Abstract
The thermodynamic profiles of target site recognition have been surveyed for homing endonucleases from various structural families. Similar to DNA-binding proteins that recognize shorter target sites, homing endonucleases display a narrow range of binding free energies and affinities, mediated by structural interactions that balance the magnitude of enthalpic and entropic forces. While the balance of ΔH and TΔS are not strongly correlated with the overall extent of DNA bending, unfavorable ΔHbinding is associated with unstacking of individual base steps in the target site. The effects of deleterious basepair substitutions in the optimal target sites of two LAGLIDADG homing endonucleases, and the subsequent effect of redesigning one of those endonucleases to accommodate that DNA sequence change, were also measured. The substitution of base-specific hydrogen bonds in a wild-type endonuclease/DNA complex with hydrophobic van der Waals contacts in a redesigned complex reduced the ability to discriminate between sites, due to nonspecific ΔSbinding.
INTRODUCTION
Site-specific DNA-binding proteins typically display tight binding to their cognate target sites, and can discriminate between those sites and potential noncognate targets under demanding physiogical conditions—including high salt, the presence of many competing DNA-binding proteins in the cell and a vast background of potential nonspecific target sites within genomic DNA (1,2). Site-specific protein–DNA recognition is driven by structural interactions and conformational changes in both molecules that occur upon binding (3,4). This includes: (i) the overall complementarity of shape and charge between the two macromolecules, (ii) generic interactions with the DNA phosphoribosyl backbone (which attenuates overall affinity), (iii) interactions between chemical groups on the protein and on the nucleotide bases and (iv) binding-induced distortions of the DNA duplex.
Interactions with the DNA bases include both nonpolar contacts that are dominated by the van der Waals (Lennard–Jones) potential and hydrogen bonds formed between individual polarizable groups (5). These latter contacts can be made directly between DNA and protein atoms, or can be mediated by water molecules trapped in the molecular interface (6). Distortion of the DNA also contributes substantially to overall specificity, through the influence of sequence-specific DNA conformational preferences, because the energetic cost of disrupting the DNA double helix (particularly by base unstacking) can be strongly dependent on the sequence of the DNA target (7).
Historical studies of specific protein–DNA-binding energetics demonstrate that the free energy change and affinity of DNA binding displayed by most proteins fall within a relatively narrow range, by balancing inversely proportional changes of enthalpy (ΔH) and entropy (TΔS) for any given binding event (1). This balance has been termed ‘isothermal enthalpy–entropy compensation’ (1,8), and allows DNA-binding proteins that must perform extensive DNA remodeling or distortion (such as the bacterial CAP protein or TATA binding protein) to function under the same physiological conditions and at similar molecular concentrations as proteins that induce almost no distortion of their DNA targets (such as the GCN4 transcriptional activator). The absolute magnitude of these opposing thermodynamic forces can vary dramatically between individual proteins (each by up to +/−30 kcal/mol).
The majority of DNA-binding proteins that have been subjected to detailed thermodynamic analyses recognize relatively short (less than 10 basepair) target sites (1). Most of those proteins display high fidelity, exhibiting a marked decrease in affinity and activity as a result of basepair substitutions in their cognate target site. For those systems, a strong correlation has been noted between the extent of DNA distortion and the relative balance of ΔH and ΔS upon protein binding. Proteins that strongly distort their DNA targets are generally observed to display unfavorable ΔH as a result of base unstacking and molecular strain (1,8,9), which is balanced by a net favorable TΔS. This can be accomplished by minimizing protein folding upon binding and by extensive release of solvent molecules and counterions when the molecules form a complex. In contrast, proteins that bind to their DNA targets with minimal distortion often display a favorable ΔH, balanced by unfavorable TΔS. This can be accomplished by coupling the binding event to additional protein folding, as well as by quenching of protein dynamic flexibility (10).
Homing endonucleases that act as mobile genetic elements (usually in concert with microbial introns and inteins) differ greatly from the previously studied DNA-binding proteins mentioned above. They are small proteins (typically less than 250 amino acids) and are among the most specific DNA-binding proteins found in nature, recognizing long target sites of 14–30 bp (11). The most specific of the homing endonucleases are under intense study as potential enzymatic reagents for various applications that require highly specific targeting of DNA sequences, including corrective gene therapy and genomic engineering (12). All homing endonucleases are able to tolerate individual polymorphisms at some target site positions, providing a mechanism to increase their overall genetic mobility. For the most specific of homing endonucleases, the length of their DNA target sites assures that any one target sequence can occur by chance at a very low frequency (for example, a single 22 bp site occurs by chance at a frequency of 1 in 1013). Even if several thousand closely related target site variants of that length can be recognized by the protein, the chance that any one of them might occur in random genomic DNA is equal to the sum of their individual probabilities—in some cases as low as 1 in 109, making them capable of acting as ‘gene specific reagents’ (13).
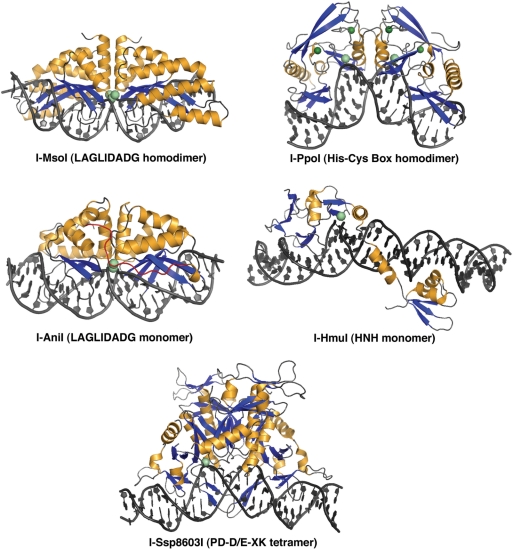
Homing endonucleases can be classified into at least five distinct structural families (Figure 1), based on their structural similarities and conservation of active site geometries(11). These families are the LAGLIDADG proteins (the most specific, found in archaebacteria and in mitochondrial and chloroplast genomes of fungi and algae), His-Cys box proteins (found in protist nuclear genomes), HNH and GIY-YIG proteins (both found in primarily in phage) and PD-(D/E)XK proteins (found in bacteria). With the exception of the His-Cys box and HNH endonucleases that share a common ββα-metal active site motif (14), each of these families has arisen from unique ancestors. The extraordinary DNA-binding specificity displayed by these proteins requires neither large tertiary structures nor the assistance of additional protein factors.
Figure 1.
Structures of homing endonuclease–DNA complexes in this study. The structures correspond to a homodimeric LAGLIDADG enzyme (I-MsoI; 2.5 Å resolution), a monomeric LAGLIDADG enzyme (I-AniI; 2.4 Å), a homodimeric His-Cys box enzyme (I-PpoI, 1.7 Å), an HNH enzyme (I-HmuI, 3.1 Å) and a tetrameric PD-D/E-XK enzyme (I-SspI; 3.1 Å). All structures shown are in complex with wild-type physiological target sites from the biological host of the corresponding homing endonuclease. Bound metal ions are shown as spheres.
