Abstract
Methylation of lysine residues on histones participates in transcriptional gene regulation. Lysine 9 methylation of histone H3 is a transcriptional repression signal, mediated by a family of SET domain containing AdoMet-dependent enzymes. G9a methyltransferase is a euchromatic histone H3 lysine 9 methyltransferase. Here, G9a is shown to methylate other cellular proteins, apart from histone H3, including automethylation of K239 residue. Automethylation of G9a did not impair or activate the enzymatic activity in vitro. The automethylation motif of G9a flanking target K239 (ARKT) has similarity with histone H3 lysine 9 regions (ARKS), and is identical to amino acids residues in EuHMT (ARKT) and mAM (ARKT). Under steady-state kinetic assay conditions, full-length G9a methylates peptides representing ARKS/T motif of H3, G9a, mAM and EuHMT efficiently. Automethylation of G9a at ARKT motif creates a binding site for HP1 class of protein and mutation of lysine in the motif impairs this binding. In COS-7 cells GFP fusion of the wild-type G9a co-localized with HP1α and HP1γ isoforms whereas the G9a mutant with K239A displayed poor co-localization. Thus, apart from transcriptional repression and regulatory roles of lysine methylation, the non-histone protein methylation may create binding sites for cellular protein–protein interactions.
INTRODUCTION
Nuclear DNA in eukaryotes is packaged with histones, a basic scaffolding protein, to form chromatin. The basic unit of the chromatin is the nucleosome which consists of 147 bp of DNA superhelically wound around a histone octamer composed of two copies of each core histones H2A, H2B, H3 and H4 (1). The N-terminal tail regions of histones in a nucleosome are unstructured in crystallography studies (2) and are accessible for enzymatic modification (3–5). Indeed, histone molecules are modified at a post-translational level by phosphorylation, ubiquitination, acetylation and methylation. Methylation was shown to occur at the ε-amino group of lysine (K) residues (6) and guanidino group of the arginine (R) residues (7) by a variety of AdoMet-dependent, enzymes (8). Both methylation and acetylation of the histone molecules are now recognized as important covalent modifications in eukaryotic chromatin, which can regulate gene expression. Histone molecules contain several K and R residues and many of them are methylated in vivo. Acting individually or in combination both methylation and/or acetylation of histones, in conjunction with DNA methylation are believed to implement an epigenetic means of gene function affecting processes such as transcriptional activation, repression, heterochromatin formation, DNA replication and repair. These modifications facilitate recruitment of various regulator proteins that can recognize different covalent modifications in a sequence-dependent manner.
Chromatin modification is dynamic, and largely dependent on gene expression profile of the cell. All the chromatin modifications are well coordinated and undertaken by a number of histone modification enzymes, and the enzymes that remove those modifications (9). The lysine 9 residue of histone H3 (H3K9) is a well-studied modification that is implicated in transcriptional repression and heterochromatin formation in Arabidopsis (10), Neurospora (11), yeast (12) and mammalian (13) cells. H3K9 methylation is also present in transcriptionally active region of the mammalian chromatin (14,15). The lysine residues are mono-, di- or trimethylated in vivo. Several lysine 9 histone H3 methyltransferases, such as SUV39H1, SUV39H2, EuHMT/GLP, SETDB1/ESET and G9a have been identified and well studied in mammals. Although the substrate specificity of these enzymes was determined by a combination of genetic and biochemical studies to be H3K9, one enzyme cannot rescue the other's function in vivo. For example, the G9a knockout mouse embryos die at 8.5 days postcoitum (16). Furthermore, G9a null embryo stem (ES) cells display altered DNA methylation in the Prader–Willi imprinted region and ectopic expression of the Mage genes (17). Similarly, the homozygous mutations of Eset resulted in periimplantation lethality between 3.5 and 5.5 days postcoitum (18). Both the mouse (Suv39h1 and Suv39h2) and human (SUV39H1 and SUV39H2) genes encode enzymes that can trimethylate histone H3 lysine 9 to create a binding site for heterochromatic protein, HP1 (19,20). Genetic disruption of Suv39h1 and Suv39h2 led to severe decrease of the H3K9me3 at the pericentric heterochromatic regions (21). Similar to Suv39h1, G9a is capable of H3K9 trimethylation in vitro (22,23). G9a is believed to be the euchromatic histone methyltransferase. Genetic disruption of G9a in cells led to reduction of H3K9me and H3K9me2 patterns in euchromatic regions (24). Therefore, it is possible that the histone methyltransferases may have roles beyond H3K9 methylation or they perform H3K9 methylation in a development specific manner. It is plausible that the known histone methyltransferases may have other substrate specificities.
In this study we have used recombinant murine G9a, and attempted to determine its substrate specificity. Surprisingly, we discovered that G9a is capable of automethylation. Mutational analyses of the target lysine residues were undertaken to determine their implication in catalysis and protein–protein interaction. Furthermore, cellular localization studies were conducted to identify the link between G9a automethylation and heterochromatin binding.
MATERIALS AND METHODS
Cell culture, constructs and protein purification
Parental HCT116 (colorectal carcinoma) cells, Hela cells, NIH3T3 cells were purchased from the American Type Culture Collection (ATCC). DNMT1−/− cells were kindly provided by Bert Vogelstein (Johns Hopkins University, Baltimore). All cells were incubated at 37°C in a 5% CO2 humidified atmosphere and propagated in Mac Coy's 5A modified medium (ATCC) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin and streptomycin. A total of 100 μg/ml hygromycin were added to the DNMT1−/− cells (25). Cell extracts were made with RIPA buffer (50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 0.02% NaN3 and 1% NP-40).
