Abstract
Previously, we reported the presence in mouse cells of a mitochondrial RNA which contains an inverted repeat (IR) of 121 nucleotides (nt) covalently linked to the 5′ end of the mitochondrial 16S RNA (16S mtrRNA). Here, we report the structure of an equivalent transcript of 2374 nt which is over-expressed in human proliferating cells but not in resting cells. The transcript contains a hairpin structure comprising an IR of 815 nt linked to the 5′ end of the 16S mtrRNA and forming a long double-stranded structure or stem and a loop of 40 nt. The stem is resistant to RNase A and can be detected and isolated after digestion with the enzyme. This novel transcript is a non-coding RNA (ncRNA) and several evidences suggest that the transcript is synthesized in mitochondria. The expression of this transcript can be induced in resting lymphocytes stimulated with phytohaemagglutinin (PHA). Moreover, aphidicolin treatment of DU145 cells reversibly blocks proliferation and expression of the transcript. If the drug is removed, the cells re-assume proliferation and over-express the ncmtRNA. These results suggest that the expression of the ncmtRNA correlates with the replicative state of the cell and it may play a role in cell proliferation.
INTRODUCTION
The mitochondrial DNA (mtDNA) is a closed-circular, double-stranded molecule that displays an exceptional economy of organization (1–3). In humans, the ∼16 500 bp of the genome encode the 12S and 16S ribosomal RNAs, 22 transfer RNAs (tRNAs) and 13 polypeptides (4–7,4–7). The H-strand encodes the 12S and 16S ribosomal RNAs, 14 tRNAs and 12 polypeptides, while the L-strand codes for 8 tRNAs and the ND6 subunit of NAD dehydrogenase (2,4–8). Between the tRNAPhe and tRNAPro genes is the D-loop that has evolved as the major control region. Besides the H-strand origin of replication, the D-loop contains the major promoters that regulate transcription of the H- (HSP) and the L-strand (LSP) (9–11). Both strands are transcribed as polycistronic RNAs, which are then processed to release the individual mRNAs, tRNAs and rRNAs (3). The human mitochondrial RNA polymerase as well as transcription factors have been extensively described (6–8).
Previously, we described the presence in mouse cells of a novel mitochondrial RNA containing an IR of 121 nt linked to the 5′ end of the 16S mtrRNA (12,13). The IR generates a perfect double-stranded structure of 121 bp and a loop of 120 nt. In situ hybridization (ISH) revealed that this ncRNA is over-expressed in spermatogenic cells, especially in mouse proliferating spermatogonia (14). Similar results were obtained with human spermatogonia using a probe complementary to the 16S mtrRNA (14).
These results suggest that human cells might contain a transcript with similar structural features to the mouse RNA, and that its expression correlates with cell proliferation. In this work, we report that the human RNA is over-expressed in several human proliferating cells but not in resting cells. The structure of this transcript of 2374 nt reveals the presence of an IR of 815 nt linked to the 5′ end of the 16S mtrRNA. Together with the 16S mtrRNA, the IR forms a long double-stranded structure or stem, which is resistant to RNase A digestion. On the other hand, in silico analysis revealed that this transcript is a new member of the expanding family of non-coding RNAs (ncRNA) (15–17), and therefore we named this molecule non-coding mitochondrial RNA or ncmtRNA. Moreover, several experimental evidences strongly suggest the mitochondrial origin of this transcript. ISH using a probe specific for the ncmtRNA, confirmed over-expression of this transcript in proliferating cells. The expression of this transcript can be induced in resting lymphocytes stimulated with phytohaemagglutinin (PHA) (18), together with DNA synthesis and the expression of the proliferation markers proliferating cell nuclear antigen (PCNA), Ki-67 and phosphohistone H3 (19–21). On the other hand, treatment of DU145 cells with aphidicolin (22,23) reversibly blocks cell proliferation as well as the expression of the ncmtRNA. These results suggest that the ncmtRNA is a new marker of cell proliferation.
MATERIALS AND METHODS
Cell culture
HeLa, SiHa, DU145, MCF/7, H441, Caco-2 and 42/95 (melanoma) cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Gibco) containing 10% fetal calf serum (FCS), 2 mM glutamine, 50 μg/ml of penicillin, 50 μg/ml of streptomycin and 0.1 mM non-essential amino acids. The human promyelocitic leukemia cell line HL-60, was cultured in Iscove's modified Dulbecco's medium (IMDM; Gibco) supplemented with 20% fetal bovine serum, 2 mM glutamine, 50 μg/ml of penicillin, 50 μg/ml of streptomycin and 0.1 mM non-essential amino acids. All human cell lines were maintained in a humidified cell culture chamber at 37°C and 5% CO2. To inhibit mitochondrial transcription, HeLa cells were cultured as described with the addition of 50 ng/ml of ethidium bromide plus 50 μg/ml of uridine and 1 mM sodium pyruvate (24). For aphidicolin treatment (22,23), DU145 cells were cultured for 16 h with 5 μg/ml of aphidicolin followed by 10 h without the drug. Then the cells were again treated with 5 μg/ml of aphidicolin for another 16 h. To release the cells from the aphidicolin block, they were cultured for 48 h in regular medium without the drug.
Human peripheral blood lymphocytes obtained from healthy donors were isolated by Ficoll gradient centrifugation as described before (18). The cells were suspended in RPMI 1640 medium supplemented with 10% FCS (18) and the number of cells per ml was determined in a Neubauer chamber. The cells were cultured for 24, 48 and 72 h at 37°C with 5% CO2, with or without 10 μg/ml of PHA (18). About 100 000 cells in 200 μl of medium were cultured in 96-wells plates and pulsed for 16 h with BrdU starting at 0, 24 and 48 h. Cells were collected at 24, 48 and 72 h of culture and BrdU incorporation was measured colorimetrically by ELISA according to manufacturer's instructions (Proliferation ELISA, Roche).
Human tissue samples
Formalin-fixed, paraffin-embedded human tissue samples were obtained from diagnostic biopsies or resection specimens from patients at the Hospital Barros Luco Trudeau (Santiago, Chile). The tissues were used in agreement with the ethical guidelines approved by the ethical committee of the hospital and our institutions.
RNA and DNA isolation
Total RNA from cells was extracted with TRIzol (Invitrogen) as described before (12,13,25). The polyA+ fraction was obtained with the Oligotex mRNA Midi kit (Qiagen) according to the manufacturer's instructions. To eliminate mtDNA contamination, RNA preparations were treated with TURBO DNA-free (Ambion) according to the manufacturer's instructions. Human mtDNA was prepared from human lymphocytes, HeLa, SiHa, Hep G2 and HL-60 cells (26).
