Abstract
Reactive oxygen species (ROS) can be induced by both endogenous and exogenous processes, and they can damage biological molecules including nucleic acids. Exposure of isolated DNA to X/γ-rays and Fenton reagents was shown to lead to the formation of intrastrand cross-link lesions where the neighboring nucleobases in the same DNA strand are covalently bonded. By employing HPLC coupled with tandem mass spectrometry (LC-MS/MS) with the isotope dilution method, we assessed quantitatively the formation of a guanine–cytosine (G[8-5]C) intrastrand cross-link lesion in HeLa-S3 cells upon exposure to γ-rays. The yield of the G[8-5]C cross-link was 0.037 lesions per 109 nucleosides per Gy, which was ∼300 times lower than that of 5-formyl-2′-deoxyuridine (0.011 lesions per 106 nucleosides per Gy) under identical exposure conditions. We further constructed a single-stranded M13 genome harboring a site-specifically incorporated G[8-5]C lesion and developed a novel mass spectrometry-based method for interrogating the products emanating from the replication of the genome in Escherichia coli cells. The results demonstrated that G[8-5]C blocked considerably DNA replication as represented by a 20% bypass efficiency, and the lesion was significantly mutagenic in vivo, which included a 8.7% G→T and a 1.2% G→C transversion mutations. DNA replication in E. coli hosts deficient in SOS-induced polymerases revealed that polymerase V was responsible for the error-prone translesion synthesis in vivo.
INTRODUCTION
Reactive oxygen species (ROS) can be generated during normal aerobic metabolism or through exogenous pathways, and excess generation of ROS in vivo results in damage to biological molecules including DNA (1–3). The DNA lesions, if not repaired properly, may give rise to mutations and induce cell death.
Several intrastrand cross-link lesions were found to form in aqueous solution of isolated DNA upon exposure to γ- or X-rays (4–11), during which hydroxyl radical (•OH) can be generated from the radiolysis of water. In this context, •OH can either couple with the C5=C6 double bond of cytosine, thymine and 5-methylcytosine or abstract a hydrogen atom from the 5-methyl group of the latter two pyrimidine bases (12). Both processes give rise to the pyrimidine base-centered secondary radicals, which may attack their neighboring purine bases to yield intrastrand cross-link lesions (4–6,8,11,13). Moreover, recent LC-MS/MS results revealed that G[8-5m]T, mC[5m-8]G, G[8-5m]mC and G[8-5]C cross-link lesions, where the C8 of guanine is covalently bonded to the methyl or C5 carbon of its vicinal pyrimidine base, were produced in DNA in aerated aqueous solution upon exposure to Fenton-type reagents (14,15). These observations suggest that this under-investigated group of oxidatively generated DNA lesions might be induced endogenously.
The oxidative intrastrand cross-link lesions result in significant destabilization to DNA double helix (16). In addition, it was found that the G[8-5m]mC, G[8-5m]T and G[8-5]C lesions could be recognized by Escherichia coli UvrABC nuclease, suggesting that these lesions might be substrates for nucleotide excision repair (NER) enzymes in vivo (16,17). On the other hand, in vitro replication studies showed that G[8-5m]T and G[8-5]C cross-link lesions either completely blocked most of the high-fidelity DNA polymerases or only allowed for the incorporation of one nucleotide opposite the 3′-thymine or cytosine portion of the two cross-link lesions (18–20). Yeast polymerase η (pol η) was able to replicate past the G[8-5]C and G[8-5m]T cross-link lesions; however, the 5′ guanine portion of the cross-links caused a marked reduction of both the efficiency and the fidelity of nucleotide incorporation (9,20). These results suggested that the oxidative intrastrand cross-link lesions, if not repaired, can be cytotoxic and mutagenic (18–20).
On the grounds that these intrastrand cross-link lesions might be substrates for NER enzymes in vivo and they can block replication and, potentially, transcription, these lesions may pose a significant challenge for people suffering from genetic diseases that are defective in NER (21). Along this line, it was shown that neurodegeneration is a major feature of people with deficiency in NER, e.g. xeroderma pigmentosum (XP) and Cockayne syndrome patients (22), though the molecular mechanisms underlying the neurodegeneration remain elusive (23). An attractive hypothesis was put forward suggesting that defective repair and the resulting accumulation of oxidative DNA lesions in the nervous systems of NER-deficient patients might be responsible for the neurodegeneration (23–25). In this regard, neurons consume a large amount of molecular oxygen and generate ROS as by-products of cellular respiration, which can cause considerable damage to DNA (25). Major DNA lesions induced by ROS were non-bulky lesions including 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), 5,6-dihydroxy-5,6-dihydrothymidine (thymidine glycol) and other single-base lesions. These lesions are, however, thought to be repaired primarily by the base excision repair (BER) pathway (24,25), though 8-oxodG- and thymidine glycol-bearing substrates could also be recognized by NER enzymes in vitro (25). Nevertheless, bulky DNA lesions like dipyrimidine photoproducts can only be removed by the NER pathway in human cells (26). Therefore, it has been proposed that neurodegeneration in NER-deficient patients is caused by ROS-induced bulky lesions (24,25).
Methods have been developed for evaluating the genotoxic effects of DNA damage with a structurally defined lesion (27). Essigmann and coworkers (28–31) introduced the ingenious restriction endonuclease and post-labeling (REAP) and competitive replication and adduct bypass (CRAB) assays to measure quantitatively the mutation frequency and lesion bypass efficiency, respectively, of a site-specifically incorporated and structurally defined DNA lesion in vivo (28–34). In these assays, the entire population of progeny, emerging from the replication of the lesion-bearing vector, was assayed for bypass efficiency and mutation frequency, which offers statistically sound conclusions about how a particular lesion is replicated in vivo (28–34). In addition, these experiments are conducted by using single-stranded vectors, which are generally not subjected to DNA repair, hence allowing for the evaluation of the efficiency and fidelity of in vivo translesion synthesis (27).