Crystallographic structures are available for representatives of all homing endonuclease families (Figure 1), showing that their mechanisms of DNA recognition and association are widely divergent. The phage endonucleases (HNH and GIY-YIG families) are completely asymmetric and are composed of separate N- and C-terminal catalytic and DNA-binding domains that are tethered together by a variety of additional domains and linkers (15–17). In nature these proteins often exchange or swap either their catalytic or their DNA-binding domains in a highly combinatorial manner. Their isolated nuclease domains typically display considerable non-specific DNA cleavage activity.
In contrast, the remaining homing endonuclease families display significant symmetry in their tertiary and/or quaternary structures, ranging from pseudosymmetric, gene-fused monomers (such as the LAGLIDADG endonucleases I-SceI and I-AniI) (18,19) to homodimers (found in many lineages of LAGLIDADG and His-Cys box proteins) (20,21) and tetramers (most recently visualized for the bacterial PD-D/E-XK endonuclease I-Ssp8603I) (22). In homing endonuclease systems involving protein dimers or tetramers, the corresponding cognate target site in the biological host is often a pseudopalindrome. In those cases, palindromic derivatives of the DNA sites are also viable substrates for their endonucleases.
In this study, we survey the free energy and DNA-binding affinity, and the balance of enthalpy and entropy, for representatives of most known families of homing endonucleases using the method of isothermal titration calorimetry (ITC) (23). In addition to a global survey of DNA-binding energetics by these proteins, we also characterize three particularly significant aspects of homing endonuclease DNA recognition: the ability of symmetric homodimers to recognize asymmetric DNA targets, the energetic costs of encorporating deleterious basepair mismatches in their cognate target sites and the effect of subsequent efforts to redesign protein contact points to accommodate those same basepair substitutions.
MATERIALS AND METHODS
Materials
All homing endonucleases used in this study were overexpressed in Escherichia coli and purified as described previously (methods found in the following citations). The proteins surveyed were: I-MsoI (a LAGLIDADG homodimer from the green algae Monomastix) (20), I-AniI (a LAGLIDADG monomer from the fungi Aspergillus nidulans) (18), I-PpoI (a His-Cys box protein from the slime mold Physarum polycephalum) (24), I-HmuI (an HNH protein from bacteriophage SPO1) (17) and I-SspI (a PD-D/E-XK protein from the cyanobacteria Synechocystis) (22). All endonucleases were wild-type constructs, with the exception of I-SspI (which was expressed and purified as a catalytically inactive point mutant, because the wild-type protein cannot be expressed in E. coli at sufficient yield to permit ITC experiments).
All DNA oligonucleotides (1 μmol scale, salt-free syntheses; see Table 1 for sequences) were purchased from Operon Biotechnologies, Inc. (Huntsville, AL, USA). Complementary strands were annealed by heating to 95°C followed by slow-cooling to room temperature to generate the double-stranded DNA target site for binding analysis by ITC. Oligonucleotide target site sequences were chosen based on previous identification of their cognate target sites and additional biochemical analyses. Asymmetric G-C anchors were added to each end of palindromic and pseudo-palindromic sites to prevent undesired hairpin formation and thus promote formation of stable heterodimers. The homogeneity of duplex constructs was verified by electrophoresis in 4% agarose.
Table 1.
DNA target site constructs
| Protein | Structural family | Site name and site type | Sequence of duplex (target site in bold) |
|---|---|---|---|
| I-PpoI | His-Cys box homodimer | WT: wild-type cognate pseudopalindrome | 5′ GGGGGAGTAACTATGACTCTCTTAAGGTAGCCAAAGGGGG 3′ 3′ CCCCCTCATTGATACTGAGAGAATTCCATCGGTTTCCCCC 5′ |
| I-PpoI | His-Cys box homodimer | LL: left–left palindrome | 5′ GGGGGTGCAGTGACTCTCTTAAGAGAGTCAGACGTGGGGG 3′ 3′ CCCCCACGTCACTGAGAGAATTCTCTCAGTCTGCACCCCC 5′ |
| I-PpoI | His-Cys box homodimer | RR: right–right palindrome | 5′ GGGGGTGCAGTGGCTACCTTAAGGTAGCCAGACGTGGGGG 3′ 3′ CCCCCACGTCACCGATGGAATTCCATCGGTCTGCACCCCC 5′ |
| I-MsoI | LAGLIDADG homodimer | WT: wild–type cognate pseudopalindrome | 5′ GGGGGGCAGAACGTCGTGAGACAGTTCGGGGGGG 3′ 3′ CCCCCCGTCTTGCAGCACTCTGTCAAGCCCCCCC 5′ |
| I-MsoI | LAGLIDADG homodimer | LL: left–left palindrome | 5′ GGGGGGCAGAACGTCGTACGACGTTCTGGGGGGG 3′ 3′ CCCCCCGTCTTGCAGCATGCTGCAAGACCCCCCC 5′ |
| I-MsoI | LAGLIDADG homodimer | RR: right–right palindrome | 5′ GGGGGGCGAACTGTCTCGAGACAGTTCGGGGGGG 3′ 3′ CCCCCCGCTTGACAGAGCTCTGTCAAGCCCCCCC 5′ |
| I-MsoI | LAGLIDADG homodimer | MIS: miscognate pseudopalindrome | 5′ GGGGGGCAGAAGGTCGTGAGACCGTTCGGGGGGG 3′ 3′ CCCCCCGTCTTCCAGCACTCTGGCAAGCCCCCCC 5′ |
| I-AniI | LAGLIDADG monomer | WT: wild–type cognate asymmetric | 5′ CCTTCCCTGAGGAGGTTTCTCTGTAAACCCTTCC 3′ 3′ GGAAGGGACTCCTCCAAAGAGACATTTGGGAAGG 5′ |
| I-AniI | LAGLIDADG monomer | WT-OPT: selected optimal target asymmetric | 5′ CCTTCCCTGAGGAGGTTACTCTGTTAACCCTTCC 3′ 3′ GGAAGGGACTCCTCCAATGAGACAATTGGGAAGG 5′ |
| I-SspI | PD-D/E-XK tetramer | WT: wild-type cognate pseudopalindrome | 5′ GGGCCTTCGGGCTCATAACCCGAAGGGACG 3′ 3′ CCCGGAAGCCCGAGTATTGGGCTTCCCTGC 5′ |
| I-HmuI | HNH monomer | WT: wild-type cognate asymmetric | 5′GGGGGGTAATGAGCCTAACGCTCAGCAATTCCCACGTAAGGGGGG3′ 3′CCCCCCATTACTCGGATTGCGAGTCGTTAAGGGTGCATTCCCCCC5′ |
Underlined bases indicate positions in target sites for homodimeric (I-PpoI, I-MsoI) or tetrameric (I-SspI) homing endonucleases that are not conserved between DNA half-sites (i.e. they contribute to asymmetry in otherwise palindromic targets).
