Abstract
BORIS, like other members of the ‘cancer/testis antigen’ family, is normally expressed in testicular germ cells and repressed in somatic cells, but is aberrantly activated in cancers. To understand regulatory mechanisms governing human BORIS expression, we characterized its 5′-flanking region. Using 5′ RACE, we identified three promoters, designated A, B and C, corresponding to transcription start sites at −1447, −899 and −658 bp upstream of the first ATG. Alternative promoter usage generated at least five alternatively spliced BORIS mRNAs with different half-lives determined by varying 5′-UTRs. In normal testis, BORIS is transcribed from all three promoters, but 84% of the 30 cancer cell lines tested used only promoter(s) A and/or C while the others utilized primarily promoters B and C. The differences in promoter usage between normal and cancer cells suggested that they were subject to differential regulation. We found that DNA methylation and functional p53 contributes to the negative regulation of each promoter. Moreover, reduction of CTCF in normally BORIS-negative human fibroblasts resulted in derepression of BORIS promoters. These results provide a mechanistic basis for understanding cancer-related associations between haploinsufficiency of CTCF and BORIS derepression, and between the lack of functional p53 and aberrant activation of BORIS.
INTRODUCTION
BORIS (Brother of the Regulator of Imprinted Sites), also designated CTCFL (CTCF-like), is the mammalian paralog of a highly conserved (1,2), multi-functional chromatin factor encoded by a candidate tumor suppressor gene, CTCF (3–6). Loukinov et al. (4,7) showed that in humans and mice, these two genes encode polypeptides of similar size that share a centrally positioned nearly identical DNA-binding domain (DBD). The DBD is composed of 11 Zn-fingers (11ZF), including ten of the classic DNA-binding C2H2-class and one (ZF 11) of the C2HC-class capable of binding both single-strand DNA and RNA (8). It is noteworthy that the 11 ZF DBD regions in BORIS and CTCF are encoded by genomic sequences, which display an accurate duplication of a region containing all ZF-coding exons of the CTCF gene from an early mammal. The accuracy of this duplication in the human genome was maintained to the extent that genomic nucleotide similarities between CTCF and BORIS at individual intron–exon junctions approach 100% identity at the single nucleotide level (7).
By virtue of this shared DNA-recognition domain, BORIS can bind specifically to the same DNA target sequences that interact with CTCF. However, the consequences of BORIS or CTCF bound to the same site would be expected to be different. This is because the amino- and carboxy-termini of BORIS and CTCF are totally unrelated such that cofactors associated with one would not partner with the other. Thus, there would be different functional outcomes depending on which one of the two paralogs occupies a CTCF/BORIS-11ZF-binding site (9,10).
Once bound to DNA, CTCF may function as a versatile component of transcriptional chromatin insulators, hormone-dependent silencers and promoter repressors or activators (3–6). CTCF-binding sites have been mapped in differentially methylated domains of imprinting control regions (ICR) that regulate a parent-of-origin-dependent mono-allelic expression within clusters of imprinted genes: IGF2/H19 (11–13), Rasgrf-1 (14) DLK1/GTL2 (15), AWT1/WT1-AS (16) and KvDMR (17) loci. In addition, several functionally distinct, important, methylation-sensitive CTCF/BORIS-binding sites involved in random and/or imprinted X-chromosome inactivation (XCI) choice have been mapped at the X-inactivation center (Xic) (18–22).
Finally, a novel class of methylation-insensitive, but CpG-containing CTCF/BORIS-target sites was found and characterized in promoter regions of genes encoding the X-linked cancer-testis antigens (CTA) MAGE-A1, NY-ESO-1 (9,23) and SPAN-X (24). Promoters of CTA genes are methylated and repressed in normal somatic cells that express CTCF and not BORIS, but are specifically demethylated and activated in testicular germ cells and in cancer cells that express BORIS (9,23). Vatolin et al. (9) and Hong et al. (23) have documented co-expression of BORIS with other CTA genes in the majority of cancer cell lines and primary breast, prostate, colorectal and lung tumors tested. They also showed that conditionally expressed BORIS can outcompete CTCF in vitro and in vivo for binding to normally methylated 11ZF-target sites present in the silenced MAGE-A1 and NY-ESO-1 promoters. In addition, within three days of BORIS expression in normal cells, they observed both demethylation and activation of these two genes. It was also demonstrated that while CTCF is bound to the repressed/methylated MAGE-A1 and NY-ESO-1 promoters in normal human fibroblasts, activation of the same promoters in cancer cell lines is associated with in vivo occupancy by BORIS of methylation-insensitive 11ZF-target sites. Moreover, 12 other CTA genes that are normally co-expressed in testis with BORIS but not in somatic cells with CTCF were activated by ectopic expression of BORIS in normal primary human fibroblasts. In contrast, promoters of genes regulated by methylation but not normally co-expressed with BORIS in male germ cells failed to respond to ectopic expression of BORIS (9). Similar findings on the important roles of CTCF/BORIS-binding sites in regulation of the germ cell specific ALF and SPAN-X genes, have been reported by DeJong (25) and Larionov (24) laboratories, respectively.
Importantly, during male germ cell development, CTA-expressing cells are also engaged in the process of re-establishing paternal gene imprinting marks. This suggests that binding of BORIS in place of CTCF to shared targets, may be responsible for two effects. The first is activation of CTA gene promoters in testis. The second is re-setting of paternal ICR marks by remethylation in mature differentiating testicular germ cells after erasure of all gene imprinting marks in primordial germ cells (7,10). Indeed, data from a recent publication have provided strong support for the concept of BORIS-mediated epigenetic regulation of paternal ICR re-methylation (10).
Taken together, these results strongly support the hypotheses that CTCF and BORIS act successively to govern epigenetic states in normal male germ cell development, while rivalry in binding to the same spectrum of DNA sites caused by aberrant activation of BORIS in somatic cells may be associated with cancer (4,7).
While the mechanisms governing regulation of CTCF by its promoter have been explored by Klenova et al. (26), data on BORIS promoter regulation has not been described. This prompted us to investigate patterns of BORIS expression in normal testicular germ cells, in cancer cell lines, and in primary tumor tissues. We characterized the 5′-flanking noncoding region of the gene with expected regulatory (promoter) function. We mapped three alternative promoters producing five alternatively spliced 5′-UTR and analyzed promoter usage in different cell types and tissues. We also showed that activity of these promoters is negatively regulated by DNA methylation and by CTCF and p53.
MATERIALS AND METHODS
Cell lines and tissue samples
A total of 293 human embryonic kidney cells; normal human dermal fibroblasts (NHDF) (Cambrex, Rockland, MD, USA), normal foreskin fibroblasts BJ and 31 tumor cell lines (Table 1) were used in this study. The majority of tumor cell lines were maintained in RPMI 1640 supplemented with 10% heat inactivated FBS, 1% PSN Antibiotic Mixture (Invitrogen, Carlsbad, CA, USA) and 0.5% fungizone (Invitrogen). The 293, HeLa and Co115 cell lines were grown in DMEM with 10% heat inactivated FBS, 1% PSN and 0.5% fungizone. Normal cells were grown in the media recommended by the ATCC and NHDF in the media recommended by Cambrex. K562 cells stably transfected with the temperature-sensitive p53Val135 mutant, which adopts a wild-type conformation at 32°C, were previously described (27). The p53−/− H1299 parental cell line and the H1299 p53wt cell line (28) were a generous gift from Dr Peter M. Chumakov (Department of Molecular Genetics Lerner Research Institute, Cleveland, Ohio, USA). Demethylation studies were performed with NHDF treated with 1 μM 5-aza-2′-deoxycytidine (5-aza-dC) (Sigma-Aldrich, St. Louis, MO, USA). Cells were harvested at different times after treatment and medium containing 5-aza-dC was replaced with the fresh medium every 48 h.
Table 1.
Cell lines used for screening of BORIS expression from the three different promoters
| Group | Cell line | Cancer type | BORIS expression | PrA% | PrB% | PrC% | Methylation MS-SSCPa | P53b | |
|---|---|---|---|---|---|---|---|---|---|
| PrB | PrC | ||||||||
| A | SNB-19 | CNSc | ++ | 95 | 0 | 5 | M | M | Mut |
| 786-0 | Kidney | ++ | 84 | 0 | 16 | M | M | Mut | |
| A498 | Kidney | + | 65 | 0 | 35 | M | M | Mut | |
| ACHN | Kidney | + | 78 | 0 | 22 | M | M | Mut | |
| CCRF-CEM | Leukemia | + | 66 | 0 | 34 | U/M | U/M | Mut | |
| RPMI-8226 | Leukemia | + | 61 | 0 | 39 | U/M | U/M | Mut | |
| HOP-92 | Lung | + | 93 | 0 | 7 | M | M | Mut | |
| SK–MEL-2 | Melanoma | ++ | 68 | 0 | 32 | M | M | WT | |
| OVCAR-3 | Ovary | ++ | 83 | 0 | 17 | M | M | WT | |
| NCCIT | Testis | + | 96 | 1 | 3 | M | M | Mut | |
| A/C | HS578T | Breast | ++ | 41 | 0 | 59 | M | M | WT |
| MCF7 | Breast | ++ | 43 | 0 | 57 | M | M | Mut | |
| MDA-MB-435 | Breast | + | 50 | 1 | 49 | M | M | Mut | |
| HCT-116 | Colon | ++ | 47 | 0 | 53 | M | M | Mut | |
| SW-620 | Colon | + | 51 | 0 | 49 | M | M | Mut | |
| SN12C | Kidney | ++ | 53 | 0 | 47 | M | M | Mut | |
| NCI-H522 | Lung | + | 48 | 0 | 52 | M | M | Mut | |
| SK-OV-3 | Ovary | ++ | 49 | 0 | 51 | M | M | WT | |
| C | MDA-MB-231 | Breast | + | 21 | 0 | 79 | M | M | Mut |
| SNB-75 | CNS | ++ | 36 | 0 | 64 | M | M | WT | |
| HCT-15 | Colon | ++ | 33 | 0 | 67 | M | M | Mut | |
| HT29 | Colon | +++ | 37 | 0 | 63 | M | M | WT | |
| HL-60 | Leukemia | + | 39 | 0 | 61 | M | M | Mut | |
| MOLT-4 | Leukemia | ++ | 25 | 0 | 74 | M | M | Mut | |
| SR | Leukemia | +++ | 31 | 0 | 69 | M | M | Mut | |
| NCI-H322 | Lung | +++ | 13 | 0 | 87 | M | M | Mut | |
| B | COLO-205 | Colon | ++ | 3 | 73 | 24 | U/M | U/M | ? |
| K-562 | Leukemia | ++++ | 3 | 80 | 18 | U | U | Mut | |
| MM-S1 | Leukemia | +++ | 2 | 85 | 13 | U | U | Mut | |
| NCI/ARD-RES | Ovary | +++ | 1 | 52 | 47 | U | U | Del | |
| OVCAR-8 | Ovary | +++ | 0 | 96 | 4 | U | U | ? | |
| Normal cells | BJ | Fibroblast | − | – | – | – | M | M | n/a |
| NHDF | Fibroblast | − | – | – | – | M | M | n/a | |
aU = unmethylated; M = methylated; U/M = partially methylated.