In this study we identified and quantified, for the first time, the formation of the G[8-5]C intrastrand cross-link lesion in HeLa-S3 cells that were exposed to γ-rays. In addition, we incorporated LC-tandem mass spectrometry (MS/MS) analysis into REAP and CRAB assays and examined how the G[8-5]C lesion affects DNA replication by hindering DNA polymerases and inducing mutations in vivo. Our results revealed that, in E. coli cells, G[8-5]C not only blocked markedly DNA replication but also caused considerable G→T and G→C mutations. Moreover, DNA polymerase V was responsible for the mutagenic bypass of the lesion in vivo. This was the first report on in vivo genotoxicity of an oxidative intrastrand cross-link lesion of DNA.
MATERIALS AND METHODS
[γ-32P]ATP, nuclease P1 and alkaline phosphatase were obtained from Amersham Biosciences Co. (Piscataway, NJ, USA), MP Biomedicals (Aurora, OH, USa) and Sigma-Aldrich (St. Louis, MO, USA), respectively. Snake venom phosphodiesterase and calf spleen phosphodiesterase were obtained from US Biological (Swampscott, MA, USA). Restriction endonucleases, DNA polymerases, T4 DNA ligase and uracil DNA glycosylase were from New England Biolabs (Ipswich, MA). Shrimp alkaline phosphatase was obtained from Roche Diagnostics (Indianapolis, IN). HeLa-S3 cells were purchased from National Cell Culture Center (Minneapolis, MN). M13mp7L2 and wild-type E. coli strains were kindly provided by Prof. John M. Essigmann, and polymerase-deficient AB1157 strains [(Δpol B1::spec (pol II-deficient), ΔdinB (pol IV-deficient) and ΔumuC::kan (pol V-deficient)] were general gifts from Prof. Graham Walker at MIT (35,36). DNA synthesis reagents were from Glen Research (Sterling, VA). 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) was purchased from TCI America (Portland, OR). Isotope-labeled d(G[8-5]C) (structure shown in Figure 1c) was synthesized previously (15).
Figure 1.
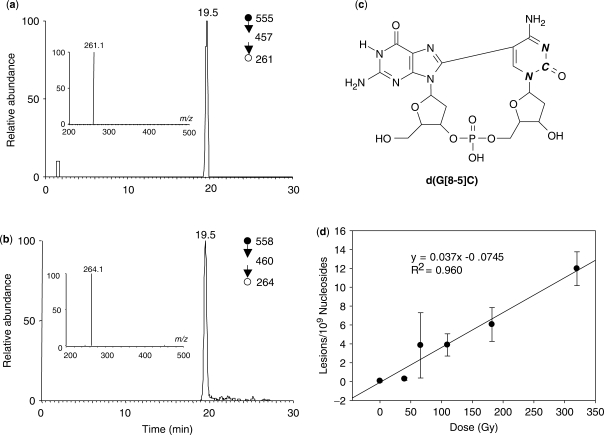
Identification of a G[8-5]C intrastrand cross-link lesion in HeLa-S3 cells. Shown are the SICs for the m/z 555→457→261 (a) and m/z 558→460→264 (b) transitions for the in vivo formed intrastrand cross-link and the isotope-labeled internal standard, respectively. Depicted in (c) is the structure for the isotope-labeled internal standard (bold italic letters designate the 15N- and 13C-labeling sites). Panel (d) reveals the dose-responsive formation of the G[8-5]C lesion.
Treatment of HeLa-S3 cells with γ-rays and enzymatic digestion of DNA
HeLa-S3 cells were centrifuged to remove the culture medium and resuspended in phosphate-buffered saline (PBS). The cell suspension (107 cells/ml) was exposed to γ-rays delivered by a Mark I 137Cs Irradiator (JL Shepherd and Associates, San Fernando, CA) at a dose rate of 2.8 Gy/min. Immediately after the exposure, the cells were harvested by centrifugation and the nuclear DNA was isolated by phenol extraction and desalted by ethanol precipitation.
The resulting DNA was digested with four enzymes following previously described procedures (14). The digestion mixture was passed through a YM-10 Centricon membrane (Millipore, Billerica, MA) to remove the enzymes. The amount of nucleosides in the mixture was quantified by UV absorbance measurements and to the mixture was added stable isotope-labeled d(G[8-5]C). The d(G[8-5]C) was enriched from the digestion mixture following previously published procedures by HPLC with a 4.6 × 50 mm C18 column (5 μm in particle size, Phenomenex®, Torrance, CA) (14). A gradient of 5-min 0–2% acetonitrile followed by a 55-min 2–5% acetonitrile in 10 mM ammonium formate (pH 6.3) was employed and the flow rate was 0.60 ml/min. The HPLC fractions eluted between 20.0 and 26.0 min were pooled, and we collected fractions in a wide retention time range to ensure that the cross-link lesion is completely collected. The collected fractions were dried in the Speed-vac, redissolved in 10 μl H2O, and injected for LC-MS/MS analysis.
LC-MS/MS identification and quantification of G[8-5]C
A 0.5 × 150 mm Zorbax SB-C18 column (5 μm in particle size, Agilent Technologies, Palo Alto, CA) was used for the separation of the DNA hydrolysis samples, and the flow rate was 8.0 μl/min, which was delivered by using an Agilent 1100 capillary HPLC pump (Agilent Technologies). A 60-min gradient of 0–30% acetonitrile in 20 mM ammonium acetate was employed. The effluent from the LC column was coupled to an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA), which was set up for monitoring the fragmentation of the [M + H]+ ions of the labeled and unlabeled d(G[8-5]C).