Isothermal titration calorimetry
Appropriate pairs of protein and DNA samples were dialyzed overnight into identical buffers, prior to calorimetric measurements, using Tube-O-Dialyzers (G Biosciences, St Louis, MO, USA). Individual buffer compositions, chosen based on previous published studies of optimal binding conditions for each endonuclease (while preventing DNA cleavage by withholding magnesium), were as follows:
I-PpoI: 20 mM sodium phosphate pH 8.0, 50 mM NaCl and 10 mM EDTA (25,26).
I-MsoI: 50 mM Tris pH 8.0, 75 mM NaCl and 10 mM CaCl2 (20).
I-AniI: 50 mM Tris, pH 7.5, 50–100 mM NaCl, 10 mM CaCl2 (27).
I-SspI: 50 mM Tris pH 7.5 and 200 mM NaCl (28).
I-HmuI: 20 mM HEPES pH 8.0, 50 mM NaCl and 1 mM EDTA (29).
Following dialysis, sample concentrations were quantified by UV spectroscopy using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA). The extinction coefficients (ε) for the proteins were calculated using ProtParam (30). The annealed DNA oligonucleotide ε-values were determined experimentally by digesting the dsDNA oligonucleotides to completion with DNAse and using known nucleotide-monophosphate ε-values (31) to calculate ε for the dsDNA complex (32). Sample concentrations ranged from 1.5 to 4.5 μM protein and from 30 to 70 μM dsDNA target.
ITC experiments were conducted on a VP-ITC MicroCalorimeter (MicroCal, LLC, Northampton, MA, USA) following manufacturer guidelines. The protein sample was placed in the cell and the DNA target in the auto-pipette. Individual runs consisted of 25–35 injections of 5–9 μl each, depending on sample concentrations, and were conducted at a temperature of 30°C and a stirring speed of 329 r.p.m. A full experiment was composed of three experimental runs with both DNA and protein components in the instrument, as well as a control run of DNA into buffer. Following integration and normalization of peaks, data from the DNA into buffer control run was subtracted to minimize the effect of DNA dilution on measured heat of mixing. Data were fit using Origin 7 SR2 software (OriginLab Corporation, Northampton, MA, USA). The data were well modeled by the one-site fitting algorithm in Origin and the resulting fit parameters were used to calculate KD.
For a ligand X binding to a single set of ‘n’ identical sites on a macromolecule, the single-site binding constant ‘KD’ = (filled sites)/(empty sites) and ΔGo = −R T lnK = ΔHo − T ΔSo, where ΔGo, Δ Ho and ΔSo are the free energy, enthalpy and entropy change for single site binding. By nonlinear least squares fit of calorimetric titration data, the parameters KD, ΔH0, and n are determined directly in a single experiment and ΔG0 and ΔS0 may then be calculated. Titration calorimetry is capable of defining all of these parameters in a single experiment, resulting in nearly complete thermodynamic characterization of the interaction (33).
DNA bending analyses
The extent of DNA bending and parameters of individual basepair steps were analyzed for the crystal structures of individual homing endonuclease-DNA complexes using program ‘ReadOut’ (34) (via a web-based server located at: http://gibk26.bse.kyutech.ac.jp/jouhou/readout/).
RESULTS AND DISCUSSION
Affinity, binding free energy and thermodynamic compensation across homing endonuclease families
The binding of 11 separate combinations of homing endonucleases and various DNA target sites, representing five major structural classes of these proteins, was analyzed by ITC. The DNA constructs used for these studies are shown in Table 1; the structure of each wild-type protein/DNA complex is shown in Figure 1. The affinities and thermodynamic signatures of these interactions are summarized in Table 2 and Figures 2 and 3.
Table 2.
Affinities and thermodynamic values of homing endonuclease–DNA-binding events
| Protein | Site | KD | ΔH | ΔS | −TΔS | ΔG |
|---|---|---|---|---|---|---|
| nM | kcal/mol | cal/mol/deg | kcal/mol | kcal/mol | ||
| I-PpoI | WT | 17 ± 1.7 | −35.5 ± 0.29 | −81.6 ± 0.87 | 24.7 | −10.8 |
| I-PpoI | LL | 87 ± 8.8 | −35.1 ± 0.57 | −83 ± 3.0 | 25.3 | −9.8 |
| I-PpoI | RR | 23 ± 1.6 | −38.8 ± 0.26 | −93.1 ± 2.4 | 28.2 | −10.6 |
| I-AniI | WT | 96 ± 10 | 11.2 ± 1.1 | 69.3 ± 3 | −21.0 | −9.8 |
| I-AniI | WT-OPT | 8 ± 1 | 6.6 ± 0.7 | 59.0 ± 3 | −17.9 | −11.3 |
| I-MsoI | WT | 21 ± 5.0 | 12.7 ± 0.27 | 77 ± 1.1 | −23.3 | −10.6 |
| I-MsoI | LL | 6 ± 1.4 | 16.4 ± 0.20 | 91.7 ± 0.36 | −27.8 | −11.4 |
| I-MsoI | RR | 17 ± 6.0 | 14.4 ± 0.40 | 83.0 ± 0.64 | −25.2 | −10.8 |
| I-MsoI | MIS | 760 ± 53 | 21.2 ± 0.42 | 98 ± 2.7 | −29.7 | −8.4 |
| I-MsoI redesigned | MIS | 46 ± 6.5 | 34.2 ± 0.42 | 146.5 ± 0.7 | −44.4 | −10.2 |
| I-MsoI redesigned | WT | 243 ± 20 | 37.1 ± 4 | 152.7 ± 6 | −46.3 | −9.2 |
| I-SspI (E11Q)a | WT | 350 ± 30 | −23.3 ± 3 | −47.3 ± 4 | 14.3 | −9.0 |
| I-HmuIb | WT | (30) | Endothermic | – | – | – |
a‘I-SspI’ corresponds to a catalytically inactive mutant (E11Q) of the I-Ssp8603I homing endonuclease, containing an alteration of an active-site putative metal-binding residue. In addition, the enzyme contains a second mutation on its surface far from the DNA-binding interface (F55K) to improve its solubility. The resulting affinity and ΔGbinding therefore may not reflect the wild-type enzyme.
bI-HmuI displays complex behavior in ITC experiments that is not reliably modeled mathematically (Figure 2). The reaction is strongly endothermic, and the Kd for that binding event has been estimated at 30 nM.
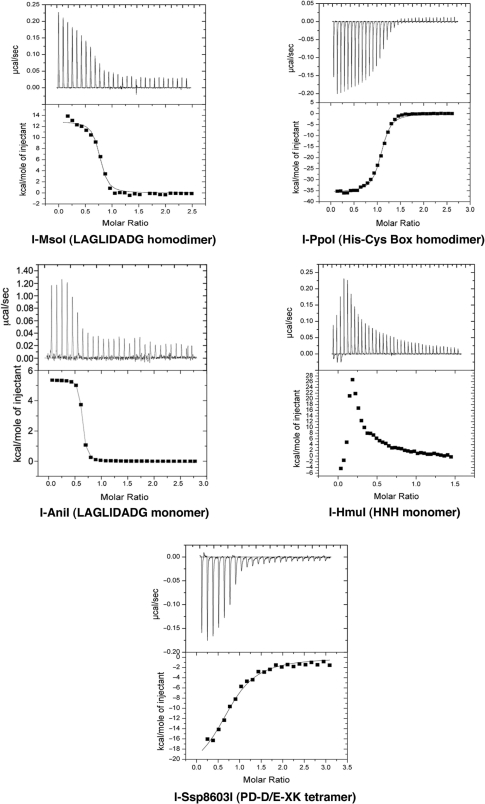
Figure 2.