The MBP-G9a▵634 construct and protein purification methods are described in Esteve et al. (26). All GST fusion constructs with various fragments of the G9a were made using pGEX5.1 vector (GE Healthcare). The specific fragments of G9a were amplified using vent DNA polymerase (New England Biolabs, NEB). Mutation(s) were generated using mutant PCR primers using vent DNA polymerase. Primer sequences used for generation of recombinant clones and mutants are available on request. The G9amut (K233/239/243A) was cloned in frame into pVIC1 (NEB) and named as pVICG9amut. The pVICG9amut was transfected into Sf9 cells. The insect cell culture and baculovirus-mediated G9afl and G9amut protein expression were performed as described before (22). Purification of the G9afl and G9amut were essentially same using NEB chitin magnetic beads (22). GST fusion G9afl (GST-G9afl) and G9afl (K167A) (GST-G9afl (K167A)) were expressed in Escherichia coli and purified using glutathione Sepharose high-performance resin (GE Healthcare).
Histone methyltransferase assay and fluorography
Histone methyltransferase assays were carried out at 25°C for 3 min in duplicate in a total volume of 25 µl. A typical reaction contained S-adenosyl-l- [methyl-3H] methionine (AdoMet) (specific activity 15 Ci/mmol, GE Healthcare), substrate peptide and enzyme in assay buffer (50 mM Tris–HCl, pH 9.0, 5 mM MgCl2, 4 mM DTT, 7 µg/ml PMSF). For kinetic analysis, 1 nM peptide with one target lysine is equal to 1 nM methyl group (22). The efficiency of [3H] incorporated methylated DNA was ∼55% and all calculations were corrected accordingly. Data obtained were plotted by regression analysis using the GraphPad PRISM program version 4c (GraphPad Software Inc.).
All the peptides used for histone methyltransferase assay were made at NEB. The peptides represented the following sequences (Wt-H3: CARTKQTARKSTGGKAPRK; G9a: CGQPKVHRARKTMPKSV; EuHMT: CADVKVHRARKTMPKSV and mAM: CPQKKVFKARKTMRVSD). G9aK167 (aa 160–175): IVLGHATKSFPSSPSK. All the histone modification peptide fragments were as follows (H3K9me2: CARTKQTARK(me2)STGGKAPRK; H3K4AK9me2: CARTAQTARK(me2)STGG; H3K4ac: CARTK(ac)QTARKSTGG; H3K4me3: CARTK(me3)QTARKSTGG; H3S10p: CARTKQTARKS(p)TGG; H3T11p: CARTKQTARKST(p)GG; Wt-H3(1–13): CARTKQTARKSTGG; H3K4A: CARTAQTARKSTGG), where p, ac and me indicate phosphorylated, acetylated and methylated amino acid residues, respectively. Peptides were purified by HPLC and the intactness was measured by mass spectroscopy. All peptides for assay were dissolved in milliQ water.
For fluorography, the radioactive reaction mixtures of 20 µl (20 µg of cell extracts or GST-fusions, 7 µM S-adenosyl-l- [methyl-3H] methionine and 80 nM recombinant G9a at 25°C for 1 h) were resolved in a 4–20% SDS/PAGE. The radioactive gel was stained with Coomassie staining for visualization followed by treatment with EN3HANCE as suggested by the supplier (Perkin Elmer). The gel was air dried between cellophane and was exposed to Kodak BioMax MS Film with a Kodak BioMax TranScreen-LE intensifying screen. Recombinant human histones were from NEB.
G9a methyltransferase protein/peptide digestion, separation and LC-MS/MS
The baculovirus expressed G9a protein (1 µg in 5 µl) was added to 2 µl of trypsin reaction buffer and digested overnight at 37°C with trypsin (ratio: 20:1; NEB). Multiple injections of 1–8 µl of this sample were carried out into an 1100 Series Nanoflow LC System (Agilent Technologies) and separated on a C18 reversed-phase column (150 µm × 150 mm, 300; Vydac), using a 40 min 5–45% B linear gradient (FA = water, 0.1% formic acid, FB = CH3CN, 0.1% formic acid) at a flow rate of 0.3 µl/min. Multiply charged peptide-ions were automatically chosen and fragmented in an XCT Ion Trap Mass Spectrometer (MS) with a Nano-Electrospray ionization (nanoESI) source (Agilent Technologies) and the data collected and stored for later analysis.
Protein identification using MS and MS/MS Data
The MS/MS fragmentation data were analyzed using Spectrum Mill (Agilent Technologies), X! Tandem and the web version of Mascot (Matrix Science) url: http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=MIS. For the Spectrum Mill, X! Tandem and Mascot analyses, data were searched against the NCBI non-redundant database and peptides generated by a tryptic digest with a maximum of four missed cleavages were considered. Spectrum Mill identified peptides were filtered by a score greater than seven and a percent scored peak intensity (% SPI) greater than 60. This Spectrum Mill identified peptides were then used to build proteins and those proteins scoring greater than 20 were considered valid identifications. Any proteins scoring less than 20 were also considered if more than five spectra were associated with one peptide. For the Mascot analysis, peptides generated by a trypsin cleavage were also considered and proteins identified with score of 60 or higher were considered valid identifications. X! Tandem was run with the default parameters. After identification of the protein(s) present in the sample, an additional analysis was performed allowing for modification of the peptides from the identified proteins.