Restriction endonuclease and mass spectrometry (REAMS) assay
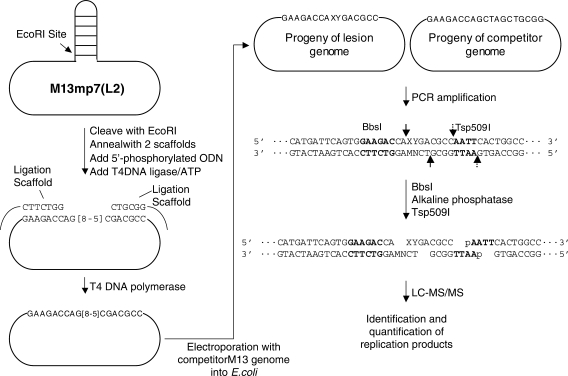
The procedures for the preparation of lesion-bearing 16-mer ODNs were described in the Supplementary Data. The lesion-carrying ODNs were ligated into the EcoRI-linearized M13mp7(L2) genome in the presence of two ligation scaffolds flanking the lesion site (30). The scaffolds were then degraded by the 3′→5′ exonuclease activity of the T4 DNA polymerase (30). To examine the bypass efficiency, we also prepared a control competitor genome by inserting a 19-mer unmodified ODN, [d(GAAGACCAGCTAGCTGCGG)], into the M13 genome. The amount of the lesion-containing genome was normalized against that of the competitor genome following the previously described method (30). The lesion-containing genome was then mixed with the competitor genome at a fixed ratio of 85/15, transformed into wild-type and DNA polymerase-deficient AB1157 E. coli cells by electroporation, and allowed for replication in the host E. coli cells for 6 h (29–33). The AB1157 cells were then pelleted and the supernatant, which contained progeny phages, was collected. Because not all copies of the lesion-containing genome were electroporated into cells, the isolated supernatant carried, other than the progeny phage, unreplicated genomes. To minimize the effect of residual lesion-containing genomes in subsequent analysis, the progeny/lesion-genome ratio was increased by several orders of magnitude via infecting SCS110 E. coli with the viable progeny phage (29–33). After culturing the cells at 37°C for 5 h, single-stranded DNA was extracted from the progeny phage.
PCR amplification of the region of interest in the resulting progeny genome was performed by using primers d(CAGCTATGACCATGATTCAGTGGAAGAC) and d(CAGGGTTTTCCCAGTCACGACGTTGTAA) (30). The PCR products were cleaned up by using QIAquick nucleotide removal kit (Qiagen, Valencia, CA) and digested with 45 U of BbsI. The terminal phosphate groups in the restriction fragments were removed by treatment with 20 U of shrimp alkaline phosphatase at 37°C for 4 h, and the shrimp alkaline phosphatase was deactivated by incubating the reaction mixture at 65°C for 20 min. The resulting DNA fragments were further digested with 40 U of Tsp509I at 65°C for 1 h. After the above restriction cleavages, the DNA fragment of interest from the full-length replication product was liberated as an 8-mer ODN, d(XYGACGCC), where ‘XY’ designate the nucleobases present at the initial G[8-5]C lesion site after DNA replication in vivo (Figure 2). On the other hand, the corresponding DNA fragment liberated from the competitor genome was an 11-mer ODN, d(GCTAGCTGCGG). In this context, it is worth noting that the removal of the 5′-phosphate groups from ODNs with shrimp alkaline phosphatase can avoid partial spontaneous dephosphorylation, which may affect the accuracy for the quantification of replication products.
Figure 2.
The REAMS assay for the monitoring of the in vivo cytotoxicity and mutagenicity of the G[8-5]C lesion. For the ligation scaffolds, only the portions of the sequences that are complementary to the 16-mer lesion insert are shown, while the remaining sequences are indicated by black lines. The PCR products and restriction fragments for the competitor genome are not listed. BbsI and Tsp509I cleavage sites are indicated by solid and broken arrows, respectively, and the recognition sites for these two restriction endonucleases are highlighted in bold.
A 0.5 × 150 mm Zorbax SB-C18 column (5 μm in particle size, Agilent Technologies) was used for the separation of the ODNs after restriction enzyme digestion, and the flow rate was 8.0 μl/min, which was delivered by using the Agilent 1100 capillary HPLC pump. A gradient of 5 min of 0–20% methanol followed by a 35 min of 20–50% methanol in 400 mM HFIP buffer (pH was adjusted to 7.0 by the addition of triethylamine) was employed for the separation (37,38). The effluent from the LC column was coupled directly to the LTQ linear ion-trap mass spectrometer, which was set up for monitoring the fragmentation of the [M-2H]2− ions of the 8-mer and 11-mer ODNs.
RESULTS
To understand the biological implications of oxidatively induced intrastrand cross-link lesions, it is important to determine whether this type of lesions can be formed in human cells. To this end, we exposed HeLa-S3 cells to a series of doses of γ-rays, isolated the genomic DNA from these cells, and digested the DNA with a combination of four enzymes (nuclease P1, calf spleen phosphodiesterase, alkaline phosphatase and snake venom phosphodiesterase). The digestion method was shown to be efficient in liberating intrastrand cross-link lesions from DNA (4,14,39,40). We then added the isotope-labeled d(G[8-5]C) (structure shown in Figure 1c) to the digestion mixture, enriched d(G[8-5]C) by HPLC, and analyzed the collected HPLC fractions by LC-MS/MS and LC-MS3. It is worth noting that the enrichment procedure is essential for the identification and accurate quantification of DNA lesions of very low abundance. In this respect, the enrichment allows for the removal of most salts employed for the enzymatic digestion and most undamaged nucleosides, thereby improving the ionization efficiency of the analyte under investigation (14).