Thermodynamic profiles of representative homing endonuclease binding events. Heat profiles of sequential injections of DNA against relevant endonucleases are shown. All endonucleases are wild-type except for a catalytically inactivated mutant of I-Ssp8603I; all DNA target sites are the physiological sequences form the corresponding biological hosts. Three of the five representative homing endonucleases (the monomeric and homodimeric LAGLIDADG enzymes and the HNH enzyme) display endothermic binding; the other two (the His-Cys box and PD-D/E-XK enzymes) display exothermic binding. All of the enzymes studied could be fit to standard saturation curves, except for I-HmuI (described in detail in the text), which displays complex multiphasic binding behavior.
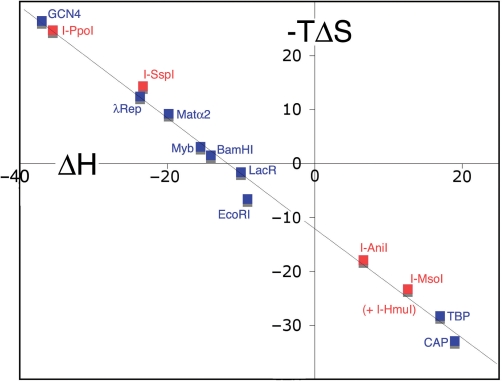
Figure 3.
Isothermal enthalpy–entropy compensation by homing endonucleases and other DNA-binding proteins. The enthalpic (ΔH) and entropic (−TΔS) contributions to site-specific DNA recognition of different protein–DNA complexes, including several representative wild-type homing endonucleases (highlighted in red) are shown. The thermodynamic values for previously studied DNA-binding proteins are shown in blue, and are taken from previous analyses by Jen-Jacobson et al. (1) and references therein.
The dissociation constants of the wild-type homing endonuclease families against their wild-type, physiological target sites range from 17 (I-PpoI) to 96 nM (I-AniI). The precise affinity and thermodynamic values for one wild-type endonuclease (I-HmuI, of the phage HNH family) could not be estimated directly from ITC data, due to multiphasic behavior in its binding isotherm (described in detail subsequently). Gel shift experiments for that endonuclease give KD values of ∼30 nM to its cognate DNA targets (B.L.S., unpublished data). Finally, the dissociation constant of a catalytically inactive point mutant (E11Q) of I-Ssp8603I (a bacterial PD-D/E-XK homing endonuclease) was 350 nM; this probably represents a compromised binding affinity due to the loss of a bound metal ion in the protein–DNA interface. For the wild-type endonucleases that could be analyzed in detail, the range of binding free energies (ΔG) to their physiological DNA targets was −10.8 kcal/mol (I-PpoI) to −9.8 kcal/mol (I-AniI).
With the exception of I-HmuI, the stoichiometries of protein–DNA interactions measured in these experiments were within 25% of 1:1 (protein functional assemblies:DNA target sites), in agreement with previously published biochemical studies and crystallographic analyses (Figure 2). The lower stoichiometry observed for I-HmuI binding may reflect a higher percentage of aggregated or misfolded protein (that may also contribute to the unusual isotherm for that endonuclease).
For two endonucleases, their physiological binding site is not the optimal substrate for that protein. I-AniI (a LAGLIDADG monomer) binds a variant target site containing two basepair substitutions with 12-fold lower KD (8 nM) than its target in Aspergillus. Similarly, I-MsoI (a LAGLIDADG homodimer) binds a palindromic DNA target that consists of its two left half-sites with a 3-fold lower KD (6 nM) than its physiological target site. Both of these observations are discussed in more detail in the following sections.
The thermodynamic DNA-binding profiles of these proteins are widely distributed across the continuum of enthalpy/entropy compensation, and obey the same linear pattern of thermodynamic compensation noted previously for a wide variety of DNA-binding proteins (1) (Figures 2 and 3). Two of the endonucleases (the His-Cys box protein I-PpoI and the PD-D/E-XK protein I-Ssp8603I) display strongly exothermic DNA binding, whereas the monomeric and homodimeric LAGLIDADG endonucleases (I-AniI and I-MsoI) and the HNH endonuclease (I-HmuI) display strongly endothermic DNA binding (Figure 2). The range of values for ΔH (from −35 to +13 kcal/mol) and TΔS (−23 kcal/mol to +25 kcal/mol) that separate these individual protein–DNA interactions are as broad as previously observed for other DNA-binding proteins (Figure 3).
DNA bending and thermodynamic signatures
All available crystal structures of homing endonuclease/DNA complexes demonstrate significant DNA bending that is associated with the mechanism by which each accomplishes recognition of long target sequences (11) (Figures 1 and 4). In the case of dimeric (LAGLIDADG and His-Cys-Box) and tetrameric (PD-D/E-XK) homing endonucleases, cleavage is executed across the minor groove of the target site (generating 3′ overhangs), thus allowing the protein to contact and ‘read out’ contiguous sets of nucleotide basepairs within the major groove of each DNA half-site, at positions that flank the cleavage sites. This strategy requires that the center of the DNA target sites be significantly distorted near the scissile phosphates, either by narrowing the minor groove and using two closely juxtaposed active sites (as seen for the LAGLIDADG enzymes) or by widening the minor groove and using two physically separated active sites (as seen for the His-Cys box and PD-D/E-XK enzymes). In contrast, the monomeric phage endonucleases (such as the HNH enzyme I-HmuI) bind even longer targets (>25 bp), and use a tandem series of protein domains to contact intermittent stretches of DNA bases within both the major and minor groove of the target site. For those complexes, DNA bending is again necessary in order to access DNA bases in the minor groove and to straddle the DNA backbone at various positions in the complex.
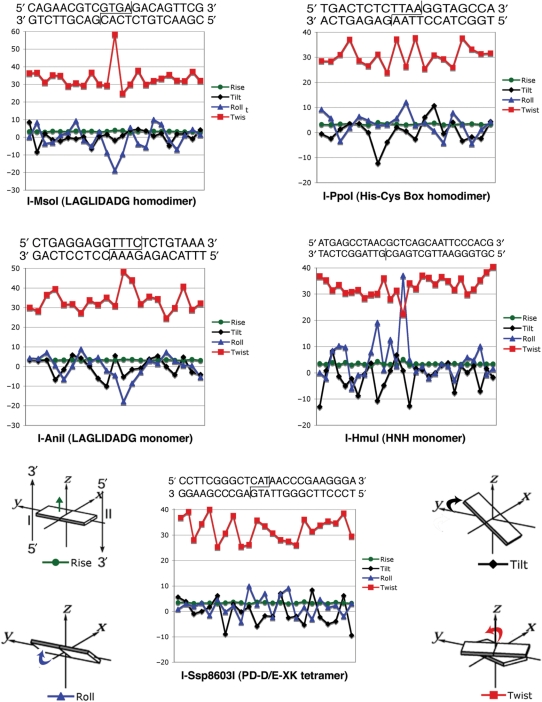
Figure 4.
