Abstract
p300 regulates the transcriptional activity of a variety of transcription factors by forming an activation complex and/or promoting histone acetylation. Here, we show a unique characteristic of orphan receptor TR3 in negatively regulating the function of p300. TR3 was found to interact with p300 and inhibited the acetylation of transcription factors induced by p300, resulting in the repression of their transcriptional activity. Further analysis revealed that both a conserved transcriptional adapter motif (TRAM) in p300 and a specific sequence FLELFIL in TR3 were critical for their interaction. TR3 binding completely covered the histone acetyltransferase (HAT) domain of p300 and resulted in suppression of the HAT activity, as the p300-induced histone H3 acetylation and transcription were inhibited with the presence TR3. Furthermore, an agonist of TR3, a natural octaketide isolated from Dothiorella sp. HTF3 of an endophytical fungus, was shown to be a potent compound for inhibiting p300 HAT activity (IC50 = 1.5 μg/ml) in vivo. More importantly, this agonist could repress the transcriptional activity of transcription factors, and proliferation of cancer cells. Taken together, our results not only delineate a novel transcriptional repressor function for TR3, but also reveal its modulation on p300 HAT activity as the underlying mechanism.
INTRODUCTION
p300 and its closely related protein CBP are thought to play a fundamental role in a variety of signal transduction pathways (1–3). They function in these pathways by acetylating multiple target proteins, including transcription factors such as p53 (4), NF-κB (5) and β-catenin (6), and nuclear receptors such as ER (7), AR (8) and GR (9). p300 and CBP are also histone acetyltransferases (HATs) (10,11) and can modulate the chromatin structure via acetylation of histones, which facilitates the interaction of histones with DNA. In addition, they can act as a bridge between transcription factors and the basal transcription machinery (12) to enhance the transcription activation.
Nuclear orphan receptor TR3 (also termed as Nur77 and NGFI-B) is a transcription factor that plays key roles in apoptosis and cell proliferation at both transcriptional and post-transcriptional levels (13). TR3 functions in the nucleus or in the mitochondria through interaction with distinct proteins. For example, by forming a heterodimer with retinoid X receptor α (RXRα), TR3 binds to the retinoic acid response element (RARE) of the retinoic acid receptor β (RARβ) promoter in the presence of RXR ligand (14,15). Once bound to DNA, the TR3/RXRα complex promotes RARβ expression, which is critical for apoptosis induction (16,17). In addition, apoptotic agents can induce nucleocytoplasmic shuttling of TR3. The translocation of TR3 to mitochondria strongly promotes apoptosis in variety of cancer cell lines (18–20). Therefore, depending on different cell context and stimuli, TR3 may employ different mechanisms to function.
Several studies have demonstrated that the activities of p300 and CBP can be modulated by their interacting partners. The first example is the viral oncoprotein E1A that can inhibit p300 and CBP acetyltransferase activities at high concentrations (21–23). Further study revealed that E1A inhibits nucleosomal histone modification by the PCAF complex and blocks p53 acetylation (21). Similar inhibition in p53 transcriptional activity through regulation of p300/CBP activity has also been observed in human papillomavirus E6 oncoprotein (24). Another example is vIRF, a viral IFN regulatory factor encoded by the K9 gene of Kaposi's sarcoma-associated herpesvirus. vIRF inhibits p300 HAT activity by direct interaction, resulting in hypoacetylation of nucleosomal histones and perturbation of cytokine gene expression (25). On the other hand, some cellular proteins with inhibitory activities on p300 and CBP have also been identified, such as Twist and the E1A-like inhibitor of differentiation (EID-1) (23,26,27). Twist interacts with the HAT domains and inhibits acetyltransferase activities of p300 and PCAF (23). E1A mimics the effect of Twist in inhibiting the p300 HAT activity (23). The HOX homeodomain proteins, acting as master developmental regulators, also inhibit CBP HAT activity and thus function as repressors of gene transcription (28). These sets of evidence strongly suggest that other cellular factors with similar regulatory activities on P300 and CBP may exist.
More recently, we have found that p300-induced p53 acetylation can be dramatically suppressed by TR3 at the transcriptional level (29). By blocking the acetylation, TR3 down-regulates p53 transcriptional activity and lead to a decrease on the transcription level of MDM2 (29). Similarly, TR3 also significantly inhibits the p300-induced RXRα acetylation, resulting in the disassociation of RXRα from DNA and the translocation of RXRα with TR3 from the nucleus to cytoplasm upon TPA stimulation (30). These results give rise to interesting questions: is that a unique characteristic of TR3 to negatively regulate transcription factors that can be acetylated by p300? Does TR3 regulate p300 acetyltransferase activity? In this study, we attempted to uncover the molecular mechanism by which TR3 controls the activity of p300-acetylated transcription factors. We first demonstrated that TR3 negatively regulates the acetylation and transcriptional activity of many p300-regulated transcription factors. Further analysis revealed that a physical interaction between TR3 and p300, which requires the recognition of a specific sequence (FLELFIL) located in the ligand-binding domain of TR3 by the transcriptional adapter motif (TRAM) of p300, is critical for the inhibitory effect of TR3 on p300. Importantly, binding of TR3 inhibits the HAT activity of p300, as the p300-induced histone H3 acetylation and transcription were repressed by the presence of TR3. Finally, we showed that an agonist of TR3 has a strong inhibitory effect on p300 HAT activity (IC50 = 1.5 µg/ml), by which the proliferation of breast cancer cells was inhibited. Therefore, repression of p300 function by TR3 may represent a unique pathway for regulating the activity of a subset of transcription factors that can be acetylated by p300.
MATERIALS AND METHODS
Cell lines and transfection
Human embryonic kidney (HEK) 293T, cervical cancer HeLa and breast cancer MCF-7 cell lines were purchased from ATCC. They were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 100 µg/ml penicillin and 100 µg/ml streptomycin. Transfection was performed using calcium phosphate precipitation method for 293T cells, and liposomal transfection reagent (Roche, Indianapolis, IN, USA, Fugene 6) for HeLa and MCF-7 cells.
Acetylation assay
Cell lysates were extracted in cell lysis buffer [20 mmol/l HEPES (pH 7.5), 0.1 mol/l KCl, 0.4 mmol/l EDTA, 0.2% NP-40, 10 mmol/l β-mercaptoethanol, 0.1 mmol/l phenylmethylsulfonyl fluoride, 10 µg of pepstatin per ml, 1 µg of Na3VO4 per ml] and immunoprecipitated with antibodies against various tags, including GFP (Santa Cruz, CA, USA) and Myc (Santa Cruz). The immunoprecipitates were then separated by SDS–PAGE and blotted with specific anti-acetyl-lysine antibody (Upstate, Lake Placid, NY).