Our LC-MS results revealed that G[8-5]C could be induced in human cells upon exposure to γ-rays. In this context, we found a strong peak in the selected-ion chromatogram (SIC) for the m/z 555→457 transition for the DNA sample obtained from γ-irradiated cells (Figure S1). The retention time for this peak is identical to the peak found in the corresponding SIC (i.e. m/z 558→460) for the internal standard. Moreover, the product-ion spectrum of the [M + H]+ ion of this fraction showed the presence of two abundant and diagnostic fragment ions, i.e. the ions of m/z 261 and 457 (Figure S1), resulting from the eliminations of the 2-deoxyribose-phosphate backbone and a 2-deoxyribose component, respectively (41). The corresponding fragment ions, that is, the ions of m/z 264 and 460, were also abundant in the product-ion spectrum of the [M + H]+ ion of the internal standard (Figure S1). The same retention time and the similar MS/MS of the analyte and the isotope-labeled internal standard support that this lesion can be induced in HeLa-S3 cells upon exposure to γ-rays. Corresponding analysis of DNA samples obtained from control cells without γ-ray exposure revealed that the G[8-5]C lesion was not detectable.
It is worth noting that, other than the two characteristic fragment ions found in the MS/MS for the internal standard and the analyte, ions from impurities were also present (Figure S1). In this respect, these impurity-related ions were also present for the control samples. However, further collisional activation of the ion of m/z 457 allowed us to obtain a product-ion spectrum, that is, MS3, of high quality (Figure 1a). Corresponding LC-MS3 analysis of control sample again revealed that the G[8-5]C was not detectable (data not shown), substantiating our conclusion that the intrastrand cross-link lesion is not produced in detectable quantity in unirradiated cells.
Because of the high specificity offered by MS3, we chose to employ LC-MS3 to quantify the G[8-5]C lesion induced in HeLa-S3 cells. The quantification results showed that the formation of the lesion is dose-responsive (Figure 1d). In addition, the yield for the formation of the G[8-5]C lesion is ∼0.037 lesions per 109 nucleosides per Gy, which is ∼300 times lower than what we recently found for the rate of the formation of 5-formyl-2′-deoxyuridine (FodU, 0.011 lesions per 106 nucleotides per Gy) under identical exposure conditions (42).
Other studies indicate that the yields for the formation of some other oxidatively generated single-base modifications, e.g. 8-oxodG and 5-(hydroxymethyl)-2′-deoxyuridine, were in a similar range as that of FodU (43). In addition, another tandem lesion, 8,5′-cyclo-2′-deoxyadenosine could be induced at a similar yield as these single-nucleobase lesions. For instance, the abundance of (5′S)-8,5′-cyclo-2′-deoxyadenosine in keratinocytes increased from about 0.23 to 0.32 lesions per 106 nucleosides after receiving 5 Gy of X-rays (44). In addition, we found that the structurally related G[8-5m]T lesion could be induced at a rate of 0.050 lesions per 109 nucleosides per Gy (20), which is slightly higher the rate for the formation of G[8-5]C. Thus, the rates for the formation of the intrastrand cross-link lesions G[8-5m]T and G[8-5]C are approximately two orders of magnitude lower than that for the formation of the (5′S)-8,5′-cyclo-2′-deoxyadenosine.
At a mechanistic point of view, the hydroxyl radical, produced from γ radiolysis of water, can add to the C6 carbon of cytosine to afford the 6-hydroxy-5-yl radical of the cytosine base, which can couple with the C8 of its neighboring 5′ guanine to render the G[8-5]C lesion. The latter radical, however, can also couple with O2, which reduces the yield for the formation of G[8-5]C. Thus, it is of interest to assess the formation of G[8-5]C in hypoxic cells in the future.
We next examined how the G[8-5]C lesion blocks DNA replication and induces mutations in vivo. We chose to develop an LC-MS/MS method based on the REAP and CRAB assays, first introduced by Essigmann and coworkers (29–32,34), because, in these assays, the entire population of progeny was assayed for bypass efficiency and mutation frequency, which offers statistically sound conclusions. In addition, these assays are high throughput and do not require phenotypic selection (30).
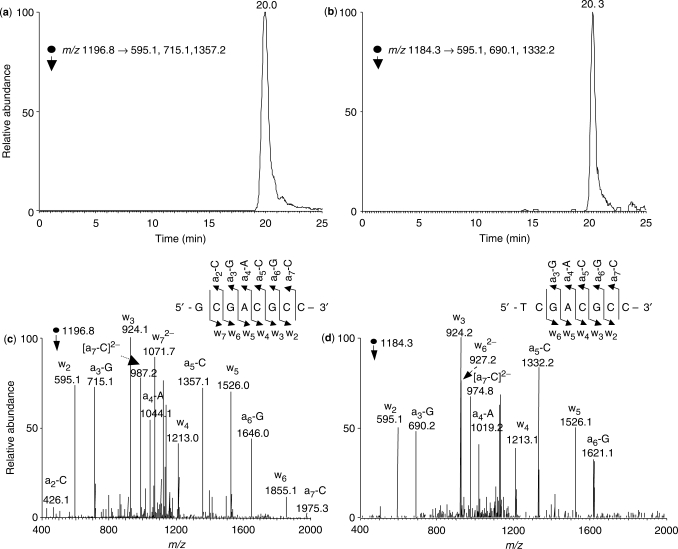
The assay begins with the insertion of the lesion under study at a defined site in a replicable single-stranded genome, i.e. M13mp7(L2) (Figure 2). After in vivo replication, the progeny genome was isolated and the DNA sequence of interest was amplified by PCR. The PCR products were digested with BbsI and Tsp509I, and the digestion products were subjected to LC-MS/MS analysis (Figure 2). Because MS was employed for interrogating the restriction fragments of replication products, we termed the method ‘restriction endonuclease and mass spectrometry’ assay, or REAMS assay. In the original REAP assay, the identity and frequency of mutation occurring at the lesion site were determined by [32P]-labeling the site that originally had the lesion and resolving at the end the 5′-[32P]dNTP by thin-layer chromatography. In the CRAB assay, the lesion bypass efficiency was calculated from the rise in the relative amount of the restriction fragment from the competitor genome over the lesion-containing genome. In our method, the restriction fragments were subjected directly to LC-MS/MS analysis under the similar conditions as described recently (38). In this context, we monitored specifically the fragmentation of the [M–2H]2− ions of the 8-mer full-length products where the nucleobase incorporated at the original 3′ cytosine site was mutated to a guanine, adenine or thymine. However, none of these products were detectable. We then monitored the fragmentation of the doubly deprotonated ions of d(XCGACGCC), where X is a A, T, C or G, it turned out all of them except d(ACGACGCC) could be found [The SICs and tandem mass spectra for d(GCGACGCC) and d(TCGACGCC) identified from the restriction digestion mixture are shown in Figure 3]. Moreover, we attempted, but failed to detect the −1 and −2 frameshift mutation products emanating from the replication of the G[8-5]C-bearing genome.