DNA distortion induced by homing endonuclease binding. DNA bend parameters were quantitated using program ‘Readout’ (34), via a web-based server located at: http://gibk26.bse.kyutech.ac.jp/jouhou/readout/). The cognate target sequence, used in the individual crystal structures from which the bend parameters were calculated, are shown; cleavage sites are indicated. All endonucleases in this study except for I-HmuI cleave both strands to produce 3' overhangs; I-HmuI nicks the lower strand only (vertical line). The individual features of basepair steps distortion shown in the graphs (roll, tilt, twist and rise) are illustrated relative to a standard coordinate frame for double-stranded DNA.
For previously examined DNA-binding proteins, unfavorable enthalpic changes upon DNA binding (corresponding to an endothermic reaction event) have been observed to correlate with increased distortion of the DNA target, which in turn corresponds to basepair unstacking and molecular strain (1). As discussed subsequently, a comparison of the binding thermodynamics of the representative homing endonuclease complexes (Figure 2, Table 2) to the distortion of their DNA target sites (Figure 4) also demonstrates the dominant role of base unstacking in promoting unfavorable changes in enthalpy during DNA binding, while conversely also demonstrating that significant DNA bending can be accomplished while still maintaining base stacking and producing a strongly exothermic-binding event.
The three homing endonucleases that display strongly ‘endothermic’ DNA binding (the two LAGLIDADG endonucleases and the I-HmuI HNH endonuclease) all display basepair roll angles and unstacking at individual basepair steps that significantly depart from undistorted B-form DNA (Figure 4). The LAGLIDADG enzymes both display a single significant negative roll angle at their central −1/+1 bp step of ∼−20°, corresponding to pronounced narrowing of the minor groove at the center of the target cleavage region. This DNA bend and unstacking is accompanied by overwinding of the central basepair step (‘twist’ rising to 50° and 60° for the I-MsoI homodimer and I-AniI monomer, respectively). In contrast, the HNH endonuclease I-HmuI displays strongly ‘positive’ roll angles at two separate basepair steps (+20° at step +9/+10 and then +37° at step +13/+14). At each position the protein ‘hurdles’ the phosphate backbone and inserts side chains into the minor groove. The latter of these bends is accompanied by widening of the minor groove and significant underwinding of the DNA (‘twist’ being reduced to 22°).
The two endonucleases that exhibit strongly ‘exothermic’ DNA binding (the His-Cys box I-PpoI endonuclease, and the PD-D/E-XK endonuclease I-SspI) also display significant DNA bending (overall bends of ∼75° and 25° across the central 8 bp of their target sites) and corresponding widening of the minor groove at the site of cleavage. However, both proteins accomplish binding and distortion of their DNA targets with roll angles that never exceed +/−12° at any single basepair step, and relatively small distortions of helical winding (‘twist’) values across their target sites (Figure 4). In the case of I-PpoI, the severe bend imparted to the DNA target is accomplished through a mixture of smaller cumulative roll angles (which act in concert to widen the minor groove at the cleavage sites) and mutually opposing tilts in a single DNA basepair step in each half-site.
Multidomain DNA recognition: I-HmuI
Whereas the eukaryotic and bacterial homing endonucleases display relatively compact structures containing single DNA-binding domains arranged in various oligomeric symmetries, the phage endonucleases (HNH and GIY-YIG families) contain separate DNA interaction domains arranged in a sequential tandem array along a single peptide chain (15,16,35). These proteins are prone to significant exchange and shuffling of their domains during evolution, and the occasional insertion of additional structural elements such as zinc fingers (36).
I-HmuI displays such a multidomain structure (Figure 1) (17). Its N-terminal, antiparallel β–sheet (contacting the 5′ end of the target site) is associated with the HNH nuclease core, which is then followed by two α-helices that intercalate in the minor groove in the center of the site, and finally a helix–turn–helix domain that generates the majority of base-specific contacts at the 3′ end of the target site. The DNA binding isotherm displayed by I-HmuI (Figure 2) is unique among the measurements described here, in that it displays complex multiphasic behavior as the total concentration of protein increases with sequential injections. Each of the first three injections (generating DNA concentrations in the sample cell rising from ∼10 to 30 nM) results in a rapid absorption of heat (+ΔH) followed by a slower, more modest release of heat (−ΔH). The overall heat signature becomes stronger with each of these early injections. Beginning with the fourth injection (resulting in a DNA concentration of ∼40 nM), the isotherm demonstrates only a single feature corresponding to heat absorption. This signal then displays a hyperbolic reduction in strength with each ensuing injection as saturation of the DNA target sites is achieved. The transition from the multiphasic injection isotherms to single peaks and subsequent return to baseline occurs at a protein concentration near the previously measured KD for the I-HmuI–DNA interaction.
While the pattern of heat absorption and release demonstrated by I-HmuI described above and shown in Figure 2 is difficult to explain to complete satisfaction, there are two likely explanations. The first is that that a small percentage of the protein is aggregated, a behavior that is eliminated by early injections of DNA (perhaps as free protein is titrated into DNA-bound complexes). The second is that the early injection isotherms might reflect to a biphasic binding event consisting of a relatively fast endothermic association of one protein domain (possibly the C-terminal helix–turn–helix domain) followed by a slower exothermic step of intramolecular association. It is possible that the N-terminal nuclease domain, which displays fewer contacts with the DNA and is known to display reduced DNA specificity on its own (37), may require a slow step of DNA distortion at the center of the target site before docking to the remaining target sequence. Recent studies of GIY-YIG endonucleases, which possess similar structural organization, have also demonstrated DNA binding and cleavage behaviors that involve initial rapid association and subsequent slower conformational changes (38).
Symmetry recognizing asymmetry
Many multimeric homing endonucleases are encoded within introns that interrupt highly conserved rDNA and tRNA genes (39–43). The preponderance of base-paired stem–loop elements in folded RNA structures increases the frequency of palindromic sequences within their coding regions, encouraging the persistence of multimeric homing endonucleases, including both homodimers (such as the His-Cys box endonuclease I-PpoI and the LAGLIDADG endonuclease I-MsoI, encoded by introns within algral rDNA genes) and tetramers (such as I-Ssp8603I, encoded by an intron within a bacterial tRNA gene). While such homing endonucleases possess the advantage of being encoded by particularly short reading frames, which are tolerated well by their intron hosts, they are presented with the challenge of maintaining activity when target site symmetry is broken. In many cases, a homodimeric protein is found to display cleavage activity against a physiological DNA target site that displays surprisingly little palindromic symmetry between left and right half-sites. For example, the LAGLIDADG homodimeric endonuclease I-CeuI acts at a target site that displays only 36% sequence identity between left and right half sites (21).
