Abstract
To initiate ϕ29 DNA replication, the DNA polymerase has to form a complex with the homologous primer terminal protein (TP) that further recognizes the replication origins of the homologous TP-DNA placed at both ends of the linear genome. By means of chimerical proteins, constructed by swapping the priming domain of the related ϕ29 and GA-1 TPs, we show that DNA polymerase can form catalytically active heterodimers exclusively with that chimerical TP containing the N-terminal part of the homologous TP, suggesting that the interaction between the polymerase TPR-1 subdomain and the TP N-terminal part is the one mainly responsible for the specificity between both proteins. We also show that the TP N-terminal part assists the proper binding of the priming domain at the polymerase active site. Additionally, a chimerical ϕ29 DNA polymerase containing the GA-1 TPR-1 subdomain could use GA-1 TP, but only in the presence of ϕ29 TP-DNA as template, indicating that parental TP recognition is mainly accomplished by the DNA polymerase. The sequential events occurring during initiation of bacteriophage protein-primed DNA replication are proposed.
INTRODUCTION
Replication of linear chromosomes raises the question how their ends are maintained through subsequent replication rounds. This dilemma, called the end replication problem (1,2), comes up from two main observations. On the one hand, all DNA polymerases synthesize a new complementary DNA strand exclusively in the 5′–3′ direction and, on the other hand, replicative DNA polymerases normally require a short RNA molecule to prime DNA synthesis. From this, it could be inferred that the DNA sequence paired to the most distal RNA primer would be lost in the daughter strand, leading to a continuous shortening of the linear chromosome, with the eventual loss of the information placed at its ends. In higher eukaryotes, telomerase prevents chromosome ends shortening by extending the 3′ end of its DNA substrate using, in a repetitive fashion, its own RNA as template, providing a single-stranded DNA (ssDNA) composed of many telomeric sequences that will be copied by the replication machinery (3). Such a quandary is not restricted to eukaryotic chromosomes, but it is also present during replication of any linear DNA. Several strategies have been developed by evolutionary distant organisms to prevent the loss of their DNA ends. Thus, organisms as phage λ recircularize their genomes by means of the complementarity of the 5′-ends (4); others, as phage T7, contain a linear double-stranded DNA (dsDNA) with a terminal direct repetition used to form concatemers during replication (5); and DNA molecules as those of poxviruses or linear plasmids of Borrelia contain a terminal covalently closed hairpin loop (6–9).
Several prokaryotic and eukaryotic viruses, as well as linear plasmids from bacteria, fungi and higher plants, and even Streptomyces spp. have solved the end replication problem by using a protein (called Terminal Protein; TP) to prime DNA synthesis from the very end of their linear genomes (10–12). In these cases, an amino acid residue of the primer TP provides the priming OH group, becoming covalently linked to the 5′- end of the DNA (parental TP). In vitro replication analyses, mainly performed with bacteriophage ϕ29, have laid the foundations for this so-called protein-priming replication mechanism. The complex formed between the replicative DNA polymerase and a free TP molecule interacts with the replication origins at both ends of the genome by specific recognition of the parental TP and DNA sequences. The DNA polymerase catalyses the incorporation of a specific dNMP onto the priming OH group of the TP, in a reaction directed by an internal dNMP in the template strand (initiation reaction). The presence of repetitive sequences at the replication origins in these genomes allows the initiation complex to recover the terminal nucleotides by sliding-back, in the case of bacteriophages ϕ29, PRD1, GA-1 and Cp1 (13–16), or jumping-back, as in adenovirus (17). The ϕ29 DNA polymerase/primer TP heterodimer does not dissociate after initiation or the sliding-back step. There is a transition stage in which the DNA polymerase synthesizes a 5-nt-long DNA molecule while complexed with the primer TP, undergoes some structural change during incorporation of nucleotides 6–9 (transition) and dissociates from the primer TP when nucleotide 10 is incorporated into the nascent DNA chain (elongation mode) (18). Finally, the same DNA polymerase catalyses chain elongation via a strand displacement mechanism to fulfil TP-DNA replication (11).
Recent crystallographic resolution of ϕ29 DNA polymerase/TP complex shows that TP forms an extended structure that is complementary to the DNA polymerase surface (19). TP is folded into an N-terminal domain, an intermediate domain that interacts with the DNA polymerase subdomain TPR-1, and a priming domain that occupies the DNA-binding site in the polymerase (20). This fact precludes the initiation at internal sites, as an upstream 3′ template would sterically clash with TP, restricting the beginning of DNA synthesis at the ends of the genome (19).
Besides DNA polymerase and TP, bacteriophage ϕ29 replication in vivo requires the presence of phage encoded ss- and dsDNA binding proteins (SSB and DBP, respectively). ϕ29 SSB covers the displaced ssDNA to prevent its degradation by cellular nucleases as well as to preclude the appearance of short palindromic DNAs that would be detrimental for phage DNA replication (21). ϕ29 DBP organizes and compacts the viral genome (22), and specifically activates the initiation of replication (23) by forming multimeric nucleoprotein complexes at the ends of the TP-DNA (24,25).
Thus, in spite of its simplicity, ϕ29 TP-DNA replication requires a number of specialized proteins to perform the reactions necessary to prime and elongate the nascent strand from both replication origins. These proteins must interact properly with each other to ensure successful DNA replication. Replication systems developed with purified proteins and DNAs from the ϕ29-related bacteriophages Nf and GA-1 have shown that the reactions catalysed by each of these three systems are similar. However, in spite of their resemblance, DNA polymerases, TPs and DBPs, cannot be interchanged individually, as they are unable to network correctly with the other components of the heterologous replication system because of the high specificity of such protein–protein interactions (26–29).
By the use of chimerical proteins, generated by swapping specific domains of the TP and DNA polymerase of both ϕ29 and GA-1 bacteriophages, we show that the DNA polymerase–TP interaction is specified mainly by the contacts established between the TP intermediate domain and the DNA polymerase TPR-1 subdomain. In addition, we show that the specific recognition of the replication origin (parental TP) and the DBP is mainly carried out by the DNA polymerase.
MATERIALS AND METHODS
Nucleotides and DNAs
Unlabelled nucleotides were purchased from Amersham Pharmacia Biochemicals. [α-32P]dATP [3000 Ci/mmol (1 Ci = 37 GBq)] was obtained from Amersham Pharmacia. Oligonucleotides were obtained from Isogen. ϕ29 and GA-1 TP-DNA were obtained as described (30). Plasmids pET-28a(+)® and pET-28b(+)® were purchased from Novagen.
Proteins
Wild-type ϕ29 DNA polymerase was purified from Escherichia coli NF2690 cells harbouring plasmid pJLPM (a derivative of pT7-4w2), as described (31). Wild-type GA-1 DNA polymerase and wild-type ϕ29 and GA-1 TPs were expressed in E. coli BL21(DE3) cells harbouring the gene cloned into plasmid pT7-4 (DNA polymerase) and pT7-3 (TPs) and further purified as described (29,31,32). ϕ29 DBP and SSB, obtained from Bacillus subtilis cells infected with phage ϕ29, were purified as described (27,33).
Construction, expression and purification of ϕ29 and GA-1 TP variants and chimerical ϕ29 DNA polymerase
Table 1 shows the DNA polymerases and TPs used in this study. For details about the construction, expression and purification of the different DNA polymerase and TP variants, see Supplementary Data.
Table I.
DNA polymerase and terminal protein variants
| DNA polymerase | ϕ29 | Wild-type ϕ29 DNA polymerase |
| GA-1 | Wild-type GA-1 DNA polymerase | |
| Chimera | ϕ29 DNA polymerase containing GA-1 DNA polymerase TPR-1 subdomain (residues 261–358) | |
| ϕ29 | Wild-type ϕ29 TP | |
| GA-1 | Wild-type GA-1 TP | |
| N-ϕ | Chimerical TP containing the ϕ29 TP N-terminal part (residues 1–173) and GA-1 TP priming domain (residues 174–265). | |
| Terminal protein | NG | Chimerical TP containing the GA-1 TP N-terminal part (residues 1–173) and ϕ29 TP priming domain (residues 174–266). |
| ϕ29-Ct | ϕ29 TP priming domain (residues 174–266) | |
| ϕ29-Nt | ϕ29 TP N-terminal part (residues 1–173) | |
| ϕ29ΔN | ϕ29 TP lacking N-terminal domain (residues 1–73) |
Protein-primed initiation assay (TP-dAMP formation)
TP-primed initiation assay performed with wild-type ϕ29 or GA-1 DNA polymerases and TP variants
The capacity to carry out the initiation step of TP-DNA replication was analysed as described (34), in the presence of either 1 mM MnCl2 or, when indicated, 10 mM MgCl2, 1.6 nM of either ϕ29 or GA-1 TP-DNA as template, 0.1 µM dATP [α-32dATP] (1 µCi), 15 nM of either ϕ29 or GA-1 DNA polymerase and the indicated amount of either wild-type, chimerical, ϕ29▵N or ϕ29-Ct mutant TPs in 25 µl of reaction volume. In the last case, when indicated, 150 nM of the TP variant ϕ29-Nt and 35 µM of ϕ29 DBP were also added. After incubation for the indicated times at 30°C, samples were processed and analysed as previously described (30). The reactions in which the TP derivative ϕ29-Ct was used as primer were analysed in Tris–tricine–SDS gels. Quantification was done by densitometric analysis of the labelled band corresponding to the TP-dAMP complex.