Figure 3.
LC-MS/MS for monitoring the restriction fragments of interest without mutation or with a G→T mutation at the original guanine portion of the lesion [i.e. d(GCGACGCC) and d(TCGACGCC)]. Shown in (a) and (b) are the SICs for the formation of indicated fragment ions of these two ODNs, and illustrated in (c) and (d) are the MS/MS of the [M–2H]2− ions of these two ODNs (m/z 1196.9 and 1184.3). Nomenclature for fragment ions follow that described by McLuckey et al. (56).
We were also able to find the restriction fragment from the progeny of the competitor genome, d(GCTAGCTGCGG) (11-mer). We then quantified the relative amounts of different replication products by LC-MS/MS with the consideration of difference in ionization efficiencies for different ODNs (calibration curves are shown in Figure S7) (38). The relative amounts of d(GCGACGCC) (8G), d(TCGACGCC) (8T), d(CCGACGCC) (8C) allowed for the calculation of mutation frequencies. The bypass efficiency was then calculated from the ratio of the total amount of 8G, 8T and 8C, which were from the replication of the lesion-containing genome, over the amount of the 11-mer, which was the replication product of the competitor genome. In this respect, the lesion/competitor genome ratio employed for the initial transformation was also considered for the calculation.
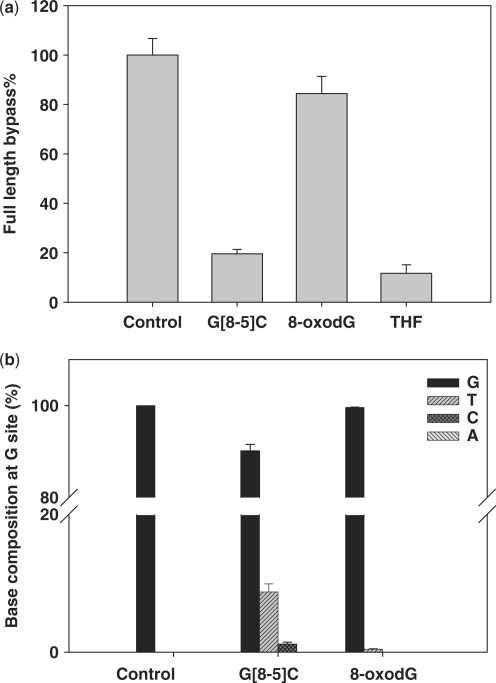
Our results with the G[8-5]C intrastrand cross-link revealed that the lesion blocked considerably DNA replication in vivo as represented by a 20% bypass efficiency in the wild-type AB1157 strain (Figure 4). The mutation frequency of the 5′ guanine moiety of G[8-5]C was significant, which included 8.7% G→T and 1.2% G→C transversion mutations (Figure 4).
Figure 4.
In vivo bypass efficiencies and mutation frequencies of DNA lesions in wild-type AB1157 E. coli cells determined by the REAMS assay. The data represent the means and standard deviations of results from three independent transformation and LC-MS/MS experiments.
The results showed that the LC-MS/MS method afforded a detection limit of 10 fmol for 8T, 8G, 8C and 8A at a signal-to-noise ratio (S/N) of greater than 10. Therefore, the method allowed for a detection of 0.2% mutation frequency if restriction fragments from 5 pmol of PCR products were injected for analysis. This detection limit is comparable to that offered by the traditional REAP assay which used [32P]-post-labeling for the quantification (28).
The LC-MS/MS method for the study of the cytotoxicity and mutagenicity of DNA lesions obviates the use of [32P]-labeling and enables the simultaneous monitoring of the bypass efficiencies and mutation frequencies of DNA lesions. In addition, the REAMS method interrogates a short piece of ODN carrying the original lesion site rather than examining only the nucleotide at the initial lesion site in the REAP assay. Therefore, the sequence information offered by MS/MS provides unambiguous identification of the replication products and facilitates the identification of replication products arising from mutations occurring at or near the original lesion site.
To compare our results with those obtained by the traditional REAP and CRAB assays, we also carried out control experiments by examining the lesion bypass and mutagenic properties of 8-oxodG and the THF analog of the abasic site. Our results revealed that 8-oxodG only inhibited weakly plasmid replication as represented by a 84% bypass efficiency and resulted in slight G→T mutation (0.5%), whereas the abasic site analog was a strong DNA replication blockade (bypass% = 11%, Figure 4) in wild-type AB1157 E. coli cells. These results were in agreement with previous findings obtained by the traditional CRAB and REAP assays (32,33), thereby validating our LC-MS/MS method for the in vivo replication studies.