RT-PCR
Reverse transcription was carried out with 50–100 ng of freshly prepared RNA, 50 ng of random hexamers or sequence-specific primers and 200 U of reverse transcriptase (M-MLV or SuperScript II, Invitrogen) (12,13). The cDNA was amplified by PCR and analyzed by electrophoresis as described before (12,13,27). The primers used to amplify the sense 16S mtrRNA were: P1 (r) 5′ AAGGTGGAGTGGGTTTGGGGC (position 11–31); P9 (f) 5′ TAGGCCTAAAAGCAGCCACCAA (position 501–522); P10 (f) 5′ ACCGTGCAAAGGTAGCATAATCAC (position 912–935); P11 (r) 5′ AATAGGATTGCGCTGTTATCCCTA (position 1260–1283); P12 (r) 5′ CTGTTCTTGGGTGGGTGTG (position 1536–1554). For the antisense 16S mtrRNA, P2 (f) 5′ GGGGTCTTAGCTTTGGCTCTCC (position 1326–1347); P3 (f) 5′ TTGGTGGCTGCTTTTAGGCCTA (position 1207–1227); P4 (f) 5′ GGTTGATTGTAGATATTGGGCT (position 833–854); P5 (f) 5′ GGTAAGATTTGCCGAGTTC (position 741–759); P6 (f) 5′ GTGATTATGCTACCTTTGCACGGT (position 626–649); P7 (r) 5′ ACCATTTACCCAAATAAAGTATAG (position 1483–1506); P8 (r) 5′ GGACCAATCTATCACCCTATA (position 942–962). To identify the position of each primer, the sequence from nt 1 to 1559 of the sense or antisense 16S mtRNA was used as reference. For the region between the IR and the 16S mtrRNA, P13 (r) 5′ AGGTTTAGCCAAACCATT (807–824). For the 12S mtrRNA, P14 (f) 5′ AGCCTATATACCGCCATCTTC (position 604–624); P15 (r) 5′ AAGTATACTTGAGGAGGGTGA (position 843–863); P16 (f) 5′ GTGTACTGGAAAGTGCACTTG (position 927–947). For COX I, P17 (f) 5′ GAACAGGTTGAACAGTCTACCCT (position 371–393) and P18 (r) 5′ TTCCGAAGCCTGGTAGGATAAGA (position 738–760). For the 18S rRNA, P19 (f) 5′ GATGCGTGCATTTATCAGATC (position 309–329) and P20 (r) 5′ AGTGGACTCATTCCAATTACA (position 652–672). For GAPDH, P21 (f) 5′ ACTCTGGTAAAGTGGATATTGT (position 131–152) and P22 (r) 5′ ATGATGTTCTGGAGAGCCC (position 662–680). For β-actin mRNA, P23 (f) 5′ AAGAGAGGCATCCTCACCCTG (position 181–202); P24 (r) 5′ GGCGACGTAGCACAGCTTCTCC (position 639–660). (f) and (r) represent forward and reverse primers, respectively. The sequence of the primers for the 16S, 12S and COX 1 transcripts were deduced from the human mtDNA (GenBank Accession No. V00662). The sequence of the primers for the 18S rRNA, GAPDH mRNA and β-actin mRNA were deduced from the GenBank Accession Nos. M10098, M33197 and NM_001101, respectively. Amplified DNA fragments were purified (Wizard SU Gel and PCR Clean-up system; Promega), cloned in pGEM®-T Easy (Promega) or pTOPO (Invitrogen) and the purified recombinant plasmids were sequenced as described before (12,13).
RNase digestion
About 1 μg of total RNA from human cells treated with TURBO DNA-free in 50 μl of 2× SSC (28) was incubated with RNase A at a final concentration of 50 μg/ml for 15 min at 25°C. The digestion products were extracted with phenol–chloroform, and after adding 10 μg of glycogen to the aqueous phase, the RNA was precipitated with ethanol and recovered by centrifugation (12,13).
S1 protection assay
S1 protection assay was carried out using a single-stranded digoxigenin-labeled DNA probe complementary to the region between the IR and the 16S mtrRNA of the ncmtRNA. First, an amplicon was obtained by RT-PCR using primers 1 and 2 and this fragment was used as a template in an asymmetric PCR reaction that contained only primer 1 and digoxigenin-11-dUTP (Roche Non-Radioactive ISH). S1 protection was carried out as described (27), using 50 μg RNA and 50 ng digoxigenin-labeled probe. After S1 digestion, reactions were ethanol-precipitated and analyzed on native 2.5% agarose gels, which were transferred onto Hybond™-XL membranes (Amersham). After blocking with 0.02% Tween 20 plus 5% skim milk in PBS, the position of the digoxigenin-labeled probe was revealed with anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche) and developing in BCIP/NBT alkaline phosphatase substrate mixture (DAKO) for 10 min.
5′ RACE
HeLa cells RNA dissolved in 2× SSC was digested with RNase A and the double-stranded structure was recovered after phenol extraction as described before. The cDNA was synthesized with primer P8 (Figure 2a) and tailed at the 3′ end with dCTP using terminal deoxynucleotidyl-transferase (TdT; Promega) (29). Amplification by PCR was carried out with the specific primer 8 (Figure 2a) and the anchor primer provided by the manufacturer. The amplicon obtained of ∼250 bp was purified and both strands were sequenced directly.
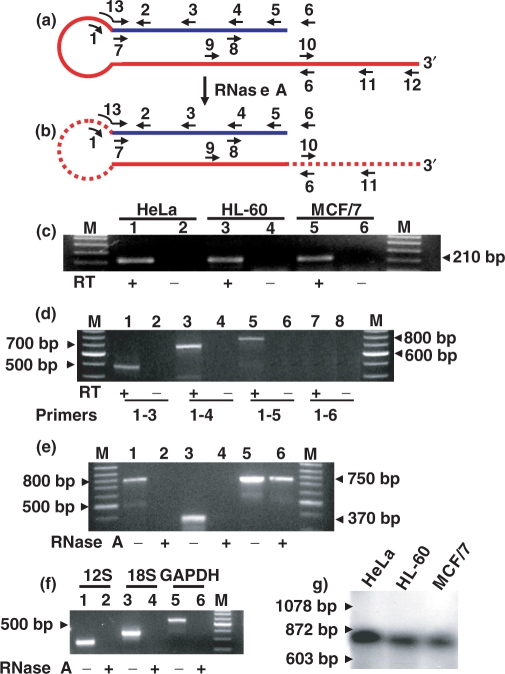
Figure 2.
The human ncmtRNA. (a) Theoretical structure of the human ncmtRNA. The 5′ end of the sense 16S mtrRNA (red line) is linked to a fragment of the antisense 16S mtrRNA or IR (blue line) forming a double-stranded structure and a loop of unknown length. The position of the reverse primers (under the lines) and the forward primers (over the lines) are indicated (see Materials and Methods section). (b) Digestion of the loop and the single-stranded region of the ncmtRNA by RNase A is also illustrated. (c) Amplification of the cDNA obtained from three tumor cell lines using primers 1 and 2. An amplicon of ∼210 bp was obtained only when the reaction was carried out with reverse transcriptase (c, odd lanes). (d) Amplification of the cDNA of HeLa cells by PCR using primers 1 in combination with primers 3 (lanes 1 and 2), 4 (lanes 3 and 4), 5 (lanes 5 and 6) and 6 (lanes 7 and 8), respectively. Amplicons of ∼500, 700 and 800 bp were obtained. No amplification products were generated with primer 1 and 6 (lanes 7 and 8) or without reverse transcriptase (d, even lanes). M, 100 bp ladder. (e) RNA from HeLa cells in 2× SSC was incubated without (odd lanes) or with 50 μg/ml of RNase A for 15 min at 25°C (even lanes). The RNA was recovered and amplified by RT-PCR using primers 1 and 5 (lanes 1 and 2), primers 10 and 11 (lanes 3 and 4) or with primers 7 and 5 (lanes 5 and 6). Amplicons of 800 and 350 bp were obtained with primer 1 and 5, and 10 and 11, respectively, only with untreated RNA (lanes 1 and 3, respectively). Amplification of the fragment of 750 bp obtained with primers 7 and 5 was not affected by the nuclease treatment (lanes 5 and 6). f) Amplification of the 12S mtrRNA (lanes 1 and 2), 18S rRNA (lanes 3 and 4) and GAPDH mRNA (lanes 5 and 6) after digestion with RNase A (mock experiment, odd lanes). (g) About 1 μg of RNA from the indicated cell lines was digested with RNase A and the digestion products were resolved by electrophoresis on a 1.5% agarose gel. After blotting, the membrane was probed with a 32P-labeled PCR fragment targeted to the double-stranded region of the ncmtRNA (see Materials Methods section). A single hybridization band corresponding to a transcript of ∼800 nt was detected.
Northern blot
Total RNA of HeLa and MCF/7 cells (3 μg each) were electrophoresed at 75 V for 90 min in a native 1.0% agarose gel prepared in TAE buffer or in an agarose gels under denaturing conditions containing 2.2 M formaldehyde (27). The RNAs were transferred in 20× SSC to a nylon membrane (Hybond™-XL; Amersham) for 18 h, and exposed to UV light for 5 min. The probe used was primer 13 (Figure 2a) labeled with 32P. Briefly, in a final volume of 20 μl, 100 ng of the oligonucleotide were mixed with 4 μl of enzyme buffer (100 mM cacodylate buffer pH 6.8, 1 mM CoCl2 and 0.1 mM DTT), 2 μl of [α-32P]-dCTP (3000 Ci/mmol) and 10 U of TdT, and incubated at 37°C for 30 min. The probe was purified using a Sephadex G-50 spin column (Amersham). Hybridization was carried out for 18 h at 37°C with 5 ml of 0.5 M sodium phosphate, pH 7.1, 2 mM EDTA, 7% SDS and 0.1% sodium pyrophosphate (30) containing 32P-labeled primer 13 at a ratio of 105 c.p.m./cm2 of membrane surface area. The blots were washed twice for 5 min at room temperature with 2× SSC plus 0.1% SDS and once with 0.5× SSC plus 0.1% SDS. Radioactivity on the membranes was visualized with a phosphor imager (Molecular™ Imager FX Phosphor Imaging System, BioRad).