bMut = mutation; WT = wild type; Del = deletion (35).
cCNS = Central nervous system.
Normal and tumor tissues were obtained from the Tissue Bank of the Institute of Pathology of Lausanne. Twenty-four human normal tissue samples from bone marrow, bladder, heart, kidney, skin, colon, testis and ovary and 26 tumor tissue samples from bladder, breast, colon, kidney, lung, testis, ovary and endometrium were analyzed. All tissue samples were examined histologically by a pathologist (R.B.). The use of human tissue samples during this study was in accordance to the guidelines of the ethical committee of the Medical Faculty of Lausanne (Switzerland).
Analysis of mRNA stability of BORIS alternative 5′UTRs
The mRNA half-life of BORIS was determined in K562 and Ovcar-8 cell lines treated with Actinomycin D (ActD). Briefly, Ovcar-8 cells were grown in 6-well tissue culture plates for 24 h to achieve ∼80% confluency. Next, cells were treated for 10 min, 30 min, 1, 2, 4, 6, 8, 12, and 24 h with 15 μg/ml of ActD dissolved in DMSO. Total RNA was extracted before and after ActD treatment, and mRNA levels were quantitated by real-time RT-PCR (qPCR). For each sample, the amount of BORIS mRNA was quantified relative to 1 μg of total RNA. First strand cDNA was synthesized using 1 μg of total RNA (DNase-treated) and alternative 5′ UTRs of BORIS mRNA were amplified using primers specific for each isoform (Table 2). Serial 10-fold dilutions (from 300 000 to 30 molecules) of cloned DNA for each alternative 5′ UTRs were used as a reference for the standard curve calculation. All qPCRs were performed using the SYBR Green fluorogenic dye and data were analyzed using ABI system software.
Table 2.
Primers and probes used in real-time PCR
| Name | Primers sequences | Probe |
|---|---|---|
| CTCF | FW 5′-TGACACAGTCATAGCCCGAAAA-3′ | 6FAMTGATTTGGGTGTCCACTTGCGAAAGC-MGB |
| REV 5′-TGCCTTGCTCAATATAGGAATGC-3′ | ||
| BORIS-MC | FW 5′-CCCATTGTGCCACCATCA-3′ | 6FAMACGGAAAAGCGACCTAC-MGB |
| REV 5′-AGCATGCAAGTTGCGCATAT-3′ | ||
| BORIS promA | FW 5′-CTTACTTCCCCCCCGGGT-3′ | 6FAMCTCCTCCCTTCCTCA-MGB |
| REV 5′-GCCTTGGGGTTGAAGTGGA-3′ | ||
| BORIS promB or | FW 5′-GCAGAGCCACAAGCCAAAG-3′ | 6FAMAGTGGGCCGAGCAT-MGB |
| BORIS | REV 5′-ATCTCAGTGGCTGCCATAATGACT-3′ | |
| BORIS promC or | FW 5′-CCCTTCTCCCCCCTATGGA-3′ | 6FAMACCGCTTGCTTATTT-MGB |
| BORIS C1 | REV 5′-CCATAATGACTTGGCCTGTTTG-3′ | |
| BORIS A1 | FW 5′-TCCCTTCCTCATCCACTTCAA-3′ | SYBR Green method |
| REV 5′-GCTCAGAAAGGACAGAGATCTCAGT-3′ | ||
| BORIS A2 | FW 5′-CATCCACTTCAACCCCAAGC-3′ | SYBR Green method |
| REV 5′-CTTTGGCTTGTGGGCTCTG-3′ | ||
| BORIS A3 | FW 5′-GCTCTCCTCCTCTCCTTATCCAC-3′ | SYBR Green method |
| REV 5′-CCATACAGGGCACTGGGAGAC-3′ | ||
| hMYC-G | FW 5′-GTGCGGGAGCCAGTGAACT-3′ | SYBR Green method |
| REV 5′-AAGATCCCAGCTCCTCAGCC-‘3 | ||
| hMYC-N | FW 5′-GGCTCTGTGAGGAGGCAAGGTG-3′ | SYBR Green method |
| REV 5′-GCTCTCTATTTGGAGTGGCGGG-3′ | ||
| CTCF ChIP PrA | FW 5′-CTCCTTATCCATTACCCACCACC-3′ | SYBR Green method |
| REV 5′CAGTATCTCAGTGCCTCCTGTGG-3′ | ||
| CTCF ChIP PrB | fw 5′-CCCTGCCCCCACAGTACAT-3′ | SYBR Green method |
| rev 5′-TTTTCCGCTCCGCGC-3′ | ||
| CTCF ChIP PrC | fw 5′-GGCCAGTCCCGGTCAAG-3′ | SYBR Green method |
| rev 5′-AGCATGGGCTGTTCTGGG-3′ |
RNA degradation curves were obtained by setting as 100% the maximum level of mRNA expression at Time 0 before treatment with ActD. mRNA levels determined at times following Time 0 are expressed as a percentage of the maximum value. The half-lives of alternatively spliced 5′ UTR mRNAs was obtained from the logarithmically transformed best-fit line by linear regression analysis (29).
RT-PCR and northern blot analyses
RNAs were extracted from cultured cells using the TRIzol LS Reagent (Invitrogen, Basel, Switzerland). One-step RT-PCR was performed with 200 ng of total RNA in 20 μl. BORIS expression was screened with the primers RT-A3 5′-AAGCCGCGAACGGAGACGAAG-3′ and RT-B3 5′-ACGCCTTCATCCACTTCCTCTTT-3′. Northern blot analyses were performed as described previously (30).
Reverse transcription and qPCR
RNA was extracted from cultured cells as described below from tumor cell lines (Table 1). Total RNA (3–4 μg) was converted to cDNA using ThermoScript reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions.
qPCR analyses were preformed using the TaqMan® Universal PCR Master Mix or the PowerSYBR® Green PCR Master Mix (Applied Biosystems, Foster city, CA, USA) and the Applied Biosystems 7900HT qPCR system. For the analyses of BORIS transcripts expressed from promoter A, primers were designed in the first noncoding exon nearest to promoter A + 1 start site. For the analyses of expression from promoter B, primers were engineered to span the first noncoding and coding exons. For the analyses of expression from promoter C, primers were selected by using the unique sequence of the first coding exon utilized only for promoter C. Primers and probes were selected using Primer Express program, and optimal experimental conditions were established for each set.
Sequences of the primers and probes are summarized in Table 2. Primer/probe mixtures for human glyceraldehyde-3-phosphate dehydrogenase (HS99999905_m1) and p53 (HS00153349_m1) were purchased as Predeveloped Assay (Applied Biosystems, Foster City, CA, USA). Data were analyzed by comparative Ct method and normalized to the untreated control.
The RNA copy number generated from each promoter was determined using a standard curve following the ABI protocols. To determine transcript copy number, standard curves were generated for each primer pair using 10-fold serial dilutions of linearized plasmids containing known copy numbers of the target PCR products. The threshold cycle of each dilution was determined and plotted against the log value of the cDNA copy number. Transcript numbers for each experimental sample were interpolated by its detection threshold value using the appropriate standard curve. Expression levels were normalized to that of the housekeeping gene, GAPDH. Standard curve coordinates and ranges are shown in Table 3.
Table 3.
Real-time PCR standard curve coordinates and ranges
| Plasmid | Curve range (copy number) | Standard curves | |
|---|---|---|---|
| Slope | R2 | ||
| GAPDH | 30–300 000 | −3.31 + 0.01 | 0.9955 + 0.02 |
| BORIS Prom A | 30–300 000 | −3.51 + 0.12 | 0.9941 + 0.0078 |
| BORIS Prom B | 30–300 000 | −3.17 + 0.02 | 0.9945 + 0.004 |
| BORIS Prom C | 30–300 000 | −3.33 + 0.03 | 0.9943 + 0.0003 |
Identification of transcription initiation sites using RNA ligase-mediated rapid amplification of 5′ cDNA ends (5′ RLM-RACE)
The GeneRacer system (Invitrogen, Carlsbad, CA, USA), based on RNA ligase-mediated and oligo-capping rapid amplification of cDNA, was carried out based on the manufacturer's instructions. The kit ensures the amplification of only full-length transcripts by eliminating truncated messages from the amplification process. Total RNA was dephosphorylated using calf intestinal phosphatase and then decapped to target full-length messenger RNAs. An RNA oligonucleotide was then ligated to the full-length decapped mRNAs. Ligated mRNA was reverse transcribed with a BORIS-specific primer located in the first exon (RACE-EX1 5′-CAGAGGTACGCTCGGCCTCCAAC-3′, +159 to +138). Then the 5′ cDNA end was amplified by nested PCR using the GC-Rich PCR system (Roche) and with BORIS-specific reversed primers (RACE-EX1N 5′-GGCCTTTTTCCGGCATCAACT-3′, +79 to +59; RACE-N 5′-TTGGGGTTGAAGTGGATGAGGAAG-3′, +1294 to +1271). Amplified products were separated by electrophoresis on 1% agarose gels and purified on SNAP columns (Invitrogen, Carlsbad, CA, USA). Purified PCR fragments were cloned in a pCR®2.1-TOPO® vector (Invitrogen), sequenced and analyzed.