In such cases, one would anticipate that a homodimeric homing endonuclease would display binding and cleavage activity towards palindromic variants of its natural target site, and that many such proteins would display energetically superior (more favorable) contacts to one half-site over the other. This was examined for homodimeric endonucleases from both the His-Cys Box (I-PpoI) and LAGLIDADG (I-MsoI) families, by measuring their binding affinity and thermodynamic profiles against palindromic DNA sequences (‘Left-Left’, or ‘LL’ and ‘Right-Right’, or ‘RR’) derived from individual half-sites of their host target sites (Table 1). The two palindromic sequences derived from the I-PpoI target site differ from one another at 4 bp out of 14 total. In contrast, the two palindromic sites derived from the I-MsoI target (which is substantially more asymmetric) differ at 12 out of 22 bp. Each of the palindromic sites differs from the wild-type target at half of those basepairs (all within a single half-site in each case).
I-PpoI and I-MsoI both display noticeable differences in affinity towards the two palindromic DNA sequences derived from their wild-type target (Table 2). I-PpoI binds its ‘RR’ target 4-fold more tightly than its ‘LL’ target; I-MsoI binds its ‘LL’ target 3-fold more tightly than ‘RR’. The corresponding differences in ΔGbinding between the palindromic targets for each enzyme (ΔΔG) are ∼0.6 and 0.8 kcal/mol, respectively. In both cases, these differences in binding affinity are attributable to more favorable ΔHbinding for the superior palindromic target (−3.7 and −2.0 kcal/mol, respectively), which is partially offset by smaller, unfavorable changes in TΔS. The differences in the binding affinities and ΔHbinding to the LL and RR palindromes is correlated to the total number of contacts made to nucleotide bases in each unique half-site. For example, I-MsoI displays 20 contacts to atoms on DNA bases in the left half-site (10 water mediated, 10 direct to side chains) versus 15 similar contacts to DNA bases in the right half-site (again, half of the contacts are water mediated). The number of contacts to DNA backbone atoms are identical in both.
Interestingly, the two enzymes behave differently towards their wild-type asymmetric target sites from the host as compared to the palindromic variants. I-PpoI binds its asymmetric cognate target site with an affinity (KD ∼17 nM) that is comparable to the more ‘tightly’ bound of the two palindromes (KD ∼23 nM). In contrast, I-MsoI binds its asymmetric cognate site with an affinity (KD ∼21 nM) that is comparable to the more ‘loosely’ bound palindromic target (KD ∼17 nM). The ability of a homodimeric homing endonuclease to recognize an asymmetric target site with an affinity that rivals a palindromic repeat of one half-site, or to even prefer the asymmetric cognate target over either palindromic variant (rather than displaying an affinity that is obviously an intermediate between the two palindromes) has also been observed for the LAGLIDADG homing endonuclease I-CeuI (21).
The LAGLIDADG family: specificity and engineering
Homing endonucleases are under intense scrutiny as potential reagents for targeted genetic applications, in which genomes of target organisms or cells are manipulated in vivo, using site-specific recombination to alter or add desired traits. This field requires the development of highly specific DNA-binding proteins that can stimulate gene conversion events at unique sites within complex genomes. Over the past several years, two different approaches to create enzymes capable of inducing site-specific DNA double-strand breaks have been developed: zinc finger nucleases (44) and engineered homing endonucleases (12,45). LAGLIDADG homing endonucleases are particularly attractive systems for the development of gene-specific reagents: they are the most specific of all known homing endonucleases, possess relatively small and highly modularized structures, and have tightly coupled mechanisms of specific DNA recognition and cleavage. A variety of studies have demonstrated that the DNA-binding specificity of LAGLIDADG endonucleases can be altered in predictable ways, ranging from the creation of artificial chimeric enzymes (46,47) to endonucleases that harbor individual amino acid substitutions in the protein–DNA interface (48–55).
Unlike many DNA-binding proteins, engineering and selection of homing endonuclease constructs that display altered DNA-binding specificity (particularly the LAGLIDADG family) is facilitated by two properties: they display relatively modest reductions in affinity when individual contacts to nucleotide bases are eliminated, and they utilize highly modularized and separable contacts between individual amino acid side chains and DNA bases. While this strategy of DNA recognition does not reduce the rules for binding specificity down to the simplicity of a one-to-one ‘code’ between protein residues and DNA bases, it does greatly simplify the complexity of DNA recognition and its redesign.
In order to investigate the exact effect of deleterious single base substitutions on binding specificity and affinity of LAGLIDADG endonucleases, we characterized the differences in binding thermodynamics for two separate LAGLIDADG homing endonucleases (monomeric I-AniI and homodimeric I-MsoI) when complexed to optimal target sites, versus nonoptimal miscognate target sites. For each study, the effect of simultaneously altering two basepairs was measured (sequences of sites shown in Table 1). We then went on to further characterize the thermodynamic effects of a ‘redesign cycle’ for I-MsoI, where amino acid substitutions in the enzyme were engineered with the goal of reacquiring high affinity, specific recognition of the formerly miscognate target sequence.
In the case of I-AniI, the optimal target site for the endonuclease differs from the wild-type, physiologic target site at two basepair positions (Table 1). Both mutations (at positions +1 and +8) consist of an inversion of an A:T basepair and were identified in an in vitro screen for hypercleavable target site variants (13). The endonuclease displays a 12-fold difference in affinity towards these two targets (KD(wild-type) = 96 nM; KD(optimal) = 8 nM), corresponding to a difference in ΔGbinding of −1.5 kcal/mol. The less favorable ΔGbinding to the wild-type site is caused by a substantial increase in the heat absorbed during binding to that site (ΔH is increased by 4.6 kcal/mol), which is partially offset by a more favorable entropic change for the formation of the miscognate complex.
In contrast, the homodimeric I-MsoI displays near optimal binding affinity to its physiological cognate target site. Two simultaneous alterations of that target sequence, consisting of a substitution of −6 C:G to −6G:C in the ‘left’ DNA half-site, and a similar change from +6 T:A to +6 G:C in the symmetry-related ‘right’ half-site, result in significant reduction of cleavage activity under standard reaction protocols. These substitutions were chosen based on structure-based computational predictions of DNA mutations that would cause a significant reduction in binding affinity of the wild-type enzyme (51). At both of these DNA positions, Lys 28 is engaged in a hydrogen bond to the purine ring and Thr 83 makes a water-mediated contact to the pyrimidine (Figure 5A). Converting either base pair to a G:C was predicted to disrupt binding by the loss of the direct hydrogen-bonding interactions and by desolvation of Lys28.