About 1 μg of RNA digested with RNAse A as described before, was subjected to electrophoresis on a 1.5% agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane (27). The membrane was probed with an amplicon of ∼250 bp corresponding to the double-stranded region of the ncmtRNA and obtained by RT-PCR using primers 8 and 5 (Figure 2a) and [α-32P]-dCTP. The probe was extracted with phenol and precipitated with isopropanol. Hybridization was carried out with a solution containing 8 million c.p.m./ml in 4× SSC, 10% dextran sulfate, 150 μg/ml yeast tRNA, 150 μg/ml herring sperm DNA, 50% formamide and 1× Denhardt's solution (27). After hybridization at 65°C overnight, the membrane was washed for 10 min at room temperature with 2× SSC and 1× SSC, 20 min at 55°C with 0.5× SSC and 20 min at room temperature with 0.2× SSC. Then the membrane was exposed to X-ray films at −70°C (12,27).
Mitochondria isolation
HeLa cells were grown as indicated and about 5 × 108 cells were harvested and recovered by centrifugation at 300g for 5 min at 4°C. The cells were resuspended in about 10 volumes of a hypotonic solution containing 10 mM KCl, 0.15 mM MgCl2 and 10 mM Tris–HCl, pH 6.8, incubated for 10 min on ice and dounce homogenized with the tight pestle (13,31). The homogenization was monitored by phase microscopy until ∼70% of the cells were broken and the mitochondrial fraction was obtained as described before (31). The final mitochondria fraction was resuspended in 2–3 ml of 0.25 M sucrose, 2 mM MgCl2 and 0.4 mM sodium phosphate buffer at pH 6.8 and treated with RNAse A at a final concentration of 50 μg/ml for 15 min at room temperature (32). The mitochondria fraction was recovered by centrifugation at 10 000g for 15 min, suspended in 100 μl of PBS containing 100 U of RNaseOut (Invitrogen) and mitochondrial RNA was extracted with TRIzol as described before.
ISH
Cells cultured in 8-well chamber slides (Lab-Tek®, NUNC) for 24–48 h, were washed three times with PBS and fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. The slides were then washed three times with PBS for 5 min and incubated with 0.2 N HCl for 10 min at room temperature. Hybridization was carried out essentially as described before (12,14). The cells were hybridized for 18 h at 37°C with 200 μl of the hybridization solution containing 3.5 pmol of the antisense probe (primer 6 or 13) or the corresponding sense probes (Figure 2a), previously labeled at the 3′ end with digoxigenin-11-dUTP (Roche Applied Science) (12,14). The slides were washed with 2× SSC and 1× SSC for 10 min at room temperature, 0.2× SSC for 30 min at 37°C and finally, with 0.2× SSC for 10 min at room temperature (14). Then the cells were incubated for 2 h at room temperature with anti-digoxigenin antibody conjugated to alkaline phosphatase (Roche Applied Science), diluted 1:500 in the blocking buffer (1% BSA, 0.3% Triton X-100 in PBS). The color reaction was carried out with a BCIP/NBT substrate mixture (DAKO) plus levamisol for 30 min (14). Resting or PHA-stimulated human lymphocytes were applied on silanized slides, air dried, fixed with 4% paraformaldehyde and subjected to the same hybridization protocol. For FISH, after hybridization the cells were incubated with anti-digoxigenin antibody conjugated to FITC (Roche Applied Science). For tissue samples, paraffin sections of ∼5 μm thick were collected in silanized slides and deparaffinized by immersion in three consecutive xylene baths, 10 min each. The sections were rehydrated with three washes in 100%, 90%, 70% and 50% ethanol, once in PBS and once in distilled water for 10 min. Afterwards the sections were fixed with 4% paraformaldehyde in PBS and incubated with 0.2 N HCl plus 4 mg/ml of pepsin for 10 min at room temperature, and subjected to ISH as described before.
Immunocytochemistry and FISH
After ISH, melanoma cells (42/95) were incubated for 30 min in blocking buffer (1% BSA, 0.3% Triton X-100 in PBS), and then incubated for 2 h at room temperature with anti-digoxigenin antibody conjugated with fluorescein (Roche) and with anti-PCNA monoclonal antibody (DAKO). The cells were washed three times with 0.05% Tween20 in PBS, incubated for 2 h at room temperature with anti-mouse IgG conjugated with rhodamin, washed again and mounted with DABCO. After hybridization, HeLa cells were incubated with anti-digoxigenin antibody conjugated with rhodamin (Roche) and with anti-cytochrome c monoclonal antibody (Promega) or anti-endonuclease G polyclonal antibody (Chemicon International). After washing with 0.05% Tween20 in PBS, the cells were incubated with either anti-mouse IgG conjugated with fluorescein or with anti-rabbit IgG conjugated with fluorescein. Fluorescence microscopy was analyzed with a Olympus BX51 microscope. Confocal microscopy was performed with a LSM 5 Zeiss microscope equipped with a 63× objective. The analysis was carried out with the Zeiss LSM 5 Image Browser software.
Fixed resting or PHA-stimulated lymphocytes were incubated with the blocking solution and then with anti-PCNA (DAKO) or anti-Ki67 antigen (DAKO) or anti-phosphohistone H3 (Upstate) for 60 min at room temperature. After washing in PBS, the sections were incubated for 30 min with anti-mouse IgG conjugated to alkaline phosphatase (KPL) diluted 1:250 in 2% BSA in PBS. The color reaction was developed for 20 min with BCIP/NBT substrate mixture (DAKO) containing levamisol. Previous to immunocytochemistry, deparaffinized tissue sections were incubated in coplin jars for 15 min at 95°C in Target Retrieval Solution (DAKO) diluted 1:10 with distilled water. The incubation with anti-PCNA was as described before.
RESULTS
The human ncmt RNA
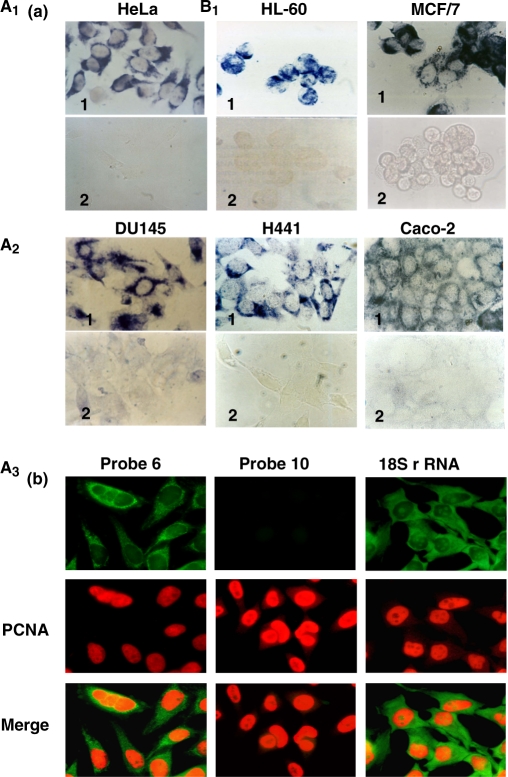
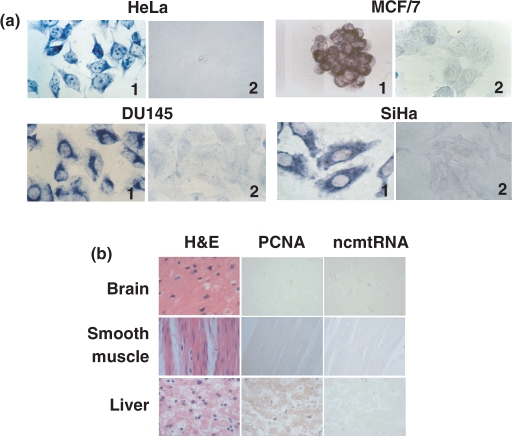
As shown in Figure 1a (panel 1), ISH with probe 6 targeted to the 16S mtrRNA (see Materials and Methods section) revealed strong hybridization signals in HeLa, HL-60, MCF/7, DU145, H441 and Caco-3 human cells. As expected, hybridization with the control sense probe 10 (see Materials and Methods section) was negative (Figure 1a, panel 2). Moreover, FISH with probe 6 in combination with immunocytochemistry of PCNA of the melanoma cell line 42/95 revealed that each cell expressing the transcript was also expressing PCNA (Figure 1b). The hybridization with probe 10 was again negative (Figure 1b). As a positive control, FISH was carried with probe 20 (see Materials and Methods section) to detect the 18S rRNA (Figure 1b).