Luciferase reporter vectors and p53 expression vectors
Fragments from the region 5′ of BORIS were prepared by PCR amplification. The amplification process introduced Asp718I and HindIII sites in the 5′ and 3′ ends, respectively. All fragments were inserted into the pGL3-basic vector (Promega, Madison, WI, USA) cut with Asp718I and HindIII. Wild type and mutant p53 expression vectors were a generous gift from Dr Peter M. Chumakov.
Transient transfection and siRNA transfection assays
For luciferase assays, 293 cells were transiently transfected using Fugene 6 (Roche, Germany) according to the manufacturer's instructions. K562 cells were transfected with a Nucleofector device (Amaxa, Inc., Cologne Germany) according to the manufacturer's recommendations with Nucleofector Solution V and program T16 generating the highest transfection efficiency with the lowest mortality. HeLa, NCCIT and OVCAR-3 cells were seeded in 12-well dishes at a concentration of 3 × 105 cells/well, one day before transfection. The cells were transiently transfected with the different constructs (0.75 μg/well) with the jetPEI Cationic Polymer Transfection reagent (4 μl/well) (PolyPlus-transfection, Illkirch, France) according to the manufacturer's instructions. All experiments were performed at least three times. The pRL-tk vector (0.25 μg/well) (Promega, Madison, WI, USA) was co-transfected as an internal control for transfection efficiency. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). To compare results, the mean values of relative luciferase activity were used. The expression levels of the different constructs were compared to the level of the pGL3-control vector containing the firefly luciferase gene under the control of the SV40 early promoter and to the level of the pGL3-basic vector.
SiRNA assays were performed in 6-well plates using the Interferin reagent (PolyPlus-transfection, Illkirch, France) according to the manufacturer's instructions. Stealth™ RNAi for CTCF 5′-GCGCUCUAAGAAAGAAGAUUCCUCU-3′ was synthesized by Invitrogen (Invitrogen, Carlsbad, CA, USA). Stealth™ RNAi Negative Control High GC siRNAs for silencing control was provided by Invitrogen (Invitrogen, Carlsbad, CA, USA). Transfected cells were harvested 48 h post-transfection.
DNA methylation analysis of promoters B and C
DNA was extracted from frozen tissue or culture cells using the DNeasy tissue kit (Qiagen, Hilden, Germany). Two microgram of DNA were modified in 40 μl of water with sodium bisulfite as previously described (31) or using the EpiTect® Bisulfite Kit (Qiagen, Hilden, Germany). After bisulfite modification, PCR on BORIS promoter B was performed with the primers 5′-CCTCCCCCAACCCTACCTAA-3′ and 5′-GTTTTTGGTTTGTGGGTTTTGTT-3′ using the master mix (Promega, Madison, WI, USA) with a final concentration of 5% DMSO, under the following PCR conditions: 35 cycles of 94°C for 30 s, 54°C for 45 s and 72°C for 50 s. The first PCR product diluted 1/50 was amplified by semi-nested-PCR with the primers 5′-CACTACCACCCTCCACTCTC-3′ (+931 to 1950) and ′-GTTTTTGGTTTGTGGGTTTTGTT-3′ (+964 to +980). Each PCR product was analyzed by MS-SSCA and MS-DBA (31,32), as previously described. In the MS-DBA approach, two 3′-end DIG-labeled probes, 5′-AACCCGACGACGACCGAC-3′ and 5′-CCAACCCAACAACAACCAAC-3′, were used to recognize the methylated and the unmethylated DNA, respectively. PCR on BORIS promoter C were performed with primers 5′-ACAAAACCCACAAACCAAAA-3′ and 5′-TTTTTGGAGGAGAGTAGGTG-3′ using the Platinum PCR supermix (Invitrogen, Carlsbad, CA, USA) with a final concentration of 5% DMSO using the following PCR conditions: 40 cycles at 94°C for 30 s, 52°C for 45 s and 72°C for 50 s. PCR products amplified from NHDF, NCCIT, OVCAR-3, K562 and OVCAR-8 cells were cloned into pCR®2.1-TOPO® vector (Invitrogen, Carlsbad, CA, USA). Ten clones for each cell lines were sequenced (Genomics Research Facility, Rocky Mountain Laboratories, NIAID/NIH, Montana, USA).
Electrophoretic mobility shift assay (EMSA)
Fragments containing BORIS promoters A, B and C were synthesized by PCR. EMSA was performed as previously described (33). Briefly, PCR fragments were end labeled using 32P-γ-ATP and T4 polynucleotide kinase (New England Biolabs). Protein–DNA complexes were allowed to form by incubation for 30 min at ambient temperature in PBS with 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM dithiothreitol, 0.1% Nonidet P-40, 10% glycerol and poly(dI-dC). Full-length CTCF and the ZF DNA-binding domain of CTCF were translated in vitro using the TnT kit (Promega, WI, USA). Protein–DNA complexes were resolved from unbound DNA probe using a 5% native polyacrylamide gel (PAAG) in 0.5× TBE.
Chromatin immunoprecipitation assay (ChIP)
Normal fibroblast BJ cells and two cancer cell lines, Ovcar-3 and Ovcar-8, were used in ChIP assays to examine the in vivo binding of CTCF on BORIS promoters. We used a ChIP Assay kit (Upstate, Charlotteville, VA, USA) and followed the manufacturers’ recommendations. One ChIP reaction used 10 μl of the anti-CTCF monoclonal antibodies previously described (20). The human CTCF-binding site N, a c-MYC insulator site (hMYC-N), was used as positive control and CTCF non-binding site G of c-MYC (hMYC-G) was used as negative control. Immunopurified DNA was used in real-time PCR using primers described in Table 2.
RESULTS
Identification of BORIS transcription initiation sites
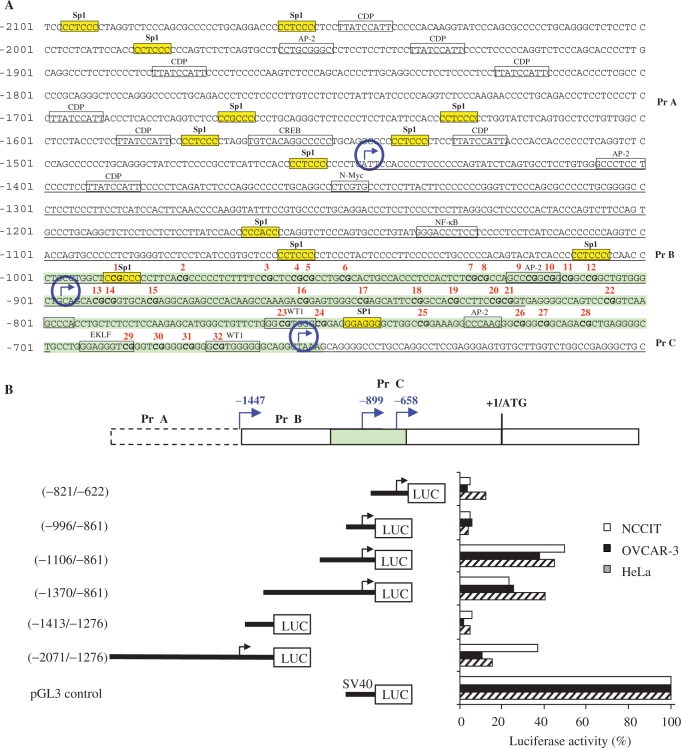
To identify transcription initiation sites, 5′ RLM-RACE was performed. The 5′ RLM-RACE method has a major advantage over other methods for mapping of transcription start sites (e.g. primer extension, nuclease protection assays or traditional 5′ RACE) in that it only detects authentic capped 5′ ends of mRNAs. The technique is based on RNA ligase-mediated (RLM-RACE) and oligo-capping rapid amplification of cDNA ends, and results in the selective ligation of an RNA oligonucleotide to the 5′ ends of decapped mRNA. This method allowed the amplification of only truly full-length transcripts via elimination of truncated messages. The 5′ RLM-RACE assays were performed on total RNA extracted from normal testis tissue, and from the NCCIT tumor cell line. Two PCR products (∼700 bp and ∼1500 bp) were obtained and, after cloning and sequencing, the first two start sites were identified at −658 and −899 bp from the ATG translational start site (Figure 1A). Using the GC-Rich PCR system and nested primers upstream of the first transcription start site, a ∼200 bp PCR product was generated. This new site mapped to −1447 bp upstream of the ATG translation start codon (Figure 1A).
Figure 1.