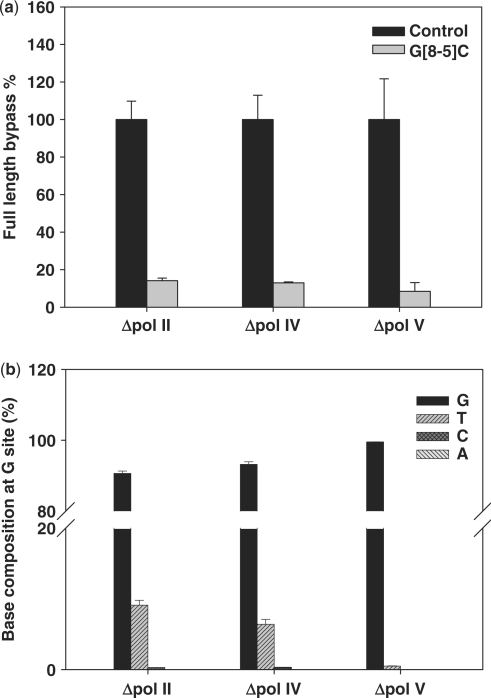
To determine which translesion synthesis DNA polymerase(s) is(are) involved in the bypass of the lesion in E. coli cells, we also examined the bypass efficiencies and mutation frequencies of the lesion in the isogenic E. coli strains (AB1157 strain, see Materials and Methods section) that are deficient in one of the three translesion synthesis DNA polymerases, pol II, pol IV and pol V (35). It turned out that the bypass efficiencies dropped from 20% for the wild-type strain to 14, 13 and 8% for the strains that are deficient in pol II, pol IV and pol V, respectively (Figure 5), though the changes are not statistically significant (P > 0.05). In addition, no significant difference in mutation frequency was found for the wild-type strain and strains that are deficient in either pol II or pol IV; however, the G→T and G→C mutations were almost completely abolished in the pol V-deficient strain (only 0.5% of G→T mutation could be detected). These results suggest that pol V was the major polymerase responsible for the error-prone translesion synthesis of G[8-5]C in E. coli cells.
Figure 5.
In vivo bypass efficiencies and mutation frequencies of G[8-5]C in pol II-, pol IV- and pol V-deficient AB1157 E. coli determined by REAMS assay. The data represent the means and standard deviations of results from three independent transformation and LC-MS/MS experiments.
DISCUSSION
We demonstrated, for the first time, the dose-dependent formation of the G[8-5]C intrastrand cross-link lesion in HeLa-S3 cells upon exposure to γ-rays. Although the formation rate for the cross-link lesion was low, that is, ∼300 times less than that of 5-FodU under identical exposure conditions (42), the formation of the bulky intrastrand cross-link lesions may have significant implications in human diseases.
First, these lesions may play an important role in the pathogenesis of human diseases associated with the aberrant accumulations of iron and copper. In this context, we demonstrated recently that iron and copper ions could induce the formation of intrastrand cross-link lesions of DNA in the presence of H2O2 via the Fenton-type reaction (14,15):
In humans, genetic hemochromatosis and Wilson's disease cause abnormal accumulation of iron and copper in various organs, and an accumulation of highly mutagenic oxidatively generated lesions in genomic DNA has been considered to be relevant to iron-induced carcinogenesis in iron-overload diseases (45). Additionally, bulky DNA lesions were found in the liver of patients with Wilson's disease and primary hemochromatosis (46), though the exact chemical structure of these lesions remains elusive. Oxidatively generated intrastrand cross-link lesions may account, in part, for the pathological conditions of these diseases. It is important to examine the formation of the intrastrand cross-link lesions in tissues from these patients in the future.
The oxidatively generated intrastrand cross-link lesions may also contribute significantly to neurodegeneration and cancer developments in patients suffering from genetic diseases with deficiencies in NER, e.g. XP, trichothiodystrophy and Cockayne's syndrome (47,48). In this respect, a hallmark of XP patients is the markedly increased risk to develop skin cancer, which is attributed to the inability of cells to repair sunlight-induced dimeric DNA photoproducts, where neighboring nucleobases, essentially dipyrimidines, are conjugated via the [2 + 2] photochemical reaction and Paternò-Büchi photocycloaddition (22,49). Because of the structure similarity between oxidatively generated intrastrand cross-link lesions and dipyrimidine photoproducts, the oxidatively generated intrastrand cross-link lesions are also expected to be repaired by the NER pathway. Consistent with this notion, we demonstrated that oxidatively generated intrastrand cross-link lesions G[8-5]C, G[8-5m]mC and G[8-5m]T could indeed be recognized by the E. coli NER machinery, and the efficiency for the cleavage of these lesion-carrying substrates were comparable as those for the cleavage of dipyrimidine photoproducts (16). Along this line, it was found that XP patients often develop neurological abnormalities due to premature neuronal death (22). Neurons are not exposed to sunlight; thus, DNA lesions formed from other endogenous mutagens (e.g. ROS) contribute to those neuron losses. The ROS-induced DNA intrastrand cross-link lesions are expected to accumulate gradually in the long-lived, non-dividing, albeit transcriptionally active neurons of XP patients thereby contributing to neurodegeneration in these patients. In this context, dipyrimidine photoproducts are known to block considerably transcription by RNA polymerases (50–52); a structurally related oxidatively generated DNA lesion, 8,5′-cyclo-2′-deoxyadenosine also inhibits transcription pronouncedly (53,54). It is important to examine how oxidatively generated intrastrand cross-link lesions affect transcription in the future.
By using the newly developed mass spectrometry-based assay, we demonstrated for the first time that the G[8-5]C cross-link lesion blocked markedly in vivo DNA replication, it also induced considerable G→T (8.7%) and G→C mutations (1.2%). Replication studies with the isogenic E. coli strains that are defective in SOS-induced bypass DNA polymerases demonstrated that pol V was responsible for the mutagenic bypass of the lesion in vivo.
This result is also in line with in vitro replication results showing that the pol V homolog in yeast, i.e. yeast pol η, is capable of synthesizing past the lesion in an error-prone fashion (9). In this context, the in vitro replication studies with yeast polymerase η showed that the cytosine portion of the G[8-5]C intrastrand cross-link is not mutagenic; the polymerase inserts predominantly a dGMP opposite the 3′ cytosine portion (9). On the other hand, the polymerase exhibits significant mis-incorporations of dAMP and dGMP opposite the guanine portion of the lesion (9), which can result in G→T and G→C mutations for the 5′ guanine portion of the lesion. In this context, it is worth noting that pol V is also important for the efficient bypass of a group of structurally diverse and highly mutagenic guanine oxidation products in vivo (36).