Co-immunoprecipitation and western blotting
Cells were transfected with various plasmids as required and harvested at 36 h post-transfection. Cell lysate preparation, immunoprecipitation and western blotting were performed as described previously (29). Briefly, cell lysates were incubated with the appropriate antibody for 1 h, and subsequently incubated with protein A-Sepharose beads for 1 h. The protein–antibody complexes that were recovered on beads were subjected to western blot analysis using different antibodies as required after separation by SDS–PAGE. The immunoreactive products were then detected by using Enhanced chemiluminescence kit (Amersham, NJ, USA). Both p300 and TR3 antibodies were purchased from Santa Cruz.
Luciferase reporter assay
Cells were transfected with different luciferase reporter genes, β-galactosidase (β-gal) and other different expression vectors as required. After transfection, luciferase activity was normalized for transfection efficiency using corresponding β-gal activity. The ratios of luciferase/β-gal activity were used as indicators for transcriptional activity of different promoters.
GST pull-down assay
Various DNA coding sequences were generated by PCR and cloned into the expression vector pGEX-4T-1. The resulting plasmids were transformed into BL-21 bacterial cells and induced to express fusion proteins at 37°C. GST-fusion proteins were purified using glutathione-agarose beads. For TR3 binding, the GST-tagged TR3 protein bound to beads were incubated with 500 µl of whole cell lysate from 293T cells that transfected with HA-p300. After incubation for 5–6 h, unbound proteins were removed by washing with lysis buffer. Bound proteins were eluted by boiling for 10 min in 1× loading buffer, resolved by SDS–PAGE and examined by immunoblot analysis with anti-HA antibody to detect p300. GST alone was used as a control.
HAT activity assay
Cells were transfected with different expression vectors, including HA-p300, Flag-TR3 or siRNA-TR3 (The DNA target sequence for silencing TR3, CAGTCCAGCCATGCTCCTC, was constructed in the pSuper vector) as required. After transfection, cell lysates were prepared and immunoprecipitated with anti-HA or anti-Flag antibody. The in vitro HAT activity was assayed by measuring the histone acetylated by p300 as described elsewhere (31). Typically, the 25 µl reaction mixture containing immunoprecipitated lysates, 10 µg histone and 10 µM acetyl-coenzyme A in HAT assay buffer (50 mM Tris–HCl, pH 8.0, 10% glycerol, 0.1 mM EDTA and 1 mM dithiothreitol) was incubated at 30°C for 30 min, and then subjected to SDS–PAGE for protein separation. Acetylated histone was analyzed by western blotting using anti-acetylated histone H3 antibody (Sigma, St. Louis, MO, USA).
For in vivo HAT activity, cells were treated with TR3 agonist at different concentration as indicated for 5 h before harvest. The cell lysates were directly subjected to analyze acetylated histone as described above.
BrdU assay
Cells were transfected with relative expression vectors as indicated, then treated with or without TR3 agonist. After incubating with 5-bromo-2′-deoxyuridine (5-BrdU, 20 μM) (Sigma) for 2 h, cells were fixed with 4% paraformaldehyde for 30 min at 4°C, and then incubated with saponin (0.1%) for another 10 min. The cells were washed twice with PBS containing 0.1% saponin, and resuspended in PBS containing 30 µg of DNase I. After incubation with anti-BrdU antibody (Santa Cruz) for 1 h, cells were given two PBS washes and then incubated with PE-linked anti-mouse antibody (Santa Cruz). Finally, cells were analyzed by flow cytometer (Backman Coulter, Fullerton, CA, USA).
Isolation of an agonist of TR3
The endophytical fungal strain Dothiorella sp. HTF3 was isolated from mangrove tree and cultured in liquid potato dextrose broth media. A bank of natural products was purified from the mycelia of the endophytic fungi and subjected to the TR3 agonist/antagonist screening designed by our group. A compound was found to function as a TR3 agonist. Structural analysis by nuclear magnetic resonance (NMR) revealed that this compound is an octaketide. Our unpublished studies demonstrated that this compound binds to TR3 ligand-binding domain to induce TR3 mRNA and protein expression, and activate its transcriptional activity (Zhan et al., unpublished data).
RESULTS
TR3 negatively regulates p300-acetylated transcriptional factors
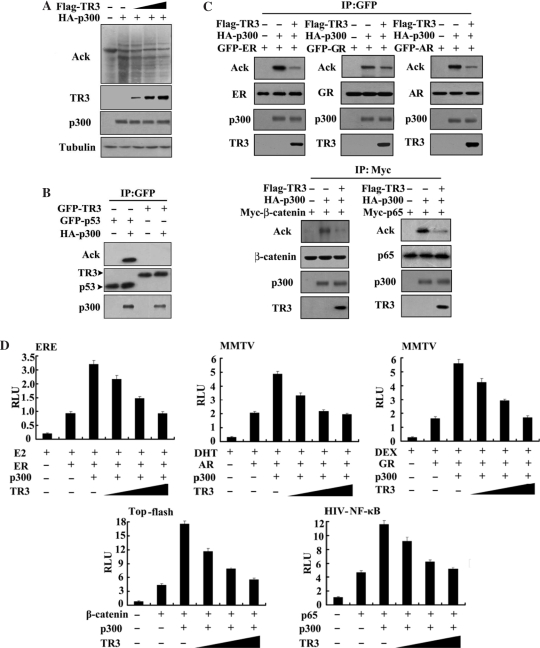
Recently, we found that TR3 negatively regulates the transcriptional activity of p53 (29) and retinoid X receptor (RXRα) (30) through the p300-dependent acetylation pathway. These results strongly suggest that TR3 may also inhibit the activity of other p300-acetylated transcription factors. To test this possibility, we first examined the effect of TR3 on the p300-induced acetylation of a broad spectrum of substrates. When whole-cell lysates were analyzed by western blotting with specific anti-acetyl-lysine antibody, a dramatic increase in total protein acetylation was observed upon the expression of p300 (Figure 1A, lane 2). As we expected, coexpression of TR3 with p300 led to a decrease in the total protein acetylation level in a dose-dependent manner (Figure 1A). We found that TR3 neither interfered with p300 expression (Figure 1A), nor was acetylated by p300 (Figure 1B). Clearly, TR3 has a unique characteristic in inhibiting p300-induced acetylation of numerous cellular proteins.
Figure 1.