Identification and activity of three BORIS promoters. (A) Nucleotide sequence of the 5′-upstream region of the human BORIS gene is shown together with a portion of the coding region. Potential binding sites of transcriptional regulatory proteins predicted by the MatInspector program are shown in boxes. Yellow boxes highlight the Sp1 factor. The A in ATG translational start site is designated as nucleotide +1 and does not appear in the figure. The main start sites for transcription, as determined by 5′-RACE, are indicated by encircled blue arrows. The transcribed sequences are underlined. Nucleotides of the CpG island are shaded in green. Red numbers (1–32) represent all CpGs within the CpG island. (B) Determination of BORIS promoters A, B and C activities by transient expression of luciferase reporter constructs in HeLa, NCCIT and OVCAR-3 cell lines. Left: schematic representation of BORIS promoter luciferase reporter constructs. Numbers indicate positions of nucleotides of the 5′-flanking region of the BORIS gene, as shown in A. Right: relative luciferase activities compared with the pGL3 control vector activity, which was considered to have 100% activity. The CpG island is shaded in green.
Identification of BORIS promoters and their transcriptional activities
To functionally characterize the three putative promoters, different fragments of the regions upstream of all three transcriptional start sites were cloned into a firefly luciferase expressing vector (Figure 1B). Transfection assays were carried out with the NCCIT, OVCAR-3 and HeLa cell lines. Two reporter constructs were used to study transcriptional regulation by promoter A (Figure 1B). The larger construct (−2071 to −1276) showed substantial transcriptional activity in all three cell lines, whereas the 5′ truncated construct (−1413 to −1276) showed only low activity. The activity of promoter A was significantly higher in NCCIT than in OVCAR-3 or HeLa cells. These results suggested that transcriptional activity of promoter A could be cell type-specific. To study transcriptional regulation by promoter B, three reporter constructs were generated. The construct containing nucleotides −1370 to −861 of the promoter B region produced an activity that was 23–40% of that generated by the SV40 promoter. A construct truncated to contain nucleotides −1106 to −861 was more active, yielding between 38% and 50% of SV40 levels, whereas the transcriptional activity of the smallest construct (−996 to −861) was very low. These results suggest that the sequences located between −1106 and −996 to the ATG translational start site defined the minimal BORIS promoter B. Promoter C is located very close to promoter B and is quite small. To study promoter C, we generated a construct containing nucleotides −821 to −622 upstream of the start site. This construct produced relatively low activity in comparison with promoters A and B. The transcriptional activity of promoter C was observed to be higher in HeLa cells corresponding to 18% of SV40 promoter activity, suggesting that promoter C may also be tissue specific (Figure 1B).
Analyses of the BORIS promoter region sequences using the MatInspector program (http://www.genomatix.de/index.html) identified several consensus sequences for transcription factor binding sites. In addition, promoters B and C are situated in a CpG island. Promoter B contained putative binding sites for Sp1, AP-2, NF-κB and N-Myc and promoter C contained Sp1, two WT1, AP-2 and EKLF putative binding sites (Figure 1A). Promoter A contained Sp1, AP-2, CREB and a large number of CAAT displacing protein (CDP) putative binding sites.
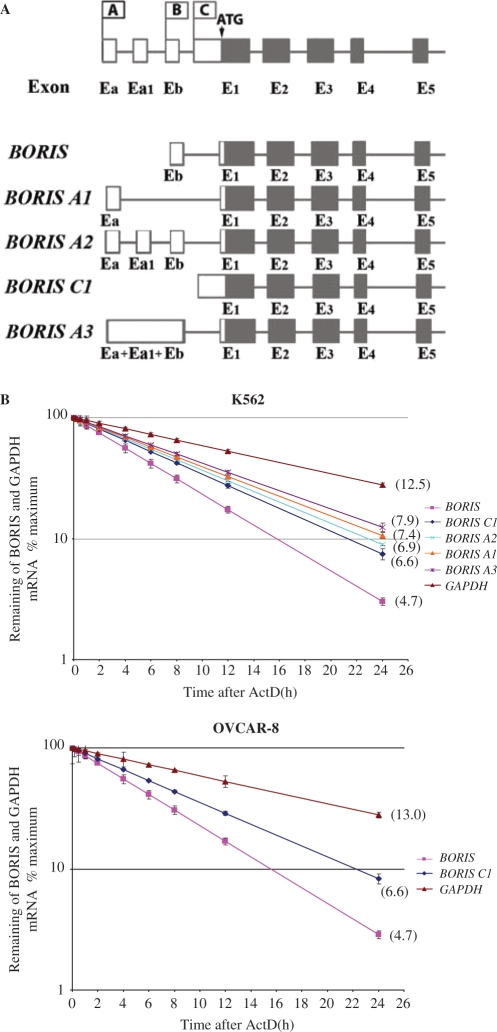
Identification of five alternatively spliced 5′-UTRs for BORIS and analyses of their stability
The fact that BORIS transcription initiated from three different promoters suggested that there would likely be a series of 5′ splice variants. Indeed, we identified five major transcripts that differed in their 5′ noncoding regions but encoded the same protein sequence (Figure 2). The BORIS A1 and BORIS A3 isoforms are expressed from promoter A and have first 180 and 548 bp noncoding exons Ea and Ea+Ea1+Eb, respectively that splice directly to the first 530 bp coding exon, E1. The BORIS A2 splice variant also originates from promoter A, but has two more noncoding exons, Ea1 and Eb, than the BORIS A1 isoform. The BORIS variant that comes from promoter B contains a first 80 bp noncoding exon, Eb, which splices to first 530 bp coding exon, E1. The BORIS C1 isoform is expressed from promoter C and is transcribed as an unspliced first 1190 bp coding exon (Figure 2A). To investigate whether the alternative 5′ UTRs affect mRNA stability, we blocked RNA synthesis by treating cells with ActD. Two cell lines were used in those experiments: K562 cells that express all five alternative 5′ UTRs and Ovcar-8 cells that express only two of five. As shown in Figure 2 (panel B), the half-lives of BORIS, BORIS A1, BORIS A2, BORIS A3 and BORIS C1 transcripts in K562 cells were 4.7, 7.4, 6.9, 7.9 and 6.4 h, respectively, indicating that BORIS mRNAs in cultured cells are relatively stable and long-lived. The alternative transcript expressed from promoter B, BORIS, was 1.5-fold less stable than BORIS transcripts expressed from promoters A and C. The relative turnover of the three alternative 5′ UTRs expressed from promoter A—BORIS A1, BORIS A2 and BORIS A3—was comparable and the rates of degradation were approximately 1.2-fold slower than for the BORIS C1 transcript expressed from promoter C. Similar half-lives of BORIS (4.7 h) and BORIS C1 (6.6 h) were obtained in Ovcar-8 cell line (Figure 2B). In both cell lines, GAPDH mRNA degradation was similar to published values for that gene (34).
Figure 2.
Detection of BORIS expression from different alternative promoters and stability of BORIS alternative transcripts. (A) Unique BORIS cDNA sequences attached to promoters A, B and C. To detect BORIS expression from different promoters, forward primer was designed on basis of unique cDNA sequence for every promoter. Reverse primer was designed on the basis of BORIS coding exon 2 sequence. Expected RT-PCR fragment was 720 bp for BORIS promoter A expression, 760 bp for BORIS promoter B expression and 810 bp for BORIS promoter C expression. All RT-PCR fragments contain splice site allowing distinguishing them from genomic DNA. (B) K562 and Ovcar-8 cells were treated with Actinomycin D for various durations to block transcription. Total RNA was extracted at the indicated time points following the addition of actinomycin D. Quantitative real-time PCR amplification were performed as described in Materials and Methods section to determine copy number of each BORIS alternative transcript. Each data point represents the average of three amplification reactions. RNA degradation curves were obtained by setting at 100% the maximum of mRNA expression at Time 0 before Act D treatment.
Analysis of BORIS transcription from promoters A, B and C in normal and cancer cells
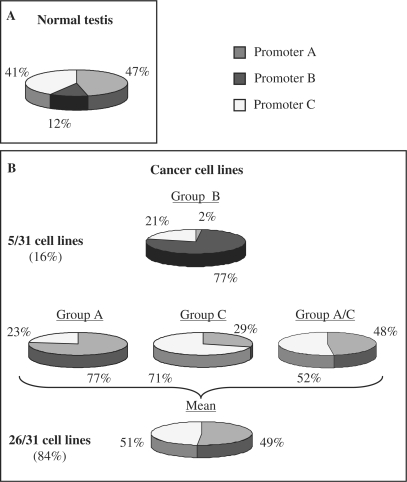
To gain further insights into the characteristics of promoters A, B and C, we studied their utilization by qPCR in a panel of normal tissues, normal cell lines and cancer cell lines. As expected from previous studies, testis was the only normal tissue to express BORIS (data not shown). The relative percent of expression from each promoter was then calculated. Figure 3A shows that in normal testis, expression comes from all three promoters. The distribution of activity is 47% from promoter A, 12% from promoter B and 41% from promoter C.
Figure 3.
BORIS expression from different alternative promoters (A, B and C) in normal testis and in multiple types of cancer cell lines by real-time PCR. (A) Relative expression of BORIS from the three different promoters in normal testis. (B) Relative expression of BORIS from the three different promoters in 31 different tumor cell lines. Tumor cell lines are divided in four groups according to the main promoter usage. Group B represents 16% of all cell lines tested and the BORIS expression is mainly originated from promoter B. In the three other groups the activity of BORIS comes from either promoter A, either promoter C or from promoters A and C. These three groups represent 84% of all cell lines tested.