Calculation of apparent Km for dATP was carried out as described (35), using increasing amounts of dATP. Reactions were stopped and analysed by SDS–polyacrylamide gel electrophoresis (PAGE). Formation of the product was plotted against dATP concentration. Apparent values for Michaelis–Menten constant (Km) for nucleotide incorporation was obtained by least squares non-linear regression to rectangular hyperbola using Kaleidagraph 3.6.4 software.
TP-primed initiation assay performed with chimerical ϕ29 DNA polymerase and TP variants
The assay was carried out as described above, in the presence of 1 mM MnCl2, 1.6 nM of either ϕ29 or GA1 TP-DNA or 5.6 µM of single-stranded oligonucleotide LOT12 (ssDNA LOT12; 5′GTGGGGGCTTACTTT), which contains the ϕ29 replication origin sequence. In the presence of TP-DNA, 120 nM of ϕ29 chimerical DNA polymerase and 240 nM of the indicated TP were used. As a control, an initiation reaction, with 15 nM of ϕ29 wild-type DNA polymerase and 30 nM of ϕ29 wild-type TP, was carried out. After incubation for 10 min at 30°C, samples were stopped and processed as described above. When the effect of ϕ29 DBP was studied, the assays were performed in the presence of 60 nM of chimerical DNA polymerase and 120 nM of the indicated TP, in the absence or presence of 35 µM of ϕ29 DBP. As a control, an initiation reaction with 30 nM of ϕ29 wild-type DNA polymerase and 60 nM of ϕ29 wild-type TP was carried out. Samples were incubated for the indicated times at 30°C and further processed as described above. When the template used was ssDNA LOT12 the assay was performed in the presence of 300 nM of either wild type or chimerical ϕ29 DNA polymerase and 600 nM of the indicated TP. In this case, samples were incubated for 10 min at 30°C.
In the case of the template-independent initiation assay, TP-DNA was omitted, 120 nM of ϕ29 DNA polymerase, 240 nM of either wild-type or mutant ϕ29▵N TP and 1 mM MnCl2 were added and the incubation was maintained for the indicated time at 30°C. The reactions were stopped by adding 10 mM EDTA and 0.1% SDS, filtered through Sephadex G-50 spin columns, and further analysed by SDS–PAGE as described (30). Quantification was done by densitometry of the labelled band corresponding to the TP-dAMP complex, detected by autoradiography.
Interference assay for DNA polymerase binding
With this assay the relative DNA polymerase binding efficiency of mutant TPs with respect to the wild-type TP is estimated by adding simultaneously both TPs and a limited amount of DNA polymerase. Reactions were carried out as described for the template-dependent initiation assay, using a limiting amount of either ϕ29 or GA-1 DNA polymerase and different proportions of a mixture of wild-type and mutant TPs. Thus, 60 nM of ϕ29 DNA polymerase was incubated with 120 nM of wild-type ϕ29 TP, 1 mM MnCl2 and increasing amounts of chimera N-ϕ (120, 240, 480, 960 and 1920 nM). Similarly, 60 nM of GA-1 DNA polymerase was incubated with 120 nM of wild-type GA-1 TP and increasing amounts of chimera N-G (120, 240, 480, 960 and 1920 nM). To analyse the competition capacity of the ϕ29 TP priming domain, 60 nM of ϕ29 DNA polymerase was incubated with 120 nM of chimerical TP N-ϕ, 1 mM MnCl2 and increasing amounts of the truncated TP ϕ29-Ct (120, 240, 480, 960 and 1920 nM). In all cases, the incubation was for 10 min at 30°C. After incubation, reactions were stopped and analysed as indicated for the protein-primed initiation assay.
Replication assay (protein-primed initiation plus elongation) using ϕ29 TP-DNA as template
The assay was performed essentially as described (36), in 25 µl of reaction volume, in the presence of the indicated metal activator, dNTPs concentration, wild-type DNA polymerase, TP, [α-32dATP] (1 µCi) and 1.6 nM of either ϕ29 or GA-1 TP-DNA as template. After incubation for the indicated time at 30°C, the reactions were stopped by adding 10 mM EDTA and 0.1% SDS and filtered through Sephadex G-50 spin columns. For size analysis, the labelled DNA was denatured by treatment with 0.7 M NaOH and subjected to electrophoresis in alkaline 0.7% agarose gels as described (37). After electrophoresis, the position of unit length TP-DNA was detected by ethidium bromide staining, and the gels were dried and autoradiographed.
TP-DNA amplification assay
The assay was performed essentially as described (38), in the presence of 16 pM ϕ29 TP-DNA, 30 nM of wild-type ϕ29 DNA polymerase, 120 nM of either wild-type ϕ29 TP or ϕ29▵N, 30 µM of ϕ29 SSB and 35 µM of ϕ29 DBP, in a final reaction volume of 25 µl. After incubation for 45 min at 30°C, the samples were processed and the amplified DNA analysed by electrophoresis in alkaline agarose gels, as described (38). After electrophoresis, the position of unit length ϕ29 TP-DNA was detected by ethidium bromide staining.
RESULTS
Modelling of GA-1 TP. Construction of chimerical TPs
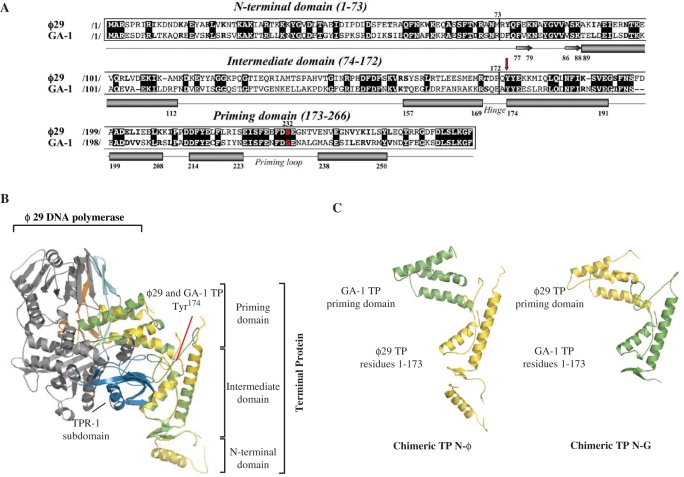
The recent structure determination of the ϕ29 DNA polymerase/TP complex has provided the first definition of the architecture of a TP (19). Thus, ϕ29 TP (266 amino acids long) is formed by a disordered N-terminal domain, followed by the intermediate domain that is constituted mainly by two long α-helices, and connected through a flexible hinge region to the priming domain, comprised of a four helix bundle (19) (Figure 1A and B). The loop in between the last two α-helices contains the Ser232 residue whose hydroxyl group is used as primer by the DNA polymerase to start initiation of phage DNA replication (10–12).
Figure 1.
(A) Alignment of the amino acid sequence of ϕ29 and GA-1 TPs. Numbers between slashes indicate the amino acid position relative to the N-terminal end of each TP. Identical residues are indicated in white letters over a black background. Other similarities are indicated by bold letters. The priming serine residue of both TPs is shown in red. The different secondary structural elements of TPs are depicted below the sequence alignment (α-helices and β-sheets are represented as cylinders and arrows, respectively), according to Ref. (19). Red arrow indicates the N-terminal residue of the swapped priming domains in chimerical TPs. (B) Ribbon representation of ϕ29 and modelled GA-1 TPs (coloured in yellow and green, respectively) complexed to ϕ29 DNA polymerase (19). Model for GA-1 TP was provided by the homology-modelling server Swiss-Model, using as template the crystallographic structure of ϕ29 TP (PDB code 2EX3). ϕ29 DNA polymerase TPR-1, TPR-2 and thumb subdomains are coloured in blue, cyan and orange, respectively. The structural domains constituting both TPs are also indicated. (C) Schematic representation of the chimerical TPs constructed for this study.