TR3 represses transcriptional activities of transcription factors through inhibiting p300-induced acetylation. (A) TR3 inhibits the acetylation of a broad spectrum of substrates. HA-p300, with increasing amount of Flag-TR3, was transfected into 293T cells. Lysates were prepared from transfected cells and subjected to western blotting. The acetylated proteins were detected by anti-acetylation-specific antibody. The levels of Flag-TR3 and HA-p300 were shown by anti-Flag and anti-HA antibodies, respectively. Tubulin was used to indicate the similar loading of proteins in each lane. (B) TR3 cannot be acetylated by p300. GFP-TR3 and HA-p300 were transfected into 293T cells. Cell lysates were prepared and GFP-TR3 was immunoprecipitated with anti-GFP antibody. The precipitated proteins were then subjected to western blotting with acetylation-specific antibody. p53, which is known to be acetylated by p300, was used as a positive control. GFP-TR3 and GFP-p53 were also detected by western blotting with anti-GFP antibody. (C) TR3 attenuates p300-induced acetylation of various transcription factors. HA-p300 and Flag-TR3 together with different tagged transcription factors as indicated were transfected into 293T cells. Acetylation for each transcription factor was determined as described above. The expression levels of various transcription factors were indicated by western blotting with antibodies corresponding to different tags. (D) TR3 represses p300-induced transcriptional activity of various transcription factors. Different transcription factors and their corresponding reporter genes, together with p300, β-gal expression vector and increasing amount of TR3, were transfected into 293T cells. Reporter gene activity was determined and normalized in relation to the cotransfected β-gal activity. The bars represent the average ± mean from three independent experiments.
We then chose several transcription factors and nuclear factors, which have been approved to be acetylated by p300, to analyze the effect of TR3 on their transcriptional activity. Consistent with previous reports, estrogen receptor (ER), glucocorticoid receptor (GR), androgen receptor (AR), p65 and β-catenin (5–9) were all acetylated by p300 (Figure 1C). Intriguingly, TR3 has a dramatically inhibitory effect on the acetylation of these factors. When TR3 was introduced into 293T cells that had been transfected with p300 and these factors respectively, the p300-induced acetylation of each factor was significantly attenuated (Figure 1C). We further tested whether TR3 could modulate p300-dependent transcriptional activity of the transcription factors. For this purpose, different reporter genes linked with luciferase, including ER response element (ERE), steroid hormone responsive elements in the mouse mammary tumor virus (MMTV-Luc), Tcf4 reporter gene (Top-flash) and NF-κB responsive elements (HIV-NF-κB) and their corresponding transcription factors, together with p300 and TR3, were transfected into 293T cells. The result showed that p300 exerted clear activation on transcriptional activities of these transcription factors (Figure 1D, bar 3). However, coexpression of TR3 rendered p300 almost completely incapable of activating transcriptional activity of these factors in a dose-dependent manner (Figure 1D), which correlates well with the result shown in Figure 1C. Together, these results demonstrate that TR3 indeed possesses a unique characteristic to repress the activity of transcription factors that are commonly regulated by p300 acetylation.
TR3 physically interacts with p300
The finding that TR3 regulates the transcriptional activity of p300-acetylated transcription factors promoted us to determine whether TR3 physically interacts with p300 in cells. To test this possibility, Myc-TR3 and HA-p300 were coexpressed in 293T cells, followed by reciprocal immunoprecipitation with anti-Myc for TR3 and anti-HA for p300. Western blotting showed that TR3 was readily detected when p300 was immunoprecipitated (upper panel); similarly, p300 was also detected in the immunoprecipitates of TR3 (lower panel), suggesting TR3 interaction with p300 at exogenous level. We carried out another parallel experiment to test whether endogenous TR3 and p300 could interact with each other using lysates of untransfected HeLa cells. The results showed that p300 could be detected in the immunoprecipitates of TR3 (Figure 2B), indicating the association of TR3 with p300 at the endogenous level. GST pull-down experiment further demonstrated that GST-TR3 can bind HA-p300 that was transfected into 293T cells and immunoblotted by HA antibody (Figure 2C). Together, these results suggest that TR3 has a physical interaction with p300 in the cells.
Figure 2.
Mapping of domains responsible for the interaction between TR3 and p300. (A) TR3 and p300 form a complex in 293T cells. Myc-TR3 and HA-p300 were transfected either alone or together into cells. Cell lysates were prepared and then subjected to immunoprecipitation, followed by western blotting analysis with anti-Myc or anti-HA antibodies as indicated. (B) Interaction of TR3 with p300 in the absence or presence of TR3 agonist in HeLa cells. Cell lysates were immunoprecipitated with anti-TR3 antibody and control IgG, respectively, followed by western blotting with anti-p300 antibody. (C) Pull-down of p300 by TR3. GST-TR3 was expressed in Escherichia coli cells and purified as described in Materials and Methods section. The beads-bound GST-TR3 was incubated with the lysate of 293T cells transfected with HA-p300. HA-p300 was indicated with anti-HA antibody. GST was used as a negative control. Lower panel indicated the amount of GST and GST-TR3 used in the assay. (D) Determination of TR3-binding sites in p300. Schematic diagrams depicting different p300 truncation mutants are shown in upper panel. Full-length Myc-TR3 and different HA-p300 mutants were transfected into 293T cells as indicated. Cell lysates were immunoprecipitated with anti-Myc antibody. The immunoprecipitates and cell lysates were analyzed by western blotting with anti-HA and anti-Myc antibodies for p300 and TR3 proteins, respectively. (E) Determination of p300-binding sites in TR3. Schematic diagrams (upper panel) depict different truncation mutants of TR3. Full-length HA-p300 and different Flag-TR3 mutants were transfected into 293T cells as indicated. Cell lysates were immunoprecipitated with anti-HA antibody. The immunoprecipitates and cell lysates were analyzed by western blotting with anti-HA and anti-Myc antibodies for p300 and TR3 proteins, respectively.
We next went on to map the regions within TR3 and p300 that are responsible for their interaction. Different truncation mutants of p300 were constructed as indicated (Figure 2D, upper panel) and tested for interaction with TR3 by co-immunoprecipitation (Co-IP) experiment. When co-expressed with the full-length TR3 in 293T cells, the p300 mutant p300/M2, but not p300/N1 and p300/C1, retained the ability to interact with TR3 (Figure 2D), indicating that the region of amino acids (aa) 1039–1874 is responsible for p300 to interact with TR3. Furthermore, we found that p300/N2 and p300/M1 also interacted with TR3 (Figure 2D), suggesting that the cysteine-histidine-rich region 2 (CH2) of p300 is sufficient for TR3 binding, but other domains, such as the cysteine-histidine-rich region CH3, may also contribute to the interaction.