The same analysis was performed on 31 cancer cell lines (Table 1, Figure 3B). Only 5 cell lines showed transcriptional activity promoter B and these were assigned to a subset designated as group B. Activity in all the other cell lines came only from promoters A or/and C. These 26 cell lines were assigned to one of three groups, based on their utilization of these two promoters. Group A is comprised of cell lines that expressed BORIS primarily from promoter A. Group C includes a series of cell lines that expresses BORIS primarily from promoter C. Cell lines in the group designated A/C use both promoters more or less equally. The patterns of promoter utilization in the cancer cell lines are distinct from that observed in normal testis. However, it must be recognized that testis tissue contains germ cells at various stages of spermatogenesis which could utilize the different promoters in a cell type-specific manner or might use all three at each stage. Moreover, based on known features of the 31 cell lines, preferential promoter utilization could not be correlated with tissue origin or with genomic characteristics that might be thought to influence BORIS expression—loss of the CTCF-containing chromosome 16; gain of chromosome 20 or amplification of 20q13 that contains the BORIS locus. In addition, there was no obvious relation between promoter utilization and patterns of p53 expression (Table 1) (35,36). These results showed that all three promoters contribute to BORIS transcription, with cancer cells preferentially using promoters A and C.
Regulation of BORIS promoters by CTCF
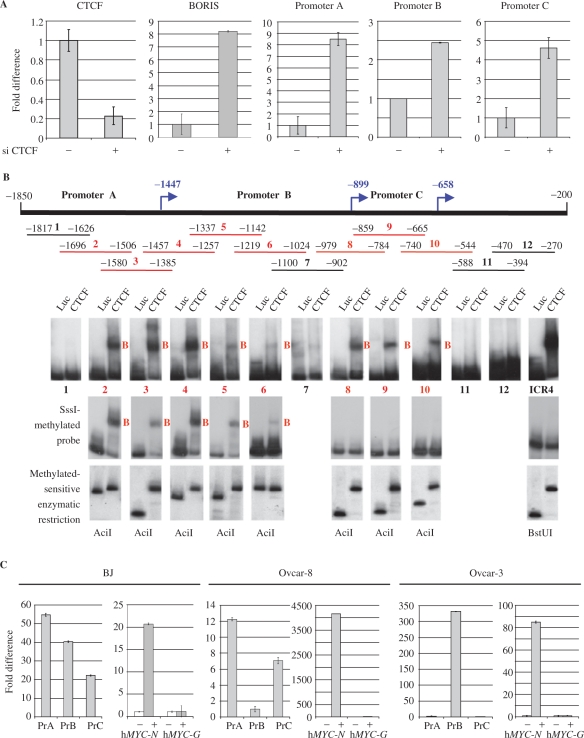
The fact that expression of CTCF and BORIS in normal testis is almost mutually exclusive raised the possibility that CTCF may negatively regulate BORIS transcription. To examine this possibility, we used siRNA to suppress CTCF expression in NHDF and then tested the cells for expression of CTCF and BORIS by qPCR (Figure 4A). We observed that the level of CTCF transcript was reduced more than 4-fold in cells expressing the siRNA while the levels of BORIS transcripts in the same cells were increased at least 8-fold (Figure 4A). To evaluate the contribution of each promoter to expression of BORIS in the siRNA-treated cells, we performed qPCR with primers and probes specific for each of the spliced isoforms. The results showed that in cells expressing the CTCF-specific siRNA, expression of BORIS from all three promoters was enhanced, but to differing extents—expression from promoter A was increased 8–fold, expression from promoter B was increased 2.5-fold, and expression from promoter C was increased 4.5-fold (Figure 4A). These results indicated that under normal circumstances, CTCF contributes to suppression of BORIS expression by acting on all three promoters.
Figure 4.
Regulation of BORIS expression by CTCF. (A) Real-time PCR analyses of RNA extracted from NHDF (Normal Human Dermal Fibroblasts) treated with siRNA against CTCF show the decrease of CTCF expression and the increase of BORIS expression coming from the three promoters at different levels. Data were analyzed by comparative Ct method and normalized to the untreated control. Results are represented for each set of primers used in real-time PCR. (B) Schematic map of the overlapping fragments used as EMSA probes for in vitro detection of CTCF-binding sites and results of EMSA analyses showing binding of CTCF within promoters A, B and C in red (first panel). Results of EMSA analyses of SssI-methylated probes showing insensitivity of CTCF binding in promoter A and B, and methylation sensitive sites in promoter B and C within the CpG island (second panel). The third panel showed the methylation-sensitive enzymatic restriction of probes used in methylation-sensitive EMSA. (C) CTCF in vivo occupancy on BORIS promoters via ChIP in normal fibroblasts BJ, Ovcar-8 and Ovcar-3 cell lines. Real-time PCR analysis of the fold difference for the presence of DNA with the CTCF-binding site in the input chromatin versus in the CTCF chromatin fraction is obtained by ChIP. The fold difference between immunoprecipitated DNA with specific CTCF antibody or rabbit serum (used as negative control) is represented for each set of primers. hMYC-N primers are used as positive control for good enrichment after ChIP and c-MYC non-binding site G is used as control for the specificity of the enrichment. Controls are represented in the right graph for each cell lines.
To obtain additional evidence that CTCF binds directly to the BORIS promoter region, we performed EMSA using recombinant CTCF with overlapping probes encompassing BORIS promoter sequences. We identified 3 binding sites in promoter A, 3 in promoter B and 2 in promoter C (Figure 4B), supporting the suggestion that CTCF is directly involved in suppressing BORIS expression. It is known that CTCF binding can be sensitive (11–13,18,20) or insensitive to methylation of its target sites (9,23,24). In order to see if CTCF binding within BORIS promoters was sensitive to DNA methylation, all fragments positive for CTCF binding were methylated in vitro using SssI DNA methylase. EMSA analysis showed that binding of the CTCF 11ZF DNA-binding domain to fragments 2, 3, 4 (promoter A), 5 and 6 (promoter B) was similar whether the fragments were methylated or not (Figure 4B). In contrast, no binding was observed to methylated fragments 8, 9 and 10 that are located in the CpG island shared by promoters B and C. The degree of probe methylation, tested using the methylation-sensitive restriction endonuclease AciI for the BORIS promoter and BstUI for ICR4 as a positive control, showed that the extent of methylation was nearly 100%. This experiment showed that some CTCF-binding sites within BORIS promoters were methylation-sensitive and others were methylation-insensitive.
To investigate CTCF binding to BORIS promoters in vivo, we performed ChIP assays in normal BJ fibroblasts, in Ovcar-8, using mostly promoter B, and in Ovcar-3, using mostly promoters A. We analyzed the promoter regions of BORIS and site N from the c-MYC locus, a well-characterized CTCF-binding site that functions as a 5′ chromatin insulator for the locus (3,20,37). Figure 4C showed that site N was occupied by CTCF in all three lines while the non-binding target used as negative control was not. In the BJ cell line, occupancy of all three promoters by CTCF correlated well with the transcriptionally repressed state of all three in this cell line. In Ovcar-8, CTCF occupancy was found only on promoter A and C. No CTCF binding was found in promoter B, which correlates with expression of BORIS coming only from promoter B in this cell line (Table 1). In contrast, CTCF binding in Ovcar-3 was identified in promoter B but not in promoter C. CTCF binding in promoter A was not significant, showing only a 2.2-fold enrichment. These results also correlate with BORIS expression coming from promoters A and C, but not from promoter B. Taken together, these results showed that there was an inverse correlation between the extent to which CTCF bound to the BORIS promoters and the activity of those promoters.
Regulation of BORIS promoters by p53
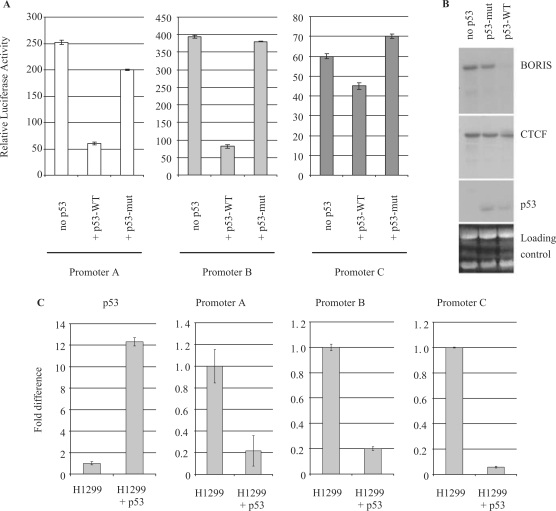
In our analyses of BORIS expression in the 31 cancer cell lines, we noted that the lines expressing BORIS at the highest levels were known to have deletions or mutations of TP53, the gene encoding the p53 tumor suppressor (data not shown). This prompted us to ask if p53 might be involved in regulating BORIS expression. To examine this possibility, we transfected K562 cells that are p53 negative with luciferase reporters for each of the three BORIS promoters together with expression plasmids for wild-type or mutant p53 (Figure 5A). The results of these studies showed that the luciferase activity of all three BORIS promoters was significantly reduced in cells transfected with wild-type p53 versus mutant p53. With wild-type p53, the activity of promoters A and B was downregulated about 3.3- and 4.6-fold, respectively, and the activity of promoter C was repressed about 1.5-fold. We then performed northern blot analysis of K562 cells transfected with a vector encoding temperature-sensitive alleles of wild-type and mutant p53. The results showed that over-expression of the mutant p53 had no effect on the levels of either BORIS or CTCF transcripts. In contrast, activation of wild-type p53 was associated with a great reduction in expression of BORIS while expression of CTCF was unaffected (Figure 5B). To further examine the impact of p53 on BORIS expression in vivo, we compared p53-negative H1299 parental cells with H1299 cells stably infected with a virus expressing wild-type p53 (28). The results (Figure 5C) showed that the level of p53 expression was 12-fold higher in the virus-infected H1299 cells than in the parental cells. Analyses of CTCF transcripts showed that they were unchanged (data not shown). In contrast, p53 had a profound effect on each of the three BORIS promoters. Expression from promoters A and B was reduced 4.6- and 4.9-fold, respectively, while that from promoter C was down 17-fold.
