, the stability of DS and LS OMe MB/RNA target duplexes were inspected using RNase H, an enzyme that selectively cleaves RNA/DNA duplexes. In the case of DS MBs, dramatic signal decrease was observed after RNase H was added to the duplexes. This means that the DS MB/RNA target duplexes are extremely vulnerable to RNase H digestion, resulting in signal fluctuation for intracellular measurements. On the contrary, LS OMe MBs/RNA target duplexes are stable and resistant to RNase H activity due to the 2′-O-Me-modified RNA loops. Even though LS O-Me MBs are resistant to DNase I and RNase H digestion, this does not mean that they are stable in true cellular environments. Thus, we tested LS OMe MBs in cell lysates. The stability of LS Ome MBs in the cell lysate clearly proves that the LS OMe MB design will be useful to eliminate many sources of false-positive and negative signals in intracellular measurements. Thus, we can conclude that the incorporation of l-DNA stems in MBs with loops composed of other non-standard nucleic acids does not alter the stability of the hairpin structure and takes full advantage of the biostability features of each component.
Figure 6.
Characterization of biostability of LS OMe MBs (green) compared to DS MBs (blue). (a) The response of LS OMe MB1 to its target and DNase I was tested. (b) Signal enhancement and stem melting temperature of each LS OMe MB were calculated. (c) Selectivity of LS OMe MB1 was tested in the same way as LS MBs using mock target sequences. (d–e) RNase H sensitivity of LS OMe MB1 (d) and DS MB1 (e) was experimented. First, each MB was incubated with its RNA target, then RNase H was added to the mixture and the fluorescence change was monitored. In the case of DS MB (e), DNA target was added to ensure the DS MB1 had conformational change after the RNA targets were digested. (f) Cell lysate sensitivity of LS OMe MB 1 (green) and DS MB 1 (blue) were tested.
CONCLUSION
In summary, we have established that the use of an l-DNA stem in a hairpin structured DNA probe like a MB eliminates unwanted intra- and intermolecular interactions. The exclusion of stem–loop interaction in an l-DNA stem MB ensures that the designed hairpin structure is dominant thereby improving the hairpin DNA probe's sensitivity and stability. The use of l-DNA stem prevents the probe from being opened by non-specific d-DNA sequences that contain short sequences complementary to the stem. Thus, stem invasion causing false-positive signals are eliminated. In addition, s l-DNA stems can be used to improve any known MB design. Because l-DNA stems can universally stabilize hairpin conformations of nucleic acid probes as long as D-type nucleic acids are used the loop. Moreover, the combination of nuclease resistant loop with l-DNA stems is a useful molecular beacon design since it removes the possibility of false-positive and negative reports coming from intracellular enzymatic activities and non-specific interactions of MBs and targets. While there are other approaches to design stable hairpin DNA probes, the l-DNA stem strategy is the simplest, most direct and most effective way to develop hairpin structured DNA probes with desired properties. Our results suggest that enantiomeric DNA (l-DNA) is useful for designing structured functional nucleic acid probes for a variety of biological and biotechnologic applications.