Figure 1.
Expression of the hypothetical human ncmtRNA. (a) ISH of the indicated cells with probe 6 (panels 1) or the control sense probe 10 (panels 2). Note cytoplasmic and perinuclear hybridization signals (×100). (b) Melanoma cell line expressing the transcript are also expressing PCNA. The cells were subjected to ISH with probe 6 or probe 10 together with immunocytochemistry of PCNA. The merge figure confirmed the co-expression of both molecules. As a positive control, the same cells were subjected to ISH for the 18S rRNA and immunocytochemistry of PCNA (×63).
The target of probe 6 might be a transcript homologous to the mouse ncRNA (12). Therefore, to characterize this transcript, a hypothetical human mitochondrial RNA was deduced (Figure 2a). The transcript contains the complete sequence of the 16S mtrRNA (Figure 2a, red line) linked at its 5′ end to a fragment of the RNA transcribed from the 16S gene corresponding to the L-strand of the mtDNA or antisense 16S mtrRNA (Figure 2a, blue line). Based on this structure, primers were designed to amplify the putative transcript by RT-PCR (Figure 2a). Primer 1 was positioned at the theoretical loop between positions 11 and 31 of the human sense 16S mtrRNA, while the forward primer 2 was at the putative IR, corresponding to position 1326–1347 of the antisense 16S mtrRNA (Figure 2a). RT-PCR carried out with RNA from HeLa, HL-60 and MCF/7 cells yielded a single amplicon of ∼210 bp (Figure 2c, odd lanes). No amplification was obtained when reverse transcriptase was omitted from the reaction (Figure 2c, even lanes). The sequence of the amplicon of each cell line revealed that an IR of 184 nt was linked to the first 31 nt at the 5′ end of the 16S mtrRNA (Supplementary Figure S1a and GenBank Accession No. DQ386868).
To determine whether the IR was longer than 184 nt, a PCR-walking strategy was utilized. The cDNA from HeLa cells was amplified between primer 1 and primers 3, 4, 5 and 6, targeted to a putative longer IR (Figure 2a). Amplification fragments of ∼500, 700 and 800 bp were obtained when primer 1 was used in combination with primers 3, 4 and 5, respectively (Figure 2d, lanes 1, 3 and 5). No amplification product was obtained with primers 1 and 6 (Figure 2d, lanes 7 and 8), suggesting that the 5′ of the IR is positioned between primers 5 and 6 (see below). The sequence of the amplicon of 800 bp revealed an IR of 769 nt linked to the first 31 nt of the 16S mtrRNA (Supplementary Figure S1b and GenBank Access No. DQ386868). The sequence of the region between the 16S mtrRNA and the IR is identical to that found in the same region of the amplicon of 215 bp (compare Supplementary Figures S1a and b), confirming that in both cases we were amplifying the same transcript. The sequences of the 500 and 700 bp fragments indicated that they were part of the 800 bp amplicon. The same results and sequences were obtained with RNA from HL-60 cells, PHA-stimulated lymphocytes and MCF/7 cells (data not shown).
The stem of the ncmtRNA is resistant to RNase digestion
The 769 nt of the IR are fully complementary to the 16S mtrRNA. Therefore, the transcript should contain a double-stranded structure or stem of at least 769 bp resistant to RNase A digestion as predicted in Figure 2b. On the other hand, the loop and the single-stranded region that extends beyond the hairpin structure should be digested by the enzyme (Figure 2b, dotted lines). To test this possibility, RNA from HeLa and HL-60 cells was digested with RNase A as described in Materials and Methods section. The cDNA from the non-digested or digested RNA was then amplified by PCR using the primers described in Figure 2a. The amplicon of 800 bp obtained with primers 1 and 5, was not amplified after RNase digestion (Figure 2e, lanes 1 and 2), and the same was true for the fragment of 360 bp obtained with primers 10 and 11 (Figure 2e, lanes 3 and 4), indicating that the single-stranded regions of the transcript were digested. Similarly, the 12S mitochondrial rRNA, 18S rRNA and GAPDH mRNA, were not amplified after digestion (Figure 2f lanes 2, 4 and 6). On the other hand, amplification of a fragment of 750 bp, corresponding to the double-stranded structure of the ncmtRNA and obtained using primers 7 and 5 (Figure 2a), was not affected by RNase A digestion (Figure 2e, lanes 5 and 6). The sequence of this fragment was identical to that of the amplicon of 800 bp obtained with primers 1 and 5, except for the absence of the first 31 nt of the 16S mtrRNA. Northern blot also confirmed the presence of the stem after RNase A digestion. A single hybridization band of ∼800 nt was detected in digested RNA of HeLa, HL-60 and MCF/7 cells (Figure 2g).
To determine the 5′ end of the transcript using 5′ RACE, HeLa cell RNA was digested with RNase A and the nuclease-resistant stem was isolated as described before, and used as template for 5′ RACE. The sequence of the amplicon of 250 bp obtained by PCR with primer 8 and the anchor primer (see Materials and Methods section) revealed that the 769 nt of the IR were extended by 46 additional nt (Supplementary Figure S1b, underlined sequence), indicating that the total length of the IR was 815 nt.
The 16S mtrRNA is contiguous with the IR
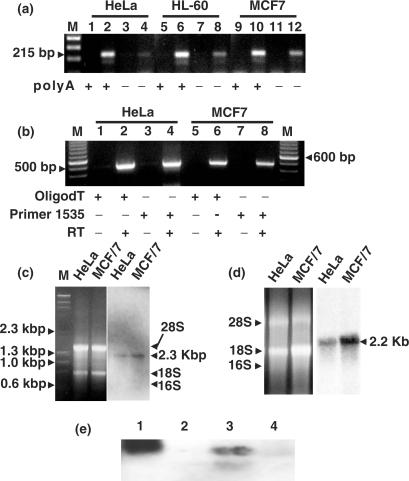
Similarly to the 12S and 16S mtrRNAs (33), the ncmtRNA is also polyadenylated. Total RNA from HeLa, HL-60 and MCF/7 cells was separated into polyA+ and polyA− fractions (see Materials and Methods section). The cDNA obtained from both fractions was then amplified with primers 1 and 2 (Figure 2a). As shown in Figure 3a, amplification of the 215 bp fragment revealed that the transcript was enriched in the polyA+ fraction (lanes 2, 6 and 10), compared to the polyA− fraction (Figure 3a, lanes 4, 8 and 12). The cDNA was synthesized from total RNA from HeLa and MCF/7 cells using either oligo dT or primer 12, which is complementary to the 3′ end of the 16S mtrRNA (Figure 2a). Then, each cDNA was amplified by PCR between primer 1 positioned at the loop and primer 3 positioned on the IR. In both cases, the expected amplicon of 500 bp was obtained (Figure 3b, lanes 2, 4, 6 and 8), indicating that the cDNA primed with oligo dT or with primer 12 comprised the complete 16S mtrRNA plus the IR. No amplification product was obtained without reverse transcriptase (Figure 3b, odd lanes).
Figure 3.