HP1 protein expression and GST pull-down assay
GST fusion HP1α, HP1β and HP1γ clones were overexpressed in E. coli using 0.3 mM IPTG overnight at 16°C. The cells were harvested, sonicated and incubated with GST Sepharose beads (GE Healthcare). The bound proteins were washed twice with 1 × PBS supplemented with 1% Triton X-100, twice more with 1 × PBS supplemented with 0.1% Triton X-100. The fusion proteins bound to beads were washed finally with PBS and stored at 4°C. The pull-down assays were performed as described before (27).
Immunofluorescence
COS-7 cells were cultured onto coverslips and transfected with a mixture of plasmid and Transpass D2 reagent (NEB) at a ratio of 1:3 µg/µl. The cells were visualized after 48 h using a Zeiss 200 M microscope with a 63 × oil objective lens at 488 nm for GFP-G9a and GFP-G9amut fusion proteins, 568 nm for HP1α and HP1γ detection and 460 nm for nuclear staining using Hoechst 33342. Briefly, cells were first fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. After three washes with PBS, slides were blocked by 5% BSA in 1 × PBS supplemented with 0.1% Tween 20 for 1 h at room temperature. For HP1α and HP1γ detection cells were incubated overnight at 4°C in 1% BSA PBS/Tween 0.1% with anti- HP1α and HP1γ rabbit polyclonal antibodies (CST). After three washes with PBS, anti-rabbit IgG conjugated with Alexa Fluor 594 (Molecular Probes) was added for 1 h before staining the slides with Hoechst 33342.
RESULTS
Wider substrate specificity of murine G9a methyltransferase
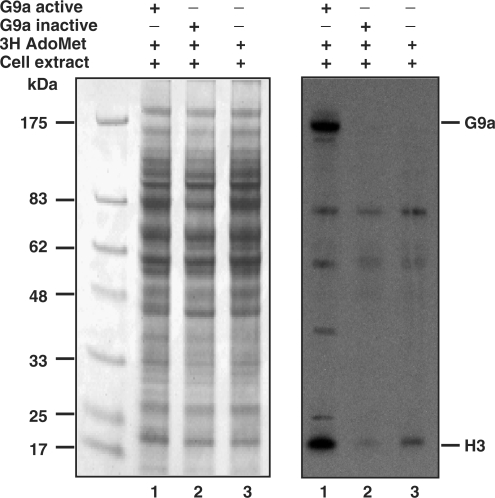
Previously, we have studied the substrate specificity and steady-state kinetic properties of the murine G9a in detail. The enzyme is capable of mono-, di- and trimethylation of the target lysine residues on the amino terminal tail of histone H3 (22). Subsequent in vitro biochemical studies suggested that the substrate recognition and sequence specificity of G9a involve seven amino acids (TARKSTG) of the histone H3 tail and that K9 methylation is impaired by S10 or T11 phosphorylation (28). In the mammalian proteome similar motifs are expected to be found in protein other than histone H3. To determine if the G9a methyltransferase is capable of methylating other cellular proteins, we incubated purified recombinant G9a enzyme with HCT116 plus tritiated AdoMet co-factor. The radioactive protein samples were separated on a SDS gel and fluorographed to visualize G9a-mediated protein methylation. Surprisingly, several prominent radioactive protein bands were visible in autoradiography after overnight exposure of the film (Figure 1). The apparent molecular weights of these bands were between 7 and 175 kDa (Figure 1, right panel, lane 1). Histone H3 molecules were radioactively labeled and visible at 17 kDa region as expected. Experiments with heat killed recombinant G9a in the reaction mixture or without additional G9a enzyme did not display strong radioactive bands (Figure 1, right panel, lanes 2 and 3). The minor bands observed in the presence of heat killed G9a and extract alone suggests the cell extracts contain other protein methyltransferases. Several signals on the autoradiograph are specific to recombinant G9a-mediated tritiated methyl group incorporation. Similar autoradiography profile was observed with HCT116 DNMT1 null, mouse 3T3 and Hela cell extracts (data not shown), suggesting G9a is capable of methylating proteins other than histone H3.
Figure 1.
Multiple substrate specificity of recombinant G9a. HCT116 cell extracts were incubated with recombinant G9a, heat killed G9a and tritiated AdoMet as indicated with a plus symbol. The radiolabeled protein mixture was separated on denaturing PAGE, stained and fluorographed. The Coomassie-stained gel is shown at the left panel and the radioactive proteins are seen as specific dark bands at the right panel. The apparent position of G9a and H3 are shown at the right. The molecular weight markers are on the left.