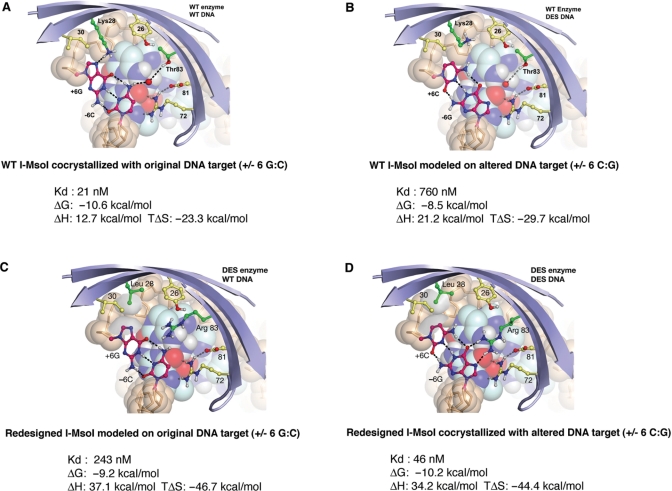
Figure 5.
Thermodynamic signature of a homing endonuclease redesign cycle. The wild-type I-MsoI endonuclease (top left; A) binds to its cognate target site with a Kd of 21 nM, driven by a favorable entropic change (TΔS) upon binding. Simultaneous alteration of a base pair in each half site (position +/− 6) results in a 30-fold increase in Kd, correlated with an unfavorable increase of 2.1 kcal/mol in the free energy of binding (ΔΔG) (top right; B). This energetic penalty is caused by a large unfavorable increase in the enthalpy of binding. Redesign of the enzyme, via two point mutations in the protein/DNA interface (bottom right; D), almost entirely restores affinity and free energy of binding (KD = 46 nM). Finally, analysis of the redesigned enzyme against the original target site (bottom left; C) indicates that the newly created LHE, while displaying a specificity switch, only displays about a 5-fold to 10-fold increase in KD.
I-MsoI displays a 36-fold increase in the dissociation constant KD against the miscognate target site relative to the wild-type target site (from 21 nM up to 760 nM) (Figure 5B). This reduction in affinity corresponds to a 2.1 kcal/mol loss of favorable ΔGbinding (from −10.6 kcal/mol for the wild-type complex to −8.5 kcal/mol for the miscognate site). Similar to I-AniI, the change in ΔGbinding to the miscognate site is caused by a significant increase in the heat absorbed during binding (ΔH rises from 12.7 to 21.2 kcal/mol), which is partially offset by a more favorable entropic change (TΔS decreases from −23.3 to −29.7 kcal/mol).
For both enzymes, the deleterious basepair mismatches are presumed to lead to the loss of hydrogen bonds between protein side chains and DNA bases, and possibly the introduction of interatomic steric clashes in the interface. The total magnitude of unfavorable thermodynamic changes induced by these mismatches is reduced, however, by increased entropy in the miscognate complex: presumably the complexes between the endonucleases and their miscognate target sites display increased torsional and vibrational disorder in both the protein and the DNA at the site of nonoptimal contacts.
The I-MsoI endonuclease was subsequently engineered to compensate for the basepair substitutions at positions +/−6 in the DNA site, by redesign of the surrounding amino acids (Figure 5D). In these studies, a double point mutation in the enzyme, consisting of K28L and T83R, was predicted to reestablish energetically favorable interactions with the DNA bases (51). This computational redesign prediction was validated by crystallographic analyses, and by functional assays for cleavage activity against the novel target site. In the structure of the redesigned cognate enzyme/DNA complex, Leu28 makes a non-polar contact with the C5 of the cytosine rings at the altered basepair positions and Arg83 makes two hydrogen bonds to the corresponding guanine base of the same basepairs.
Armed with crystal structures of the WT:WT and Mutant:Mutant complexes of I-MsoI bound to DNA (Figure 5A and D), the thermodynamic profiles of the redesigned enzyme to its corresponding ‘cognate’ target and to the original wild-type DNA sequence were determined. As described above, the wild-type enzyme displays dissociation constants of 21 and 760 nM to its cognate and noncognate target sites (a 36-fold difference in affinity). In comparison, the redesigned enzyme displays a binding affinity to the mutated DNA target that is almost entirely restored as compared to the original wild-type complex (KD = 46 nM) and a 5.3-fold higher dissociation constant for the original wild-type site (KD = 243 nM). The amount of discrimination between cognate and noncognate sites displayed by the redesigned enzyme is therefore somewhat lower than the wild-type protein, corresponding to a ΔΔGbinding (cognate versus noncognate) of ∼1 kcal/mol.
The near wild-type affinity and ΔGbinding between the redesigned I-MsoI and the altered DNA target site is driven entirely by a significant improvement in the already favorable entropic change upon binding. TΔSbinding is increased by over 20 kcal/mol as compared to the original wild-type binding event, compensating for a significant unfavorable increase in heat absorption (+ΔH) upon binding. This dramatic alteration in the thermodynamic profile of the redesigned protein–DNA pair is probably a result of the introduction of a hydrophobic leucine side chain at residues 28 and 28′ in place of lysine. In both enzyme constructs, residue 28 is solvent-exposed in the unbound enzyme and buried in the DNA complex. In the case of the wild-type protein, the lysine side chain simply exchanges hydrogen bonds with solvent for similar interactions with the DNA. In contrast, binding of the redesigned endonuclease is presumably accompanied by significant desolvation of the solvent-exposed leucine side chain. When the original DNA target sequence is bound by the same redesigned enzyme construct, the less favorable enthalpy of binding is partially negated by the still strongly favorable entropic changes that accompany burial of the hydrophobic leucine side chains. Therefore, the enzyme does not display as large a difference in affinity between cognate and miscognate DNA targets as is observed for the wild-type enzyme. An important take-home message of this analysis is that the replacement of hydrogen-bond contacts with hydrophobic van der Waals contacts in the protein–DNA interface, during protein engineering, may lead to an undesirable loss of specificity and discrimination.