We reason that the presence of the cross-link lesion may lead to a structural distortion to duplex DNA, which may render the hydrogen-bonding property of the 5′-guanine portion not to be recognized by translesion synthesis DNA polymerases during nucleotide insertion. Indeed, molecular modeling results showed that the N-glycosidic linkage of the 3′ nucleoside of the cross-link assumes an ‘anti’ configuration, whereas that of the 5′ nucleoside exhibits a syn configuration (9). Under such circumstances, purine nucleotides can conceivably be incorporated opposite the 5′-G more efficiently than pyrimidine nucleotides because the former nucleotides exhibit a stronger stacking interaction with the 3′ nucleotide of the primer than the latter nucleotides (55).
Taken together, we reported for the first time the in vivo formation and the cytotoxic and mutagenic properties of the G[8-5]C lesion. In addition, the REAMS assay introduced here can be generally applicable for the examination of the replication of other lesions in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Prof. John M. Essigmann and Prof. Graham Walker at MIT for providing E. coli strains and M13 shuttle vector and the National Cell Culture Center for providing the HeLa-S3 cells. The authors also want to thank Dr James C. Delaney in the Essigmann group for the generous help with the in-vivo mutagenesis assay. This work was supported by the National Institutes of Health (R01 CA96906 and R01 CA101864 to Y.W.) and the University of California Toxic Substances Research and Teaching Program (to H.H.). Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. DNA lesions generated in vivo by reactive oxygen species, their accumulation and repair. NATO ASI Ser., Ser. A: Life Sci. 1999;302:251–257. [Google Scholar]
- 3.Marnett L.J. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 4.Bellon S., Ravanat J.L., Gasparutto D., Cadet J. Cross-linked thymine-purine base tandem lesions: synthesis, characterization, and measurement in gamma-irradiated isolated DNA. Chem. Res. Toxicol. 2002;15:598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- 5.Box H.C., Budzinski E.E., Dawidzik J.B., Gobey J.S., Freund H.G. Free radical-induced tandem base damage in DNA oligomers. Free Radic. Biol. Med. 1997;23:1021–1030. doi: 10.1016/s0891-5849(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 6.Box H.C., Budzinski E.E., Dawidzik J.B., Wallace J.C., Iijima H. Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res. 1998;149:433–439. [PubMed] [Google Scholar]
- 7.Box H.C., Budzinski E.E., Dawidzik J.D., Wallace J.C., Evans M.S., Gobey J.S. Radiation-induced formation of a crosslink between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA) Radiat. Res. 1996;145:641–643. [PubMed] [Google Scholar]
- 8.Budzinski E.E., Dawidzik J.B., Rajecki M.J., Wallace J.C., Schroder E.A., Box H.C. Isolation and characterization of the products of anoxic irradiation of d(CpGpTpA) Int. J. Radiat. Biol. 1997;71:327–336. doi: 10.1080/095530097144210. [DOI] [PubMed] [Google Scholar]
- 9.Gu C., Wang Y. LC-MS/MS identification and yeast polymerase η bypass of a novel γ-irradiation-induced intrastrand cross-link lesion G[8-5]C. Biochemistry. 2004;43:6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- 10.Romieu A., Bellon S., Gasparutto D., Cadet J. Synthesis and UV photolysis of oligodeoxynucleotides that contain 5-(phenylthiomethyl)-2′-deoxyuridine: a specific photolabile precursor of 5-(2′-deoxyuridilyl)methyl radical. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q., Wang Y. The reactivity of the 5-hydroxy-5,6-dihydrothymidin-6-yl radical in oligodeoxyribonucleotides. Chem. Res. Toxicol. 2005;18:1897–1906. doi: 10.1021/tx050195u. [DOI] [PubMed] [Google Scholar]
- 12.von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor & Francis; 1987. [Google Scholar]
- 13.Zhang Q., Wang Y. Independent generation of 5-(2′-deoxycytidinyl)methyl radical and the formation of a novel cross-link lesion between 5-methylcytosine and guanine. J. Am. Chem. Soc. 2003;125:12795–12802. doi: 10.1021/ja034866r. [DOI] [PubMed] [Google Scholar]
- 14.Hong H., Cao H., Wang Y., Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao H., Wang Y. Quantification of oxidative single-base and intrastrand cross-link lesions in unmethylated and CpG-methylated DNA induced by Fenton-type reagents. Nucleic Acids Res. 2007;35:4833–4844. doi: 10.1093/nar/gkm497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu C., Zhang Q., Yang Z., Wang Y., Zou Y., Wang Y. Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry. 2006;45:10739–10746. doi: 10.1021/bi060423z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z., Colis L.C., Basu A.K., Zou Y. Recognition and incision of gamma-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chem. Res. Toxicol. 2005;18:1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellon S., Gasparutto D., Saint-Pierre C., Cadet J. Guanine-thymine intrastrand cross-linked lesion containing oligonucleotides: from chemical synthesis to in vitro enzymatic replication. Org. Biomol. Chem. 2006;4:3831–3837. doi: 10.1039/b609460k. [DOI] [PubMed] [Google Scholar]
- 19.Gu C., Wang Y. Thermodynamic and in vitro replication studies of an intrastrand G[8-5]C cross-link lesion. Biochemistry. 2005;44:8883–8889. doi: 10.1021/bi050036+. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y., Hong H., Wang Y. In-vivo formation and in-vitro replication of a guanine-thymine intrastrand cross-link lesion. Biochemistry. 2007;46 doi: 10.1021/bi7012195. doi: 10.1021/bi7012195, in press. [DOI] [PubMed] [Google Scholar]
- 21.Cleaver J.E., Kraemer K.H. Xeroderma pigmentosum. In: Scriver C., Beaudet A.L., Sly W.S., Vale D., editors. The Metabolic Basis of Inherited Disease. Vol. 2. New York: McGraw-Hill; 1989. pp. 2949–2971. [Google Scholar]