Murine G9a is automethylated in amino terminus lysine residues
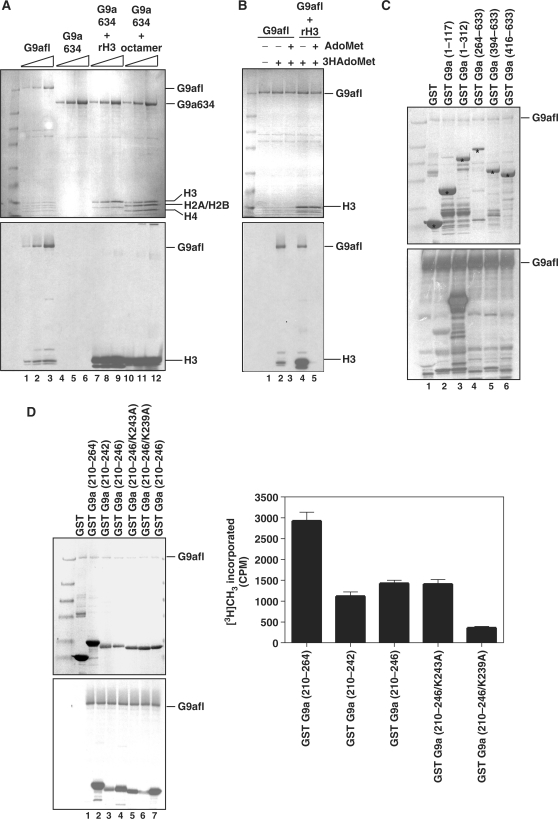
The highest molecular mass radioactive bands appeared ∼175 kDa, the apparent molecular weight of purified murine recombinant G9a. To determine if G9a is automethylated, we incubated full-length G9a (G9afl) or MBP-G9a▵634 (G9a▵634) alone with tritiated AdoMet, or G9a▵634 with recombinant H3 and tritiated AdoMet, and G9a▵634 with in vitro reconstituted recombinant octamer plus tritiated AdoMet. If G9a is capable of methylating itself, the tritiated methyl group will be incorporated to the protein and can be detected by fluorography. The fluorography showed a dark band coinciding with the G9afl in the SDS gel at an apparent molecular weight of 175 kDa (Figure 2A, lanes 1–3). A small amount of insect cell histone H3 that co-purifies with G9a also was methylated, as viewed in the bottom of the gel. G9a enzyme lacking the first 633 amino acids, G9a▵634, did not display any dark band implying that the truncated enzyme is incapable of automethylation (Figure 2A, lanes 4–6). The G9a▵634 with rH3 or recombinant nucleosome showed methylation of the H3 (Figure 2A, lanes 7–12). To demonstrate that methylation is dependent on AdoMet cofactor, we incubated G9afl with tritiated AdoMet or a combination of tritiated AdoMet and 100 fold excess cold AdoMet. If the automethylation reaction is AdoMet dependent, excess cold AdoMet will compete for the AdoMet-binding site of the enzyme and will result in incorporation of the cold AdoMet. Incorporation of cold AdoMet will render either a poor or no G9a signal on autoradiography film. Indeed, in the presence of excess cold AdoMet the autoradiography signal for G9a (Figure 2B, lanes 2 versus 3) or H3 was not apparent (Figure 2B, lanes 4 versus 5).
Figure 2.
Automethylation of G9a. (A) Coomassie-stained gel showing all the components in the in vitro automethylation assay on the top panel. G9afl indicates the full-length recombinant murine G9a, G9a634 is the 633 amino acids deletion MBP fusion murine G9a (G9a▵634). Recombinant histones were used for in vitro assembly of the octamer. Histones and enzymes are indicated on the right of the gel or fluorography image shown in the bottom panel. Lanes 1, 2 and 3 show co-purified insect histones that eluted with the enzyme preparation. The enzyme concentration was 20, 40, 80 nM. (B) AdoMet dependency of automethylation by recombinant G9a. Cold AdoMet or tritiated AdoMet [3H AdoMet] are shown on the top along with the reaction components. The top panel is the Coomassie-stained gel and the bottom shows its autoradiograph. (C) Mapping of the automethylation site in the amino terminus of G9a. The GST fusion constructs representing various fragments of G9a are shown on top along with amino acid numbers in parenthesis. The top panel is the Coomassie-stained gel of the reaction mix along with G9afl. The fusion proteins are marked with an asterisk. The bottom panel is the fluorography image of the gel. (D) Mutational analysis of the lysine amino acid residues of the amino terminus region of G9a. The GST fusion constructs representing various fragments and point mutations of G9a fragments are shown on top along with amino acid numbers in parenthesis. The top panel is the Coomassie-stained gel of the reaction mix along with G9afl. The bottom panel corresponds with the fluorography image of the gel. The right panel shows relative radioactivity of the fusion fragments. GST-G9a (210–264) is plotted as 100%, and its fusion derivatives are shown.
To dissect the exact sequence for automethylation, we constructed several GST fusion deletion mutants of the first 633 amino acids of G9a (Figure 2C). Each GST-fusion G9a peptide fragment was incubated with the full-length G9a in the presence of the tritiated AdoMet and the reaction mixtures were separated on a gel and fluorographed. G9afl did not methylate GST alone or fusions containing 1–117 or 264–633 amino acids (Figure 2C, lanes 2 and 4). This suggests that the methylated lysine residue/(s) reside between amino acids 118–263. Further deletion analysis demonstrated amino acids between 210 and 264 are methylated in vitro. We cloned amino acids 210–246 as GST fusions and generated point mutants (K243A and K239A). The wild type and point mutant GST fusions were purified, in vitro methylated by G9afl, and fluorographed to determine the amino acids crucial for methylation (Figure 2D). Indeed K239A reduced the methylation significantly in fluorography (Figure 2D, lane 7). Furthermore, we measured the radioactive incorporation of each band. The GST-G9a (210–264) displayed strong radioactive incorporation and the incorporation was impaired in GST-G9a (210–264/K239A) suggesting K239 may be the methylated amino acids residue of G9a (Figure 2D, right panel). Deletion or mutation of amino acids between 243 and 264 also resulted in poor methylation of the GST fusion fragment. For example mutation of K243 residue [(GST-G9a (210–246/K243A)] resulted in decrease in K239 methylation. Therefore, the surrounding amino acids at K239 may be important for automethylation.