CONCLUDING REMARKS
Homing endonucleases are unique in that they all must arrive at a successful balance of two somewhat contradictory properties: high-enough specificity to avoid toxicity to their biological host, and low-enough fidelity to maximize opportunities for evolutionary transfer and mobility. Each unique structural family of homing endonuclease is largely constrained to a certain biological host range, and has arrived at the optimal balance of these properties relative to the average genomic size of those hosts. The comparative analysis of multiple lineages of these proteins offers many insights into the constraints placed on DNA recognition by such proteins. Additionally, the characterization of thermodynamic profiles of engineered homing endonucleases promises to add detail to this field for future applications in genomic engineering and medicine.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 2.vonHippel PH. From ‘Simple’ DNA-Protein interactions to the macromolecular machines of gene expression. Ann. Rev. Biophys. Biomol. Struct. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arauzo-Bravo MJ, Fujii S, Kono H, Ahmad S, Sarai A. Sequence-dependent conformational energy of DNA derived from molecular dynamics simulations: toward understanding the indirect readout mechanism in protein-DNA recognition. J. Am. Chem. Soc. 2005;127:16074–16089. doi: 10.1021/ja053241l. [DOI] [PubMed] [Google Scholar]
- 4.Michael Gromiha M, Siebers JG, Selvaraj S, Kono H, Sarai A. Intermolecular and intramolecular readout mechanisms in protein-DNA recognition. J. Mol. Biol. 2004;337:285–294. doi: 10.1016/j.jmb.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Heyningen PV, Berman HM, Thornton JM. Protein-DNA interactions: a structural analysis. J. Mol. Biol., 1999:287. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 6.Jayarum B, Jain T. The role of water in protein-DNA recognition. Ann. Rev. Biophys. Biomol. Struct. 2004;33:343–361. doi: 10.1146/annurev.biophys.33.110502.140414. [DOI] [PubMed] [Google Scholar]
- 7.Koudelka GB, Mauro SA, Ciubotaru M. Indirect readout of DNA sequence by proteins: the roles of DNA sequence-dependent intrinsic and extrinsic forces. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:143–177. doi: 10.1016/S0079-6603(06)81004-4. [DOI] [PubMed] [Google Scholar]
- 8.Searle MS, Williams DH. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 1993;21:2051–2056. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayaram B, McConnell KJ, Dixit SB, Beveridge DL. Free energy analysis of protein-DNA binding: the EcoRI endonuclease-DNA complex. J. Comp. Phys. 1999;151:333–357. [Google Scholar]
- 10.Spolar RS, Record M.T.,, Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 11.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 12.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 13.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of homing endonuclease specificity and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlmann UC, Moore GR, James R, Kleanthous C, Hemmings AM. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- 15.VanRoey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanRoey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat. Struct. Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 17.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–696. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 20.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 21.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: Evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L, Bonocora RP, Shub DA, Stoddard BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 2007;26:2432–2442. doi: 10.1038/sj.emboj.7601672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem. Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 24.Flick KE, McHugh D, Heath JD, Stephens KM, Monnat R.J., Jr., Stoddard BL. Crystallization and preliminary X-ray studies of I-PpoI: a nuclear, intron-encoded homing endonuclease from Physarum polycephalum. Protein Sci. 1997;6:2677–2680. doi: 10.1002/pro.5560061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellison EL, Vogt VM. Interaction of the intron-encoded mobility endonuclease I-PpoI with its target site. Mol. Cell. Biol. 1993;13:7531–7539. doi: 10.1128/mcb.13.12.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittmayer PK, Raines RT. Substrate binding and turnover by the highly specific I-PpoI endonuclease. Biochemistry. 1996;35:1076–1083. doi: 10.1021/bi952363v. [DOI] [PubMed] [Google Scholar]
- 27.Ho Y, Kim SJ, Waring RB. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. PNAS USA. 1997;94:8994–8999. doi: 10.1073/pnas.94.17.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonocora R, Shub DA. A novel group I intron-encoded endonuclease specific for the anticodon region of tRNA-fMet genes. Mol. Microbiol. 2001;39:1299–1306. doi: 10.1111/j.1365-2958.2001.02318.x. [DOI] [PubMed] [Google Scholar]
- 29.Landthaler M, Lau NC, Shub DA. Group I intron homing in Bacillus phages SP01 and SP82: a gene conversion event initiated by a nicking homing endonuclease. J. Bacteriol. 2004;186:4307–4314. doi: 10.1128/JB.186.13.4307-4314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker JM, editor. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 31.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallansrud G, Ward B. A comparison of measured and calculated single- and double-stranded oligodeoxynucleotide extinction coefficients. Anal. Biochem. 1996;236:134–138. doi: 10.1006/abio.1996.0141. [DOI] [PubMed] [Google Scholar]
- 33.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 34.Ahmad S, Kono H, Arauzo-Bravo MJ, Sarai A. ReadOut: structure-based calculation of direct and indirect readout energies and specificities for protein-DNA recognition. Nucleic Acids Res. 2006;34:W124–127. doi: 10.1093/nar/gkl104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen BW, Dyer DH, Huang JY, D’Ari L, Rabinowitz J, Stoddard BL. The crystal structure of a bacterial, bifunctional 5,10 methylene- tetrahydrofolate dehydrogenase/cyclohydrolase. Protein Sci. 1999;8:1342–1349. doi: 10.1110/ps.8.6.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean AB, Stanger MJ, Dansereau JT, VanRoey P, Derbyshire V, Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc. Natl Acad. Sci. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landthaler M, Shen BW, Stoddard BL, Shub DA. I-BasI and I-HmuI: two phage intron-encoded endonucleases with homologous DNA recognition sequences but distinct DNA specificities. J. Mol. Biol. 2006;358:1137–1151. doi: 10.1016/j.jmb.2006.02.054. [DOI] [PubMed] [Google Scholar]
- 38.Mueller J, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemieux B, Turmel M, Lemieux C. Unidirectional gene conversions in the chloroplast of Chlamydomonas inter-specific hybrids. Mol. Gen. Genet. 1988;212:48–55. [Google Scholar]
- 40.Turmel M, Boulanger J, Schnare MN, Gray MW, Lemieux C. Six group I introns and three internal transcribed spacers in the chloroplast large subunit ribosomal RNA gene of the green alga Chlamydomonas eugametos. J. Mol. Biol. 1991;218:293–311. doi: 10.1016/0022-2836(91)90713-g. [DOI] [PubMed] [Google Scholar]
- 41.Turmel M, Gutell RR, Mercier J.-P, Otis C, Lemieux C. Analysis of the chloroplast large subunit ribosomal RNA gene from 17 Chlamydomonas taxa: three internal transcribed spacers and 12 group I intron insertion sites. J. Mol. Biol. 1993;232:446–467. doi: 10.1006/jmbi.1993.1402. [DOI] [PubMed] [Google Scholar]
- 42.Biniszkiewicz D, Cesnaviciene E, Shub DA. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turmel M, Cote V, Otis C, Mercier JP, Gray MW, Lonergan KM, Lemieux C. Evolutionary transfer of ORF-containing group I introns between different subcellular compartments (chloroplast and mitochondrion) Mol. Biol. Evol. 1995;12:533–545. doi: 10.1093/oxfordjournals.molbev.a040234. [DOI] [PubMed] [Google Scholar]
- 44.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Stoddard BL, Monnat RJJ, Scharenberg AM. Advances in engineering homing endonucleases for gene targeting: ten years after structures. Prog. Gene Ther. 2007 in press. [Google Scholar]
- 46.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat R.J., Jr, Stoddard BL. Design, activity and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 48.Arnould S, Perez C, Cabaniols J.-P, Smith J, Gouble A, Grizot S, Epinat J.-C, Duclert A, Duchateau P, Paques F. Engineered I-CreIderivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 49.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 51.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat R.J., Jr., Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 53.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sussman DJ, Chadsey M, Fauce S, Engel A, Bruett ARJ, Monnat J, Stoddard BL, Seligman LM. Isolation and characterization of new homing endonuclease specificities at individual target site positions. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 55.Gruen M, Chang K, Serbanescu I, Liu DR. An in vivo selection system for homing endonuclease activity. Nucleic Acids Res. 2002;30:29–34. doi: 10.1093/nar/30.7.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]