Different deletion mutants of TR3 were also generated (Figure 2E, upper panel) to test the interaction with p300. As shown in Figure 2E, TR3 deletion mutant D4 but not D3 showed the ability to interact with p300, suggesting that the N-terminal region of TR3 is not required for p300 binding. Further analysis revealed that deletion mutants TR3/D1, TR3/D5 and TR3/D6 all interacted with p300, but TR3/D2 failed to do so (Figure 2E). These results suggest that at least two regions, the DNA-binding domain (DBD) and the region of aa 439–521 of TR3, are capable of binding to p300.
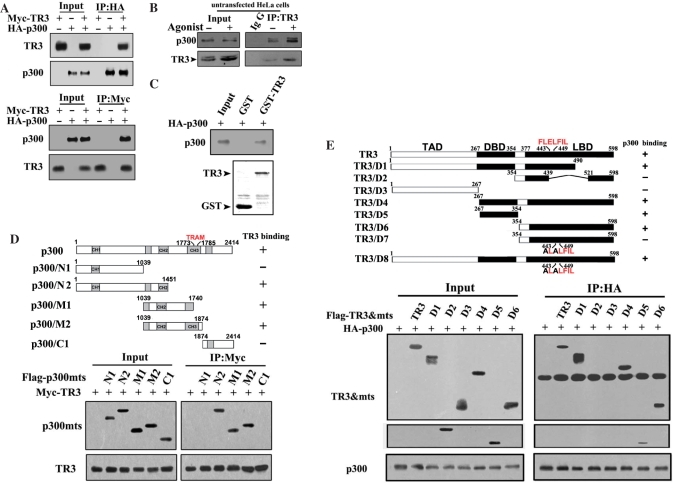
TR3 binding inhibits the HAT activity of p300
The HAT domain of p300 is located at the middle region of the protein that shows a physical interaction with TR3 (Figure 2D), indicating that the HAT activity of p300 may also be impaired by TR3 binding. We therefore assayed the p300 HAT activity by utilizing previously described systems (28,32). We first examined the acetylation level of histone H3, as Histone H3 acetylation is usually used to reflect p300 HAT activity (25,32). In 293T cells that were cotransfected with TR3 and p300, TR3 indeed exhibited a strong inhibition on p300 HAT activity, as ∼74% of such activity had lost when 2 µg of TR3 was introduced (Figure 3A). We further examined the effect of different TR3 deletion mutants on the HAT activity of p300. Similar to full-length TR3, TR3/D1, TR3/D4 and TR3/D6 could efficiently inhibit p300 HAT activity, reaching to 60% inhibition (Figure 3B). A common feature for these deletion mutants is that they all contain the region of aa 439–521 that interacts with p300 (Figure 2E). In contrast, TR3/D2, TR3/D3 and TR3/D5 showed no effect on p300 HAT activity (the activity was remained at almost 100% in all the tested mutants except TR3/D5) (Figure 3B), even though TR3/D5 could interact with p300 via the DBD (Figure 2E). Thus, these results suggest that the region of aa 439–521, rather than the DBD, is essential for TR3 to inhibit p300 HAT activity.
Figure 3.
TR3 inhibits p300 HAT activity. (A) Effect of TR3 on p300 HAT activity. HA-p300 and increasing amount of Falg-TR3 were transfected into 293T cells. Cell lysates were prepared and HA-p300 was immunoprecipitated with anti-HA antibody. The precipitates were then incubated with histone (10 µg) and Lys-CoA (10 µM) at 30°C for 30 min. The reaction products were subjected to western blotting with anti-Ack-H3 antibody to show the HAT activity. The levels of TR3, p300 and histone H3 were indicated by western blotting with anti-Flag, -HA and -histone antibodies. The percentage of Ack-H3 inhibition was calculated by determining the Ack-H3 protein amount of individual bands using densitometry. (B) Different TR3 mutants impair p300 HAT activity. Flag-TR3 mutants together with full-length HA-p300, were transfected into 293T cells. The HAT activity of p300 was determined as described above. Ack-H3 inhibition was determined by using densitometry. (C and D) TR3 inhibits p300 HAT-mediated transcription. Gal4-p300/HAT, together with a luciferase reporter gene pGAL4 and increasing amount of TR3 (C) or TR3 deletion mutants (D), was cotransfected into 293T cells. Reporter gene activity was determined as described in Figure 1D. The bars represent the average ± mean from three independent experiments.
Since p300 HAT activity is thought to stimulate transcription by acetylating Histones (25,33), we next examined whether the repression of p300 acetyltransferase activity by TR3 will result in inhibition of p300-activated gene transcription. For this end, a truncated p300 fragment containing the HAT domain (aa 1039–1874) was fused to Gal4 protein (Gal4-p300/HAT), and cotransfected into 293T cells together with a reporter construct containing the luciferase gene controlled by five GAL4 response elements (pGAL4) (34). As shown in Figure 3C, Gal4-p300/HAT obviously activated the transcription of the reporter gene. However, cotransfection of TR3 abrogated the effect of Gal4-p300/HAT in a dose-dependent manner (Figure 3C), suggesting that p300-mediated gene transcription is regulated by TR3. In the parallel experiment, a decrease in transcription activation was observed when TR3 mutants D1, D4 and D6 but not D2, D3 and D5 were used (Figure 3D). Considering the varied interactions between p300 and TR3 mutants (Figure 2E), it is likely that TR3 binding to p300 inhibits both p300 HAT activities and activation of transcription.
TRAM in p300 and FLELFIL sequence in TR3 are critical to modulate HAT activity
Interestingly, when TR3 was cotransfected into 293T cells with either p300/M1 or p300/M2, both of which retain the p300 HAT activity fully, it only inhibited the HAT activity of p300/M2 with 78% approximately, but not that of p300/M1 (the approximate inhibition was 6%) (Figure 4A). Compared to p300/M1, p300/M2 contains extra sequences in which a motif (RKTNGGCPICKQ) called TRAM can be found (Figure 2D). TRAM has been shown to be conserved in all members of the CBP/p300 family of proteins and is responsible for specific recognition of target proteins (35). In p300, TRAM is located within the CH3 domain just after the HAT domain (36,37). Our result clearly demonstrated that TRAM plays an important role in mediating the inhibition of TR3 on p300 HAT activity.
Figure 4.