Mass spectroscopic analysis of post-translational modification of G9a
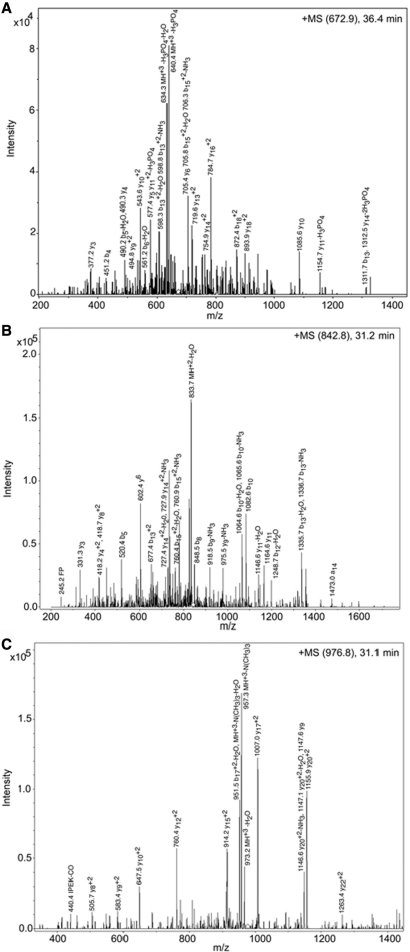
To establish the authenticity of the methylated lysine residue of G9a, the tryptic fragments of the purified G9a were subjected to mass-spectroscopy analysis. Mascot, Spectrum Mill and X! Tandem (29) analysis identified the major protein present in the sample as the murine G9a methyltransferase. All three programs identified a serine (residue S194) as being in both a phosphorylated and unmodified state in multiple peptides. Figure 3A shows a spectrum obtained from a phosphorylated peptide. Mascot, X! Tandem and Spectrum Mill also identified a dimethyllysine at residue 167 (K167me2) only in the modified form (Figure 3B). Mascot and Spectrum Mill also identified a trimethyllysine at residue 239 (K239me3) only in the modified form (Figure 3C). The spectra of the peptide containing a mass 42 modification has a characteristic loss of mass 59 known to be associated with trimethyllysine (30) hence distinguishing it from an acetylation.
Figure 3.
Analysis of the post-translational modifications of G9a by mass spectroscopy. Three post-translationally modified peptides were observed from digestion and nano-ESI-MS/MS analysis. MS/MS Ion Trap spectra: The major ions are labeled with the corresponding m/z value, a or b or y ion fragments and neutral loss species where they could be assigned. The observed b and y ions are indicated by \ and / and where both were present. Any position that gave rise to a neutral loss species is indicated in bold. (A) Phosphoserine-containing peptide. The MS/MS spectrum of an ion with m/z value of 672.9, corresponding to a G9a tryptic peptide that eluted at 36.4 min. M S M/TG/AG/K Sp/P/P/S VQ/SL/A/M/R where Sp = phosphoserine (B) Dimethyllysine-containing peptide. The MS/MS spectrum of an ion with m/z value of 842.8, corresponding to a G9a tryptic peptide that eluted at 31.2 min. IVLGH\ATKdS/FPS/S/P/S/K where Kd = dimethyllysine. (C) Trimethyllysine-containing peptide. The MS/MS spectrum of an ion with m/z value of 976.8, corresponding to a G9a tryptic peptide that eluted at 31.1 min. Kt T M S K P S NG/Q/P P/I P E/K R/P P E/V Q/H F R where Kt = Trimethyllysine.
K167 is surrounded by amino acids that are similar to K239 (K167: ATKS; K239: ARKT). Therefore, we performed a methyltransferase assay with a peptide spanning amino acids 160–175 of G9a (G9aK167). We did not observe methyl group incorporation to the peptide even after 1 h of incubation. This result demonstrates that G9a automethylation involves K239 residue (Supplementary Figure 1).
Mutation of methylated lysine and G9a enzymatic activity
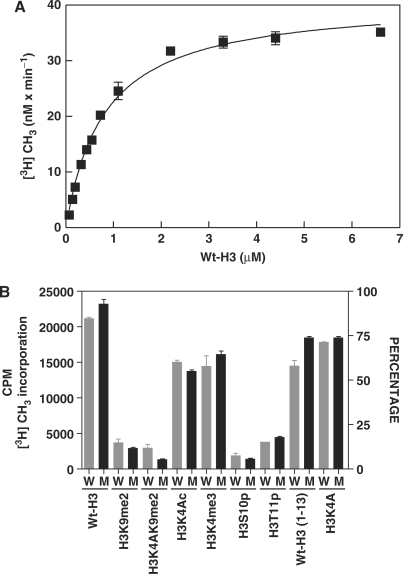
Recently, it was shown that lysine methylation regulates the function of non-histone proteins such as p53 (31,32). Thus, to study if there is a functional role of lysine automethylation on methyltransferase activity of G9a, we generated point mutations at the K233A, K239A and K243A of G9a. The mutant G9a (G9amut) was expressed in the baculovirus expression system. The enzyme was purified and steady-state kinetic properties were studied with a peptide representing the amino terminus (1–17 amino acids) of histone H3 (Wt-H3) (Figure 4A). The Michaelis–Menten kinetic constants were determined for the G9a mutant (G9amut) by using non-linear regression analysis and compared with that of the G9a wild-type enzyme and reported in Table 1. The turnover numbers, Km and catalytic efficiency values for G9amut were comparable to that of the wild-type enzyme. These results suggest that mutation of the lysine cluster does not affect the catalysis of the enzyme. However, deletion of the first 633 amino acids resulted in significant loss of catalytic activity (Table 1, 24).