The IR is contiguous with the 16S mtrRNA. (a) Total RNA from HeLa (lanes 1 to 4), HL-60 (lanes 5 to 8) and MCF/7 (lanes 9 to 12) cells were separated into polyA+ and polyA− fractions. A total of 100 ng of each fraction was used to synthesize cDNA, which was then amplified by PCR using primers 1 and 2, to generate the 215 bp amplicon as indicated. Odd lanes correspond to reactions carried out in the absence of RT. (b) cDNA was synthesized from the polyA+ fraction of HeLa or MCF/7 cells, using oligo dT (lanes 1, 2, 5 and 6) or primer 12 (lanes 3, 4, 7 and 8). A single amplicon of 500 bp was obtained after PCR amplification of the cDNAs with primers 1 and 3 only when reverse transcriptase was included in the reaction mixture (even lanes). (c) About 3 μg of HeLa and MCF/7 cells RNA was resolved by electrophoresis on a 1% native agarose gel and subjected to northern blot. The membrane was probed with 32P-labeled primer 13 (Figure 2a). The probe hybridized with a single transcript, which migrated below the 1353 bp DNA marker (M = λDNA/HindIII and φDNA/HaeIII). The size of this transcript, deduced from the dsDNA ladder, corresponds to 2280 nt. (d) Northern blot was carried out with RNA from the indicated cells, under denaturing electrophoretic conditions. In this case, probe 13 hybridized with a single band that migrated on top of the 18S rRNA and corresponding to a transcript of 2200 nt. (e) For S1 protection assay, an asymmetric PCR fragment 215 nts containing the 31 nt of the sense 16S mtrRNA plus 184 nt of the IR was synthesized and labeled with digoxigenin (see Materials and Methods section). After denaturation at 100°C for 5 min, the probe was incubated overnight at 50°C either alone (lane 2), with 20 μg of HeLa RNA (lane 3) or with 20 μg of yeast RNA (lane 4). After hybridization, the products were digested with S1 nuclease and the products resolved by 2.5% native agarose gel electrophoresis and blotted to a nylon membrane. The products of digestions were reveled with anti-digoxigenin antibody conjugated to alkaline phosphatase (see Materials and Methods section). Lane 1 represents the probe alone without treatment.
If the IR of 815 nt is linked to the 1559 nt of the 16S mtrRNA, one would expect a transcript of 2374 nt. Total RNA from HeLa and MCF/7 cells was subjected to northern blot under native electrophoretic conditions using as a probe oligonucleotide 13 labeled with 32P. Probe 13 contains 9 nt complementary to the 5′ end of the 16S mtrRNA followed by 9 nt complementary to the 3′ end of the IR (Figure 2a), and therefore is specific for the ncmtRNA. A single hybridization band that migrates just below the 1353 bp DNA marker was observed (Figure 3c). It is pertinent to mention that dsDNA and dsRNA have the same behavior on native electrophoretic conditions (34). The size of this component corresponds to a transcript of ∼2280 nt, close to the expected size of the ncmtRNA. Northern blot with probe 13 under denaturing electrophoretic conditions revealed a single band on top of the 18S rRNA and corresponding to a transcript of 2200 nt (Figure 3d).
S1 protection assay also confirmed that the IR was contiguous with the 16S mtrRNA. An asymmetric PCR product labeled with digoxigenin (see Materials and Methods section) and complementary to a region of the transcript comprising the first 31 nt of the 16S mtrRNA linked to 184 nt of the IR (Supplementary Figure S1a), was protected from S1 digestion after hybridization with HeLa cell RNA (Figure 3e, lane 3) but not when hybridization was carried out without RNA or with yeast RNA (Figure 3e, lanes 2 and 4, respectively).
Synthesis of the ncmtRNA requires mitochondrial transcription
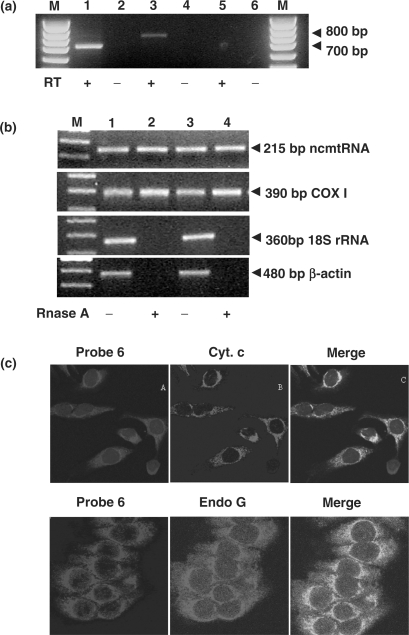
To determine whether the ncmtRNA was present in mitochondria, the organelles were isolated from HeLa cells and treated with RNase A to eliminate cytoplasmic RNA contamination (32). RNA was extracted from the isolated mitochondria and then amplified by RT-PCR using primer 1 in combination with primers 4 or 5. The expected amplicons of 700 and 800 bp, respectively, were obtained (Figure 4a, lanes 1 and 3), and their sequences were identical to those described before (Supplementary Figure S1b). No amplification was obtained with primers 1 and 6 (Figure 4a, lane 5), similarly to the results described before (Figure 2d). To ascertain that RNase treatment eliminated cytoplasmic RNA contamination, total RNA was extracted from two different preparations of HeLa cell mitochondria treated with the nuclease or untreated. As shown in Figure 4b, treatment with RNase did not affect the amplification of the amplicon corresponding to the ncmtRNA (215 bp) or to the mRNA of COX I (390 bp). In contrast, amplification of the 18S rRNA and β-actin mRNA was abolished after nuclease-treatment (Figure 4b, lanes 2 and 4). To confirm the presence of the ncmtRNA in mitochondria, co-localization studies were carried out. HeLa cells were subjected to FISH with digoxigenin-labeled probe 6 and anti-digoxigenin antibody conjugated to rhodamin to detect the ncmtRNA, and to immunocytochemistry with fluorescein-labeled anti-mouse antibody to detect the mitochondrial marker cytochrome c or with fluorescein-labeled anti-rabbit antibody to detect endonuclease G (35). As shown in Figure 4c, confocal microscopy revealed co-localization of the transcript with both mitochondrial markers.
Figure 4.
Mitochondrial localization of the ncmtRNA. (a) The mitochondrial fraction from HeLa cells was treated with RNase A, previously to RNA extraction. Amplification of the mitochondrial RNA with primer 1 in combination with primer 4 (lanes 1 and 2) or primer 5 (lanes 3 and 4) yielded the expected amplicons of 700 and 800 bp, respectively. No amplification was obtained with primers 1 and 6 (lanes 5 and 6) or when the reaction was carried out without reverse transcriptase (RT−, lanes 2, 4 and 6). (b) Total RNA extracted from two different preparations of HeLa mitochondria (lanes 1and 2, and 3 and 4) treated without (lanes 1 and 3) or with RNase A (lanes 2 and 4) was used to amplify the 215 bp fragment of the ncmtRNA, and the indicated amplicons of COX I mRNA, 18S rRNA and β-actin mRNA. Note that RNase A treatment abolished contamination with cytoplasmic transcripts. (c) Co-localization of the ncmtRNA with the mitochondrial markers cytochrome c and endonuclease G. HeLa cells were subjected to FISH to detect the ncmtRNA and immunocytochemistry to detect cytochrome c or endonuclease G (see Methods section) and analyzed by confocal microscopy.
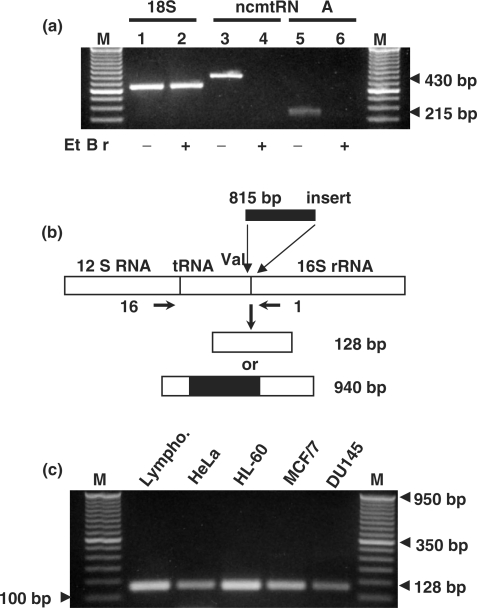
To determine whether mitochondrial transcription is required for the synthesis of this transcript, HeLa cells were cultured with EtBr (24,36,37). Total RNA from HeLa cells cultured with or without EtBr for 28 days was RT-PCR-amplified using primers 1 and 2 or 9 and 6. The expected amplicons of 215 and 430 bp, respectively, were obtained with RNA from control cells, but not from the EtBr-treated cells (Figure 5a, lanes 3 and 4, and 5 to 6, respectively). This treatment also inhibited the expression of other mitochondrial transcripts (data not shown). In contrast, EtBr did not affect the nuclear expression of the 18S rRNA (Figure 5a, lanes 1 and 2).