Effects of different mutants of p300 and TR3 on regulation of HAT activity and transcriptional activity of transcription factors. (A and C) Different p300 or TR3 mutants impair TR3 inhibition on p300 HAT activity. Different Flag-p300 mutants (A) or Myc-TR3 mutants (C), together with full length of Myc-TR3 (A) or HA-p300 (C), were transfected into 293T cells. The HAT activity of p300 was determined as described above. The percentage of Ack-H3 inhibition was calculated by using densitometry. (B) Interaction of p300 with different TR3 mutants. HA-p300 and Myc-TR3 mutants were transfected into 293T cells. Cell lysates were prepared and then subjected to Co-IP/western blotting as described in Figure 2A. (D) Effect of different TR3 deletion mutants on transcriptional activity of various transcription factors. p300, together with different TR3 mutants, transcription factors and reporter genes, was cotransfected into 293T cells. The transcriptional activity was determined as described in Figure 1D.
It has been shown that CBP/p300 can specifically recognize a small sequence (FXE/DXXXL) on E1A, p53, E2F and TFIIB through its TRAM (35). Interestingly, we found that TR3 contains a FLELFIL sequence (aa 443–449), which is similar to the FXE/DXXXL motif, within the region of TR3/D6 (Figure 2E). To determine whether this sequence facilitates p300 TRAM recognition, we constructed a new mutant (TR3/D7) based on the structure of TR3/D6 mutant, where the Phe (F) in aa 443 and the Glu (E) in aa 445 were replaced with Ala (A) simultaneously (Figure 2E). When this mutant was cotransfected into 293T cells with p300, it failed to bind p300 (Figure 4B), and exhibited no inhibitory effect on p300 HAT activity (the inhibitory rate was only 1%), but TR3/D6 exerted higher inhibition (68%) on HAT activity (Figure 4C), suggesting that the FLELFIL sequence is critical for TR3 to inhibit the HAT activity of p300. To exclude out the possibility that such inhibitory effect of TR3/D7 is due to the deleted sequences of TR3 instead of the mutated sequence, we introduced the same mutations into full-length TR3 to create TR3/D8 (Figure 2E). Although TR3/D8 still interacted with p300 (probably through its DBD) (Figure 4B), it could not repress the HAT activity of p300 anymore (Figure 4C, the inhibitory rate was only 3%) as compared to the full-length TR3 (Figure 3A, the inhibitory rate was 74%). This result further confirmed that the FLELFIL sequence is necessary to ensure the suppression of TR3 on p300 HAT activity.
As p300 HAT activity is associated with its ability to activate transcription factors (4–8,38), we further determined whether TR3 modulation on p300 HAT activity impairs the transcriptional activity of various transcription factors, and whether this regulation requires the FLELFIL sequence of TR3. When different TR3 mutants were tested, we found that TR3/D1, TR3/D4 and TR3/D6 repressed the reporter gene activities of all the transcription factors induced by p300 as effectively as the full-length TR3 (Figure 4D). In contrast, mutants lacking the FLELFIL sequence, such as TR3/D2, TR3/D3 and TR3/D5, did not show any inhibition on activities of reporter genes for all transcription factors (Figure 4D). Similar result was observed when the TR3/D7 and TR3/D8 mutants, of which the FLELFIL sequence had been mutated, was used (Figure 4D). Taken together, these results all suggest that TR3 may inhibit the transcriptional activity of transcription factors through repression of p300 HAT activity.
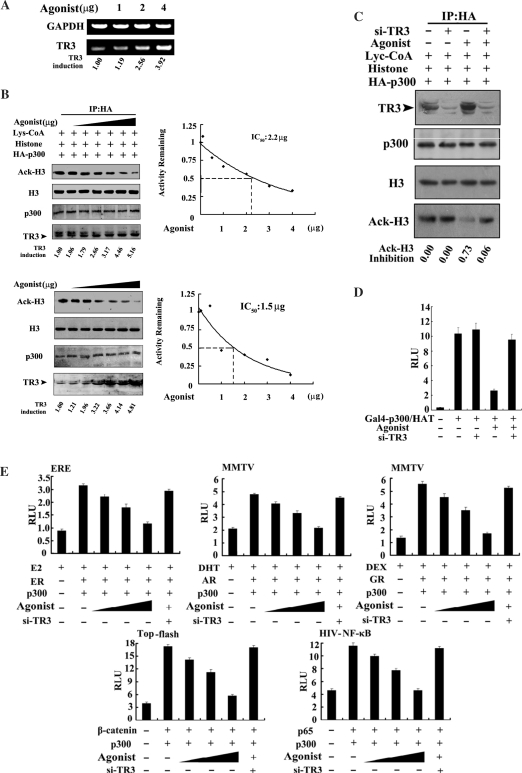
Roles of a TR3 agonist in regulating p300 functions
Recently, we screened a bank of the natural products from fungi and found an agonist of TR3. This agonist specifically activated TR3 transactivation through targeting to the TR3 ligand-binding domain (Zhan et al., unpublished data), and promoted the expression of TR3 mRNA and protein (Figure 5A–C). Co-IP assay showed that agonist could enhance the TR3–p300 interaction at endogenous level (Figure 2B). If TR3 is a genuine factor for inhibition of p300 HAT activity, it is expected that this agonist would also have such an inhibitory effect on p300 HAT activity as well as the transcriptional activity of p300-induced transcription factors. We first analyzed the role of TR3 agonist in p300-dependent HAT activity. As shown in Figure 5B, agonist did not impair the expression levels of p300 and Histone H3, but indeed down-regulated p300 HAT activity in a concentration-dependent manner; meanwhile, the expression levels of endogenous TR3 were up-regulated with increase of agonist dose (Figure 5B, left panels). The IC50 (inhibitory concentration of agonist for 50% reduction of the HAT activity) was 2.2 µg/ml in the in vitro assay and 1.5 µg/ml in the in vivo assay (Figure 5B, right panels). To confirm that such inhibition was directly mediated by TR3, a siRNA against TR3 was introduced into cells to inhibit endogenous TR3 expression. When endogenous TR3 was inhibited by its RNA interfere, the inhibitory effect of agonist on p300 HAT activity was not detected (Figure 5C). TR3 agonist also repressed p300-activated transcription of GAL4 response elements linked luciferase gene, and such repression could be diminished by siRNA-TR3 (Figure 5D). In addition, while introduction of p300 into 293T cells activated the expression of luciferase reporter gene for a series of transcription factors (Figure 5E, lane 2), agonist exerted an obvious inhibition on reporter gene activities with a dose-dependent fashion (Figure 5E), but failed to do so in the presence of siRNA-TR3 (Figure 5E, lane 6). Clearly, the repression of p300 HAT activity and activities of reporter genes by agonist is mediated by TR3.