Figure 4.
Steady-state kinetic analysis and methylation properties of mutant G9a methyltransferase. (A) Substrate (Wt-H3) concentration is on X-axis and the reaction velocity on Y-axis. The reaction was performed at 25°C with 25 nM G9amut enzyme using filter-binding assay. (B) Comparison of methylation by G9afl and G9amut on various substrates. The substrates and their sequences along with modifications are indicated at the bottom and in the Materials and Methods section. W is G9afl and M is G9amut enzyme. Twenty-five nanomolar of either G9afl or G9amut was used in the assay.
Table 1.
Comparison of steady-state kinetics of G9amut with wild-type and amino terminus deletion enzyme
| Substrate | kcat (h−1) |
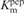 (µM) (µM) |
kcat/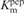 (106 × M−1 h−1) (106 × M−1 h−1) |
Reference | |
|---|---|---|---|---|---|
| G9afl | Wt-H3 | 88 ± 4 | 0.9 ± 0.1 | 98 | 20 |
| G9amut | Wt-H3 | 98 ± 2 | 0.8 ± 0.05 | 122.5 | Present work |
| G9a▵634 | Wt-H3 | 24 ± 1.4 | 0.3 ± 0.05 | 80 | 24 |
We also performed methyltransferase assay with GST-G9afl and GST-G9afl (K167A) along with Wt-H3 to determine if the K167 has any role in enzymatic activity. Under steady-state conditions both GST-G9afl and GST-G9afl (K167A) displayed similar properties. The kcat values for GST-G9afl and GST-G9afl (K167A) were 78 h−1 and 66 h−1, respectively (Supplementary Figure 2).
Since modification of proximal amino acids of the H3 tail affected catalysis of the wild-type G9a (28), we further examined if the G9amut will be affected by similar substrate modifications. Both wild-type G9a and G9amut were incubated separately with the same substrates such as H3K9me2, K4 acetylated (H3K4Ac), S10 phosphorylated (H3S10p) and T11 phosphorylated (H3T11p) substrates. Both enzymes incorporated methyl group in a similar efficiency, and S10 or T11 phosphorylated peptide inhibited both enzymes (Figure 4B). These data confirm that K239me3 does not impair or activate enzymatic catalysis of G9a.
G9a methylates other H3K9 methyltransferase and methyltransferase activator protein
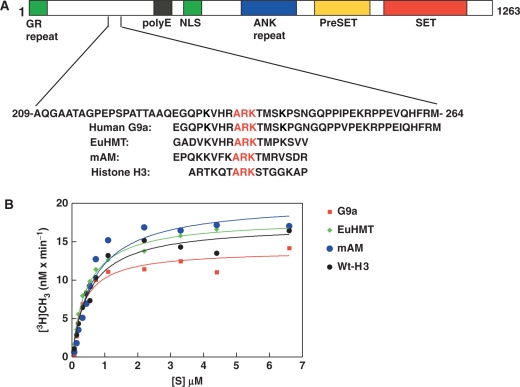
The methylated lysine of G9a is conserved between human and mouse G9a. A homology search of automethylation motif of G9a in the database yielded matches with histone H3, EuHMT and mAM, a SetDB1 methyltransferase activator protein (33) (Figure 5A). Both EuHMT and mAM displayed similar consensus (ARKT) motifs with histone H3 and G9a. Since G9a can methylate histone H3, and is automethylated in the ARK motif, it is possible that G9a may methylate EuHMT and mAM. To validate this hypothesis, we performed steady-state kinetic analysis of peptides representing ARK motif of G9a, EuHMT, mAM and control histone H3 peptide (Wt-H3) (Figure 5B). The steady-state kinetic parameters are presented in Table 2. The turnover numbers for peptides representing G9a, EuHMT, mAM and histone H3 were 66, 86, 98 and 82 h−1. The Km value remained between 0.4 and 0.7 µM. These data suggest that G9a enzyme is capable of automethylation and methylation of other H3K9 methyltransferases.
Figure 5.
G9a is capable of methylation of the other H3K9 methyltransferases. (A) A schematic diagram showing various domains of murine G9a methyltransferase. Repeated GR repeats at the extreme amino terminus region in green, poly E residues in black, nuclear localization signal (NLS) in green, ANK repeats in blue, preSET domain in yellow and SET domain in red are shown. The amino acids surrounding the automethylated K239 is shown along with conserved ARKT/S motifs of histone H3K9 methyltransferase EuHMT, mAM and histone H3. The conserved motifs are underlined. (B) Steady-state kinetic analysis of peptides representing the conserved ARKT/S motif of G9a (red square), EuHMT (green diamond), mAM (blue filled circle) and Wt-H3 (black filled circle) are shown. The peptide concentration [S] is shown on the X-axis and the incorporation at the Y-axis.
Table 2.