- 22.Cleaver J.E. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 23.Cleaver J.E. Xeroderma pigmentosum: the first of the cellular caretakers. Trends Biochem. Sci. 2001;26:398–401. doi: 10.1016/s0968-0004(01)01833-3. [DOI] [PubMed] [Google Scholar]
- 24.Satoh M.S., Jones C.J., Wood R.D., Lindahl T. DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical-induced base lesions. Proc. Natl Acad. Sci. USA. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reardon J.T., Bessho T., Kung H.C., Bolton P.H., Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl Acad. Sci. USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd R.S. Investigations of pyrimidine dimer glycosylases—a paradigm for DNA base excision repair enzymology. Mutat. Res. 2005;577:77–91. doi: 10.1016/j.mrfmmm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C–>T.A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaney J.C., Essigmann J.M. Context-dependent mutagenesis by DNA lesions. Chem. Biol. 1999;6:743–753. doi: 10.1016/s1074-5521(00)80021-6. [DOI] [PubMed] [Google Scholar]
- 29.Delaney J.C., Essigmann J.M. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl Acad. Sci. USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaney J.C., Essigmann J.M. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 31.Delaney J.C., Smeester L., Wong C., Frick L.E., Taghizadeh K., Wishnok J.S., Drennan C.L., Samson L.D., Essigmann J.M. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 32.Henderson P.T., Delaney J.C., Gu F., Tannenbaum S.R., Essigmann J.M. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 33.Henderson P.T., Delaney J.C., Muller J.G., Neeley W.L., Tannenbaum S.R., Burrows C.J., Essigmann J.M. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 34.Delaney J.C., Essigmann J.M. Effect of sequence context on O(6)-methylguanine repair and replication in vivo. Biochemistry. 2001;40:14968–14975. doi: 10.1021/bi015578f. [DOI] [PubMed] [Google Scholar]
- 35.Jarosz D.F., Beuning P.J., Cohen S.E., Walker G.C. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Neeley W.L., Delaney S., Alekseyev Y.O., Jarosz D.F., Delaney J.C., Walker G.C., Essigmann J.M. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 37.Apffel A., Chakel J., Fischer S., Lichtenwalter K., Hancock W. Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal. Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 38.Gu C., Wang Y. In-vitro replication and thermodynamic studies of methylation and oxidation modifications of 6-thioguanine. Nucleic Acids Res. 2007;35:3693–3704. doi: 10.1093/nar/gkm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong H., Wang Y. Formation of intrastrand cross-link products between cytosine and adenine from UV irradiation of d(BrCA) and duplex DNA containing a 5-bromocytosine. J. Am. Chem. Soc. 2005;127:13969–13977. doi: 10.1021/ja0531677. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q., Wang Y. Independent generation of the 5-hydroxy-5,6-dihydrothymidin-6-yl radical and its reactivity in dinucleoside monophosphates. J. Am. Chem. Soc. 2004;126:13287–13297. doi: 10.1021/ja048492t. [DOI] [PubMed] [Google Scholar]
- 41.Zeng Y., Wang Y. Facile formation of an intrastrand cross-link lesion between cytosine and guanine upon Pyrex-filtered UV light irradiation of d(BrCG) and duplex DNA containing 5-bromocytosine. J. Am. Chem. Soc. 2004;126:6552–6553. doi: 10.1021/ja049760q. [DOI] [PubMed] [Google Scholar]
- 42.Hong H., Wang Y. Derivatization with Girard reagent T combined with LC-MS/MS for the sensitive detection of 5-formyl-2′-deoxyuridine in cellular DNA. Anal. Chem. 2007;79:322–326. doi: 10.1021/ac061465w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pouget J.P., Frelon S., Ravanat J.L., Testard I., Odin F., Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat. Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 44.D'Errico M., Parlanti E., Teson M., Degan P., Lemma T., Calcagnile A., Iavarone I., Jaruga P., Ropolo M., et al. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene. 2007;26:4336–4343. doi: 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- 45.Toyokuni S. Iron-induced carcinogenesis: the role of redox regulation. Free Radic. Biol. Med. 1996;20:553–566. doi: 10.1016/0891-5849(95)02111-6. [DOI] [PubMed] [Google Scholar]
- 46.Carmichael P.L., Hewer A., Osborne M.R., Strain A.J., Phillips D.H. Detection of bulky DNA lesions in the liver of patients with Wilson's disease and primary haemochromatosis. Mutat. Res. 1995;326:235–243. doi: 10.1016/0027-5107(94)00177-7. [DOI] [PubMed] [Google Scholar]
- 47.Petit C., Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 48.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 49.Taylor J.-S. DNA, sunlight and skin cancer. Pure Appl. Chem. 1995;67:183–190. [Google Scholar]
- 50.Mei Kwei J.S., Kuraoka I., Horibata K., Ubukata M., Kobatake E., Iwai S., Handa H., Tanaka K. Blockage of RNA polymerase II at a cyclobutane pyrimidine dimer and 6-4 photoproduct. Biochem. Biophys. Res. Commun. 2004;320:1133–1138. doi: 10.1016/j.bbrc.2004.06.066. [DOI] [PubMed] [Google Scholar]
- 51.Tornaletti S., Reines D., Hanawalt P.C. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J. Biol. Chem. 1999;274:24124–24130. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donahue B.A., Yin S., Taylor J.S., Reines D., Hanawalt P.C. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brooks P.J., Wise D.S., Berry D.A., Kosmoski J.V., Smerdon M.J., Somers R.L., Mackie H., Spoonde A.Y., Ackerman E.J., et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 54.Marietta C., Gulam H., Brooks P.J. A single 8,5′-cyclo-2′-deoxyadenosine lesion in a TATA box prevents binding of the TATA binding protein and strongly reduces transcription in vivo. DNA Repair. 2002;1:967–975. doi: 10.1016/s1568-7864(02)00148-9. [DOI] [PubMed] [Google Scholar]
- 55.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 56.McLuckey S.A., Van Berkel G.J., Glish G.L. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.