Figure 5.
Inhibition of TR3 agonist on p300 HAT activity. (A) Agonist up-regulates TR3 mRNA expression levels. HeLa cells were treated with different concentrations of agonist as indicated for 5 h, and total RNA was then prepared. The levels of TR3 mRNA were determined by RT-PCR. Representatives of at least two independent experiments with similar results are shown. The level of TR3 mRNA induced by agonist was determined by using densitometry. (B) Effect of TR3 agonist on p300 HAT activity. 293T cells transfected with HA-p300 (upper panel) or HeLa cells (lower panel) were treated with different concentrations of agonist (1–4 µg) as indicated for 5 h, then the p300 HAT activity was determined as described in Figure 3A. The expression levels of HA-p300, endogenous TR3 and histone H3 were determined by western blotting with anti-HA, -TR3 and -histone H3 antibody, respectively. The level of TR3 protein induced by agonist was determined by using densitometry. Right panels indicated IC50 values that were calculated by determining the concentration of agonist needed to promote 50% inhibition of agonist activity. (C) Effect of agonist on p300 HAT activity is attenuated by siRNA-TR3. siRNA-TR3 with p300 were transfected into 293T cells. Transfected cells were treated with agonist (4 µg) for 5 h. The HAT activity was determined as described in Figure 3A. (D) Effect of agonist on transcription induced by p300 HAT. Gal4-p300/HAT and a luciferase reporter gene pGAL4, with or without siRNA-TR3, was cotransfected into 293T cells. Transfected cells were treated with agonist (4 µg) for 5 h. Reporter gene activity was determined as described in Figure 1D. The bars represent the average ± mean from three independent experiments. (E) Effects of agonist and siRNA-TR3 on p300-induced transcriptional activity. p300 with different transcription factors and reporter genes was transfected into 293T cells, and cells were treated with agonist at different concentration (1, 2 and 5 µM). The transcriptional activity was determined as described in Figure 1D. In addition, siRNA-TR3 was introduced into cells to abolish agonist function as required. (F) TR3 agonist inhibits p300 and AR-mediated mitogenic activity. Different expression vectors, including HA-p300, GFP-AR and siRNA-TR3, were transfected into breast cancer MCF-7 cells as indicated, and the transfected cells were treated with or without agonist (4 µg) for 5 h. Cells were maintained in BrdU containing medium for 2 h and then identified by flow cytometry.
Finally, we addressed the biological significance of TR3 agonist in DNA synthesis by performing the BrdU assay in breast cancer cells MCF-7. As shown in Figure 5F (left panel), transfection of AR along caused the curve right-moved (green curve) as compared with the control (red curve). Cotransfection of p300 with AR displayed an obvious increase in BrdU incorporation (black curve), consistent with the fact that p300 can acetylate AR to enhance the growth of prostate cancer cell (8). We further examined whether TR3 agonist has an inhibitory effect on p300-mediated and AR-dependent proliferation of MCF-7 cells (right panel). Compared with the control (transfection of p300 and AR only) (red curve), a left-moved curve was seen after treatment with agonist (green curve). Once siRNA against TR3 was introduced into cells, the curve converted from the left to right and almost overlapped with the control (black curve). These results demonstrate an important role of TR3 in p300-mediated cell growth.
DISCUSSION
p300 and the closely related protein CBP have been shown to interact with a large number of transcription factors (39). In the present study, we showed that orphan nuclear receptor TR3 physically interacts with p300 in the cells and consequently inhibits the HAT activity of p300. Analysis of a structural basis for the mechanism by which TR3 controls the p300-dependent HAT activity revealed that the sequence FLELFIL in TR3 is required for p300 recognition, and the TRAM motif in p300 is critical for TR3 binding to repress p300 HAT activity. Furthermore, we demonstrated a unique characteristic of TR3 in negative regulation of a broad range of transcription factors that were acetylated by p300. TR3 was found to exert its inhibitory effect on the transcriptional activity of transcription factors through repressing the p300-induced acetylation. In addition, we used a TR3 agonist that mimics TR3 function to verify the findings mentioned above. The results indicated that the agonist indeed represses transcriptional activities of transcription factors through inhibiting p300 HAT activity, and suppresses the DNA synthesis and the proliferation of breast cancer cells by regulating p300. Together, our results uncover a novel mechanism by which the orphan receptor exerts its effects on fundamental aspects of cell functions.
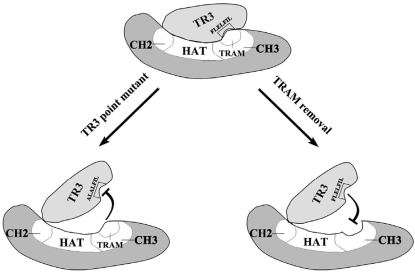
E1A and Twist have been shown to bind the CH3 domain of p300/CBP (23,36). Within CH3 domain there exists a conserved 12-residue transcriptional adapter motif named TRAM. TRAM is a hot spot to be bound by various transcription factors, such as E1A (35) and numerous cellular transcription factors (24,35). TRAM functions as a transcriptional adapter to provide HAT activity and/or to bridge the gap between transcription factors and components of the basal transcription machinery (4,35). In the present study, p300 could activate transcriptional activity of various transcription factors (Figure 1D) through the acetylation pathway (Figure 1C). On the other hand, TR3 displayed a pronounced preference for inhibiting p300-induced transcriptional activity (Figure 1D). It is very likely that this unique property of TR3 is associated with its interaction with p300 (Figure 2). TR3 binding to TRAM of p300 resulted in the inhibition of p300 HAT activity (Figure 4A). When TRAM was removed, TR3 still bound to p300 at the CH2 domain (Figure 2D), but failed to inhibit the HAT activity (Figure 4A). Therefore, TRAM is a necessary binding target for TR3 to inhibit p300 HAT activity. Here, we propose a possible model to illustrate how TR3 negatively regulates p300 function (Figure 6). According to this model, TRAM is a key site for TR3 binding to p300 (Figure 6, lower right panel). Once seated on TRAM, TR3 might completely cover the region of CH3 (Figure 6, upper panel), hence abrogates p300 HAT activity (Figure 3B). In addition to TRAM, another key factor is the FLELFIL sequence of TR3, as it is crucial for TRAM recognition (Figure 6, lower left panel).
Figure 6.