Figure 5.
Synthesis of the ncmtRNA requires mitochondrial transcription. (a) Total RNA was extracted from HeLa cells incubated without (odd lanes) or with (even lanes) 1 μg/ml of ethidium bromide for 28 days. The RNA was amplified by RT-PCR using specific primers for 18S rRNA (lanes 1 and 2) (see Methods section), primers 9 and 11 (lanes 3 and 4) and primers 1 and 2 for the ncmtRNA (lanes 5 and 6). (b) Theoretical structure of an anomalous mtDNA containing an insert of 815 bp between the tRNAval and the 16S mtrRNA genes. (c) mtDNA from the indicated cell lines was amplified between primer 16 positioned close to the 3′ end of the 12S gene and primer 1 positioned on the 16S mtrRNA gene as shown in (b). All samples yielded a single amplicon of 128 nt. (a) M, 100 bp ladder. (c) 50 bp ladder.
The presence of the long IR in the ncmtRNA might be explained if some anomalous molecules of mtDNA contains the IR of 815 bp inserted between the genes of the tRNAVal and the 16S mtrRNA (Figure 5b). Transcription of this mtDNA followed by processing of the H-strand polycistronic RNA would then generate the ncmtRNA. However, PCR amplification of the human mtDNA from five different cell types between primer 16 (positioned at the 3′ end of the 12S mtrRNA, see Materials and Methods section) and primer 1 (Figure 2a) yielded only the expected amplicon of 128 bp (Figure 5c), corresponding to the normal sequence of the mtDNA.
The ncmtRNA is expressed in proliferating cells
As shown in Figure 1, seven different human cell lines over-expressed a transcript that was complementary to probe 6. However, this probe does not distinguish between the 16S mtrRNA and the ncmtRNA. On the other hand, we demonstrated that probe 13 (Figure 2a), targeted to the region between the IR and the 16S mtrRNA, was specific for the ncmtRNA. As described before, probe 13 hybridized only with the ncmtRNA in northern blots carried out under native or denaturing conditions. Moreover, probe 13 supported the amplification of the transcript between the target region and the IR. This amplification was abolished after digestion with RNase A (Supplementary Figure S2). Therefore, ISH with probe 13 was carried out with HeLa, MCF/7, Du145 and SiHa cells. The hybridization signals were the same as that observed with probe 6, revealing strong cytoplasmic and perinuclear staining (Figure 6a), and confirming that the ncmtRNA is expressed in proliferating cells. Then we asked whether this transcript is expressed in resting cells. As shown in Figure 6b, no staining of human brain, smooth muscle and liver cells was observed after ISH with probe 13 or with probe 6 (data not shown). The absence of proliferating cells in these tissues was confirmed by the null expression of PCNA (Figure 6b).
Figure 6.
Expression of the ncmtRNA in proliferating cells. (a) The indicated cells were subjected to ISH with probe 13 (panels 1) or with corresponding control sense probe (panels 2). Strong cytoplasmic and perinuclear hybridization signals were found (×100). No hybridization was found with the control sense probe. (b) ISH and immunocytochemistry to determine the expression of the ncmtRNA and PCNA, respectively, in brain, smooth muscle and liver cells. Each panel of tissues was also stained with hematoxylin-eosin (×40).
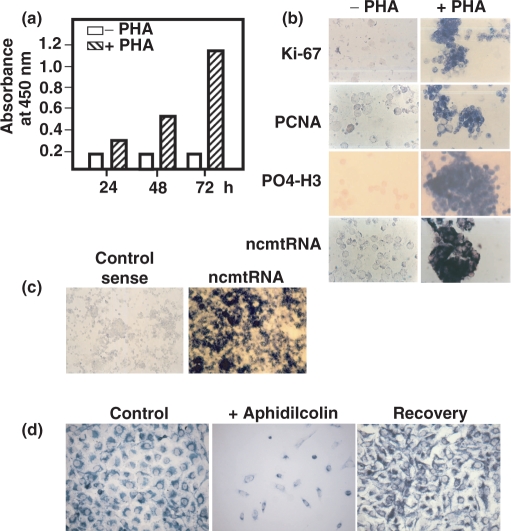
Since human circulating lymphocytes, as resting cells, do not express the ncmtRNA (Figure 7b, −PHA), we asked whether the expression of this transcript can be induced if lymphocytes are stimulated to enter the S phase. Peripheral human lymphocytes stimulated with PHA for 72 h were actively engaged in DNA synthesis as measured by BrdU incorporation (Figure 7a). As expected, the stimulated cells were also expressing the proliferating antigens Ki-67, PCNA and phosphohistone H3 (Figure 7b, +PHA). At the same time, ISH with probe 13 revealed that the PHA-stimulated lymphocytes were also over-expressing the ncmtRNA (Figure 7b, +PHA). At low magnification, hybridization of PHA-stimulated lymphocytes with probe 13 show a strong signal in most of the cells (Figure 7c, ncmtRNA), while no staining was observed with the corresponding control sense probe (Figure 7c, Control sense). Similar results were obtained with lymphocytes obtained from 11 different healthy donors (data not shown).
Figure 7.
Reversibility of the ncmtRNA expression. (a) Human lymphocytes were incubated without (white bars) or with PHA (hatched bars) for 24, 48 and 72 h. At each time period, the cells were incubated with BrdU for 16 h and the incorporation of the nucleoside was determined by ELISA (see Materials and Methods section). The incorporation of BrdU is expressed as the absorbance at 450 nm. (b) About 100 000 lymphocytes incubated without (−PHA) or with PHA (+PHA) for 72 h were subjected to immunocytochemistry to determine the expression of Ki-67, PCNA and phospho-histone H3 (PO4-H3), and to ISH with probe 13 to determine the expression of the ncmtRNA (×40). (c) PHA-stimulated lymphocytes for 72 h were hybridized with probe 13 (ncmtRNA) or with the corresponding sense control probe (Control sense) (×20). (d) DU145 cells were incubated twice with aphidicolin (see Methods section) and subjected to ISH together with a control culture. The blocked cells were changed to normal medium for 48 h and then subjected to ISH (Recovery) (×40).
Next we asked whether the arrest of the cell cycle at G1 with aphidicolin would affect the expression of the ncmtRNA. As shown in Figure 7d, treatment of DU145 cells with the drug induces both, a block on cell proliferation and a marked down regulation of this transcript when compared with the untreated control cells. When the DU145 cells are removed from the drug-containing medium to normal culture medium for 2 days, cell proliferation was re-initiated and the level of expression of the transcript is recovered and become similar to the expression rate of the control cells (Figure 7d).
DISCUSSION
The ncmtRNA
This is the first report on a novel non-coding mitochondrial RNA over-expressed in human proliferating cells but not in resting cells. Based on the hypothetical structure shown in Figure 2a, we established that the human ncmtRNA contains an IR of 815 nt covalently linked to the first 865 nt of the 16S mtrRNA. To confirm the existence of the ncmtRNA additional experiments were carried out. First, northern blot with probe 13 revealed a transcript of 2280 nt (native conditions) or 2200 nt (denaturing conditions), which are close to the expected size of 2374 nt of the ncmtRNA. Second, amplification of the region comprising the loop and the IR of the ncmtRNA was possible after using a cDNA primed with oligo dT or with a primer targeted to the 3′ end of the transcript. This experiment was similar to the strategy used to establish that different regions of the long ncRNA Tsix were contiguous (38). Third, the long double-stranded structure of 815 bp was isolated after RNase A digestion, and its presence in the digestion products was demonstrated by amplification between the 5′ and 3′ ends of the stem or by northern blot analysis. Fourth, S1 protection assay demonstrated the presence in the ncmtRNA of the sequence region between the IR and the 16S mtrRNA. Altogether, these results support the conclusion that the ncmtRNA contains an IR of 815 nt contiguous with the 16S mtrRNA.