Figure 5.
p53 effect on BORIS promoters A, B and C activities. (A) Luciferase assays were performed by transient transfection of reporter constructs and p53 wild-type (WT) or mutant (mut) expression vectors, in the p53−/− K562 cell line. (B) Northern blot showing the expression of BORIS, CTCF and p53 in the K562 p53 thermo-inducible cell line. (C) Real-time PCR on RNA extracted from p53−/− H1299 parental cells or H1299 cells expressing p53 wild type. Real-time PCR data were analyzed using the comparative Ct method and normalized to the non-induced cells.
Regulation of BORIS promoters by DNA methylation
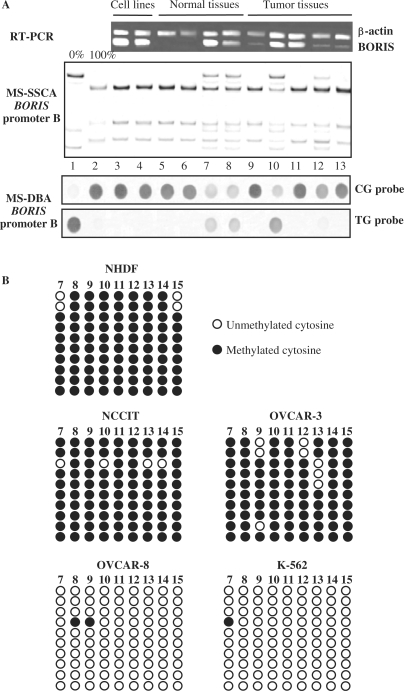
To further analyze the molecular mechanisms involved in regulating the activity of the BORIS promoters, we examined the effect of CpG methylation. The fact that promoters B and C co-localize within a CpG island raised the possibility that their activities might be affected by methylation (Figure 1A). DNA samples from 2 cell lines, 24 human normal tissue samples and 26 tumor tissue samples were extracted and analyzed by methylation-sensitive single-strand conformation analysis (MS-SSCA) and methylation-sensitive dot blot assay (MS-DBA) after sodium bisulfite modification (31,32). The CpG island was found to be completely methylated in samples of all normal tissues with the exception of testis, the tissue in which BORIS is normally expressed. Studies of the CpG island in tumors revealed demethylation in testicular tumors (6/6), ovarian tumors (1/3), breast cancers (1/6) and endometrial tumors (1/3) (Table 4). Representative examples of results from studies of the cell lines and tissues are shown in Figure 6A. The partial methylation observed in normal testis tissue is likely to be due to the presence of both germ cells and non-germ cells in this tissue. As might be expected, BORIS was expressed in almost all the tumors and cell lines that contained a hypomethylated CpG island. Moreover, it has been shown that demethylation of BORIS promoter B in tumor tissues, versus methylated BORIS promoter B in normal tissues, is correlated with the expression of BORIS in tumors (Table 4). However, among the 26 cell lines which belong to group A/C, only 2 showed partial demethylation of promoters B and C. Full methylation of these promoters in all the other lines was correlated with inhibition of BORIS transcription from promoter B (Table 1). In the case of cell lines from group B, promoters B and C were found to be unmethylated or partially methylated in the case of COLO-205. Nine out of 32 CpGs within promoters B and C were analyzed by genomic bisulfite sequencing in NHDF, 2 cell lines from group A/C (NCCIT and Ovcar-3) and 2 cell lines from group B (Ovcar-8 and K562) (Figure 6B). In normal fibroblasts, 98% of the CpGs analyzed were found to be methylated and BORIS expression could not be detected. In NCCIT and Ovcar-3, 98% and 96% of CpGs were methylated (Figure 6B). In these cells, BORIS was expressed primarily from promoter A, in association with minimal expression from promoter C and no expression from promoter B (Table 1). In Ovcar-8 and K562, sequencing showed that 0.9% and 0.45% of CpGs were methylated (Figure 6B). In these two lines, expression of BORIS comes almost exclusively from promoter B, 96% in Ovcar-8 and 80% in K562 (Table 1).
Table 4.
BORIS expression and BORIS promoter B methylation in normal and tumor tissues
| Normal tissues | Tumor tissues | ||
|---|---|---|---|
| BORIS expression | Bladder | − | − |
| Breast | − | + (1/6) | |
| Colon | − | − | |
| Kidney | − | − | |
| Testis | + | + | |
| Ovary | − | + | |
| BORIS promoter B methylation | Bladder | + | + |
| Breast | + | − (1/6) | |
| Colon | + | + | |
| Kidney | + | + | |
| Testis | − | − | |
| Ovary | + | − (1/3) |
Figure 6.
Analyses of DNA methylation of BORIS promoter B. (A) BORIS expression and methylation patterns of BORIS promoter B in human tissues and cell lines. Lanes 1–2: MS-SSCA and MS-DBA unmethylated and fully methylated controls respectively, obtained from plasmids containing BORIS promoter B sequences; lanes 3–4: NCCIT and OVCAR-3 cell lines; lanes 5–8: normal tissues, respectively colon, skin, and two testis; lanes 9–13: tumor tissues, respectively bladder, testis, ovary, breast, colon. BORIS mRNA was detected by RT-PCR, with β-actine as internal control. (B) Genomic bisulfite sequencing of 16/32 CpGs within the CpG island covering promoters B in normal fibroblast NHDF, NCCIT (group A), Ovcar-3 (group A), Ovcar-8 (group B) and K562 (group B) cell lines.
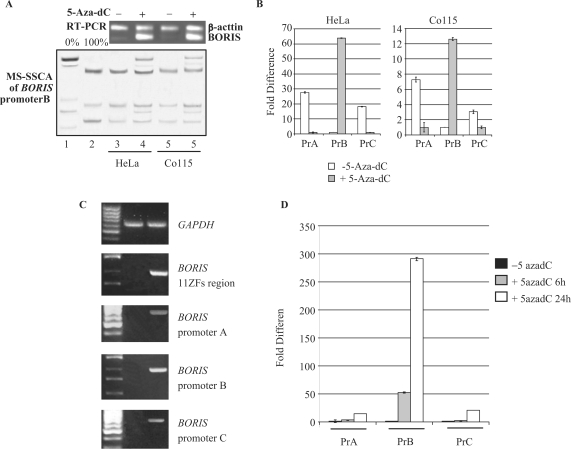
To confirm the regulation of promoter B by DNA methylation, we treated two weakly BORIS-positive cancer cell lines (HeLa and Co115) with 5-aza-dC. Treatment resulted in demethylation of the CpG island and strong activation of BORIS expression in both lines (Figure 7A). Moreover, the real-time PCR analyses showed that BORIS expression detected before 5-aza-dC treatment was coming from promoters A and C, but after 5-aza-dC treatment the BORIS expression switched to promoter B in both cell lines (Figure 7B). The decrease of expression from promoters A and C after 5-aza-dC treatment might be explained by the effect of 5-aza-dC itself on other transcriptional factors that could also in their turn downregulate BORIS expression through promoters A and C. Expression of BORIS in normal cells was also induced after treatment with 5-aza-dC (Figure 7C and D) with BORIS transcripts originating from all three promoters. qPCR analyses showed that BORIS transcripts from promoter B were readily detected in cells treated for just 6 h and that transcripts from all three promoters could be detected in cells treated for 24 h with promoter B being the strongest (Figure 7D). Remarkably, we were not able to detect any demethylation of the CpG island at the 24 h time point, suggesting that a mechanism other than demethylation was responsible for the effects of 5-aza-dC on the BORIS promoters.
Figure 7.
Effect of demethylating agent on BORIS expression in cancer cell lines and normal fibroblasts. (A) BORIS expression and methylation patterns of BORIS promoter B in two BORIS-negative cell lines treated with 5-aza-dC. Lanes 1–2: MS-SSCA unmethylated and fully methylated controls respectively; lane 3: HeLa cells; lane 4: HeLa cells after 5-aza-dC treatment; lane 5: Co115 cells; lane 6: Co115 cells after 5-aza-dC treatment. BORIS mRNA was detected by RT-PCR, with β-actine as internal control. (B) Real-time PCR on HeLa and Co115 before and after 5-aza-dC treatment. Real-time PCR data were analyzed using the comparative Ct method between treated and untreated cells and for each set of primers. (C) and (D) Determination of BORIS promoters A, B and C activities after treatment with 5-aza-dC of NHDFs by RT-PCR (C) and real-time PCR (D). Real-time PCR data were analyzed using the comparative Ct method between treated and untreated cells and for each set of primers.
DISCUSSION
In previous studies, it was shown that BORIS is normally expressed only in male germ cells where it is involved in epigenetic reprogramming (7,10). BORIS was also shown to be a CTA and its transcription is abnormally activated in varying proportions of a wide variety of cancers (9,25,38–40). In some cancers, BORIS is the most frequently activated CTA (40). Moreover, it has been suggested that expression of BORIS may have diagnostic implications (39), and recent work has identified BORIS as an attractive candidate target for anti-cancer immunotherapy (41,42). The present study significantly extends these foundations by developing molecular understandings of the mechanisms that tightly restrict BORIS expression in normal tissues but are permissive for expression in many malignancies.
Here, we first identified three transcriptional start sites at −1447, −899 and −658 bp upstream of the first ATG of the BORIS ORF. Regions upstream of those putative transcriptional starts were tested in reporter assays and were shown to exhibit promoter activities. These three promoters designated A, B and C in association with the start sites at −1447, −899 and −658, respectively induced the synthesis of five transcripts that are different from one another in their 5′-noncoding regions. Numerous studies have documented the importance of 5′-noncoding regions for both mRNA stability (43–45) and translational efficiency (46), as well as for having an impact on the specificity of tissue expression (47–49). Moreover, promoter regions and 5′-UTRs may determine alternative splicing of downstream exons, thus influencing the function of resulting products (50,51). We suggest that similar regulatory mechanisms might well be involved in the regulation of BORIS expression. In our study, we estimated the half-life of BORIS transcripts with alternative 5′ UTRs and found that the stability of the mRNAs differed significantly. The variance ranged from 4.7 to 7.9 h, with the BORIS transcript having the shortest half-life and the BORIS A3 transcript the longest. mRNA stability plays a major role in gene expression in mammalian cells, affecting the rates at which mRNAs disappear following transcriptional repression and accumulate following transcription induction (29). Thus, expression of BORIS alternative transcripts with different stabilities in testis and cancer cells suggests that these isoforms may have unique physiological functions, possibly related to different stages of spermatogenesis or events involved in cancer development and progression.