A working model to illustrate the mechanism by which TR3 negatively regulates p300. TR3 binding to p300 TRAM may cover the HAT domain and consequently inhibits HAT activity (upper panel). Point mutations of FLELFIL sequence in TR3 (lower left panel) or removal of TRAM in p300 (lower right panel) may render TR3 fail to block the HAT domain, and therefore p300 HAT activity is not repressed.
Although few signature motifs have been identified in transcription regulators that would predict with certainly their ability to bind to a given domain within p300/CBP, a short sequence FXE/DXXXL has been identified in E1A, p53 and E2F that forms specific contacts with the TRAM motif in the CH3 domain of p300/CBP (35). Within TR3, two p300-interacting domains were determined, one is the DBD, the other is the FLELFIL sequence located in the ligand-binding domain (Figure 2E). Functional analysis indicated that p300 binding by DBD of TR3 did not impair its HAT activity (Figure 3B), whereas binding by the FLELFIL sequence of TR3 led to p300 HAT activity lost (Figure 4C). When the F and E in FLELFIL sequence were substituted with A simultaneously in full-length TR3 or TR3 deletion mutant, the effect of TR3 on HAT activity was attenuated (Figure 4C). These results suggest that TR3 inhibition on p300 HAT activity may be dependent on an FLELFIL–TRAM interaction. In addition, we also demonstrated that this FLELFIL sequence is critical for TR3 to regulate transcriptional activities of different transcription factors (Figure 4D). When this sequence was deleted, or two sites in this sequence were mutated, TR3 showed no effect on the transcriptional activity of all the tested transcription factors (Figure 4D). The physiological significance of this inhibition is further strengthened by the observation that an agonist for TR3 mRNA and protein (Figure 5A–C), inhibited transcriptional activity of various transcription factors (Figure 5E) through repression of p300 HAT activity (Figure 5B). Furthermore, we noted that transfection of si-TR3 did not influence p300 HAT activity (Figure 5C, lane 2), suggesting a possibility that the physiological levels of TR3 protein in cells are important in setting the threshold for inhibiting p300 HAT activity. It may be only under the circumstances that TR3 was overexpressed (like Figure 3A, transfection of TR3) or activated (like Figure 5B, activation of TR3 by its agonist), TR3 would exert its inhibition on p300 HAT activity. Taken together, these in vivo and in vitro results support a potentially global role of TR3 in regulation of p300 HAT activity. In future, it will be of importance to determine whether other coactivation/HAT complexes, such as the SAGA, CBP and TFIID complexes, as well as other nonhistone substracts, are also sensitive to TR3 inhibition.
In fact, the interaction of TR3 with p300 has been demonstrated previously by in vitro pull-down assay, where 35S-radiolabeled full-length p300 interacted efficiently with full-length TR3 and the N-terminus of TR3, but poorly with the C-terminus of TR3 (40). Our Co-IP assay showed that endogenous TR3 could bind p300 (Figure 2B), and the in vivo complex formation of TR3 with p300 appeared to be TR3 agonist-dependent (Figure 2B). However, our results revealed that the binding to p300 occurred in both the DBD and the C-terminus, rather than N-terminus of TR3 (Figure 2E). This discrepancy might be derived from the difference of assay methods and conditions: in vivo Co-IP assay versus in vitro pull-down assay, and/or proteins extracted from cell lysates versus GST fusion protein expressed in bacterial and/or 35S-radiolabeled protein. Such discrepancy has also been reported in KSHV vIRF targeting to p300 (25,41,42). Alternatively, other factors, coactivators or corepressors, which are likely absent in the in vitro systems, might be involved in the formation of p300–TR3 complex in vivo.
HATs and histone deacetylases (HDAC) are thought to be potent targets for cancer therapy. In the past years, a number of HDAC inhibitors that can induce apoptosis in tumor cells have been identified (43). Some of them, such as SAHA, are already used in human trial as antineoplastic drug (44). Compared with HDAC inhibitors, little is known about HAT inhibitors. Recently, garcinol and curcumin were found to be p300 HAT inhibitors (45,46). They all repress p300 HAT-dependent chromatin transcription, but not transcription from DNA template. Garcinol alters global gene expression related to apoptosis, cell cycle, oncogene (45) and curcumin inhibits the multiplication of HIV1. Here we showed that a TR3 agonist, a natural octaketide isolated from Dothiorella sp. HTF3 of endophytical fungus, possesses the ability to inhibit p300 HAT activity (Figure 5B). More importantly, we found that TR3 could be the potential target of this agonist, as siRNA-TR3 attenuated agonist function on repression of p300 HAT activity and transcriptional activity of transcription factors (Figure 5C–E). It has been reported that in breast cancer cells, p300 binding to AR alters expression of specific growth control genes and promotes aberrant cellular growth in vivo (8), which is closely associated with p300 acetylation for AR. We found that when breast cancer cells were introduced with TR3 or treated with agonist, the cell proliferation mediated by AR via p300 acetylation (Figure 1C) was abolished through blockage of DNA synthesis in the cells (Figure 5F). Thus, our study reveals a novel function of TR3 to inhibit breast cancer cell growth through repression of p300 HAT activity, and indicates that the TR3 agonist may be used as a lead compound for anticancer drug.
In summary, our study demonstrates the regulation of p300 acetyltransferase activity by orphan receptor TR3, and suggests that TR3 may function as a negative regulator of a set of transcription factors whose transcriptional activities are controlled by p300-mediated acetylation. We have conformed in current study that TR3 would inhibit p300 acetylation-dependent transcription regulation, since we have detected that p300-regulated Histone acetylation was impaired by TR3 (Figure 4) or its agonist (Figure 5B–D). However, we cannot preclude the possibility that TR3 would affect general transcription machinery through chromatin remodeling. Histone acetylation plays an important role in the modulation of chromatin structure associated with signal-dependent transcriptional activation (25,47). Previous studies have demonstrated a dependence on acetyltransferase activity for in vivo p300 modulation of gene transcription (28,48). In this study, we have found that GAL4-p300/HAT could activate the transcription of reporter gene (Figure3C), and cotransfected TR3, which would bind to p300 (Figure3E and F), greatly abrogated the p300-mediated activation of transcription (Figure. 3C and D). These results demonstrate that TR3 not only diminishes the HAT activity of p300, but also inhibits p300 HAT domain-induced transcription activation. Therefore, TR3 may regulate the transcription of a broader set of genes than we previously thought.