Translation in silico of the 2374 nt of the ncmtRNA (GenBank Accession No. DQ386868) using either the standard or the vertebrate mitochondrial codes, yielded several open reading frames corresponding to peptides ranging from 15 to 60 amino acids. These results agree with the definition of ncRNAs, in which open reading frames greater than 100 amino acids are rarely present (15–17).
A search of the ncmtRNA was conducted in human EST database (www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Although several mitochondrial cDNA entries were present, including the 16S mtrRNA, the sequence of the ncmtRNA or at least part of the sequence containing the region between the IR and the 16S mtrRNA was not found. We believe that the absence of this sequence is due to both, the double-stranded structure of the transcript and replication slippage of the polymerases commonly used to construct EST libraries. Escherichia coli DNA polymerases I, II and III as well as polymerases of phages T4 and T7, have poor strand separation activity resulting in replication slippage of DNA templates carrying hairpin structures (39,40). As a consequence, deletion mutations of hairpin sequences have been observed in vitro and in vivo (41,42). Thermophilic DNA polymerases, including Taq polymerase also exhibit replication slippage with templates containing hairpin structures (43), with fateful consequences for amplification and DNA sequencing (44–46).
Synthesis of the ncmtRNA requires mtDNA transcription
Several results strongly suggest that the ncmtRNA is synthesized in mitochondria. The sequence of the transcript exhibits a minimum of 99.9% identity with several haplotypes of the human mtDNA (see http://www.ncbi.nlm.nih.gov/gquery/). Moreover, we showed that the transcript was present in isolated mitochondria and confocal microscopy demonstrated that the ncmtRNA co-localized with cytochrome c and endonuclease G, two specific markers of the organelle (35). On the other hand, treatment of HeLa cells with EtBr abolished the presence of the ncmtRNA as well as other mitochondrial transcripts, indicating that the synthesis of the ncmtRNA requires mitochondrial transcription (24,36,37). These results also indicate that the ncmtRNA is not a transcription product of a mitochondrial pseudogene (47). Searching for the sequence of the ncmtRNA in the human genome (www.ncbi.nlm.nih.gov/BLAST/Blast), yielded only fragments of the 16S mtrRNA (sense and antisense orientations) having an identity of 82–94% with contigs corresponding to several human chromosomes. However, the sequence of the ncmtRNA was not found (data not shown). This result is consistent with the results of amplification of the transcript by RT-PCR. As shown here, we have never observed amplification between the loop and the IR when the reactions were carried out without reverse transcriptase.
Since there is not a contiguous sequence in the mtDNA encoding the ncmtRNA, it is reasonable to hypothesize that the synthesis of this transcript involves post-transcriptional reactions. Although other possibilities cannot be discarded, the synthesis of the ncmtRNA might involve either a trans splicing reaction or a RNA ligase reaction as reported in other organisms (48–50). Although the synthesis of the ncmtRNA warrants future work, it is interesting to mention the similarity between the structures of this transcript with the presence of double-stranded RNAs in HeLa cell mitochondria. This is a heterogeneous fraction sedimenting from 4S to 17S and resistant to RNase A digestion (28,51). Unfortunately, no further sequence analysis of these transcripts was reported, and perhaps one of these double-stranded structures corresponds to the ncmtRNA.
Besides the ribosomal RNAs 16S and 12S, and the tRNAs, the ncmtRNA is not the only ncRNA present in mitochondria. Recently, a new family of small ncRNAs was reported to be present in isolated mouse mitochondria, consisting of six transcripts ranging in size between 23 and 68 nt (52). The authors also reported the presence of similar molecules in isolated chloroplast of Nicotiana tabacum. Unfortunately, no information on the biological function was provided (52).
Expression of the ncmtRNA and cell proliferation
The most striking results of the present work are the close correlations between expression of the ncmtRNA and cell proliferation. Besides the cell lines reported here, over-expression of the ncmtRNA was observed in 16 additional tumor and normal cell lines (data not shown). In contrast, this transcript was not expressed in non-proliferating cells including resting circulating lymphocytes. However, the expression of this transcript can be induced in PHA-stimulated lymphocytes together with DNA synthesis and the expression of the proliferation markers Ki-67, PCNA and phosphohistone H3 (19–21). Moreover, treatment of DU145 cells with aphidicolin confirmed the previous results. After treatment with the drug, the cells stop proliferating as described before (22,23). At the same time, the expression of the ncmtRNA was down-regulated. However, after removal of the drug the expression of this transcript was re-established together with cell proliferation. Altogether, these results strongly suggest that the ncmtRNA is a new marker of cell proliferation. However, the precise function that this transcript may play in the cell cycle remains unclear and warrants future studies.
Similarly to the present work, other ncRNAs have been reported to be involved in cell proliferation. ncRNAs are involved in different cellular and molecular events, including differentiation and development, imprinting, regulation of X chromosome silencing and human diseases (17). Moreover, microRNA, an important group within the expanding family of ncRNAs (53), are also involved in cell proliferation and the regulation of the cell cycle. In Drosophila melanogaster, germline stem cell division is regulated by microRNAs. This conclusion was reached after observing a reduction of the germline stem cell proliferation in the dicer-1 mutant, the RNase III essential for microRNA production (54). Although the microRNAs involved were not identified, the authors proposed that the function of these molecules is to inhibit Dacapo, which in turn negatively regulates the G1/S transition (54). In human lymphoma, c-Myc up-regulates the expression of six microRNAs encoded by the c13orf24 cluster of chromosome 13 (55). Two of these molecules, microR-17-5p and microR-20a, negatively regulate the translation of the transcription factor E2F1, which in turn is also up-regulated by c-Myc. The authors propose that the microRNAs play a fine-tuning role to regulate proliferative signals (55). MicroRNAs also regulate cell proliferation or differentiation. Thus, microRNA-133 enhances proliferation of the mouse myoblast C2C12 cells, while microRNA-1 promotes differentiation and myogenesis (56).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Giuseppe Attardi, California Institute of Technology, Pasadena, CA, for suggesting the treatment of HeLa cells with ethidium bromide. We also thank Dr Marc Shuman from UCSF for providing several human cell lines and for his enthusiastic support. We thank Dr Bernardita Mendez, Dr Pablo Valenzuela, Dr Arturo Yudelevich and Dr Mario Rosemblatt for their continuous support and enriched discussions. This work was supported by Millennium Scientific Initiative N° P04-071-F, Grant D04I1338, FONDEF, CONICYT, Santiago, Chile, and Grants DID-32-03, DID-26-04 and DID-57-04, Universidad Nacional Andrés Bello. Funding to pay the Open Access publication charges for this article was provided by MIFAB P04-071-F.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nass MM, Nass S, Afzelius BA. The general occurrence of mitochondrial DNA. Exp. Cell Res. 1965;37:516–539. doi: 10.1016/0014-4827(65)90204-1. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, de Brujin MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 4.Chomyn A, Attardi G. Recent advances on mitochondrial biogenesis. In: Ernster L, editor. Molecular Mechanisms in Biogenesis. Amsterdam: Elsevier Science Publishers B. V.; 1992. pp. 483–509. [Google Scholar]