Bacteriophage GA-1 TP is a 265 amino acids long polypeptide that shares 40% of sequence identity with ϕ29 TP (15) (Figure 1A). Based on the high degree of identity between these two TPs, the protein structure homology-modelling server Swiss-Model (39,40) supplied a model for GA-1 TP (coloured in green in Figure 1B), obtained by using the crystallographic structure of ϕ29 TP as template [in yellow in Figure 1B; PDB 2EX3 (19)]. The predicted structure exhibits both the intermediate and priming domains folded as in ϕ29 TP, enabling us to model GA-1 TP complexed with the DNA polymerase (Figure 1B). The absence of a model for the GA-1 TP N-terminal domain is due to the fact that the two helices within this domain in ϕ29 TP were originally built as poly-alanine. As previously described (19), the intermediate domain of TP would pack against the TPR-1 subdomain of the DNA polymerase (coloured in dark blue in Figure 1B) while the TP priming domain would be encircled by the DNA polymerase TPR-2, thumb and TPR-1 subdomains [in cyan, orange and dark blue, respectively, in Figure 1B; PDB 2EX3 (19)].
Previous studies showed that both, ϕ29 and GA-1 DNA polymerases display a great specificity for their corresponding TP, as the heterologous systems did not give any detectable initiation product (29). Similar results were obtained by interchanging the DNA polymerase and TP from the closely related bacteriophages ϕ29 and Nf, in spite of the high amino acid sequence identity shared by the two TPs (62.4%) and DNA polymerases (81.8%) (28), revealing also the existence of a specific recognition by the DNA pol/TP complex of parental TP, which forms part of the replication origin (28).
In order to assess the importance of the TP domains in relation with its interaction with the homologous DNA polymerase and parental TP, we have made chimerical TPs by swapping the priming domains of both ϕ29 and GA-1 TPs (see Materials and Methods section). The conserved sequence ‘YYE’ at the beginning of the first α-helix of the priming domain in both TPs (first Tyr174 is indicated with a red arrow in Figure 1A) denotes the N-terminal limit of such exchanged regions to guarantee the proper folding of the heterologous domains in the resulting chimeras N-ϕ [ϕ29 TP N-terminal part containing the N-terminal and intermediate domains (residues 1–173) linked to the GA-1 TP priming domain] and N-G (GA-1 TP N-terminal part connected to the ϕ29 TP priming domain) (Figure 1C and Table 1). Chimeras were overexpressed and purified as described in Materials and Methods section, and their specific recognition of the DNA polymerase and TP-DNA were evaluated in in vitro assays corresponding to different stages of the TP-primed DNA replication process.
The N-terminal part of TP specifies the interaction with the DNA polymerase
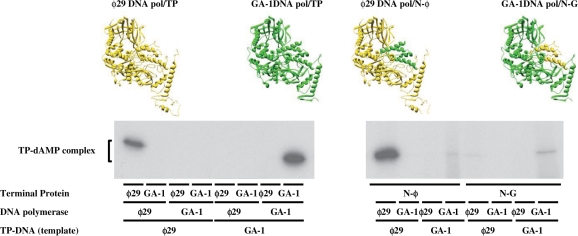
Replication of ϕ29 and GA-1 TP-DNA starts at both terminal origins with the DNA polymerase-catalysed template-directed insertion of 5′-dAMP onto the hydroxyl group of ϕ29 TP Ser232 and, most likely, of GA-1 TP Ser231 (initiation reaction), with the subsequent elongation (via strand displacement) of the initiation complex to produce full-length TP-DNA (10–12). As expected, each wild-type TP solely primed the initiation reaction catalysed by the homologous DNA polymerase, such a reaction being exclusively templated by the homologous TP-DNA (Figure 2; left part).
Figure 2.
In vitro protein-primed initiation with chimerical TPs. Reaction mixtures contained, in addition to the indicated wild-type DNA polymerases (15 nM), and wild-type (30 nM) or chimerical (120 nM) TPs, 1.6 nM of the indicated TP-DNA. Reactions were started by adding 1 mM MnCl2. After incubation for 1 min (wild-type systems) or 10 min (with chimerical TPs) at 30°C, the reactions were stopped, processed and analysed by SDS–PAGE and autoradiography (see Materials and Methods section for details). The various TP-DNAs, DNA polymerases and TP variants, as well as the mobility of the TP-dAMP complexes are indicated. For clarity, the active complexes are depicted on top of the figure. ϕ29 DNA polymerase and TP are coloured in yellow, whereas GA-1 DNA polymerase and the homologous TP are in green. The different parts of chimerical TPs are coloured according to such a colour code.
The reactions primed by the TP chimeras were less efficient than the ones primed by the wild-type TPs, higher protein doses and longer reaction times being required to observe initiation reaction (see Materials and Methods section), indicating that the high specificity between DNA polymerase and TP depends on both, the priming domain and the N-terminal part of the TP. Nonetheless, chimera N-ϕ, which holds the N-terminal part of ϕ29 TP, specifically primed the initiation reaction carried out by the ϕ29 DNA polymerase mainly when ϕ29 TP-DNA was used as template (Figure 2; right part). Only a very faint band was obtained when the GA-1 DNA polymerase and the GA-1 TP-DNA were used. These results would suggest that specific recognition between the DNA polymerase and the TP is mainly contributed through the N-terminal part of the TP, and that the TP priming domain is not involved in the recognition of the parental TP. In addition, ϕ29 DNA polymerase seems to show a less stringent requirement in placing the GA-1 TP priming domain in a catalytically proficient form. This argument was further supported from the fact that the apparent Km for the initiating dATP of chimera N-ϕ (0.9 µM) was only slightly higher than that of ϕ29 TP (0.6 µM). Although the amount of initiation product was much lower with chimera N-G (containing the N-terminal part of GA-1 TP) than with chimera N-ϕ, it displayed a similar specificity pattern, as it was mainly used as primer by GA-1 DNA polymerase in GA-1 TP-DNA templated reactions (Figure 2; right part). As before, a very faint band was obtained when the ϕ29 DNA polymerase and the ϕ29 TP-DNA were used. In the case of the chimera N-G, while the N-terminal part of the GA-1 TP confers specificity to the interaction with the DNA polymerase, the latter shows a high stringency in the placement of the non-homologous ϕ29 TP priming domain. In fact, this chimerical TP displayed a 14-fold increase in the Km for dATP (10 µM) with respect to GA-1 TP (0.7 µM). These results could indicate that GA-1 DNA polymerase/TP interaction displays a higher dependence on the priming domain than the ϕ29 DNA polymerase/TP heterodimer.
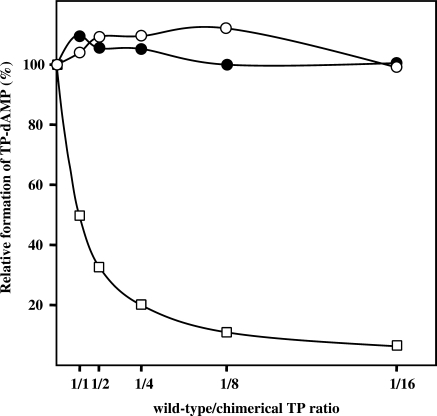
The differences in the apparent Km between the two chimerical TPs would justify their relative activity. However, the low activity exhibited by both chimeras in comparison with the wild-type TPs cannot be attributed only to Km differences, but it could also be contributed by an impaired DNA polymerase binding stability. Thus, the ability of chimeras N-ϕ and N-G to interact with ϕ29 and GA-1 DNA polymerase, respectively, was tested by using an interference assay in which, on the one hand ϕ29 TP and chimera N-ϕ and, on the other hand GA-1 TP and chimera N-G, compete for the ϕ29 and GA-1 DNA polymerase, respectively (see Materials and Methods section). As shown in Figure 3, none of the wild-type TPs were competed by the corresponding chimera, even at the highest chimera/wild-type TP ratio assayed. This result points to a weak interaction between DNA polymerase and chimerical TP.
Figure 3.