Comparison of steady-state kinetics of G9afl on various ARKT/S motifs from other enzymes or histone H3
| Motif | kcat (h−1) |
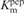 (µM) (µM) |
kcat/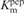 (106 × M−1 h−1) (106 × M−1 h−1) |
|
|---|---|---|---|---|
| G9afl | ARKT | 66 ± 2 | 0.4 ± 0.04 | 165 |
| EuHMT | ARKT | 86 ± 2 | 0.5 ± 0.05 | 172 |
| mAM | ARKT | 98 ± 4 | 0.7 ± 0.05 | 140 |
| H3 | ARKS | 82 ± 2 | 0.6 ± 0.05 | 137 |
Methylated ARK motif of G9a acts as an anchor to HP1
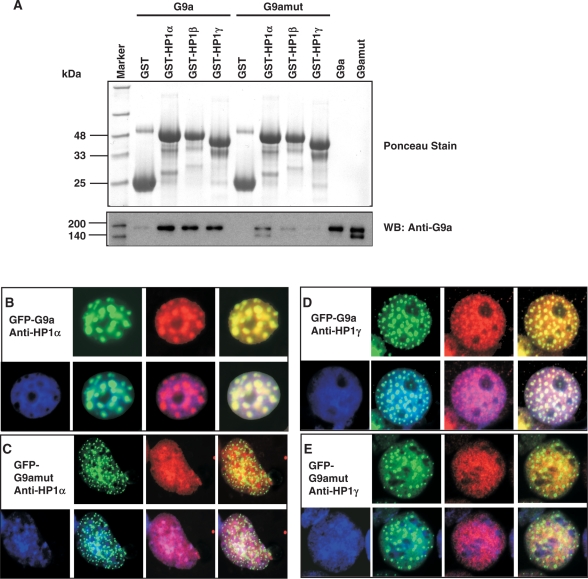
Since G9a automethylates K239, it creates a motif very similar to methylated H3 tail (G9a: ARKme3T; H3: ARKme3S). The similarity of these amino acids residues between G9a and H3 led us to investigate if G9aK239me3 would act like H3K9me3 and bind to HP1. To examine the HP1 interaction with G9a, we performed GST pull-down assay with immobilized GST-HP1α, GST-HP1β and GST-HP1γ. A fixed amount of G9a wild type or G9amut was added to the beads and incubated for a fixed time followed by three washes to remove unbound enzymes. The bound proteins were separated on a SDS/PAGE and western blotted with anti-G9a antibody. Indeed, all the three isoforms of HP1 were able to bind strongly to the wild-type G9a that predominately contained K239me3, as compared to the G9amut (Figure 6A). This suggests that the K239 trimethylation creates an anchoring site for HP1α, HP1β and HP1γ, similar to H3K9me3-HP1 binding.
Figure 6.
Automethylated G9a binds to HP1 family of proteins. (A) GST pull-down assay with G9afl (automethylated) or G9amut containing K239A (unmethylated) enzyme. The top panel shows ponceau stain of the western blotted membrane demonstrating similar loading of GST-HP1α, GST-HP1β and GST-HP1γ. The blot was probed with anti-G9a as indicated. The molecular weight markers are on the left. Immunofluorescence detection of co-localization events are as follows (B) GFP-G9a and anti-HP1α; (C) GFP-G9amut and anti-HP1α; (D) GFP-G9a and anti-HP1γ; (E) GFP-G9amut and anti-HP1γ. Both HP1α and HP1γ are shown in red.
To validate whether the wild-type G9a associates with HP1 class of proteins in cells and if the association will be impaired by mutation of K239, we transfected GFP-G9afl or GFP-G9amut constructs into COS-7 cells and after 48 h, the cells were fixed and probed with either anti-HP1α or HP1γ. HP1α and HP1γ have different chromatin-binding properties. HP1α binds predominantly to pericentric heterochromatin, whereas HP1γ binds to both heterochromatin and euchromatin. The HP1α appeared as red punctate nuclear spots throughout the nucleus corresponding to the densely stained heterochromatic region except the nucleolus, which remained dark. A similar green punctate nuclear spot appeared in the nuclei of the GFP-G9afl transfected cells. Superimposition of the GFP-G9afl with HP1α resulted in yellow nuclear spots throughout the nucleus except the nucleolus, which remained dark (Figure 6B). However, the GFP-G9amut displayed much smaller green punctate pattern and did not co-localize with HP1α. Similar sets of co-localization patterns between GFP-G9afl and HP1γ (Figure 6C) and GFP-G9amut and HP1γ (Figure 6D) were observed. We have also performed co-localization studies with GFP-G9a and HP1β, and GFP-G9amut and HP1β, and have seen similar co-localization (data not shown). These data suggest automethylation of K239 is an effective mediator for HP1 and G9a interaction.
DISCUSSION
Lysine methylation is one of the robust histone modifications in eukaryotic chromatin. Various lysine modifications take central roles in conferring epigenetic control to the chromatin template during cellular processes. Recently, several histone lysine demethylases have been identified (34), and it thus appears that the dynamic balance between histone lysine methyltransferases and lysine demethylases regulates the abundance and stability of histone lysine methylation, thus, regulating gene expression.