To understand these three promoters in greater depth, we first analyzed their utilization in a panel of normal tissues and in 31 cancer cell lines representing different types of human tumors. We demonstrated that the extent to which the promoters are employed is very different for normal testis and the cancer cell lines. In testis, all three promoters are expressed but to differing extents. Further studies will be required to determine if this pattern is common to all expressing germ cells or if promoter utilization varies depending on the state of differentiation with all being used at one point or another. In contrast, the patterns of promoter utilization in BORIS-positive cancer cell lines indicated that the lines fell into two main subsets: group B, comprising 16% of the lines, in which activity came primarily from promoter B; and group A/C, with 84% of the cases, in which activities come from promoters A and/or C. These results suggest that the normally stringent restriction of BORIS expression to testis requires repression of all three promoters in somatic cells. These data also demonstrate that release of BORIS expression in cancers does not reflect relaxation of promoter control to a state like that in testis but occurs in a more selective mode of promoter deregulation. Understanding if there are specific functional consequences to selective regulation of each promoter and variant transcript is a subject on ongoing studies.
As CTCF appears to be expressed in a complementary fashion with BORIS during spermatogenesis, we evaluated a potential inhibitory role for CTCF in BORIS transcription. We demonstrated that downregulation of CTCF by siRNA results in upregulation of BORIS in NHDF. We also showed that all three BORIS promoters contain CTCF-binding sites, suggesting that CTCF acts directly to regulate the BORIS promoters. The demonstration that CTCF is involved in the negative regulation of BORIS expression is likely to be important for understanding the uniform expression of BORIS in normal rat cells transformed due to a retroviral disruption of one CTCF allele (52) and in tumors of mice heterozygous for a null allele of CTCF (unpublished data).
Previous research convincingly demonstrated that binding of CTCF to many of its targets is sensitive to CpG methylation. This is exemplified by studies of the IGF2/H19 ICR, which showed that CTCF binds to the unmethylated maternal allele but not the methylated paternal allele (11–13). We thus hypothesized that regulation of BORIS transcription would be dependent on the methylation status of CTCF target sites in the promoters. Consistent with this suggestion, BORIS promoters B and C were found to be located in a shared CpG island. We therefore proceeded to determine how methylation status would affect the activities of these promoters. EMSAs performed using SssI methylated probes showed that CTCF-binding sites in promoter A were insensitive to methylation. In promoter B, however, CTCF binding is insensitive to methylation outside of the CpG island, but sensitive within the island. Likely, CTCF binding in promoter C was sensitive to methylation. In addition, ChIP studies showed a reverse correlation between CTCF binding and expression of BORIS from promoter B. Indeed, CTCF did not bind in promoter B in Ovcar-8 cells that expressed BORIS from promoter B. In contrast, CTCF bound strongly in promoter B in Ovcar-3 cells that expressed BORIS from promoter A. Analyses of methylation status of the CpG island in the Ovcar-3 and BJ cell lines showed that promoters B and C were fully methylated, although CTCF binding was found in both promoters in vivo. It seems likely that these results can be explained by the limitations of ChIP resolution as promoter B also contains CTCF sites insensitive to methylation and promoters B and C are only ∼250 bp apart. However, the possibility of indirect CTCF recruitment by another factor that can bind these methylated sequences cannot be formally excluded. Analyses of normal cells, tumor tissues and tumor cell lines showed that the activity of promoter B is downregulated by DNA methylation. Regulation of BORIS transcriptional activity by methylation was suggested but not conclusively demonstrated by other recent studies (9,53). As shown here, treatment of cancer cell lines that express BORIS at low levels with the demethylating agent, 5-aza-dC, resulted in demethylation of the promoter CpG island and substantial increases in BORIS transcription. However, treatment of BORIS-negative normal fibroblasts with 5-aza-dC also activated BORIS transcription, this time from all three promoters. The rapidity with which transcription was induced, and the lack of demethylation of the CpG island that contains promoters B and C, indicated that molecular mechanisms by which 5-aza-dC activated expression of BORIS in NHDF was very different from that governing activation in the tumor cells. Based on this, we conclude that although the methylation status of BORIS promoters is likely to be the determining regulatory feature in many cellular contexts, methylation-insensitive regulatory mechanisms exist that may well be cell type-specific.
The tumor suppressor protein, p53, is well known as a transcriptional activator of genes involved in cell cycle arrest, apoptosis, DNA repair and senescence (54,55). Activation of transcription by p53 occurs mostly through direct binding to DNA at a consensus target site. Nevertheless, in addition to gene activation through direct binding to DNA, p53 has been shown to repress transcription by indirect interactions with a target promoter through a promoter-bound transcriptional activator as the intermediary. For example, the interaction of p53 with Sp1 inhibits hTERT (56), CCNB1 (57) and IGF1 (58) promoters, while the interaction of p53 with NF-Y inhibits CDC2 (59), FN1 (fibronectin) (60) and COX2 (61) promoters. In this study, we showed that expression of wild-type p53 protein resulted in repression of all three BORIS promoters, but strongest forcefully for promoters A and B. However, ChIP assays showed that p53 may not bind directly to BORIS promoters (data not shown) or that if bound, it may be masked by multiple p53-binding co-factors from interactions with antibodies used in our assay. It is also possible that the strong inhibitory effects of p53 on BORIS promoters could be explained by interaction of p53 with the transcription factor Sp1 since consensus-binding sites for Sp1 are present in several copies in each BORIS promoter. Additionally, a recent study showed that p53 is required for the maintenance of DNA methylation patterns (62). Thus, it is possible that the maintenance of DNA methylation by p53 (62) could contribute to keeping BORIS silent.
In summary, we characterized three promoters for the BORIS gene that lead to the transcription of five mRNA differing in their 5′-UTRs. All three promoters were involved in BORIS expression during normal spermatogenesis; however, promoters A and C are preferentially used upon activation of BORIS during tumorigenesis. Interestingly, a few tumors that exhibited no activity from any of these three promoters still expressed BORIS transcripts (data not shown), suggesting the possible existence of additional regulatory sequences that act upstream of promoter A. Thus, DNA methylation, and expression of CTCF and p53 represent three mechanisms involved in the negative regulation of BORIS transcription, although it seems likely that additional levels of control also exist. These results showed that regulation of BORIS activity is complex, being both promoter- and cell type-dependent. Varying promoter usage driving multiple BORIS transcripts could influence many aspects of epigenetic reprogramming in normal development and in tumorigenesis.