Competition for DNA polymerase between wild-type and chimerical TPs. The assay of formation of TP-dAMP initiation product by the wild-type ϕ29 and GA-1 DNA polymerase/TP heterodimers was performed in the presence of increasing amounts of chimeras N-ϕ (open circles) and N-G (filled circles), respectively (the reaction conditions are described under Materials and Methods section). Reactions were started by adding 1 mM MnCl2 and, after incubation for 10 min at 30°C, reactions were stopped and analysed as indicated for the protein-primed initiation assay. The TP-dAMP formed in the different competition conditions relative to that formed in the absence of competition (100%), as well as the theoretical inhibition profile (open squares) that would be obtained if chimerical TPs showed a wild-type interaction with the corresponding DNA polymerase are indicated. Graphic is representative of three independent experiments.
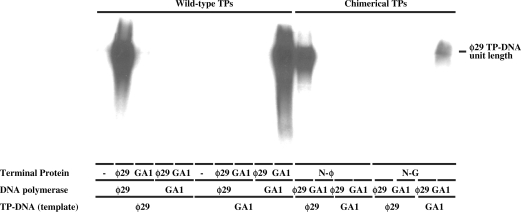
The second phase of bacteriophages ϕ29 and GA-1 TP-DNA replication consists in the elongation of the initiation product. Thus, to analyse whether the chimerical TP-dAMP products could be elongated by the DNA polymerase, we made use of a minimal replication system based on TP-DNA, DNA polymerase, and either wild-type or chimerical TP, as described (29,36). As shown in Figure 4, ϕ29 and GA-1 DNA polymerases elongated the (N-ϕ)-dAMP and (N-G)-dAMP products, respectively, rendering full-length TP-DNA with an efficiency parallel to that observed in the initiation reaction. As expected, each DNA polymerase elongated the homologous wild-type TP-dAMP using the homologous TP-DNA as template. The lack of replication products with ϕ29 DNA polymerase in the absence of TP (Figure 4) guarantees that the observed products come from a bona fide TP-DNA replication.
Figure 4.
In vitro TP-DNA replication. The assays were carried out in the presence of 1 mM MnCl2, 60 nM of the indicated DNA polymerase, 120 nM of wild-type or chimerical TP and 80 µM of the four dNTPs. After incubation for 8 min (wild-type TPs) and 30 min (with chimerical TPs) at 30°C, samples were stopped and processed as described in Materials and Methods section. The labelled DNA was denatured and subjected to electrophoresis in alkaline agarose gels to determine the lengths of the synthesized DNAs. The position of unit length TP-DNA is indicated.
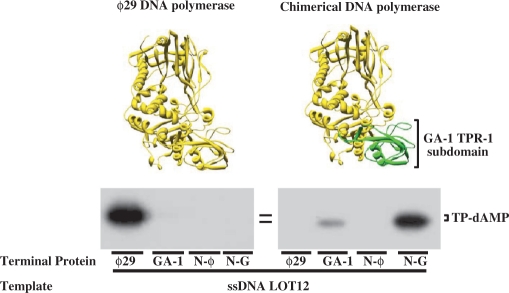
Parental TP is mainly recognized by the corresponding DNA polymerase
From the above results, the relative importance of the DNA polymerase with respect to the TP in the specific recognition of the parental TP cannot be established, as the DNA polymerase and the N-terminal part of chimerical TP in the active complexes belong to the same phage as the parental TP. To address this point, we made a chimerical DNA polymerase by substituting the ϕ29 DNA polymerase TPR-1 subdomain (residues 261–358, coloured in blue in Figure 1B) (41) by that of GA-1 DNA polymerase (29) (see Materials and Methods section), as this region of the ϕ29 DNA polymerase packs against the TP intermediate domain (19). We firstly evaluated the activity of the complexes formed by the chimerical DNA polymerase and the different TPs by performing initiation assays in which the formation of the TP-dAMP product was templated by a linear ssDNA containing the sequence of the ϕ29 left replication origin (ssDNA LOT12, see Materials and Methods section), to avoid the effects of the parental TP on the activity. As expected, the wild-type ϕ29 DNA polymerase could make an active complex only with its homologous TP (Figure 5). Interestingly, the complex formed by the chimerical DNA polymerase and the chimera N-G was only slightly less active than the wild-type ϕ29 heterodimer (Figure 5), in great contrast to the low activity displayed by this chimerical TP with wild-type GA-1 DNA polymerase in the presence of GA-1 TP-DNA (Figure 2). On the other hand, the chimerical DNA polymerase could also use GA-1 TP as primer, although the activity exhibited by this heterodimer was 10-fold lower than with chimera N-G. In contrast, chimerical DNA polymerase could not use either ϕ29 TP or chimera N-ϕ as primer, supporting the results presented previously that suggested that the main interaction with the DNA polymerase was that established between the TP N-terminal part and the DNA polymerase TPR-1 subdomain.
Figure 5.
Use of wild-type and chimerical TPs by the chimerical DNA polymerase with ssDNA as template. The TP-dAMP formation assay was carried out as described in Materials and Methods section, in the presence of 1 mM MnCl2, and using 300 nM of either wild-type or chimerical DNA polymerase, 600 nM of the indicated TP and 5.6 µM of ssDNA LOT12 as template. After incubation for 10 min at 30°C, the reactions were stopped, processed, and analysed by SDS–PAGE and autoradiography. The position of TP-dAMP is indicated. For clarity, both ϕ29 wild-type and chimerical DNA polymerase are depicted on top of the figure, following the same colour code as in Figure 2.
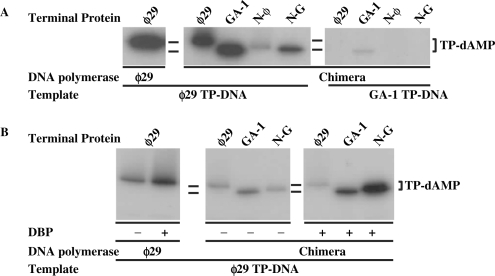
Interestingly, when TP-DNA was used as template, chimerical DNA polymerase displayed a stringent specificity with respect to the parental TP (covalently linked to the phage genome and forming part of the replication origin) as it was almost exclusively active in the presence of ϕ29 TP-DNA (Figure 6A), indicating that recognition of parental TP is mainly accomplished by the DNA polymerase. It should also be noted that, when ϕ29 TP-DNA was used as template, the complex formed with ϕ29 TP was only two-fold lower than that formed with GA-1 TP (note that when ssDNA, lacking parental TP, was used as template, chimerical DNA polymerase did not give any detectable reaction with ϕ29 TP, see above and Figure 5). The great improvement shown by the chimerical DNA polymerase/ϕ29 TP complex in the presence of ϕ29 TP-DNA, as well as the faint band observed with the chimerical DNA polymerase/N-ϕ TP (which possesses the N-terminal part of ϕ29 TP, see Figure 1 and Table 1) could indicate that the poor interaction displayed by both TPs with the chimerical DNA polymerase in the presence of ssDNA is partially overcome by a specific recognition of the parental ϕ29 TP by the ϕ29 N-terminal part of both ϕ29 and N-ϕ primer TPs. This proposal would also explain the formation of a faint initiation band only with the chimerical DNA polymerase/GA-1 TP complex in the GA-1 TP-DNA templated reaction (Figure 6A). The relative activity with chimera N-G (containing the N-terminal part of GA-1 TP) using ϕ29 TP-DNA as template, decreased with respect to that of the wild-type ϕ29 DNA polymerase/TP heterodimer (Figure 6A). The lack of a specific recognition between the N-terminal part (belonging to GA-1 TP, see Table 1 and Figure 1) of the N-G primer TP and the parental ϕ29 TP (just described) cannot justify such decrease in the activity, as a similar behaviour should have been expected for GA-1 TP. To ascertain whether this unforeseen behaviour was the consequence of an impaired opening of the replication origin by the chimerical DNA polymerase/N-G complex that would hinder its proper placement to carry out the initiation reaction, we performed the same kind of assays in the presence of ϕ29 DBP, as this protein assists the necessary initial unwinding of the DNA (23,24,42), specifically activating the initiation reaction. As shown in Figure 6B, the presence of DBP stimulated the activity of chimerical DNA polymerase complexed with GA-1 TP to a similar extent as in the case of the wild-type ϕ29 DNA polymerase/TP heterodimer. Interestingly, such improvement was much higher in the case of the complex formed with chimera N-G, suggesting that the lower activity displayed by this heterodimer in the absence of DBP could be due to an impaired opening of the DNA. On the other hand, DBP had essentially no effect on the reaction carried out by the chimerical DNA polymerase/ϕ29 TP complex.
Figure 6.