ACKNOWLEDGEMENT
This work was supported by NIH grants and ONR glant. Funding to pay the Open Access publication charges for this article was provided by NIH NIGMS and ONR grants.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR. Molecular beacons - Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 3.Yang CJ, Lin H, Tan W. Molecular Assembly of Superquenchers in Signaling Molecular Interactions. Journal of the American Chemical Society. 2005;127:12772–12773. doi: 10.1021/ja053482t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokol DL, Zhang XL, Lu PZ, Gewitz AM. Real time detection of DNA RNA hybridization in living cells. Proc. Natl Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuo T. In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta. 1998;1379:178–184. doi: 10.1016/s0304-4165(97)00090-1. [DOI] [PubMed] [Google Scholar]
- 6.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leal NA, Sukeda M, Benner SA. Dynamic assembly of primers on nucleic acid templates. Nucleic Acids Res. 2006;34:4702–4710. doi: 10.1093/nar/gkl625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsourkas A, Behlke MA, Rose SD, Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JWJ, Tan WH. A real-time assay for DNA sticky-end pairing using molecular beacons. Anal. Biochem. 2003;312:251–254. doi: 10.1016/s0003-2697(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH. Locked nucleic acid molecular beacons. J. Am. Chem. Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 11.Browne KA. Sequence-specific, self-reporting hairpin inversion probes. J. Am. Chem. Soc. 2005;127:1989–1994. doi: 10.1021/ja046369w. [DOI] [PubMed] [Google Scholar]
- 12.Crey-Desbiolles C, Ahn DR, Leumann CJ. Molecular beacons with a homo-DNA stem: improving target selectivity. Nucleic Acids Res. 2005;33:e77. doi: 10.1093/nar/gni076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urata H, Shinohara E, Ogura K, Ueda Y, Akagi M. Mirror-Image DNA. J. Am. Chem. Soc. 1991;113:8174–8175. [Google Scholar]
- 14.Damha MJ, Giannaris PA, Marfey P, Reid LS. Oligodeoxynucleotides containing unnatural L-2¢-deoxyribose. Tetrahedron Lett. 1991;32:2573–2576. [Google Scholar]
- 15.Urata H, Ogura E, Shinohara K, Ueda Y, Akagi M. Synthesis and Properties of Mirror-Image DNA. Nucleic Acids Res. 1992;20:3325–3332. doi: 10.1093/nar/20.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashley GW. Modeling, synthesis, and hybridization properties of (L)-ribonucleic acid. J. Am. Chem. Soc. 1992;114:9731–9736. [Google Scholar]
- 17.Garbesi A, Capobinanco ML, Colonna FP, Tondelli L, Arcamone F, Manzini G, Hilbers CW, Aelen JME, Blommers MJJ. L-DNAs as potenital antimessenger oligonucleotides: a reassessment. Nucleic Acids Res. 1993;21:4159–4165. doi: 10.1093/nar/21.18.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan W. Simultaneous Monitoring of the Expression of Multiple Genes Inside of Single Breast Carcinoma Cells. Anal. Chem. 2005;77:4713–4718. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 19.Hauser NC, Martinez R, Jacob A, Rupp S, Hoheisel JD, Matysiak S. Utilizing the left-helical conformation of L-DNA for analyzing different marker types on a single universal microarray platform. Nucleic Acids Res. 2006;34:5101–5011. doi: 10.1093/nar/gkl671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JJ, Geyer R, Tan W. Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res. 2000;28:e52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn H, Demidov VV, Gildea BD, Fiandaca MJ, Coull JC, Frank-Kamenetskii MD. PNA beacons for duplex DNA. Antisense Nucleic Acid Drug Dev. 2001;11:265–270. doi: 10.1089/108729001317022269. [DOI] [PubMed] [Google Scholar]
- 22.Petersen K, Vogel U, Rockenbauer E, Nielsen KV, Kolvraa S, Bolund L, Nexo B. Short PNA molecular beacons for real-time PCR allelic discrimination of single nucleotide polymorphisms. Mol. Cell. Probes. 2004;18:117–122. doi: 10.1016/j.mcp.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Seitz O. Solid-phase synthesis of doubly labeled peptide nucleic acids as probes for the real-time detection of hybridization. Angew. Chem. Int. Ed. Engl. 2000;39:3249–3252. doi: 10.1002/1521-3773(20000915)39:18<3249::aid-anie3249>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 24.Koshkin AA, Rajwanshi VK, Wengel J. Novel convenient syntheses of LNA [2.2.1]bicyclo nucleosides. Tetrahedron Lett. 1998;39:4381–4384. [Google Scholar]
- 25.Koshkin AA, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 26.Wengel J. Synthesis of 3′-C- and 4′-C-branched oligodeoxynucleotides and the development of locked nucleic acid (LNA) Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 27.Vester B, Wengel J. LNA (Locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 28.Kehlenbach RH. In vitro analysis of nuclear mRNA export using molecular beacons for target detection. Nucleic Acids Res. 2003;31:e64. doi: 10.1093/nar/gng063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marras SAE, Gold B, Kramer FR, Smith I, Tyagi S. Real-time measurement of in vitro transcription. Nucleic Acids Res. 2004;32:e72. doi: 10.1093/nar/gnh068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molenaar C, Marras SA, Slats JCM, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:e89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan W, Wang K, Drake TJ. Molecular beacons. Current Opinion in Chemical Biology. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]