ACKNOWLEDGEMENTS
We are grateful to Dr Herbert C. Morse III for critical reading of the manuscript. We thank Dr Peter Chumakov for kindly providing us the H1299 p53wt cell line and p53 expression vectors. This work was supported in part by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases and by Swiss National Science Foundation Grant number 3100AO-101732 (to J.B.). Funding to pay the Open Access publication charges for this article was provided by NIAID intramural funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pugacheva EM, Kwon YW, Hukriede NA, Pack S, Flanagan PT, Ahn JC, Park JA, Choi KS, Kim KW, et al. Cloning and characterization of zebrafish CTCF: developmental expression patterns, regulation of the promoter region, and evolutionary aspects of gene organization. Gene. 2006;375:26–36. doi: 10.1016/j.gene.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 3.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 4.Klenova EM, Morse H.C., III, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 5.Dunn KL, Davie JR. The many roles of the transcriptional regulator CTCF. Biochem. Cell Biol. 2003;81:161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- 6.Recillas-Targa F, De La Rosa-Velazquez IA, Soto-Reyes E, Benitez-Bribiesca L. Epigenetic boundaries of tumour suppressor gene promoters: the CTCF connection and its role in carcinogenesis. J. Cell Mol. Med. 2006;10:554–568. doi: 10.1111/j.1582-4934.2006.tb00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl Acad Sci. USA. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ladomery M, Dellaire G. Multifunctional zinc finger proteins in development and disease. Ann. Hum. Genet. 2002;66:331–342. doi: 10.1017/S0003480002001215. [DOI] [PubMed] [Google Scholar]
- 9.Vatolin S, Abdullaev Z, Pack SD, Flanagan PT, Custer M, Loukinov DI, Pugacheva E, Hong JA, Morse H., III, et al. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005;65:7751–7762. doi: 10.1158/0008-5472.CAN-05-0858. [DOI] [PubMed] [Google Scholar]
- 10.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 12.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 13.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 14.Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, West AG, Chang Y, Stablewski A, et al. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol. Cell. Biol. 2005;25:11184–11190. doi: 10.1128/MCB.25.24.11184-11190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock AL, Brown KW, Moorwood K, Moon H, Holmgren C, Mardikar SH, Dallosso AR, Klenova E, Loukinov D, et al. A CTCF-binding silencer regulates the imprinted genes AWT1 and WT1-AS and exhibits sequential epigenetic defects during Wilms’ tumourigenesis. Hum. Mol. Genet. 2007;16:343–354. doi: 10.1093/hmg/ddl478. [DOI] [PubMed] [Google Scholar]
- 17.Fitzpatrick GV, Pugacheva EM, Shin JY, Abdullaev Z, Yang Y, Khatod K, Lobanenkov VV, Higgins MJ. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 2007;27:2636–2647. doi: 10.1128/MCB.02036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 19.Lee JT. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol. 2003;13:R242–254. doi: 10.1016/s0960-9822(03)00162-3. [DOI] [PubMed] [Google Scholar]
- 20.Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, Quitschke WW, Loukinov DI, Ohlsson R, Lobanenkov VV. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum. Mol. Genet. 2005;14:953–965. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- 21.Boumil RM, Ogawa Y, Sun BK, Huynh KD, Lee JT. Differential methylation of Xite and CTCF sites in Tsix mirrors the pattern of X-inactivation choice in mice. Mol. Cell. Biol. 2006;26:2109–2117. doi: 10.1128/MCB.26.6.2109-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43––56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, Fischette MR, Adnani MT, Loukinov DI, Vatolin S, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763––7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 24.Kouprina N, Noskov VN, Pavlicek A, Collins NK, Schoppee Bortz PD, Ottolenghi C, Loukinov D, Goldsmith P, Risinger JI, et al. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PLoS ONE. 2007;2:e359. doi: 10.1371/journal.pone.0000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M, Li D, Cui Y, Mueller K, Chears WC, DeJong J. Regulatory factor interactions and somatic silencing of the germ cell-specific ALF gene. J. Biol. Chem. 2006;281:34288––34298. doi: 10.1074/jbc.M607168200. [DOI] [PubMed] [Google Scholar]
- 26.Klenova EM, Fagerlie S, Filippova GN, Kretzner L, Goodwin GH, Loring G, Neiman PE, Lobanenkov VV. Characterization of the chicken CTCF genomic locus, and initial study of the cell cycle-regulated promoter of the gene. J. Biol. Chem. 1998;273:26571––26579. doi: 10.1074/jbc.273.41.26571. [DOI] [PubMed] [Google Scholar]
- 27.Ceballos E, Delgado MD, Gutierrez P, Richard C, Muller D, Eilers M, Ehinger M, Gullberg U, Leon J. c-Myc antagonizes the effect of p53 on apoptosis and p21WAF1 transactivation in K562 leukemia cells. Oncogene. 2000;19:2194––2204. doi: 10.1038/sj.onc.1203541. [DOI] [PubMed] [Google Scholar]
- 28.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306––1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423––450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klenova EM, Chernukhin IV, El-Kady A, Lee RE, Pugacheva EM, Loukinov DI, Goodwin GH, Delgado D, Filippova GN, et al. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol. Cell. Biol. 2001;21:2221––2234. doi: 10.1128/MCB.21.6.2221-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benhattar J, Clement G. Methylation-sensitive single-strand conformation analysis: a rapid method to screen for and analyze DNA methylation. Methods Mol. Biol. 2004;287:181––193. doi: 10.1385/1-59259-828-5:181. [DOI] [PubMed] [Google Scholar]
- 32.Clement G, Benhattar J. A methylation sensitive dot blot assay (MS-DBA) for the quantitative analysis of DNA methylation in clinical samples. J. Clin. Pathol. 2005;58:155––158. doi: 10.1136/jcp.2004.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33:6850––6860. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287––4293. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- 35.O’Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285––4300. [PubMed] [Google Scholar]
- 36.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634––8647. [PubMed] [Google Scholar]
- 37.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, et al. Chromatin architecture near a potential 3. Mol. Cell. Biol. 2005;25:1511––1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, Gorlick R, Meyers P, Healey J, et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 2003;12:535––549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 39.D’Arcy V, Abdullaev ZK, Pore N, Docquier F, Torrano V, Chernukhin I, Smart M, Farrar D, Metodiev M, et al. The potential of BORIS detected in the leukocytes of breast cancer patients as an early marker of tumorigenesis. Clin. Cancer Res. 2006;12:5978––5986. doi: 10.1158/1078-0432.CCR-05-2731. [DOI] [PubMed] [Google Scholar]
- 40.Risinger JI, Chandramouli GV, Maxwell GL, Custer M, Pack S, Loukinov D, Aprelikova O, Litzi T, Schrump DS, et al. Global expression analysis of cancer/testis genes in uterine cancers reveals a high incidence of BORIS expression. Clin. Cancer Res. 2007;13:1713––1719. doi: 10.1158/1078-0432.CCR-05-2569. [DOI] [PubMed] [Google Scholar]
- 41.Loukinov D, Ghochikyan A, Mkrtichyan M, Ichim TE, Lobanenkov VV, Cribbs DH, Agadjanyan MG. Antitumor efficacy of DNA vaccination to the epigenetically acting tumor promoting transcription factor BORIS and CD80 molecular adjuvant. J. Cell. Biochem. 2006;98:1037––1043. doi: 10.1002/jcb.20953. [DOI] [PubMed] [Google Scholar]
- 42.Ghochikyan A, Mkrtichyan M, Loukinov D, Mamikonyan G, Pack SD, Movsesyan N, Ichim TE, Cribbs DH, Lobanenkov VV, et al. Elicitation of T cell responses to histologically unrelated tumors by immunization with the novel cancer-testis antigen, brother of the regulator of imprinted sites. J. Immunol. 2007;178:566––573. doi: 10.4049/jimmunol.178.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gauss KA, Bunger PL, Crawford MA, McDermott BE, Swearingen R, Nelson-Overton LK, Siemsen DW, Kobayashi SD, Deleo FR, et al. Variants of the 5. Gene. 2006;366:169––179. doi: 10.1016/j.gene.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE. Differential inhibition of mRNA degradation pathways by novel cap analogs. J. Biol. Chem. 2006;281:1857––1867. doi: 10.1074/jbc.M509121200. [DOI] [PubMed] [Google Scholar]
- 45.Suay L, Salvador ML, Abesha E, Klein U. Specific roles of 5. Nucleic Acids Res. 2005;33:4754––4761. doi: 10.1093/nar/gki760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou Z, Eibl C, Koop HU. The stem-loop region of the tobacco psbA 5'UTR is an important determinant of mRNA stability and translation efficiency. Mol. Genet. Genomics. 2003;269:340––349. doi: 10.1007/s00438-003-0842-2. [DOI] [PubMed] [Google Scholar]
- 47.De Jaco A, Camp S, Taylor P. Influence of the 5. Chem. Biol. Interact. 2005;157, 158:372––373. doi: 10.1016/j.cbi.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 48.Li WD, Reed DR, Lee JH, Xu W, Kilker RL, Sodam BR, Price RA. Sequence variants in the 5. Ann. Hum. Genet. 1999;63(Pt 3):227––234. doi: 10.1046/j.1469-1809.1999.6330227.x. [DOI] [PubMed] [Google Scholar]
- 49.Ning B, Nowell S, Sweeney C, Ambrosone CB, Williams S, Miao X, Liang G, Lin D, Stone A, et al. Common genetic polymorphisms in the 5. Pharmacogenet Genomics. 2005;15:465––473. doi: 10.1097/01.fpc.0000166823.74378.79. [DOI] [PubMed] [Google Scholar]
- 50.Gendra E, Colgan DF, Meany B, Konarska MM. A sequence motif in the Simian Virus 40 (SV40) early core promoter affects alternative splicing of transcribed mRNA. J. Biol. Chem. 2007;282:11648–11657. doi: 10.1074/jbc.M611126200. [DOI] [PubMed] [Google Scholar]
- 51.Russcher H, Dalm VA, de Jong FH, Brinkmann AO, Hofland LJ, Lamberts SW, Koper JW. Associations between promoter usage and alternative splicing of the glucocorticoid receptor gene. J. Mol. Endocrinol. 2007;38:91––98. doi: 10.1677/jme.1.02117. [DOI] [PubMed] [Google Scholar]
- 52.Sheng J, Organ EL, Hao C, Wells KS, Ruley HE, Rubin DH. Mutations in the IGF-II pathway that confer resistance to lytic reovirus infection. BMC Cell Biol. 2004;5:32. doi: 10.1186/1471-2121-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann MJ, Muller M, Engers R, Schulz WA. Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem. Pharmacol. 2006;72:1577––1588. doi: 10.1016/j.bcp.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 54.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594––604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 55.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431––442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 56.Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin. Cancer Res. 2000;6:1239––1247. [PubMed] [Google Scholar]
- 57.Innocente SA, Lee JM. p53 is a NF-Y- and p21-independent,Sp1-dependent repressor of cyclin B1 transcription. FEBS Lett. 2005;579:1001––1007. doi: 10.1016/j.febslet.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 58.Ohlsson C, Kley N, Werner H, LeRoith D. p53 regulates insulin-like growth factor-I (IGF-I) receptor expression and IGF-I-induced tyrosine phosphorylation in an osteosarcoma cell line: interaction between p53 and Sp1. Endocrinology. 1998;139:1101––1107. doi: 10.1210/endo.139.3.5832. [DOI] [PubMed] [Google Scholar]
- 59.Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, Shin DY. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 1999;274:29677––29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 60.Iotsova V, Stehelin D. Down-regulation of fibronectin gene expression by the p53 tumor suppressor protein. Cell Growth Differ. 1996;7:629––634. [PubMed] [Google Scholar]
- 61.Subbaramaiah K, Altorki N, Chung WJ, Mestre JR, Sampat A, Dannenberg AJ. Inhibition of cyclooxygenase-2 gene expression by p53. J. Biol. Chem. 1999;274:10911––10915. doi: 10.1074/jbc.274.16.10911. [DOI] [PubMed] [Google Scholar]
- 62.Park IY, Sohn BH, Choo JH, Joe CO, Seong JK, Lee YI, Chung JH. Deregulation of DNA methyltransferases and loss of parental methylation at the insulin-like growth factor II (Igf2)/H19 loci in p53 knockout mice prior to tumor development. J. Cell. Biochem. 2005;94:585––596. doi: 10.1002/jcb.20263. [DOI] [PubMed] [Google Scholar]