- 5.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Ann. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 6.Taanman J.-W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 8.Falkenberg M, Larsson N.-G, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Ann. Rev. Biochem. 2007;76:30.1–30.21. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 9.Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36:635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- 10.Bogenhagen DF, Applegate EF, Yoza BK. Identification of a promoter for transcription of the heavy strand of human mtDNA: in vitro transcription and deletion mutagenesis. Cell. 1984;36:1105–1113. doi: 10.1016/0092-8674(84)90061-8. [DOI] [PubMed] [Google Scholar]
- 11.Shuey DJ, Attardi G. Characterization of an RNA polymerase activity from HeLa cell mitochondria, which initiates transcription at the heavy strand rRNA promoter and the light strand promoter in human mitochondrial DNA. J. Biol. Chem. 1985;260:1952–1958. [PubMed] [Google Scholar]
- 12.Villegas J, Zarraga AM, Müller I, Montecinos L, Werner E, Brito M, Meneses AM, Burzio LO. A novel chimeric mitochondrial RNA localized in the nucleus of mouse sperm. DNA Cell Biol. 2000;19:579–588. doi: 10.1089/104454900439809. [DOI] [PubMed] [Google Scholar]
- 13.Villegas J, Müller I, Arredondo J, Pinto R, Burzio LO. A putative RNA editing from U to C in a mouse mitochondrial transcript. Nucleic Acids Res. 2002a;30:1895–1901. doi: 10.1093/nar/30.9.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villegas J, Araya P, Bustos-Obregon E, Burzio LO. Localization of the 16S mitochondrial rRNA in the nucleus of mammalian spermatogenic cells. Mol. Hum. Reprod. 2002b;8:977–983. doi: 10.1093/molehr/8.11.977. [DOI] [PubMed] [Google Scholar]
- 15.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 16.Pang KC, Stephen S, Engström PG, Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS. RNAdb – a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Dergunova NN, Bulycheva TI, Artemenko EG, Shpakova AP, Pegova AN, Gemjian EG, Dudnik OA, Zatsepina OV, Malashenko OS. A major nucleolar protein B23 as a marker of proliferation activity of human peripheral lymphocytes. Immunol. Lett. 2002;83:67–72. doi: 10.1016/s0165-2478(02)00085-8. [DOI] [PubMed] [Google Scholar]
- 19.Fuss J, Linn S. Human DNA polymerase epsilon colocalizes with proliferating nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt MH, Broll R, Bruch HP, Duchrow M. Proliferation marker pKi-67 affects the cell cycle in a self-regulated manner. J. Cell. Biochem. 2002;87:334–341. doi: 10.1002/jcb.10302. [DOI] [PubMed] [Google Scholar]
- 21.Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblast. J. Biol. Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- 22.O’Connor PM, Ferris DK, Pagano M, Draetta G, Pines J, Hunter T, Longo DL, Kohn KW. G2 delay induced by nitrogen mustard in human cells affects cyclinA/cdk2 and cyclin B1/cdc2-kinase complex differently. J. Biol. Chem. 1993;268:8298–8308. [PubMed] [Google Scholar]
- 23.Wetzler M, Talpaz M, Yee G, Stass SA, Van Etten RA, Andreeff M, Goodacre AM, Kleiner HD, Mahadevia RK, et al. Cell cycle-related shifts in subcellular localization of BCR: association with mitotic chromosomes and with heterochromatin. Proc. Natl Acad. Sci. USA. 1995;92:3488–3492. doi: 10.1073/pnas.92.8.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Walsh PS, Metzger DA, Higuchi R. Chelex 199 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch ET, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Young PG, Attardi G. Characterization of double-stranded RNA from HeLa cell mitochondria. Biochem. Biophys. Res. Commun. 1975;65:1201–1207. doi: 10.1016/s0006-291x(75)80357-3. [DOI] [PubMed] [Google Scholar]
- 29.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Church GM, Gilbert W. Genomic sequencing. Proc. Natl Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse B, Murdter NN, Attardi G. Transcription system using a HeLa cell mitochondrial lysate. Methods Mol. Biol. 1995;37:179–197. doi: 10.1385/0-89603-288-4:179. [DOI] [PubMed] [Google Scholar]
- 32.Gaines G, Attardi G. Highly efficient RNA-synthesizing system that uses isolated human mitochondria: new initiation events and in vivo-like processing patterns. Mol. Cell. Biol. 1984;4:1605–1617. doi: 10.1128/mcb.4.8.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Etten RA, Bird JW, Clayton DA. Identification of the 3′-ends of the two mouse mitochondrial ribosomal RNAs. The 3′-end of 16 S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J. Biol. Chem. 1983;258:10104–10110. [PubMed] [Google Scholar]
- 34.Rodriguez-Cousiño N, Esteban LM, Esteban R. Molecular cloning and characterization of W double-stranded RNA, a linear molecule present in Saccharomyces cerevisiae. J. Biol. Chem. 1991;266:12772–12778. [PubMed] [Google Scholar]
- 35.Coté J, Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi J, Takemitsu M, Nonaka I. Recovery of the missing tumorigenicity in mitochondrial DNA-less HeLa cells by introduction of mitochondrial DNA from normal cells. Somat. Cell. Mol. Genet. 1992;18:123–129. doi: 10.1007/BF01233159. [DOI] [PubMed] [Google Scholar]
- 37.Hayakawa T, Noda M, Yasuda K, Yorifuji H, Taniguchi S, Miwa I, Sakura H, Terauchi Y, Hayashi J, et al. Ethidium bromide-induced inhibition of mitochondrial gene transcription suppresses glucose-stimulated insulin release in the mouse pancreatic β-cell line βHC9. J. Biol. Chem. 1998;273:20300–20307. doi: 10.1074/jbc.273.32.20300. [DOI] [PubMed] [Google Scholar]
- 38.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 39.Canceill D, Ehrlich D. Copy-choice recombination mediated by DNA polymerase III haloenzyme from Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:6647–6652. doi: 10.1073/pnas.93.13.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canceill D, Viquera E, Ehrlich SD. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 1999;274:27481–27490. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 41.Glickman BW, Ripley LS. Structural intermediates of delation mutagenesis: a role for palindromic DNA. Proc. Natl Acad. Sci. USA. 1984;81:512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madsen CS, Ghivizzani SC, Hauswirth WW. In vivo and in vitro evidence for slipped mispairing in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 1993;90:7671–7675. doi: 10.1073/pnas.90.16.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viguera E, Canceill D, Ehrlich SD. In vitro replication slippage by DNA polymerases from thermophilic organisms. J. Mol. Biol. 2001;312:323–333. doi: 10.1006/jmbi.2001.4943. [DOI] [PubMed] [Google Scholar]
- 44.Hew Y, Grzelczak Z, Lau C, Keeley FW. Identification of a large region of secondary structure in the 3′-unstranslated region of chicken elastin mTNA with implications for the regulation of mRNA stability. J. Biol. Chem. 1999;274:14415–14421. doi: 10.1074/jbc.274.20.14415. [DOI] [PubMed] [Google Scholar]
- 45.Viswanathan VK, Krcmarik K, Cianciotto NP. Template secondary structure promotes polymerase jumping during PCR amplification. Biotechniques. 1999;27:508–511. doi: 10.2144/99273st04. [DOI] [PubMed] [Google Scholar]
- 46.Geiszt M, Lekstrom K, Leto TL. Analysis of mRNA transcripts from the NAD(P)H oxidase 1 (Nox1) gene. Evidence against production of the NADPH oxidase homolog-1 short (NOH-1S) transcript variant. J. Biol. Chem. 2004;279:51661–51668. doi: 10.1074/jbc.M409325200. [DOI] [PubMed] [Google Scholar]
- 47.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanc V, Alfonzo JD, Aphasizhev R, Simpson L. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 1999;274:24289–24296. doi: 10.1074/jbc.274.34.24289. [DOI] [PubMed] [Google Scholar]
- 49.Agorio A, Chalar C, Cardozo S, Salinas G. Alternative mRNAs arising from trans-splicing code for mitochondrial and cytosolic variants of Echinococcus granulosus thioredoxin glutathione reductase. J. Biol. Chem. 2003;278:12920–12928. doi: 10.1074/jbc.M209266200. [DOI] [PubMed] [Google Scholar]
- 50.Simpson L, Sbisego S, Aphasizhev R. Uridine insertation/delation RNA editing in trypanosome mitochondria: a complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc. Natl Acad. Sci. USA. 1971;68:1757–1761. doi: 10.1073/pnas.68.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Hüttenhofer A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartel DP. Micro RNAs: genomics, biogenesis, mechanisms and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 54.Hatfield SD, Sccherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 55.O’Donell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 56.Chen J.-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.