Specific recognition of replication origins by chimerical DNA polymerase. (A) TP-primed initiation assays were carried out in the presence of 1 mM MnCl2, 1.6 nM of the indicated TP-DNA, 120 nM of chimerical DNA polymerase and 240 nM of the indicated TP. As control, 15 nM and 30 nM of wild-type ϕ29 DNA polymerase and TP, respectively, were used. After incubation for 10 min at 30°C, samples were stopped and processed as described in Materials and Methods section. (B) Effect of ϕ29 DBP on the activity of chimerical DNA polymerase. The assay was performed in the presence of 1 mM MnCl2, 1.6 nM of ϕ29 TP-DNA as template, in the absence (−) or presence (+) of 35 µM of ϕ29 DBP. Under these conditions, 60 nM of chimerical DNA polymerase and 120 nM of the indicated TP were incubated for 20 min at 30°C. As control, 30 and 60 nM of wild-type ϕ29 DNA polymerase and TP, respectively, were incubated for 2 min at 30°C. Further processing of the samples was performed as described under Materials and Methods section.
The ϕ29 TP N-terminal part is required to allow the TP priming domain to initiate replication
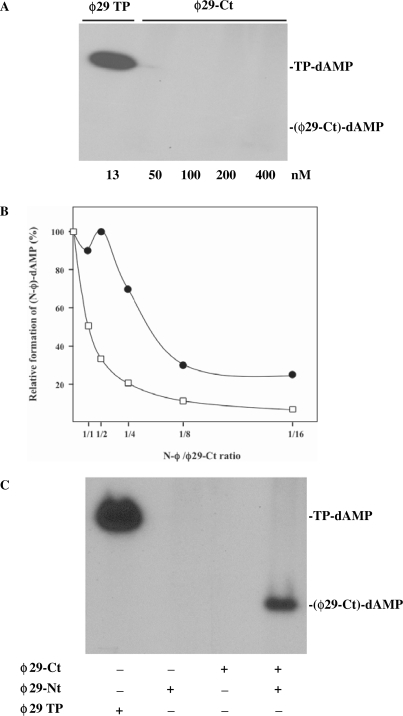
Results presented above show that the DNA polymerase cannot place properly at its catalytic site the homologous TP priming domain when there is no structural complementarity between the DNA polymerase TPR-1 subdomain and the TP N-terminal part (residues 1–173). To find out whether placement of the TP priming domain in a catalytically competent form depends on the TP N-terminal part, we cloned, expressed and purified independently both parts of the ϕ29 TP (see Materials and Methods section), rendering ϕ29 TP variants ϕ29-Ct (priming domain; residues 174–266) and ϕ29-Nt (residues 1–173). As it can be observed in Figure 7A, ϕ29-Ct was not able to prime the initiation reaction by the ϕ29 DNA polymerase even at the highest dose assayed (400 nM). However, interference assays, similar to the ones described above, in which ϕ29-Ct and chimera N-ϕ compete for the ϕ29 DNA polymerase, showed that at the higher doses of the priming domain there was interference with the binding of the chimera N-ϕ to the DNA polymerase (Figure 7B), suggesting that the lack of reaction at such doses was the consequence of a non-suitable placement of the priming domain at the DNA polymerase active site. This led us to hypothesize that the TP N-terminal part was assisting the priming domain orientation by promoting a conformational change in the DNA polymerase. Thus, as it can be seen in Figure 7C, addition of ϕ29-Nt variant allowed the priming domain to act as primer, giving rise to a labelled band which migrates at a position corresponding with the priming domain molecular weight.
Figure 7.
ϕ29 TP N-terminal part–DNA polymerase interaction is required to allow TP priming domain to prime the initiation reaction. (A) ϕ29 TP priming domain cannot accomplish TP-dAMP formation. The initiation reaction (TP-dAMP formation) was carried out in the presence of 1 mM MnCl2, 1.6 nM of ϕ29 TP-DNA, 60 nM ϕ29 DNA polymerase and the indicated amounts of either wild-type ϕ29 TP or ϕ29-Ct variant. After incubation for 10 min at 30°C, samples were stopped and processed as described. (B) ϕ29 TP priming domain binds the DNA polymerase active site in a non-productive fashion. Formation of TP-dAMP initiation product by 60 nM wild-type ϕ29 DNA polymerase and 120 nM of chimerical TP N-ϕ was carried out in the presence of 1 mM MnCl2 and increasing amounts of TP ϕ29-Ct variant (filled circles). The TP-dAMP formed in the different competition conditions relative to that formed in the absence of competition (100%) is represented. The theoretical inhibition profile that would be obtained if the TP ϕ29-Ct variant displayed a wild-type interaction with ϕ29 DNA polymerase (open squares) is indicated. Graphic is representative of three independent experiments. (C) ϕ29 N-terminal part assists the priming domain to act as primer. The assay was performed in the presence of 1 mM MnCl2, 60 nM ϕ29 DNA polymerase and 150 nM of either ϕ29-Ct, ϕ29-Nt or both, during 30 min at 30°C. As control, 15 nM and 30 nM of wild-type ϕ29 DNA polymerase and TP, respectively, were allowed to react for 1 min at 30°C. After incubation, samples were processed as described.
Role of the ϕ29 TP N-terminal domain during in vitro ϕ29 TP-DNA replication
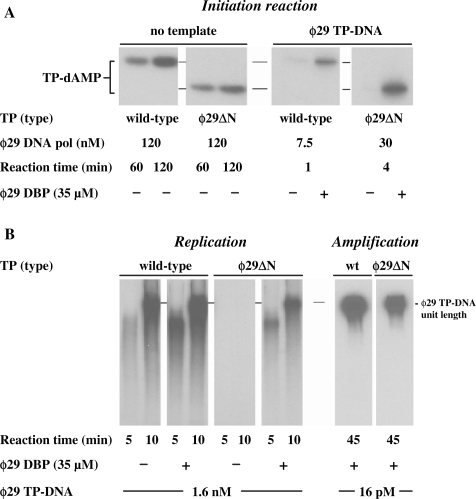
The crystallographic structure of ϕ29 TP shows the N-terminal domain (residues 1–73) as a protein segment that contains disordered sequence and, contrary to the rest of the protein, is not interacting with the polymerase (19) (Figure 1B). To determine whether this TP domain plays some role during in vitro TP-DNA replication, we constructed the ϕ29 TP deletion mutant ϕ29▵N, lacking amino acid residues 1–73. We firstly evaluated the ability of ϕ29▵N mutant to interact with ϕ29 DNA polymerase by analysing the primer capacity of the mutant TP, taking advantage of the fact that ϕ29 DNA polymerase can catalyse the deoxynucleotidilylation of TP in the absence of template (43). Under these conditions, the activity of the ϕ29▵N derivative was slightly lower (50%) with respect to that displayed by the wild-type TP (Figure 8A), indicating that deletion of the TP N-terminal domain does not preclude the interaction with the DNA polymerase. Conversely, in the presence of TP-DNA, the ϕ29▵N mutant TP required the presence of DBP to give a detectable initiation reaction. Thus, under these conditions DBP stimulated ϕ29▵N mutant TP priming activity 100-fold, in comparison to the 10-fold stimulation observed with the wild-type TP (Figure 8A). Accordingly, although a defective interaction with the parental TP cannot be ruled out, the results strongly point again to an impaired ability of the mutant TP to open the replication origins.
Figure 8.
ϕ29 TP N-terminal domain role during in vitro ϕ29 TP-DNA replication. (A) Formation of TP-dAMP complex with either wild-type or ϕ29▵N mutant TP. Non-templated initiation reactions were carried out in the presence of 1 mM MnCl2, 240 nM of either wild-type or mutant TP and the indicated concentration of wild-type ϕ29 DNA polymerase. In the templated reactions, 1.6 nM of ϕ29 TP-DNA, 10 mM MgCl2, 60 nM of either wild-type or mutant TP and the indicated concentration of ϕ29 DNA polymerase were used, in the absence (−) or presence (+) of the indicated amount of ϕ29 DBP. After incubation at 30°C for the indicated times, samples were analysed by SDS–PAGE and autoradiography. The position of the TP-dAMP initiation complexes is indicated. Quantification was by densitometry of the band corresponding to the labelled TP-dAMP complex, detected by autoradiography. (B) ϕ29 TP-DNA replication and amplification with either wild-type or ϕ29▵N mutant TP. Reactions were carried out with 10 mM MgCl2, 60 nM of either wild-type or mutant ϕ29 TP, 15 nM wild-type ϕ29 DNA polymerase, 20 µM the four dNTPs, the indicated amount of ϕ29 TP-DNA, in the absence (−) or presence (+) of ϕ29 DBP. After incubation for the indicated times at 30°C, relative activity values were calculated, and the length of the synthesized DNA was analysed by alkaline agarose gel electrophoresis. The amplification assay was performed in the presence of 120 nM of either wild-type or mutant ϕ29 TP, 30 nM of wild-type ϕ29 DNA polymerase, 80 µM the four dNTPs, the indicated amount of ϕ29 TP-DNA and 35 µM of ϕ29 DBP and 30 µM of ϕ29 SSB. After 45 min of incubation at 30°C, the reaction was stopped with 10 mM EDTA. The relative activity values were calculated, and the length of the synthesized DNA was analysed by alkaline agarose gel electrophoresis. The migration position of unit length ϕ29 DNA is indicated.