G9a in both human and mouse are large proteins with very distinct domains. For example, the murine G9a is ∼1263 amino acids long with amino terminus poly E repeat followed by nuclear localization signal, ANK repeat, preSET and SET domain. The SET domain is the catalytic center of the enzyme and point mutation of the conserved R1097, W1103, Y1138, R1162 and C1168 makes it catalytically impaired (26). The biological function of the entire region of the amino terminus is unknown. Deletion of amino terminus 634 amino acid residues makes the enzyme catalytically impaired but not inactive (26). Thus, it is plausible that a cooperation between amino and carboxy terminus dictates the catalytic activation status of the enzyme. Lysine residues in protein undergo autoacetylation and automethylation. It is demonstrated that autoacetylation of HAT p300 facilitates its dissociation from the promoter to allow TFIID promoter occupancy (35). In this study, we have reported automethylation activity of murine G9a. Till date there is one other automethylation example of histone methyltransferase, PRMT6 (36). The biological significance of PRMT6 automethylation is unknown. So why did a histone methyltransferase automethylate itself? The answer may lie in the domain that is methylated, and how a methylated and unmethylated protein interacts with their neighbors during cellular processes. We postulate that the biological consequence of G9a automethylation alone or in context of other modifications (phosphorylation), might be dictated by other proteins or physiological status of the cell. G9a automethylation at ARKT motif makes it an ideal candidate for interaction with HP1. Binding of G9a to HP1 perhaps creates a platform for recruitment of transcriptional repressor complexes such as DNMT1 and HDAC1 (37). Indeed, DNMT1 is generally found in the pericentric heterochromatin of mammalian cells and can be loaded onto DNA between G2-M phase in a replication independent manner (38). We have also demonstrated that DNMT1 is capable of binding to G9a for coordinated histone and DNA methylation during DNA replication (39) and transcriptional repression of survivin gene (40). A plausible scenario would be that the condensed pericentric heterochromatin maintains its silenced status by recruiting DNMT1-G9a-HP1 trimeric complex to maintain residual H3K9 methylation and insulating from unwanted events that may alter the repressive status. Once the H3K9me3 marks are fully established by G9a alone, or in combination with SUV39H1, it falls off and HP1 remain recruited to heterochromatin, presumably before the next cell cycle. The HP1-H3K9me3 binary complex may further recruit the de novo methyltransferase DNMT3B to direct DNA methylation of the major satellites in the endogenous C-type retrovirus (41). A recent report also demonstrates that HP1 can mediate gene silencing via direct interaction with DNMT1 (42). Therefore, HP1 may be a key silencing element that would be necessary to maintain intact epigenetic silencing status from generation to generation.
We have also demonstrated that G9a is capable of methylation of other proteins such as EuHMT and mAM at the amino terminus with a similar ARKT motif. Therefore, it is possible that ARKT motif methylation of other histone methyltransferases may have similar affinity for HP1 interaction. Furthermore, other cellular proteins may recognize methylated lysine on G9a. Cellular proteins with chromo, tudor, MBD and PHD domain can recognize and bind to Kme3 proteins and modulate their function(s). For example G9a and EuHMT form functional heterodimeric complex in vivo and steady-state level of G9a decreases in cells lacking EuHMT (43). G9a appears to be more stable in G9a/EuHMT heterodimeric complex than when expressed alone. One of the possible explanations of the mechanism of dimer formation may start from enzyme–substrate interaction between G9a and EuHMT. If G9a methylate EuHMT, then both proteins will be in the close proximity of each other facilitating a heterodimer formation via interaction of their SET domain. This heterodimeric complex may direct H3K9 methylation of the euchromatic region. Another implication of G9a-mediated methylation of mAM may be to influence SETDB1 methylation. It is known that mAM facilitates ESET catalysis from di- to trimethyl at H3K9 both in vitro and in vivo (33). Since most of our in vitro studies were carried out with peptide segments representing EuHMT and mAM, further validation with full-length mAM and EuHMT is needed.
In summary, we show that the euchromatic histone methyltransferase G9a automethylates and methylates other H3K9 methyltransferases. The methylation events by G9a create a binding site for HP1, and thus, creating another layer of silencing effector molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The HP1α, HP1β and HP1γ expression constructs were obtained from Dr Naoko Tanese, for which we are thankful. We thank Dr Jim Samuelson and Dr Jeong Kyong Kim for discussion and critical reviews. We thank Dr D. G. Comb and Dr Rich Roberts of New England Biolabs, Inc. for their support and encouragement. Funding to pay the Open Access publication charges for this article was provided by New England Biolabs, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 4.Imhof A. Histone modifications: an assembly line for active chromatin? Curr. Biol. 2003;13:R22–R24. doi: 10.1016/s0960-9822(02)01383-0. [DOI] [PubMed] [Google Scholar]
- 5.Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat. Struct. Mol. Biol. 2005;12:110–112. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 6.Murry K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 7.Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem. Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr. Opin. Struct. Biol. 2006;16:753–760. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I. Histone modifications in Arabidopsis- high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003;33:471–480. doi: 10.1046/j.1365-313x.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- 11.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 13.Horn PJ, Peterson CL. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- 14.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J. Role of histone methyltransferase G9a in CpG methylation of the Prader–Willi syndrome imprinting center. J. Biol. Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- 18.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 22.Patnaik D, Chin HG, Esteve PO, Benner J, Jacobsen SE, Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J. Biol. Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 23.Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 25.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 26.Esteve PO, Patnaik D, Chin HG, Benner J, Teitell MA, Pradhan S. Functional analysis of the N- and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic Acids Res. 2005;33:3211–3223. doi: 10.1093/nar/gki635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin HG, Pradhan M, Esteve PO, Patnaik D, Evans T.C., Jr, Pradhan S. Sequence specificity and role of proximal amino acids of the histone H3 tail on catalysis of murine G9A lysine 9 histone H3 methyltransferase. Biochemistry. 2005;44:12998–13006. doi: 10.1021/bi0509907. [DOI] [PubMed] [Google Scholar]
- 29.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 30.Hirota J, Satomi Y, Yoshikawa K, Takao T. Epsilon-N,N,N-trimethyllysine-specific ions in matrix-assisted laser desorption/ionization-tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:371–376. doi: 10.1002/rcm.924. [DOI] [PubMed] [Google Scholar]
- 31.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 37.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 38.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteve PO, Chin HG, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J. Biol. Chem. 2007;282:2615–2625. doi: 10.1074/jbc.M606203200. [DOI] [PubMed] [Google Scholar]
- 41.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–2000. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 42.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.