ACKNOWLEDGEMENTS
We thank for Dr Sheng-cai Lin for plasmids of p300/M1 and p300/C1; Dr Chun-dong Yu for plasmids of ERE, MMTV-Luc, HIV-NF-κB reporter genes; Dr Chawn-shang Chang for plasmids of AR and GR; Dr Hans Clevers for plasmid of Top-flash reporter gene. This work was supported by the National Natural Science Foundation of China (30630070 and 30425014 to Q.W.); grants from the Ministry of Education (706036 to Q.W., IRT0649 to Q.W. and Y.-M.S.); National Foundation for Fostering Talents of Basic Science (J0630649 to Q.W.); Fujian Commission of Science & Technology (2007Y0033 to Q.W.); ‘973’ project (2007CB914402 to Q.W.); ‘863’ project (2006AA09Z410 to Z.-H.Z.). Funding to pay the Open Access publication charges for this article was provided by 30630070.
Conflict of interest statement. None declared.
REFERENCES
- 1.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 2.Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 3.Nordheim A. Transcription factors. CREB takes CBP to tango. Nature. 1994;370:177–178. doi: 10.1038/370177a0. [DOI] [PubMed] [Google Scholar]
- 4.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 6.Levy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, Neuveut C. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol. Cell. Biol. 2004;24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, et al. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 2001;276:18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 8.Fu M, Rao M, Wang C, Sakamaki T, Wang J, Di VD, Zhang X, Albanese C, Balk S, et al. Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol. Cell. Biol. 2003;23:8563–8575. doi: 10.1128/MCB.23.23.8563-8575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. J. Exp. Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 11.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson S, Pillus L. Modifying chromatin and concepts of cancer. Curr. Opin. Genet. Dev. 1999;9:175–184. doi: 10.1016/S0959-437X(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 13.Dequiedt F, Kasler H, Fischle W, Kiermer V, Weinstein M, Herndier BG, Verdin E. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity. 2003;18:687–698. doi: 10.1016/s1074-7613(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 14.Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 15.Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q, Dawson MI, Zheng Y, Hobbs PD, Agadir A, Jong L, Li Y, Liu R, Lin B, et al. Inhibition of trans-retinoic acid-resistant human breast cancer cell growth by retinoid X receptor-selective retinoids. Mol. Cell. Biol. 1997;17:6598–6608. doi: 10.1128/mcb.17.11.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Q, Li Y, Liu R, Agadir A, Lee MO, Liu Y, Zhang X. Modulation of retinoic acid sensitivity in lung cancer cells through dynamic balance of orphan receptors nur77 and COUP-TF and their heterodimerization. EMBO J. 1997;16:1656–1669. doi: 10.1093/emboj/16.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q, Liu S, Ye XF, Huang ZW, Su WJ. Dual roles of Nur77 in selective regulation of apoptosis and cell cycle by TPA and ATRA in gastric cancer cells. Carcinogenesis. 2002;23:1583–1592. doi: 10.1093/carcin/23.10.1583. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Wu Q, Chen ZM, Su WJ. The effect pathway of retinoic acid through regulation of retinoic acid receptor alpha in gastric cancer cells. World J. Gastroenterol. 2001;7:662–666. doi: 10.3748/wjg.v7.i5.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD, Lin B, Chen G, Lu J, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 22.Perissi V, Dasen JS, Kurokawa R, Wang Z, Korzus E, Rose DW, Glass CK, Rosenfeld MG. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl Acad. Sci. USA. 1999;96:3652–3657. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 24.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung JU. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell. Biol. 2000;20:8254–8263. doi: 10.1128/mcb.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyake S, Sellers WR, Safran M, Li X, Zhao W, Grossman SR, Gan J, DeCaprio JA, Adams PD, et al. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol. 2000;20:8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen WF, Krishnan K, Lawrence HJ, Largman C. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 2001;21:7509–7522. doi: 10.1128/MCB.21.21.7509-7522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao BX, Chen HZ, Lei NZ, Li GD, Zhao WX, Zhan YY, Liu B, Lin SC, Wu Q. p53 mediates the negative regulation of MDM2 by orphan receptor TR3. EMBO J. 2006;25:5703–5715. doi: 10.1038/sj.emboj.7601435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao WX, Tian M, Zhao BX, Li GD, Liu B, Zhan YY, Chen HZ. Orphan receptor TR3 attenuates the p300-induced acetylation of retinoid X receptor. Mol. Endocrinol. 2007;00 doi: 10.1210/me.2007-0107. (doi:10. 1210/me. 2007-0107) [DOI] [PubMed] [Google Scholar]
- 31.Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell. Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao F, Carrick-Walmsley R, Akerblad P, Sigvardsson M, Kadesch T. Inhibition of p300/CBP by early B-cell factor. Mol. Cell. Biol. 2003;23:3837–3846. doi: 10.1128/MCB.23.11.3837-3846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine M, Hoey T. Homeobox proteins as sequence-specific transcription factors. Cell. 1988;55:537–540. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 34.Chintharlapalli S, Burghardt R, Papineni S, Ramaiah S, Yoon K, Safe S. Activation of Nur77 by selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J. Biol. Chem. 2005;280:24903–24914. doi: 10.1074/jbc.M500107200. [DOI] [PubMed] [Google Scholar]
- 35.O’Connor MJ, Zimmermann H, Nielsen S, Bernard HU, Kouzarides T. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J. Virol. 1999;73:3574–3581. doi: 10.1128/jvi.73.5.3574-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 37.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Su A, Chen J, Lefebvre YA, Hache RJ. Attenuation of glucocorticoid signaling through targeted degradation of p300 via the 26S proteasome pathway. Mol. Endocrinol. 2002;16:2819–2827. doi: 10.1210/me.2002-0154. [DOI] [PubMed] [Google Scholar]
- 39.Janknecht R, Hunter T. Versatile molecular glue. Transcriptional control. Curr. Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 40.Wansa KD, Harris JM, Muscat GE. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J. Biol. Chem. 2002;277:33001–33011. doi: 10.1074/jbc.M203572200. [DOI] [PubMed] [Google Scholar]
- 41.Burysek L, Yeow WS, Lubyova B, Kellum M, Schafer SL, Huang YQ, Pitha PM. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayachandra S, Low KG, Thlick AE, Yu J, Ling PD, Chang Y, Moore PS. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc. Natl Acad. Sci. USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 44.Richon VM, Zhou X, Rifkind RA, Marks PA. Histone deacetylase inhibitors: development of suberoylanilide hydroxamic acid (SAHA) for the treatment of cancers. Blood Cells Mol. Dis. 2001;27:260–264. doi: 10.1006/bcmd.2000.0376. [DOI] [PubMed] [Google Scholar]
- 45.Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, Kundu TK. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 2004;279:33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 46.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 47.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Balbas MA, Bannister AJ, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]