By making use of the minimal replication system described above, in the presence and absence of DBP, similar replication properties were observed with the ϕ29▵N mutant. Thus, in the absence of DBP, replication products were not detected, while the activity displayed in the presence of DBP (34% with respect to the wild-type TP) paralleled the initiation reaction (Figure 8B). In addition, the velocity of the reaction was the same when either the mutant or the wild-type TP was used, implying that the TP N-terminal domain (residues 1–73) does not play any role during transition from the protein-primed to the DNA-primed elongation mode. The replication assays described above give rise to only one replication round, as described (44). Thus, the possible effects of mutant ϕ29▵N on its function as parental TP cannot be studied with this kind of assay. For this, we carried out amplification assays under more physiological conditions in which appropriate amounts of the four ϕ29 DNA replication proteins, TP, DNA polymerase, DBP, and SSB are required to amplify limited amounts of ϕ29 TP-DNA molecules (38) (see Materials and Methods section, Amplification assay). In these amplification reactions, primer TP that becomes covalently attached to the newly synthesized DNA strand during the first round of replication will act as parental TP in further replication rounds. The relative levels of amplification obtained with the mutant TP with respect to the wild-type TP (39%) were close to those displayed in the initiation and replication assays (both in the presence of DBP), indicating that the N-terminal domain (residues 1–73) of the parental TP is not required to interact with the DNA polymerase/TP complex during initial recognition of the replication origins (Figure 8B).
DISCUSSION
Extensive studies performed both in vitro and in vivo, mainly using bacteriophage ϕ29 and Adenovirus, have provided the general insights about the mechanism of protein-primed DNA replication (10–12). In the first instance, replicative DNA polymerase forms, with the corresponding TP, a heterodimer that further recognizes the replication origins, located at both ends of the linear genome, that comprise a 5′ covalently linked TP and a specific DNA sequence. The DNA polymerase then catalyses both, the formation of the covalent complex between a free TP molecule and the initial dAMP and its further elongation coupled to strand displacement. Similarly, TP-DNA replication of bacteriophages PRD1 (14,45), Cp1 (16), GA-1 (15,29) and Nf (28,29) was also shown to occur by a similar protein-primed mechanism, involving its corresponding DNA polymerase and TP. Previous results obtained by interchanging the DNA polymerases, TPs and TP-DNAs coming from ϕ29, GA-1 and Nf have revealed a high specificity among the DNA polymerase, the TP and the replication origin (27–29).
Specific recognition between the DNA polymerase and the free TP
The recently published structure of the ϕ29 DNA polymerase/TP heterodimer shows that TP is composed of an N-terminal domain making no interactions with the polymerase, an intermediate one that extensively interacts with the TPR-1 subdomain of the polymerase, and a third domain (priming domain) containing the priming serine that occupies the same binding cleft in the polymerase as duplex DNA does during elongation (19,20). In order to elucidate the relative importance of the different TP domains in the specific recognition of the homologous DNA polymerase, we have made hybrid TPs by swapping the structurally independent priming domains of both ϕ29 and GA-1 TPs. In addition, we have cloned and expressed individually the ϕ29 TP priming domain (residues 174–266) and the TP N-terminal part (residues 1–173), as well as a ϕ29 TP deletion mutant lacking the N-terminal domain (residues 1–73). Besides that, we have generated a chimerical DNA polymerase by substituting the ϕ29 DNA polymerase TPR-1 subdomain with the corresponding one from GA-1 DNA polymerase.
Biochemical analyses of the above constructions have shown that the interaction between the TP N-terminal part (that includes the TP N-terminal and intermediate domains) and the DNA polymerase TPR-1 subdomain confers specificity to the recognition between both proteins, as DNA polymerase can form a catalytically active heterodimer exclusively with the chimerical TP that contains the N-terminal part (residues 1–173) of the homologous TP. In addition, deletion of the ϕ29 TP N-terminal domain (residues 1–73) does not have drastic consequences on such interaction. From this, a major importance of the TP intermediate domain in the specific interaction with the DNA polymerase could be inferred. Such ϕ29 TP domain buries 575 Å2 of surface area against the TPR-1 subdomain of ϕ29 DNA polymerase, both domains fitting into each other. This interaction has been shown to be stabilized mainly by TP intermediate domain residues R158 and R169 that make salt bridges with the DNA polymerase TPR-1 residues E291 and E322, respectively (19). Structural alignment of both, the ϕ29 and the modelled GA-1 heterodimers, demonstrates the conservation of these four residues. However, modelling of the interaction between ϕ29 DNA polymerase TPR-1 subdomain and GA-1 TP intermediate domain shows the presence of many steric clashes that are likely responsible for the hampered formation of this heterodimer. These arguments favour the hypothesis that the specificity of the interaction between the DNA polymerase and the TP is essentially dependent on an intimate structural complementarity between both protein domains.
Chimerical TPs N-ϕ and N-G displayed an activity much lower than the ϕ29 and GA-1 TPs with ϕ29 and GA-1 DNA polymerases, respectively. This suggested the requirement of a structural complementarity between the TP priming domain and the DNA polymerase. This requirement was also demonstrated as chimerical ϕ29 DNA polymerase (containing the GA-1 DNA polymerase TPR-1 subdomain) recovered a nearly wild-type activity when assayed with the complementary TP N-G chimera. Thus, the TP priming domain should also give specificity and affinity to the interaction with the DNA polymerase, although to a lesser extent than the TP intermediate domain, as the results presented here indicate that both ϕ29 and, to a lower degree, GA-1 DNA polymerases can properly allocate the non-homologous TP priming domain-containing chimerical TPs. The crystal structure of the ϕ29 DNA polymerase/TP heterodimer shows a looser interaction between the TP priming domain and the DNA polymerase, in which only part of the C-terminal helix of the priming domain packs against the TPR-2 subdomain of polymerase, establishing hydrogen bonds between residues E252, Q253 and R256 of TP and L416, G417 and E419 of polymerase (19). Modelling of GA-1 TP priming domain into the structure of ϕ29 DNA polymerase, by structural fitting on the complexed ϕ29 TP, demonstrates that those residues are not conserved either in the polymerase or in the TP. In addition, only minor steric clashes can be observed. This fact would explain a better tolerance of each DNA polymerase with respect to their non-homologous TP priming domains than the ones observed in the case of the TP-intermediate domains.
The ϕ29 TP priming domain could not prime the initiation reaction by itself, because of an impaired productive binding to the DNA polymerase active site. Interestingly, addition of the ϕ29 TP N-terminal part assisted the function of the TP priming domain. Since both TP parts were not structurally linked, this result lead us to propose that the N-terminal part of TP induces a conformational change in the DNA polymerase that allows the TP priming domain to be correctly positioned at the polymerase catalytic site. Conformation of the ϕ29 DNA polymerase forming a complex with the TP is very similar to that of the apo enzyme, the main conformational changes being restricted to TPR-1 residues 304–315 (19). Such residues form a loop with a high degree of flexibility in the apo enzyme. On the contrary, the ϕ29 heterodimer structure shows that this loop moves out to allow TP to access the polymerase active site. Altogether, these results lead us to propose a model on how the DNA polymerase-TP interaction could take place (Figure 9A). Thus, the TP intermediate domain would recognize and interact with the DNA polymerase TPR-1 subdomain (coloured in blue in Figure 9A). Such an interaction would promote the TPR-1 loop change from a flexible (coloured in magenta) to the stable moved out conformation (coloured in orange) that would now allow the proper (prone to catalysis) placement of the TP priming domain into the DNA polymerase structure.
Figure 9.
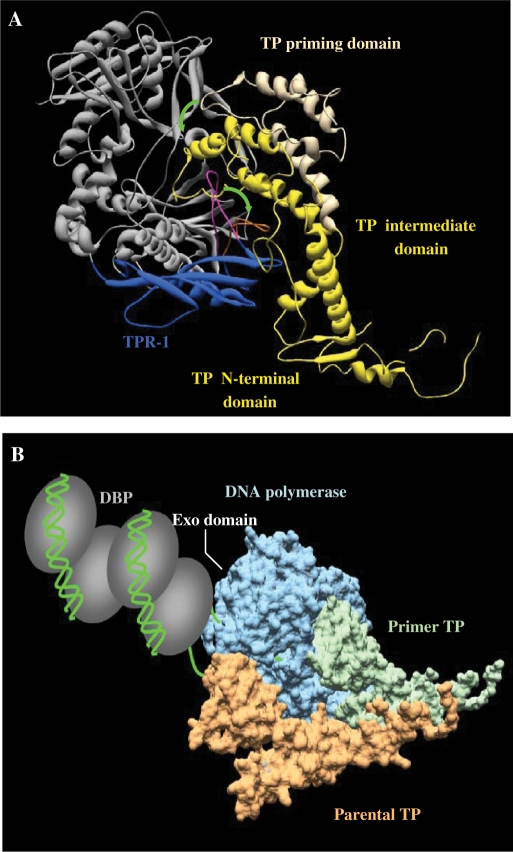
Modelling of the temporal sequence of interactions that take place during initiation of TP-DNA replication. (A) Proposed conformational changes occurring during DNA polymerase/TP heterodimer formation. ϕ29 DNA polymerase is coloured in grey and the TPR-1 subdomain in dark blue. Flexible orientation of TPR-1 loop in the apoenzyme [PDB code 1XHX; (41)] and its stable and moved out structural conformation shown in the DNA polymerase/TP complex [PDB code 2EX3; (19)] is shown in magenta and orange, respectively. TP is coloured in yellow (TP priming domain in the proposed non productive orientation is indicated with light yellow). Green arrows indicate the suggested conformational changes of both, the DNA polymerase TPR-1 loop and the TP priming domain to allow the formation of a stable heterodimer. (B) Model of the DNA polymerase/TP recognition of replication origin. DNA polymerase molecular surface is coloured in blue, priming TP in green and parental TP in orange. The nucleoprotein complex at the replication origin, formed by DNA (depicted in green) and DBP (depicted as grey ovals) is mainly contacted by the DNA polymerase.
The different contribution to the strength of the DNA polymerase–TP interaction by the priming and intermediate domains would also support the model proposed for the transition from the protein-primed initiation to the DNA-primed elongation modes. Previous biochemical studies demonstrated that the DNA polymerase/TP heterodimer is not dissociated immediately after initiation (18). There is a transition stage in which the heterodimer undergoes structural changes during replication of nucleotides 6–9, and finally the DNA polymerase dissociates from the TP when it inserts the 10th nucleotide (18). Thus, the TP intermediate domain would be in a fixed orientation on the polymerase by means of stable contacts with the TPR-1 subdomain. The weak interaction observed with the DNA polymerase would facilitate the TP priming domain to rotate following the helicoidal pathway as DNA is synthesized. The relative motion of the TP priming domain with respect to the fixed TP intermediate domain would be possible due to the flexibility of the hinge region that connects both domains. After incorporation of 6–7 nucleotides the proximity of the priming Ser to the hinge region would impede a further priming domain rotation, promoting complex dissociation (19).
Recognition of replication origins by the DNA polymerase/TP heterodimer
The ϕ29 DNA polymerase/TP heterodimer recognizes the replication origins at the genome ends. The presence of parental TP, which is covalently linked to the 5′ end of the non-template strand by a previous cycle of replication, is the main signal to be recognized by the polymerase/TP complex for initiation of replication, as when terminal DNA fragments lacking parental TP were used as templates, the initiation reaction fell 6- to 10-fold with respect to the activity obtained with TP-DNA (28,46). In addition, ϕ29 DBP forms a multimeric complex at the origins of replication (24,25), activating in vitro the initiation of ϕ29 TP-DNA replication (23). Detection of initiation activity by using heterologous systems in which DNA polymerase, TP and TP-DNA came from ϕ29 and Nf-related phages, showed that initiation was selectively enhanced when DNA polymerase and TP-DNA were from the same phage, implying a specific interaction between DNA polymerase and parental TP (28). Similarly, mutations introduced at several TP-intermediate domain residues rendered TP mutants that could not support DNA amplification when they acted as parental TP, suggesting also a contribution of the primer TP in the specific recognition of the replication origins (47,48). Furthermore, measurement of the ability of the different DBPs coming from ϕ29, Nf and GA-1 bacteriophages to activate homologous and heterologous replication origins showed also a specific recognition of each nucleoprotein complex by the homologous DNA polymerase/TP heterodimer (27).
The results presented here show that the chimerical ϕ29 DNA polymerase containing the GA-1 DNA polymerase TPR-1 subdomain is capable of catalysing the initiation reaction primed by GA-1 TP but solely in the presence of ϕ29 TP-DNA, indicating that the major contribution to the parental TP recognition is carried out by the DNA polymerase, likely through its exonuclease domain (Figure 9B). The fact that ϕ29 DBP stimulates the initiation activity of the heterodimer formed by chimerical DNA polymerase and GA-1 TP to a similar extent as in the ϕ29 wild-type system, also favours the hypothesis of a main and specific recognition of the DBP by the DNA polymerase. On the other hand, the relative improvement of the priming function of ϕ29 TP complexed to chimerical DNA polymerase, when ϕ29 TP-DNA was used as template with respect to that displayed with ssDNA LOT12, could also indicate an involvement of primer TP in parental TP detection, as it has been suggested (47,48). Interestingly, the lack of stimulation of the priming activity displayed by ϕ29 TP in the presence of ϕ29 DBP could suggest that, under physiological conditions, and once the replication origin is opened by the DBP, the interactions between the primer and parental TP are precluded, at least partially. Intriguingly, TP chimera N-G displayed a singular behaviour when complexed to chimerical ϕ29 DNA polymerase, as it was the most active of all TPs in the presence of opened origins [ssDNA LOT12 and DBP/TP-DNA], but worse than ϕ29 and GA-1 TPs with TP-DNA (closed origins). The results could suggest that in vitro opening of the replication origins by DNA polymerase/TP heterodimer requires a structurally intact primer TP. This could account for the recovery of the template-directed initiation activity displayed by mutant ϕ29▵N in the presence of DBP, even though a direct contact between the priming TP N-terminal domain and parental TP intermediate domain cannot be ruled out.
Two ϕ29 encoded proteins, p1 and p16.7, have been proposed to anchor ϕ29 TP-DNA replication to the membrane. The early expressed protein p1 assembles into large multimeric structures that are associated with the bacterial membrane through the C-terminal part of the protein (49,50), whereas the N-terminal (soluble) part is able to interact with the primer TP (51). These observations led to the proposal of a model for the role of protein p1 in targeting the DNA polymerase/TP complex to the membrane through a p1–TP interaction (49). In addition, ϕ29 gene p16.7 codes for an early expressed integral membrane protein that is also involved in the organization of membrane-associated ϕ29 DNA replication (52). Interestingly, the soluble C-terminal portion of p16.7 interacts with free TP (53). The TP region bound by these two proteins is unknown. It is tempting to speculate that TP regions bound by p1/p16.7 and DNA polymerase should not overlap, in order to ensure membrane associated TP-DNA replication. The biochemical results presented here together with the crystallographic data would preclude a function of the N-terminal domain (residues 1–73) of TP during in vitro TP-DNA replication. Thus, an attractive hypothesis is that the TP N-terminal domain, which does not interact with the DNA polymerase, could be bound by membrane proteins p1 and/or p16.7 to recruit phage replication in the cell membrane, without interfering with the replication machinery.
Supplementary Data
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
P.P-A. and E.L. were predoctoral fellows of the Spanish Ministry of Education and Science. Spanish Ministry of Education and Science (BFU 2005-00733 to M.S.); Institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular ‘Severo Ochoa’. Funding to pay the Open Access publication charges for this article are provided by research grant BFU 2005-00733 from the Spanish Ministry of Education and Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD. Origin of concatemeric T7 DNA. Nat. New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg A, Baker T. DNA Replication. 2nd. NY: W.H. Freeman; 1992. edn. [Google Scholar]
- 4.Taylor K, Wegrzyn G. Replication of coliphage lambda DNA. FEMS Microbiol. Rev. 1995;17:109–119. doi: 10.1111/j.1574-6976.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 5.Schlegel RA, Thomas CA., Jr Some special structural features of intracellular bacteriophage T7 concatemers. J. Mol. Biol. 1972;68:319–345. doi: 10.1016/0022-2836(72)90216-1. [DOI] [PubMed] [Google Scholar]
- 6.Barbour AG, Garon CF. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 7.Baroudy BM, Venkatesan S, Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982;28:315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- 8.Garon CF, Barbosa E, Moss B. Visualization of an inverted terminal repetition in vaccinia virus DNA. Proc. Natl Acad. Sci. USA. 1978;75:4863–4867. doi: 10.1073/pnas.75.10.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geshelin P, Berns KI. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J. Mol. Biol. 1974;88:785–796. doi: 10.1016/0022-2836(74)90399-4. [DOI] [PubMed] [Google Scholar]
- 10.Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 11.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet. Eng. (NY) 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 12.Salas M, Miller J, Leis J, DePamphilis M. Mechanisms for Priming DNA Synthesis. NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 13.Méndez J, Blanco L, Esteban JA, Bernad A, Salas M. Initiation of phi29 DNA replication occurs at the second 3' nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl Acad. Sci. USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldentey J, Blanco L, Bamford DH, Salas M. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993;21:3725–3730. doi: 10.1093/nar/21.16.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Illana B, Blanco L, Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J. Mol. Biol. 1996;264:453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- 16.Martín AC, Blanco L, García P, Salas M, Méndez J. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3' nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 1996;260:369–377. doi: 10.1006/jmbi.1996.0407. [DOI] [PubMed] [Google Scholar]
- 17.King AJ, van der Vliet PC. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994;13:5786–5792. doi: 10.1002/j.1460-2075.1994.tb06917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Méndez J, Blanco L, Salas M. Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase: protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman AJ, Kamtekar S, Goodman JL, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteban JA, Blanco L, Villar L, Salas M. In vitro evolution of terminal protein-containing genomes. Proc. Natl Acad. Sci. USA. 1997;94:2921–2926. doi: 10.1073/pnas.94.7.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutiérrez C, Freire R, Salas M, Hermoso JM. Assembly of phage phi29 genome with viral protein p6 into a compact complex. EMBO J. 1994;13:269–276. doi: 10.1002/j.1460-2075.1994.tb06257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco L, Gutiérrez J, Lázaro JM, Bernad A, Salas M. Replication of phage phi29 DNA in vitro: role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 1986;14:4923–4937. doi: 10.1093/nar/14.12.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieto I, Serrano M, Lázaro JM, Salas M, Hermoso JM. Interaction of the bacteriophage phi29 protein p6 with double-stranded DNA. Proc. Natl Acad. Sci. USA. 1988;85:314–318. doi: 10.1073/pnas.85.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serrano M, Gutiérrez J, Prieto I, Hermoso JM, Salas M. Signals at the bacteriophage phi29 DNA replication origins required for protein p6 binding and activity. EMBO J. 1989;8:1879–1885. doi: 10.1002/j.1460-2075.1989.tb03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bravo A, Hermoso JM, Salas M. In vivo functional relationships among terminal proteins of Bacillus subtilis phi29-related phages. Gene. 1994;148:107–112. doi: 10.1016/0378-1119(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 27.Freire R, Serrano M, Salas M, Hermoso JM. Activation of replication origins in phi29-related phages requires the recognition of initiation proteins to specific nucleoprotein complexes. J. Biol. Chem. 1996;271:31000–31007. doi: 10.1074/jbc.271.48.31000. [DOI] [PubMed] [Google Scholar]
- 28.González-Huici V, Lázaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J. Biol. Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 29.Longás E, de Vega M, Lázaro JM, Salas M. Functional characterization of highly processive protein-primed DNA polymerases from phages Nf and GA-1, endowed with a potent strand displacement capacity. Nucleic Acids Res. 2006;34:6051–6063. doi: 10.1093/nar/gkl769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peñalva MA, Salas M. Initiation of phage phi29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc. Natl Acad. Sci. USA. 1982;79:5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage phi29 DNA polymerase. Meth. Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 32.Zaballos A, Lázaro JM, Méndez E, Mellado RP, Salas M. Effects of internal deletions on the priming activity of the phage phi29 terminal protein. Gene. 1989;83:187–195. doi: 10.1016/0378-1119(89)90104-2. [DOI] [PubMed] [Google Scholar]
- 33.Soengas MS, Gutiérrez C, Salas M. Helix-destabilizing activity of phi29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J. Mol. Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 34.de Vega M, Blanco L, Salas M. phi29 DNA polymerase residue Ser122, a single-stranded DNA ligand for 3′-5′ exonucleolysis, is required to interact with the terminal protein. J. Biol. Chem. 1998;273:28966–28977. doi: 10.1074/jbc.273.44.28966. [DOI] [PubMed] [Google Scholar]
- 35.Dufour E, Méndez J, Lázaro JM, de Vega M, Blanco L, Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- 36.Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 37.McDonell MW, Simon MN, Studier FW. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 1977;110:119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- 38.Blanco L, Lázaro JM, de Vega M, Bonnin A, Salas M. Terminal protein-primed DNA amplification. Proc. Natl Acad. Sci. USA. 1994;91:12198–12202. doi: 10.1073/pnas.91.25.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 40.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Serrano M, Salas M, Hermoso JM. A novel nucleoprotein complex at a replication origin. Science. 1990;248:1012–1016. doi: 10.1126/science.2111580. [DOI] [PubMed] [Google Scholar]
- 43.Blanco L, Bernad A, Esteban JA, Salas M. DNA-independent deoxynucleotidylation of the phi29 terminal protein by the phi29 DNA polymerase. J. Biol. Chem. 1992;267:1225–1230. [PubMed] [Google Scholar]
- 44.Martín G, Lázaro JM, Méndez E, Salas M. Characterization of the phage phi29 protein p5 as a single-stranded DNA binding protein. Function in phi29 DNA-protein p3 replication. Nucleic Acids Res. 1989;17:3663–3672. doi: 10.1093/nar/17.10.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caldentey J, Blanco L, Savilahti H, Bamford DH, Salas M. In vitro replication of bacteriophage PRD1 DNA. Metal activation of protein-primed initiation and DNA elongation. Nucleic Acids Res. 1992;20:3971–3976. doi: 10.1093/nar/20.15.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutiérrez J, García JA, Blanco L, Salas M. Cloning and template activity of the origins of replication of phage phi29 DNA. Gene. 1986;43:1–11. doi: 10.1016/0378-1119(86)90002-8. [DOI] [PubMed] [Google Scholar]
- 47.Illana B, Zaballos A, Blanco L, Salas M. The RGD sequence in phage phi29 terminal protein is required for interaction with phi29 DNA polymerase. Virology. 1998;248:12–19. doi: 10.1006/viro.1998.9276. [DOI] [PubMed] [Google Scholar]
- 48.Serna-Rico A, Illana B, Salas M, Meijer WJ. The putative coiled coil domain of the phi29 terminal protein is a major determinant involved in recognition of the origin of replication. J. Biol. Chem. 2000;275:40529–40538. doi: 10.1074/jbc.M007855200. [DOI] [PubMed] [Google Scholar]
- 49.Bravo A, Salas M. Initiation of bacteriophage phi29 DNA replication in vivo: assembly of a membrane-associated multiprotein complex. J. Mol. Biol. 1997;269:102–112. doi: 10.1006/jmbi.1997.1032. [DOI] [PubMed] [Google Scholar]
- 50.Serrano-Heras G, Salas M, Bravo A. In vivo assembly of phage phi29 replication protein p1 into membrane-associated multimeric structures. J. Biol. Chem. 2003;278:40771–40777. doi: 10.1074/jbc.M306935200. [DOI] [PubMed] [Google Scholar]
- 51.Bravo A, Illana B, Salas M. Compartmentalization of phage phi29 DNA replication: interaction between the primer terminal protein and the membrane-associated protein p1. EMBO J. 2000;19:5575–5584. doi: 10.1093/emboj/19.20.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meijer WJ, Serna-Rico A, Salas M. Characterization of the bacteriophage phi29-encoded protein p16.7: a membrane protein involved in phage DNA replication. Mol. Microbiol. 2001;39:731–746. doi: 10.1046/j.1365-2958.2001.02260.x. [DOI] [PubMed] [Google Scholar]
- 53.Serna-Rico A, Muñoz-Espín D, Villar L, Salas M, Meijer WJ. The integral membrane protein p16.7 organizes in vivo phi29 DNA replication through interaction with both the terminal protein and ssDNA. EMBO J. 2003;22:2297–2306. doi: 10.1093/emboj/cdg221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.